Abstract
Disease named entity recognition (NER) is a critical task for most biomedical natural language processing (NLP) applications. For example, extracting diseases from clinical trial text can be helpful for patient profiling and other downstream applications such as matching clinical trials to eligible patients. Similarly, disease annotation in biomedical articles can help information search engines to accurately index them such that clinicians can easily find relevant articles to enhance their knowledge. In this paper, we propose a domain knowledge-enhanced long short-term memory network-conditional random field (LSTM-CRF) model for disease named entity recognition, which also augments a character-level convolutional neural network (CNN) and a character-level LSTM network for input embedding. Experimental results on a scientific article dataset show the effectiveness of our proposed models compared to state-of-the-art methods in disease recognition.
Introduction
Deep learning techniques have demonstrated superior performance over traditional machine learning (ML) techniques for various general-domain NLP tasks e.g. named entity recognition (NER), parts-of-speech (POS) tagging, language modeling, paraphrase identification, sentiment analysis etc. However, clinical documents pose unique challenges for NER compared to general-domain text due to widespread use of acronyms and non-standard clinical jargons by healthcare providers and inconsistent document structure and organization, and the demand for effective de-identification and anonymization of patient data for research. Specifically, addressing gaps in NER for the clinical domain can foster more research and innovation for meaningful clinical applications including patient cohort identification, patient engagement support, population health management, pharmacovigilance, personalized medicine, and clinical trial matching.
Disease named entity recognition1–3 is an important task in many biomedical natural language processing applications. This task is particularly challenging due to the critically large impact of a possible misrecognition. For example, cancer has both disease diagnosis and many complex histologies that need to be delineated. Thus, accurate identification of disease mentions can be beneficial for the optimal patient outcome e.g. extracting disease names from a clinical trial text may be helpful for patient profiling, which would further enable efficient matching of clinical trials to eligible patients. Similarly, disease recognition in biomedical articles can help information search engines to accurately index them such that clinicians can easily find relevant articles to enhance their knowledge.
The main contribution of this paper is to perform disease named entity recognition by encoding clinical domain knowledge via various types of embeddings into different layers of a deep neural network architecture consisting of LSTM and CNN. Our experiments show the positive impact of clinical domain knowledge on the performance of the model whilst adding such knowledge at different parts of the neural network. Our proposed model achieves the new state-of-the-art results in disease named entity recognition on a scientific article dataset.
Task Description and Data
Disease named entity recognition from free text can be cast as a sequence tagging problem. A BIO schema4 can be used for tagging the input sequence. For example, Figure 1 denotes a tag for each word from the input text. “B-disease” represents the beginning word of a disease mention, “I-disease” represents the other intermediate word(s) in a disease name, and “O” represents a word not belonging to a disease name.
Figure 1:
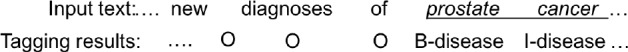
An example input and output for disease named entity recognition task.
Existing rule-based systems or traditional machine learning methods for disease named entity recognition heavily depend on hand-crafted features5, such as syntactic, lexical, n-gram etc. Although neural network-based methods do not depend on hand-crafted features, large labeled datasets are required for training the models. In this paper, we propose a domain knowledge-enhanced neural network architecture for improved accuracy in disease named entity recognition.
Existing clinical NLP systems e.g. MetaMap6, cTAKES7 etc. have been used for disease name recognition in various studies8,9. We opt to take advantage of one such existing system instead of training a neural network-based model from scratch. Thus, we explore to encode output from the clinical NLP system to improve the state-of-the-art performance for disease name recognition. To this end, we use a hybrid clinical NLP engine10 to generate the BIO tagging output. Although the engine generates tags for diseases and other biomedical concepts, we only use the disease tags for our experiments.
Domain Knowledge Sources: Our main source of clinical domain knowledge are clinical ontologies. Significant research efforts have been dedicated to build dictionaries/ontologies that facilitate biomedical NLP tasks2. MEDIC (http://ctd.mdibl.org/voc.go?type=disease) is a disease vocabulary, which includes 9,700 unique diseases and 67,000 unique terms in total. MEDIC is derived from a combination of concepts from the Online Mendelian Inheritance in Man (OMIM, https://www.omim.org/) and Medical Subject Headings (MeSH, https://www.nlm.nih.gov/mesh/) gazetteers. We use the MEDIC vocabulary in integrating lexicon features into our proposed model. In addition to the output from the clinical NLP engine, the MEDIC vocabulary constitutes the domain knowledge in our proposed neural network-based model for disease recognition.
Dataset: We use the publicly available NCBI dataset for our experiments and evaluation. The NCBI disease corpus (https://www.ncbi.nlm.nih.gov/CBBresearch/Dogan/DISEASE/)11 is a collection of 793 PubMed abstracts fully annotated at the mention and concept level. The public release of the NCBI disease corpus contains 6892 disease mentions, which are mapped to 790 unique disease concepts. The detailed statistics of the NCBI dataset is presented in Table 1. For our experiments, the dataset is split into three subsets for training, validation, and testing.
Table 1:
Dataset statistics.
| number of sentences | average sentence length | number of tokens | number of unique tokens | number of annotations | |
|---|---|---|---|---|---|
| Train-NCBI | 5,576 | 23 | 132,584 | 9,805 | 2,911 |
| Valid-NCBI | 918 | 25 | 23,456 | 3,580 | 487 |
| Test-NCBI | 941 | 25 | 24,019 | 3,679 | 535 |
Methodology
In this section we first describe a generic architecture for NER task. Then we explain our proposed methods of encoding domain knowledge into this architecture.
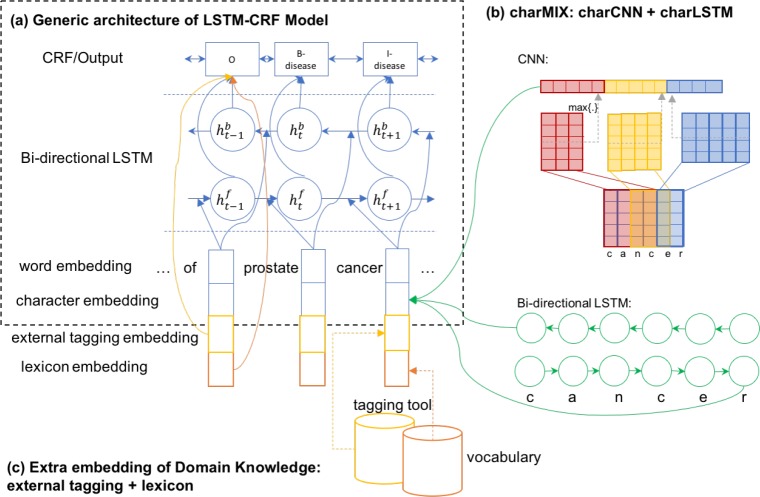
The generic neural network architecture for entity recognition task is a bidirectional LSTM with a sequential conditional random field layer (LSTM-CRF, Figure 2(a)). It takes as input a sequence of vectors (x1, x2,…,xn) and returns another sequence (y1, y2, yn) that represents the corresponding tagging information for the input sequence. LSTM-CRF models have been shown to achieve state-of-the-art performances on general domain NER tasks12–15. The LSTM-CRF has also been applied successfully for biomedical NER tasks16.
Figure 2:
The proposed architecture for disease name recognition.
The LSTM-CRF model contains the following layers: a character embedding layer, a word embedding layer, a bi-directional LSTM layer, and a CRF tagging layer. For a given sentence (x1, x2, … , xn) containing n words, each word is represented as a d—dimensional vector. The d— dimensional vector is concatenated from two parts: a d1-dimensional vector Vchar from the character embedding layer and a d2–dimensional vector Vword from the word embedding layer. The bi-directional LSTM layer reads the vector representations of the input sentence (x1, x2, xn) to produce two sequences of hidden vectors, i.e., the forward sequence () and the backward sequence (). Both vectors are concatenated into .
Methods for encoding the character embedding layer include: using a bi-directional LSTM layer (charLSTM), or Convolutional Neural Network (charCNN). charLSTM14 and charCNN13,17 have been studied separately for various NLP tasks. As displayed in Figure 2(b), we explore a combination of both methods in our architecture.
In Figure 2(c), domain knowledge either from the domain vocabulary or from the external tagging engine are introduced through a lexicon embedding layer and an external tagging embedding layer respectively. Moreover, external tagging embedding can be encoded either before or after the bi-directional LSTM layer.
LSTM-CRF-charMIX: The charCNN architecture generates the character embedding for each word in sentence. First, we define a vocabulary of characters C. Let d be the dimensionality of character embeddings, and Q ∈ Rdx|C| is the matrix character embeddings. As an example, charCNN takes the current word “cancer” as input and performs a lookup of Q ∈ Rdx|C| and stacks them to form the matrix Ck. The convolution operations are applied between Ck and multiple filter/kernel matrices. Then a max-over-time pooling operation is applied to obtain a fixed-dimensional representation of the word, which is denoted as Vcnn.
charLSTM is similar to the bi-directional LSTM layer in the generic architecture of the LSTM-CRF model. Instead of taking a sequence of words as input, it takes a sequence of characters in a word as input. It then outputs the concatenation of the forward and backward hidden states , which we denote as Vlstm.
As mentioned above, we consider both charCNN and charLSTM for learning the character embeddings. The charMIX architecture concatenates into , which is the same d1–dimensional vector Vchar for character embedding layer.
LSTM-CRF-DK: As mentioned previously, we consider the domain knowledge (DK) from the clinical vocabulary and tagging from the external clinical NLP engine10. We encode the DK into lexicon embedding and external tagging embedding correspondingly as discussed below.
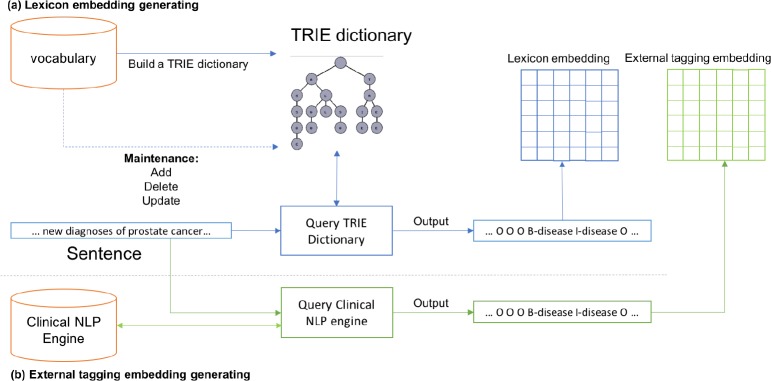
As displayed in Figure 3(a), we build a TRIE dictionary for the vocabulary (MEDIC), which can be easily maintained. TRIE is an efficient data structure for frequent word/phrase matching18. A sentence is used to query the TRIE dictionary. The TRIE dictionary then outputs a sequence of BIO tags. For example, in the sentence “… new diagnoses of prostate cancer…”, the phrase “prostate cancer” is tagged as “B-disease I-disease”. The tagging results are then used to generate the lexicon embedding Vlex accordingly.
Figure 3:
Lexicon embedding and external tagging embedding generation.
The external tagging embedding is generated similarly as the lexicon embedding as shown in Figure 3(b). In place of the clinical vocabulary, we use the clinical NLP engine (which leverages a syntactic parser and other clinical ontologies). The same sentence/input for the lexicon embedding is processed by the engine to generate another set of tagging results. Then the external tagging embedding Vtag is generated based on the tagging results. These extra embeddings essentially serve as two additional avenues to enhance the knowledge encoding of the neural network architecture.
We integrate Vlex and Vtag into the LSTM-CRF model (Figure 2) in two ways: (1) before the Bi-directional LSTM layer, by concatenating them with word embedding and character embedding, which results in a concatenated vector [Vword; Vchar; Vlex; Vtag] and acts as the input for the Bi-directional LSTM layer, and (2) after the Bi-directional LSTM layer: by concatenating them with the output from the Bi-directional LSTM layer, which generates a concatenated vector of and acts as the input for the final CRF layer.
Experimental Setup
We implement our proposed models using TensorFlow (https://www.tensorflow.org/) and conduct extensive experiments to assess their effectiveness for disease name recognition. Below we discuss the experimental setup.
Word Embeddings: We use the publicly available GloVe (https://nlp.stanford.edu/projects/glove/) 300-dimensional embeddings trained on 6 billion tokens from a large corpus of text19.
Character Embeddings: Character embeddings are initialized randomly for both charCNN and charLSTM. We set the embedding dimension size to 100. charLSTM’s state size is set to 100. For charCNN, we set 7 filters with dimensions of: {25,50, 75,100,100,100,100} and with window sizes of {l, 2, 3,4,5, 6, 7}. The maximum word length (number of characters) is set to 40; padding with a constant value of –1 is used to generate uniform inputs.
Generic Architecture: The hidden unit size of bi-directional LSTM in the generic neural network architecture is set to 300. To mitigate overfitting, we apply a dropout20 of 0.3 before the bi-directional LSTM layer.
Optimization: Parameter optimization is performed with minibatch stochastic gradient descent (SGD) with the batch size of20. We use Adam optimizer21 with the learning rate initially set to 0.001. The learning rate is decayed in every epoch using an exponential rate of 0.95. We use early stopping22 if the performance (F1 value) on the validation set does not improve for more than 5 training epochs.
Evaluation Results and Discussion
Metrics and Study Design: We evaluate the performance of our proposed models and compare with various methods from the existing work using accuracy, precision, recall, and F1-score on the test set. Accuracy is computed as the exact matching at the word level. Precision, recall and F1-score are computed at the phrase (i.e. concept) level. We conduct experiments for evaluation of our proposed models based on the following aspects:
(1) Character embeddings: we analyze the importance of various kinds of character embeddings for disease named entity recognition. For this purpose, we compare the performance of charCNN, charLSTM, and charMIX.
(2) Domain knowledge: we conduct a thorough ablation study to verify the contribution of lexicon embedding and external tagging embedding in different layers of the network.
The following subsections present detailed results and analyses based on the aforementioned aspects.
Variation of Character Embeddings: The test set results for using different types of character embeddings are shown in Table 2. We can see that the use of various character embeddings can significantly (p < 0.05) improve the model performance over the model that does not leverage character embeddings (LSTM-CRF-NOchar). The use of charCNN yields slightly better scores than using charLSTM. Ultimately, the concatenation of charCNN and charLSTM improves the overall performance as LSTM-CRF-charMIX achieves the best F1 value of 0.838. So, we use LSTM-CRF-charMIX as our strong baseline model for the domain knowledge experiments reported in the next section.
Table 2:
Results for using different character embeddings in LSTM-CRF (test set).
| Accuracy | Precision | Recall | F1 | |
|---|---|---|---|---|
| LSTM-CRF-NOchar | 0.962 | 0.755 | 0.583 | 0.658 |
| LSTM-CRF-charCNN | 0.978 | 0.841 | 0.811 | 0.826 |
| LSTM-CRF-charLSTM | 0.979 | 0.830 | 0.808 | 0.819 |
| LSTM-CRF-charMIX | 0.980 | 0.852 | 0.824 | 0.838 |
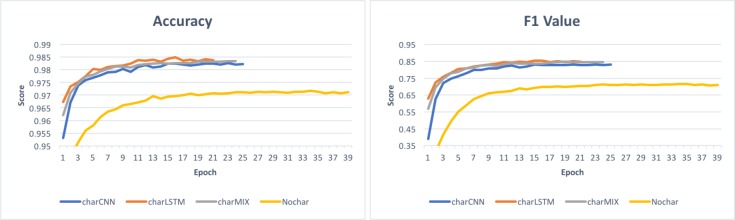
In Figure 4, we show the trend of accuracy and F1 scores over epochs on the validation set during the training process. We can observe that the models with character embeddings converge faster than the model with no character embeddings by requiring less number of epochs for training. We also notice that both accuracy and F1 scores are higher when character embeddings are used in the LSTM-CRF model.
Figure 4:
Accuracy and F1 scores on validation set during training.
Impact of Domain Knowledge: Table 3 shows the results of our model (LSTM-CRF-charMIX) in comparison with various other baselines and existing state-of-the-art systems reported in the literature. The ablation study results are also presented to outline the contribution of each fragment of introducing the domain knowledge into various layers of the LSTM-CRF model.
Table 3:
Comparative performance measures.
| Accuracy | Precision | Recall | F1 | |
|---|---|---|---|---|
| Our simple baselines | ||||
| Vocabulary only (MEDIC) | 0.939 | 0.347 | 0.517 | 0.415 |
| Clinical NLP engine10 | 0.922 | 0.316 | 0.458 | 0.374 |
| Existing systems | ||||
| Sahu and Anand (2016)3 | - | 0.849 | 0.741 | 0.791 |
| Doĝan and Lu (2012)23 | - | - | - | 0.818 |
| Habibi et al. (2017)16 | - | 0.853 | 0.836 | 0.844 |
| Zhao et al. (2017)2 | - | 0.851 | 0.853 | 0.852 |
| Our strong baseline | ||||
| LSTM-CRF-charMIX | 0.980 | 0.852 | 0.824 | 0.838 |
| Our strong baseline + lexicon embeddings | ||||
| before LSTM layer | 0.980 | 0.857 | 0.842 | 0.849 |
| after LSTM layer | 0.980 | 0.834 | 0.837 | 0.836 |
| both | 0.980 | 0.856 | 0.849 | 0.852 |
| Our strong baseline + external tagging embeddings | ||||
| before LSTM layer | 0.980 | 0.823 | 0.825 | 0.824 |
| after LSTM layer | 0.981 | 0.841 | 0.827 | 0.834 |
| both | 0.981 | 0.843 | 0.842 | 0.843 |
| Our strong baseline + both embeddings | ||||
| before LSTM layer | 0.980 | 0.847 | 0.834 | 0.841 |
| after LSTM layer | 0.980 | 0.836 | 0.834 | 0.835 |
| both | 0.981 | 0.868 | 0.839 | 0.853 |
We use vocabulary matching (MEDIC) and the clinical NLP engine as mentioned previously to generate tagging results as our simple baselines. We can see that their scores are relatively lower than the LSTM-CRF based models. Our LSTM-CRF-charMIX model (shown in Table 2) is also presented here as another baseline as it does not encode any additional embeddings (i.e. lexicon and external tagging). The models presented in the later rows demonstrate the contribution of lexicon embedding and extra tagging embedding in different layers of the network. As shown in the results, the use of both lexicon and external tagging embeddings enable the models to have better F1 scores than the baseline LSTM-CRF-charMIX model, which confirms the effectiveness of domain knowledge in our proposed neural network architecture.
We also compare our models with various existing state-of-the-art systems on the NCBI dataset.3 used CNN based character embeddings induced in a RNN architecture for disease name recognition, which achieved the F1 score of 0.791. Their model structure is similar to our proposed model structure, however, they did not have an integrated character embedding approach using both CNN and LSTM. Moreover, they did not use any domain knowledge, which further justifies the better performance of our models.
Doğan and Lu (2012)23 reported a F1 score of 0.818 on the NCBI dataset using BANNER system24, which is based on conditional random fields with hand-crafted features. By contrast, neural network-based methods do not use handcrafted features, and our proposed domain knowledge-enhanced models only require minimal efforts to generate extra lexicon and external tagging embeddings.
Habibi et al. (2017)16 used a similar LSTM-CRF model architecture as we discussed in Figure 2 (with charLSTM) with different data preprocessing and parameters settings yielding the F1 value of 0.844. By contrast, our models with combined charMIX and encoded domain knowledge improve this model by a considerable margin.
Zhao et al. (2017)2 used CNN based character embeddings with stacked convolutional layers for disease name recognition, and also leveraged lexicon features. Compared to their method, we use additional external tagging embeddings and the generic LSTM-CRF model architecture to achieve better precision and F1 scores.
For our ablation study, we encode lexicon embeddings or external tagging embeddings before or after the bi-directional LSTM layer in the generic architecture. For both lexicon embeddings and external tagging embeddings, adding the embeddings to both before and after the LSTM layer yields better results than just adding to either before or after the LSTM layer. Adding lexicon embeddings before the LSTM layer has better performance than adding to after the LSTM layer, while adding external tagging embeddings after the LSTM layer has better performance than adding to before the LSTM layer. Since lexicon embedding is more like the basic features for training the model, it has better performance of adding it before the LSTM layer, however, external tagging embeddings has better performance when added after the LSTM layer.
We further encode both lexicon and external tagging embeddings, and our best model with addition of both embeddings to both before and after the LSTM layer has the best accuracy of 0.981, precision of 0.868, and F1 value of 0.853, establishing the new state-of-the-art performance compared with existing methods on the NCBI dataset. All improvements (F1 values) of our best model are computed to be statistically significant (p < 0.05) with respect to our baselines.
In Figure 5, we compare the labels from lexicon and external tagging with gold standard labels at the word level using the normalized confusion matrix. The y-axis represents gold standard labels, and the x-axis represents lexicon labels (the confusion matrix on the left) or external tagging labels (the confusion matrix on the right). In the left matrix, 0.58 denotes the fraction of “B-disease” in gold labels are correctly labeled as “B-disease” in lexicon labels; 0.31 denotes the fraction of “I-disease” in gold labels are correctly labeled as “I-disease” in lexicon labels; 0.96 denotes the fraction of “O” in gold labels are correctly labeled as “O” in lexicon labels. In the right matrix, 0.47 denotes the fraction of “B-disease” in gold labels are correctly labeled as “B-disease” in external tagging labels; 0.4 denotes the fraction of “I-disease” in gold labels are correctly labeled as “I-disease” in external tagging labels; 0.96 denotes the fraction of “O” in gold labels are correctly labeled as “O” in external tagging labels. In terms of “B-disease” labels, lexicon labels are better than external tagging labels.
Figure 5:
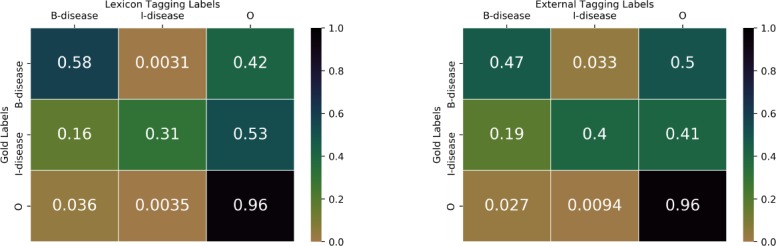
Normalized confusion matrix of gold labels with lexicon labels and external tagging labels.
Similar to Figure 5, in Figure 6 we compare gold standard labels with predicted results from our two models: one is the LSTM-CRF-charMIX model without domain knowledge, and the other is our best model using domain knowledge (lexicon embeddings and external tagging embeddings). In the left matrix, 0.84 denotes the fraction of “B-disease” in gold labels are correctly predicted as “B-disease”; 0.84 denotes the fraction of “I-disease” in gold labels are correctly predicted as “I-disease”; 0.99 denotes the fraction of “O” in gold labels are correctly predicted as “O”. In the right matrix, 0.84 denotes the fraction of “B-disease” in gold labels are correctly predicted as “B-disease”;0.85 denotes the fraction of “I-disease” in gold labels are correctly predicted as “I-disease”; 0.99 denotes the fraction of “O” in gold labels are correctly predicted as “O”. In terms of “I-disease” labels, the model with domain knowledge is better than the model without using domain knowledge.
Figure 6:
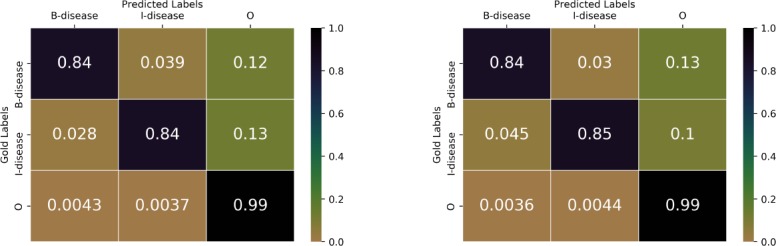
Normalized confusion matrix of gold labels with predicted labels (Left: model without using domain knowledge; Right: Model using domain knowledge).
Related Work
Our work has been motivated by the recent success of deep learning models that effectively incorporate external domain knowledge for various biomedical NLP tasks25,26. CNNs have been successfully applied to a variety of biomedical NLP tasks in the literature as well as for named entity recognition. For example, CNNs are successfully used to recognize named entities from biomedical text recently27. Some recent works explore the use of RNN architectures for the task of clinical event detection such as disease, treatment, test, adverse drug event etc. from free text EHR notes28–30.
Bidirectional RNNs are used for the task of biomedical events trigger identification31 and with combination of CNNs to learn disease name recognition models with word- and character-level embedding features3.
Bidirectional LSTMs are used to model relational and contextual similarities between the named entities in biomedical articles to understand meaningful insights towards providing appropriate treatment suggestions32, to extract clinical concepts from EHR reports33, and for named entity recognition from clinical text34,35. Both CNNs and LSTMs have been used in generating character embeddings for a variety of NLP tasks. Hence, we explore an integration of CNN and LSTM for generating character embeddings to improve performance.
Zhao et al. (2017)2 used CNN based character embeddings with stacked convolutional layers for disease name recognition, and also leveraged lexicon features. Compared to their method, we used additional external tagging embeddings and the generic LSTM-CRF model architecture to obtain better scores. The model architecture of Sahu and Anand (2016)3 is similar to our proposed model structure, however, they did not have an integrated character embedding approach using both CNN and LSTM, and they did not use any domain knowledge. Habibi et al.(2017)16 used a similar LSTM-CRF model architecture as we discussed in Figure 2 (with charLSTM) with different data preprocessing and parameters settings. By contrast, our models with combined charMIX and encoded domain knowledge improve this model by a considerable margin.
Conclusion
In this paper, we proposed a domain knowledge-enhanced LSTM-CRF model for disease named entity recognition. Firstly, we proposed charMIX, an approach to generate character embeddings by integrating embeddings from both charCNN and charLSTM. Then we integrated domain knowledge from a vocabulary and a clinical NLP engine into different layers of the LSTM-CRF model architecture. Our experiments show the impact of domain knowledge on the performance of the models when added at different parts of the network. Our proposed models achieved new state-of-the-art results in disease named entity recognition on the NCBI scientific article dataset.
References
- 1.Qikang Wei, Tao Chen, Ruifeng Xu, Yulan He, Lin Gui. Disease named entity recognition by combining conditional random fields and bidirectional recurrent neural networks. Database: The Journal of Biological Databases and Curation. 2016. [DOI] [PMC free article] [PubMed]
- 2.Zhehuan Zhao, Zhihao Yang, Ling Luo, Lei Wang, Yin Zhang, Hongfei Lin, Jian Wang. Disease named entity recognition from biomedical literature using a novel convolutional neural network. BMC medical genomics. 2017;10(5):73. doi: 10.1186/s12920-017-0316-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sunil Kumar Sahu, Ashish Anand. Recurrent neural network models for disease name recognition using domain invariant features. In ACL. 2016.
- 4.Lance A. Ramshaw, Mitchell P. Marcus. Text chunking using transformation-based learning. CoRR. 1995.
- 5.Robert Leaman, Ritu Khare, Zhiyong Lu. Challenges in clinical natural language processing for automated disorder normalization. Journal of biomedical informatics. 2015;57:28–37. doi: 10.1016/j.jbi.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alan R Aronson. Effective mapping of biomedical text to the umls metathesaurus: the metamap program. In Proceedings of the AMIA Symposium; American Medical Informatics Association; 2001. p. 17. [PMC free article] [PubMed] [Google Scholar]
- 7.Savova G.K, Masanz J.J, Ogren P.V, Zheng J, Sohn S, Kipper-Schuler K.C, Chute C.G. Mayo clinical text analysis and knowledge extraction system (ctakes): architecture, component evaluation and applications. Journal of the American Medical Informatics Association. 2010;17(5):507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ritu Khare, Jiao Li, Zhiyong Lu. Labeledin: Cataloging labeled indications for human drugs. Journal of biomedical informatics. 2014;52:448–56. doi: 10.1016/j.jbi.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nigam Shah, Nipun Bhatia, Clement Jonquet, Daniel Rubin, Annie P Chiang, Mark Musen. Comparison of concept recognizers for building the open biomedical annotator. BMC bioinformatics. 2009;10(Suppl 9) doi: 10.1186/1471-2105-10-S9-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vivek V Datla, Sadid A Hasan, Ashequl Qadir, Kathy Lee, Yuan Ling, Joey Liu, Oladimeji Farri. Automated clinical diagnosis: The role of content in various sections of a clinical document. In IEEE International Conference on Bioinformatics and Biomedicine, BIBM; 2017. pp. 1004–1011. [Google Scholar]
- 11.Rezarta Islamaj Doĝan, Robert Leaman, Zhiyong Lu. Ncbi disease corpus: a resource for disease name recognition and concept normalization. Journal of biomedical informatics. 2014;47:1–10. doi: 10.1016/j.jbi.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhiheng Huang, Wei Xu, Kai Yu. Bidirectional lstm-crf models for sequence tagging. arXiv preprint arXiv:1508.01991. 2015.
- 13.Xuezhe Ma, Eduard Hovy. End-to-end sequence labeling via bi-directional lstm-cnns-crf. arXiv preprint arXiv:1603.01354. 2016.
- 14.Guillaume Lample, Miguel Ballesteros, Sandeep Subramanian, Kazuya Kawakami, Chris Dyer. Neural architectures for named entity recognition. arXiv preprint arXiv:1603.01360. 2016.
- 15.Nils Reimers, Iryna Gurevych. Optimal hyperparameters for deep lstm-networks for sequence labeling tasks. arXiv preprint arXiv:1707.06799. 2017.
- 16.Maryam Habibi, Leon Weber, Mariana Neves, David Luis Wiegandt, Ulf Leser. Deep learning with word embeddings improves biomedical named entity recognition. Bioinformatics. 2017;33(14):i37–i48. doi: 10.1093/bioinformatics/btx228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoon Kim, Yacine Jernite, David Sontag, Alexander M Rush. Character-aware neural language models. In AAAI. 2016. pp. 2741–2749.
- 18.Ferenc Bodon, Lajos Rónyai. Trie: an alternative data structure for data mining algorithms. Mathematical and Computer Modelling. 2003;38(7-9):739–751. [Google Scholar]
- 19.Jeffrey Pennington, Richard Socher, Christopher D. Manning. Glove: Global vectors for word representation. In Empirical Methods in Natural Language Processing (EMNLP) 2014. pp. 1532–1543.
- 20.Nitish Srivastava, Geoffrey Hinton, Alex Krizhevsky, Ilya Sutskever, Ruslan Salakhutdinov. Dropout: A simple way to prevent neural networks from overfitting. The Journal of Machine Learning Research. 2014;15(1):1929–1958. [Google Scholar]
- 21.Diederik P Kingma, Jimmy Ba. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980. 2014.
- 22.Rich Caruana, Steve Lawrence, Giles C Lee. In Advances in neural information processing systems. 2001. Overriding in neural nets: Backpropagation, conjugate gradient, and early stopping; pp. 402–408. [Google Scholar]
- 23.Rezarta Islamaj Doĝan, Zhiyong Lu. In Proceedings of the 2012 workshop on biomedical natural language processing. Association for Computational Linguistics; 2012. An improved corpus of disease mentions in pubmed citations; pp. 91–99. [Google Scholar]
- 24.Robert Leaman, Graciela Gonzalez. In Biocomputing 2008. World Scientific; 2008. Banner: an executable survey of advances in biomedical named entity recognition; pp. 652–663. [PubMed] [Google Scholar]
- 25.Yuan Ling, Yuan An, Sadid A. Hasan. In Proceedings of the 1st Workshop on Sense, Concept and Entity Representations and their Applications. Association for Computational Linguistics; 2017. Improving clinical diagnosis inference through integration of structured and unstructured knowledge; pp. 31–36. [Google Scholar]
- 26.Yuan Ling, Yuan An, Mengwen Liu, Sadid A. Hasan, Ye-tian Fan, Xiaohua Hu. Integrating extra knowledge into word embedding models for biomedical NLP tasks. In 2017 International Joint Conference on Neural Networks, IJCNN2017, Anchorage, AK, USA, May 14-19, 2017; 2017. pp. 968–975. [Google Scholar]
- 27.Gamal Crichton, Sampo Pyysalo, Billy Chiu, Anna Korhonen. A neural network multi-task learning approach to biomedical named entity recognition. BMC bioinformatics. 2017;18(1):368. doi: 10.1186/s12859-017-1776-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abhyuday N Jagannatha, Hong Yu. Bidirectional rnn for medical event detection in electronic health records. In Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies; 2016. pp. 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abhyuday Jagannatha, Hong Yu. In EMNLP. 2016. Structured prediction models for RNN based sequence labeling in clinical text; pp. 856–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adyasha Maharana, Meliha Yetisgen. In BioNLP. 2017. Clinical Event Detection with Hybrid Neural Architecture; pp. 351–355. [Google Scholar]
- 31.Rahul V S S Patchigolla, Sunil Sahu, Ashish Anand. In BioNLP. 2017. Biomedical event trigger identification using bidirectional recurrent neural network based models; pp. 316–321. [Google Scholar]
- 32.Hua He, Kris Ganjam, Navendu Jain, Jessica Lundin, Ryen White, Jimmy Lin. An insight extraction system on biomedical literature with deep neural networks. In Proceedings of the 2017 Conference on Empirical Methods in Natural Language Processing, EMNLP 2017; Copenhagen, Denmark: 2017 2017. Sep 9-11, pp. 2691–2701. [Google Scholar]
- 33.Raghavendra Chalapathy, Ehsan Zare Borzeshi, Massimo Piccardi. In ClinicalNLP@COLING. 2016 2016. Bidirectional LSTM-CRF for clinical concept extraction; pp. 7–12. [Google Scholar]
- 34.Inigo Jauregi Unanue, Ehsan Zare Borzeshi, Massimo Piccardi. Recurrent neural networks with specialized word embeddings for health-domain named-entity recognition. Journal of Biomedical Informatics. 2017;76:102–109. doi: 10.1016/j.jbi.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Zengjian Liu, Ming Yang, Xiaolong Wang, Qingcai Chen, Buzhou Tang, Zhe Wang, Hua Xu. Entity recognition from clinical texts via recurrent neural network. BMC Medical Informatics and Decision Making. 2017;17(2) doi: 10.1186/s12911-017-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]