Abstract
Developing high-throughput and high-performance phenotyping algorithms is critical to the secondary use of electronic health records for clinical research. Supervised machine learning-based methods have shown good performance, but often require large annotated datasets that are costly to build. Simulation studies have shown that active learning (AL) could reduce the number of annotated samples while improving the model performance when assuming that the time of labeling each sample is the same (i.e., cost-insensitive). In this study, we proposed a cost- sensitive AL (CostAL) algorithm for clinical phenotyping, using the identification of breast cancer patients as a use case. CostAL implements a linear regression model to estimate the actual time required for annotating each individual sample. We recruited two annotators to manual review medical records of 766 potential breast cancer patients and recorded the actual time of annotating each sample. We then compared CostAL, AL, and passive learning (PL, aka random sampling) using this annotated dataset and generated learning curves for each method. Our experimental results showed that CostAL achieved the highest area under the curve (AUC) score among the three algorithms (PL, AL, and CostAL are 0.784, 0.8501, and 0.8673 for user 1 and 0.8006, 0.8806 and 0.9006 for user 2). To achieve an accuracy of 0.94, AL and CostAL could save 36% and 60% annotation time for user 1 and 53% and 70% annotation time for user 2, when they were compared with PL, indicating the value of cost-sensitive AL approaches.
Introduction
In the past decade, data from electronic health records (EHR) systems has become a source for clinical and translational research1. The potential for mining this data to support evidence-based practice and population health initiatives is immense. However, one of the challenges of harvesting the information within the EHR is efficiently and accurately identifying patients with a specific phenotype (e.g., disease or drug exposure) from the heterogeneous clinical data, including coded data and clinical narratives. Manual review of patients’ medical records by domain experts, although accurate, is costly and time-consuming. Physical review is also not scalable given the large number of medical records involved2–4. Therefore, developing high-throughput and high-performance phenotyping algorithms is of great interest to the clinical and informatics communities1.
Many efforts have devoted to EHR phenotyping tasks, including knowledge base development and rule-based phenotyping algorithms5. Recently, a number of studies applied supervised machine learning (ML) algorithms to high-throughput phenotyping tasks2,4,6–8. Wei et al.4 developed support vector machine (SVM) classifiers to identify type 2 diabetes using extracted concepts, achieving F-scores over 0.95. Carroll et al.2 also applied SVM classifiers to identify rheumatoid arthritis (RA) cases using billing codes, medication exposures, and extracted concepts, achieving a precision of 0.94 and a recall of 0.87. Some studies applied logistic regression to identify rheumatoid arthritis (RA)6, Crohn’s disease (CD) and ulcerative colitis (UC)7 patients using coded data and clinical narratives, achieving accuracy over 0.94. Kawaler el al.8 showed that Naïve Bayes, along with random forests and SVM, are the best learners for predicting post-hospitalization venous thromboembolism (VTE) risk from EHR records. However, to achieve such high performance, these supervised ML-based methods often require a large number of annotated samples. There is a need to develop methods to achieve high-performance phenotyping classification models while minimizing annotation cost, as the time and expertise required to annotate samples may otherwise be prohibitive.
Active learning (AL)9, which actively selects the most informative samples for annotation (compared with random sampling or passive learning - PL), is one way to minimize annotation cost while achieving high-accuracy machine learning models. In seminal work, Chen et al.10 and Dligach et al.11 successfully applied AL to phenotyping tasks. Chen et al.10 integrated an uncertainty-based AL algorithm with SVM-based phenotyping classifiers and showed that AL could reduce the number of annotated samples required to achieve an area under the curve (AUC) score of 0.95 by up to 68%. Dligach et al.11 employed an uncertainty sampling AL method with Naïve Bayes-based phenotyping methods and demonstrated that AL could achieve a two-thirds reduction in the amount of manual labeling, as compared to its passive counterpart. Both of these studies showed that AL reduces the number of annotated samples in simulation studies, which is promising. However, this type of simulation assumes that the cost of labeling each sample is the same in terms of time, which does not hold in real-world annotation tasks.
A number of studies12–15 have shown that in order to reduce the overall burden of annotating a corpus, AL algorithms need to consider annotation cost (i.e., time) in the model. Settles et al.13 conducted an extensive empirical study of AL with annotation costs in four open domain tasks, including named entity recognition (NER), relation extraction, sentence classification and image retrieval. They concluded that AL without a cost model might perform no better than random sampling when evaluated for time saved rather than a reduction in the number of annotated examples, but that improved learning curves could be achieved if AL took the variable costs of annotation into account appropriately. Haertel et al.14 proposed a cost-sensitive heuristic Return On Investment (ROI) based active learner and showed a 73% reduction in hourly cost over random sampling on a part-of-speech (POS) tagging task. Tomanek and Hahn12 proposed three different methods to incorporate costs into AL and revealed that cost-sensitive AL could reduce up to 54% of annotation time compared to random sampling on an NER task. In the medical domain, Chen et al.16 also showed that simple uncertainty-based AL algorithms do not guarantee a reduction of annotation time for different users in a clinical NER task, and argued that if the querying algorithm took into account the real annotation time, the AL algorithms could perform better.
Despite promising work on cost-sensitive AL algorithms for NLP tasks such as NER and POS tagging, no existing work has investigated the cost model for AL in the context of a clinical phenotyping task. This task is very different from above NLP tasks – annotators make decisions at patient-level rather than sentence-level. They have larger amounts of material to review and may choose to review only certain segments of it. Consequently, development of a cost model for this task requires understanding how experts review chart to determine phenotypes from EHRs when building the actual cost model. As a preliminary study, here we proposed to develop cost-sensitive AL algorithms for clinical phenotyping, using the identification of breast cancer patients as a use case.
Methods
Overview
In this study, we used an existing dataset containing 766 potential breast cancer patients, each of them was labeled with its breast cancer status (yes vs. no) by a previous study17. The dataset was annotated again by two recruited annotators so that the actual annotation time of each sample was recorded and a cost model was trained on a subset of the newly-annotated data for each annotator. We then conducted simulation experiments following the pool-based active learning framework18 and compared three querying algorithms: passive learning (PL, aka random sampling), active learning without a cost model (AL), and active learning with our proposed cost model (CostAL) by generating learning curves (accuracy against time or number of samples) and reporting area under the curve scores.
Dataset
We used an existing dataset for which gold standard decisions regarding the presence or absence of breast cancer were previously determined17. This dataset was composed of clinical records for 766 female patients, of which 492 patients (~64%) have had, or currently have breast cancer. Each patient’s clinical records contain the patient’s coded data (i.e., diagnostic codes, medication orders, procedure orders and lab tests) and clinical notes. For clinical phenotyping, several studies3,19–21 shown that coded data such as the International Classification of Disease (ICD) codes were not sufficient or accurate enough. Therefore, many studies2,10,11 also involved a set of Unified Medical Language System (UMLS)22 concept identifiers (CUIs) extracted from patients’ clinical notes using NLP tools. In this study, we used both the coded data and clinical notes of each patient’s clinical records for clinical phenotyping. We randomly separated these 766 samples into four parts for different purposes as shown in Table 1. More specifically, the first part (66 patients) was used to train the annotators to get familiar with the task and the annotation system (described in the next subsections). The second part (250 patients) was used for collecting the actual annotation time of each sample to train the cost model for each annotator. The third part (250 patients) was used for evaluating the cost model and also used as the pool for different querying algorithms. Note that, this part of data was seen as labeled data if it was used for evaluating the cost model and unlabeled data if it was used as the algorithm pool. The last part (200 patients) was used for evaluating the phenotyping algorithms with different querying strategies. The size and distribution of each part are shown in the table, which illustrates a similar distribution of positive and negative examples across the partitions.
Table 1.
Statistics of the dataset used in this study.
| Part | #Positive Samples (Percentage) | #Negative Samples (Percentage) | Total | Purpose | Annotation Phase |
|---|---|---|---|---|---|
| 1 | 43 (65.2%) | 23 (34.8%) | 66 | Annotator Training | 1st Phase |
| 2 | 158 (63.2%) | 92 (36.8%) | 250 | Cost Model Training | 2nd Phase |
| 3 | 169 (67.6%) | 81 (32.4%) | 250 | Cost Model Evaluation Algorithm Pool | |
| 4 | 122 (61%) | 78 (39%) | 200 | Evaluation Set | --- |
| Total | 492 (64.2%) | 274 (35.8%) | 766 | --- | --- |
Annotation
Since most of the active learning research for phenotyping10,11 did not consider the real annotation cost (i.e., annotation time or labeling time), there was no available dataset that recorded the annotation cost of each example Therefore, we recruited two annotators with medical background to annotate the above dataset and recorded the time spent annotating each sample. The TURF software23 was used to record annotators’ keystrokes and mouse clicks. Within the annotation system and TURF software, we recorded the time of each annotator spend reviewing each patient’s clinical records to make their decision.
Annotation Interface
We developed a web-based annotation system for the annotators to manually review the clinical records of each patient. Figure 1 shows a screenshot of the home screen, which provided a patient list including patients’ age, gender, and other statistical information. By clicking on any line, the annotator entered the patient’s clinical records. Figure 2 shows a screenshot of the annotation system showing part of the patient’s clinical records. The interface had limited interactivity allowing the annotator to expand/contract details regarding the major components of the records. Each segment of the records (i.e. Diagnostic codes, Medications, Procedures, Labs, and Notes) was marked to indicate the number of available records. A filter/search function was available for each segment within the records. Thus, annotators had the capacity to explore the records to identify those aspects of it most pertinent to the classification task.
Figure 1.
Home screen of the annotation system showing a patient list.
Figure 2.
The annotation system showing part of a patient’s clinical records.
Annotation Decision and Confidence
Annotators were tasked with making a binary decision as to whether or not each patient on their list (1) currently has breast cancer or has had a history of breast cancer or (2) the patient does not have evidence of breast cancer at any point current or past. Additionally, for each decision, they were asked to indicate their confidence level in their determination. We defined the confidence levels from 1 to 5, which denotes “very low confidence”, “low confidence”, “average confidence”, “high confidence” and “very high confidence”, respectively.
Annotation Phase
Two phases were taken to carry out the annotation process as shown in Table 1. In the first phase, 66 samples were used to train the annotators to get familiar with the task and the annotation system. During this phase, each annotator was provided with annotation guidelines and feedback on their annotations of the 66 samples. Annotators and researchers discussed their decisions between the two annotators, and the annotators were provided with protocols to support adherence to the guidelines. In the second phase, 500 samples were divided across 10 sessions (50 samples one session) to allow for regular periods of rest. In total, we obtained the real annotation cost (i.e., annotation time or labeling time) of 500 samples.
ML-based Phenotyping Method
We treated phenotyping as a classification task - to predict whether a patient has a medical condition given the patient’s clinical records. In this study, we used a logistic regression model provided by LIBLINEAR24 to build a classifier to predict the probability of a patient having a specific medical condition. In the logistic regression model, we compute the probability with , where is a vector of features extracted from the patient’s clinical records, y is a binary variable representing whether or not a patient x has a specific medical condition, is a vector of weights associated with the features, and c is a constant. For each patient x, we extracted all the diagnostic codes (ICD codes), medications, procedure orders (CPT billing codes), lab tests, and UMLS CUIs extracted from clinical notes, resulting in 12,225 features in total.
AL and Cost-sensitive AL for ML-based phenotyping
AL9 applies a querying algorithm to actively select the most informative samples for annotation. For example, uncertainty sampling selects the lowest confidence samples based on the current model18,25. This type of active learning algorithm assumes that the cost (i.e., annotation time) is identical for different samples (i.e., cost-insensitive) and that reducing the number of annotated examples leads to reduced annotation cost. Although the assumption is partially true, in practice AL algorithms that operate without considering annotation cost do not guarantee reduced annotation time13,16. Cost-sensitive active learning (CostAL) algorithms 12,13 consider differences in annotation cost between different samples, by building models to estimate the cost for each sample. In this study, we employed a pool-based AL framework18, which is shown in Figure 3. The querying algorithm in line 6 is the key component and is implemented using the following three different strategies:
PL: We used random sampling as the querying algorithm.
AL: We employed the uncertainty sampling algorithm18,25 as the querying algorithm, which is to query the sample whose predicted label from the classifier is the least confident (aka the most informative): , where is the information content of an unlabeled example x and is the prediction with the highest posterior probability given the current classifier.
CostAL: We queried the sample by balancing informativeness against annotation cost, to maximize the informativeness/cost ratio, or return on investment (ROI)13,14: where info(x) is the information content of an unlabeled example x, and cost(x) is the annotation cost when acquiring a label for the unlabeled example x. We developed a linear regression model, which was trained using the collected dataset with real annotation time (i.e., the 2nd part of the dataset as shown in Table 1), to predict the annotation cost on the unlabeled samples (i.e., the 3rd part of the dataset as shown in Table 1) as , where is a vector of features extracted from the patient’s clinical records, is a vector of weights associated with the features, and c’ is a constant. We extracted 6 features to represent each patient x, including features about the complexity of patient’s records: the number of (1) diagnostic codes (ICD codes), (2) medication orders, (3) procedure orders, (4) lab tests, and (5) clinical notes; and (6) whether a record contains obvious evidence about breast cancer (i.e., a set of ICD codes concerning breast cancer identified by the two annotators).
Figure 3.
A pool-based active learning algorithm.
Evaluation
We measured the accuracy of each annotator in both the two annotation phases. Higher accuracy indicates the high quality of the annotation. We reported both Root Mean Square Error (RMSE) and coefficient of determination (R2) of the linear regression-based cost model26 on the training data. RMSE is used to measure the difference between values predicted by a model (i.e., the predicted time by the learned cost model) and the values actually observed (i.e., the real annotation time). R2 is a statistical measure of how well the regression model approximates the data. An R2 of 1 indicates that the regression model perfectly fits the data. Since phenotyping is treated as a classification task, we used accuracy to evaluate the performance of different phenotyping algorithms. We generated learning curves by plotting the accuracy of a classifier on the test data (i.e., the 4th part of the dataset as shown in Table 1) as a function of the number of the annotated samples or the time spent on the annotated samples. The area under the learning curve (AUC) score was used as the primary measure to compare different learning curves of PL, AL, and CostAL. More specifically, we repeated PL with random sampling 10 times and plot the averaged learning curve with variation, and we also computed its averaged AUC score with variation.
Results
Annotation Accuracy
In the first phase of annotation, which was to train the annotators with 66 samples to get familiar with the task and the annotation system, the accuracy was 98% (65/66) for user 1 and 100% (66/66) for user 2. In the second phase of annotation, which was to collect real annotation cost of 500 samples, the accuracy was 99% (496/500) for both user 1 and 2. The high annotation accuracies indicated that the two annotators were well trained and fit for the task.
Statistics of the Annotation Confidence
Table 2 shows the statistics of the annotation confidence across the 500 samples for each annotator. From the table, we can find that (1) most of the samples can be decided with very high confidence, and (2) most of the samples with confidence level less than 5 fall into the negative samples.
Table 2.
Statistics of the annotation confidence across the 500 samples for each annotator.
| Confidence Level | #Positive Samples | #Negative Samples | Total | |
|---|---|---|---|---|
| User 1 | 1 | 0 | 0 | 0 |
| 2 | 0 | 1 | 1 | |
| 3 | 0 | 5 | 5 | |
| 4 | 1 | 27 | 28 | |
| 5 | 326 | 140 | 466 | |
| User 2 | 1 | 0 | 0 | 0 |
| 2 | 0 | 2 | 2 | |
| 3 | 0 | 2 | 2 | |
| 4 | 0 | 6 | 6 | |
| 5 | 327 | 163 | 490 |
Statistics of the Annotation Time
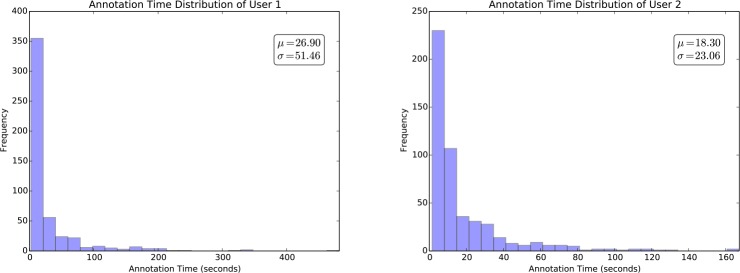
Figure 4 shows the distribution of the annotation time across the 500 samples for each annotator. The mean and standard deviation were 26.90 and 51.46 for user 1 and 18.30 and 23.06 for user 2, respectively. User 1 had much higher variance than user 2, which showed the difference between the two annotators. The variance of annotation time for each annotator among different samples shows that it is necessary to consider the real annotation cost of each sample in order to truly reduce the time for annotating a corpus when applying active learning algorithms to phenotyping.
Figure 4.
Distribution of the annotation time across the 500 samples for each annotator.
To better understand what kind of samples were more time-consuming or effort-intensive for annotation, we further made statistics to the samples that took much more time than the average. For user 1, there were totally 128 samples that took much more time than 26.90 seconds. Among them, 23 samples were positive and 105 samples were negative. For user 2, there were totally 143 samples that took much more time than 18.30 seconds. Among them, 55 samples were positive and 88 samples were negative. These statistics showed that negative samples were more time- consuming for annotation.
Accuracy of the Cost Model
The RMSE of the learned linear regression-based cost models from two annotators are 16.97 and 20.04 respectively. The R2 scores are 0.3204 and 0.1389 respectively. The lower RMSE and higher R2 of the first user indicates that the proposed cost model (i.e., selected features) fits much better on data for user 1 than that for user 2.
Effectiveness of the Cost Model for Active Learning
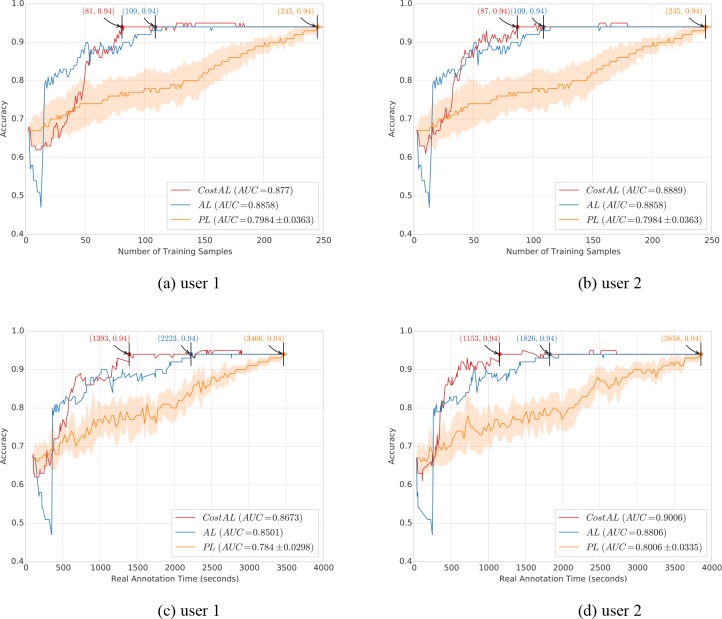
Figure 5 illustrates the learning curves and AUC scores of different query algorithms. More specifically, the yellow curve is the averaged learning curve for PL by repeating random sampling 10 times, and the light-yellow area shows the variation of each point in the averaged learning curve. Similarly, the AUC score for PL is the averaged AUC score by repeating random sampling 10 times, and the symbol ± indicates the variation. Figure 5 (a) and (b) show the learning curves between accuracy and the number of annotated training samples for user 1 and 2 respectively.
Figure 5.
Learning curves (in terms of accuracy vs. number of annotated samples or time of annotated samples) and AUC scores (shown in the legend of each subfigure) for different query algorithms. CostAL: Cost-sensitive Active Learning; AL: Active Learning without cost model; PL: Passive Learning. (a)(b) Learning curves between accuracy and the number of annotated training samples for user 1 and 2 respectively. (c)(d) Learning curves between accuracy and the annotation time spent on the annotated training samples for user 1 and 2 respectively. (a)(c) CostAL using the cost model trained with user 1’s annotation time. (b)(d) CostAL using the cost model trained with user 2’s annotation time.
For AL and PL, the learning curves are the same for two users because they did not consider actual time. According to the learning curves, PL achieved an accuracy of 0.94 by training on 245 annotated samples, while AL needed 109 annotated samples to achieve the same performance, indicating a 56% reduction. The corresponding AUC scores of AL and PL were 0.8858 and 0.7984 ± 0.0363 respectively. When CostAL was used, user 1 needed 81 annotated samples and user 2 needed 87 annotated samples to achieve an accuracy of 0.94, indicating a 67% reduction and a 64% reduction, respectively. The AUC scores were 0.877 and 0.8889 for user 1 and 2.
Figure 5 (c) and (d) show the learning curves between accuracy and the annotation time spent on the annotated training samples. For user 1 (Figure 5 (c)), to achieve an accuracy of 0.94, PL required 3,466 seconds of annotation. However, AL and CostAL required 2,223 and 1,393 seconds of annotation respectively, indicating a 36% and a 60% reduction of time compared to that of PL. Furthermore, CostAL shows a 37% reduction of time compared to AL. The AUC scores of PL, AL, and CostAL were 0.784 ± 0.0298, 0.8501, and 0.8673 respectively. Figure 5 (d) shows results from user 2. To achieve an accuracy of 0.94, PL required 3,858 seconds. However, AL and CostAL required 1,826 and 1,153 seconds of annotation respectively, indicating a 53% and 70% reduction of time compared to that of PL. Moreover, CostAL shows a 37% reduction of time compared to AL. The AUC scores of PL, AL, and CostAL were 0.8006 ± 0.0335, 0.8806 and 0.9006, respectively.
When cost is evaluated using the number of annotated samples (Figure 5 (a) and (b)), AUC scores of CostAL were not better than AL. However, when actual annotation time was used in evaluation (Figure 5 (c) and (d)), AUC scores of CostAL outperformed AL by at least 2%. The utility of cost-sensitive AL algorithms is clearly demonstrated when real annotation times are used in the evaluation. Overall, AL performed better than PL, reducing the number of annotated samples by up to 56% and the time by up to 53%. CostAL outperformed PL by reducing the number of annotated samples by up to 67% and the time by up to 70%. Furthermore, CostAL performs better than AL by reducing the number of annotated samples by up to 26% and the time by up to 37%.
Discussion
In this study, we investigated the impact of cost-sensitive AL algorithms for clinical phenotyping, using the identification of breast cancer patients as a use case. CostAL effectively reduced the annotation time that was needed to achieve a high-accuracy model, with reductions of up to 70% as compared with passive learning, which would constitute a substantive saving in clinician resources required to produce an annotated reference set. To the best of our knowledge, this is the first study to apply and evaluate CostAL for ML-based clinical phenotyping.
Since there was no available dataset that recorded the annotation cost of each example for clinical phenotyping, we recruited two annotators with a medical background to re-annotate an existing breast cancer dataset and recorded the time spent annotating each sample. The high accuracy and confidence of the annotation shown that (1) the two annotators were well trained and fit for the task, and the annotation quality was sufficient for our task, and (2) the identification of breast cancer cohort was an easy phenotyping task. Although, we achieved successful results on CostAL for ML-based clinical phenotyping, how much we can generalize findings in this study to phenotyping tasks of other diseases are not assessed yet. Moreover, the sample size of the breast cancer dataset used here is relatively small. In the future, we will expand and evaluate CostAL for other disease cohorts, and use many more samples.
The statistics of the annotation time shown that negative samples were more time-consuming for annotation. In this work, we used both the coded data and clinical notes of each patient for clinical phenotyping, as suggested by previous work2,3,10,11,19–21. If the diagnosis of breast cancer is clearly mentioned in the diagnosis section of the records, it is not hard to determine that the patient currently has breast cancer or has had a history of breast cancer. However, if the diagnosis is not clearly mentioned in the diagnosis section, it doesn’t mean that the patient does not have breast cancer. The annotator still has to read through the other sections of the patient chart to firmly say that the patient does not has breast cancer. Sometimes, the patient chart contains many clinical notes and it takes more time to read through all the notes.
For CostAL, an accurate cost model is important. In this work, we used linear regression with some heuristic features to build the cost model, which is very preliminary. Although the CostAL-based phenotyping was successful in the simulation study, the 𝑅G scores of the cost model suggest room for improvement. In practice, annotation scenarios may be more complicated than the simulation environment. For example, modeling the effects of fatigue over time would require more sophisticated models considering additional features. In addition, we noticed differences between the two users, e.g., annotation speed, which obviously affects annotation time. Currently, our cost models are user-specific, which requires training for each user. It would be advantageous to develop generalizable models that can be user independent, as this would eliminate the need for user-specific training.
From the four subfigures in Figure 5, we noticed that AL and CostAL did not perform well at the early stage when the number of training samples was small. AL was even worse than CostAL at the beginning (i.e., less than 15 training examples or 250 seconds. One possible explanation is that the classification models were not well-trained due to the small training sample size and high feature dimensionality, thus resulting in poor sample selection by the uncertainty sampling algorithms. This could be addressed by better initial sample selection strategies and we will address this issue in the future work. From the subfigures, we also found that AL and CostAL reached an accuracy of 0.94 very quickly and then kept a stable performance, while PL achieved an accuracy of 0.94 very slow. AL and CostAL could save up to 53% and 70% annotation time compared with PL, indicating the value of cost-sensitive AL approaches. Note that, the accuracy of 0.94 in our experiments was the best accuracy the classifiers could achieved on the evaluation set (i.e., the 4th part of the dataset as shown in Table 1) when the classifiers were trained using some or all of the data in the algorithm pool (i.e., the 3rd part of the dataset as shown in Table 1).
At this time, we employed the uncertainty-based querying algorithm for AL and CostAL only. We will further investigate other querying algorithms such as query-by-committee27, density-based sampling28, etc. To fully demonstrate the use of CostAL in real-world annotation tasks, we are also planning to develop a CostAL-enabled annotation system and conduct a user study to compare CostAL, AL, and PL in real-time annotation tasks.
Conclusion
In this study, we applied cost-sensitive active learning for clinical phenotyping, using the identification of breast cancer patients as a use case. Preliminary results showed that CostAL effectively reduced the annotation time compared to its cost-insensitive and passive counterparts.
Acknowledgement
We thank Harish R Siddhanamatha and Alokananda Ghosh for their time and expertise during the annotation of this dataset, and Deevakar Rogith for his help with TURF software. This study is supported by the grant interactive machine learning methods for clinical natural language processing (NLM 2R01LM010681-05).
Figures & Table
References
- 1.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. JAMIA. 2013;20(1):117–121. doi: 10.1136/amiajnl-2012-001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll RJ, Eyler AE, Denny JC. Naive Electronic Health Record phenotype identification for Rheumatoid arthritis. AMIA. 2011;Vol 2011:189–196. [PMC free article] [PubMed] [Google Scholar]
- 3.Xu H, Fu Z, Shah A, et al. Extracting and integrating data from entire electronic health records for detecting colorectal cancer cases. AMIA. 2011;Vol 2011:1564–1572. [PMC free article] [PubMed] [Google Scholar]
- 4.Wei W-Q, Tao C, Jiang G, Chute CG. A high throughput semantic concept frequency based approach for patient identification: a case study using type 2 diabetes mellitus clinical notes. AMIA. 2010;Vol 2010:857. [PMC free article] [PubMed] [Google Scholar]
- 5.Kirby JC, Speltz P, Rasmussen L V, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. JAMIA. 2016;23(6):1046–1052. doi: 10.1093/jamia/ocv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll RJ, Thompson WK, Eyler AE, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. JAMIA. 2012;19(e1):e162–e169. doi: 10.1136/amiajnl-2011-000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ananthakrishnan AN, Cai T, Savova G, et al. Improving case definition of Crohn’s disease and ulcerative colitis in electronic medical records using natural language processing: a novel informatics approach. Inflamm Bowel Dis. 2013;19(7):1411–1420. doi: 10.1097/MIB.0b013e31828133fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawaler E, Cobian A, Peissig P, Cross D, Yale S, Craven M. Learning to predict post-hospitalization VTE risk from EHR data. AMIA. 2012;Vol 2012.:436–445. [PMC free article] [PubMed] [Google Scholar]
- 9.Settles B. Active Learning. Synth Lect Artif Intell Mach Learn. 2012;6(1):1–114. [Google Scholar]
- 10.Chen Y, Carroll RJ, Hinz ERM, et al. Applying active learning to high-throughput phenotyping algorithms for electronic health records data. JAMIA. 2013;20(e2):e253. doi: 10.1136/amiajnl-2013-001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dligach D, Miller T, Savova G. Active learning for phenotyping tasks. Workshop on NLP for Medicine and Biology Associated with RANLP 2013. 2013:1–8. [Google Scholar]
- 12.Tomanek K, Hahn U. A comparison of models for cost-sensitive active learning. COLING. 2010:1247–1255. [Google Scholar]
- 13.Settles B, Craven M, Friedland L. Active Learning with Real Annotation Costs. NIPS Workshop on Cost-Sensitive Learning. 2008:1–10. [Google Scholar]
- 14.Haertel RA, Seppi KD, Ringger EK, Carroll JL. Return on investment for active learning. NIPS Workshop on Cost-Sensitive Learning. 2008;Vol 72 [Google Scholar]
- 15.Donmez P, Carbonell JG. Proactive learning: cost-sensitive active learning with multiple imperfect oracles. CIKM. 2008:619–628. [Google Scholar]
- 16.Chen Y, Lask TA, Mei Q, et al. An active learning-enabled annotation system for clinical named entity recognition. BMC Med Inform Decis Mak. 2017;17(2):82. doi: 10.1186/s12911-017-0466-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joffe E, Pettigrew EJ, Herskovic JR, Bearden CF, Bernstam E V. Expert guided natural language processing using one-class classification. JAMIA. 2015;22(5):962–966. doi: 10.1093/jamia/ocv010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis DD, Gale WA. A sequential algorithm for training text classifiers. SIGIR. 1994:3–12. [Google Scholar]
- 19.Schmiedeskamp M, Harpe S, Polk R, Oinonen M, Pakyz A. Use of international classification of diseases, ninth revision clinical modification codes and medication use data to identify nosocomial clostridium difficile infection. Infect Control Hosp Epidemiol. 2009;30(11):1070–1076. doi: 10.1086/606164. [DOI] [PubMed] [Google Scholar]
- 20.Kern EFO, Maney M, Miller DR, et al. Failure of ICD-9-CM codes to identify patients with comorbid chronic kidney disease in diabetes. Health Serv Res. 2006;41(2):564–580. doi: 10.1111/j.1475-6773.2005.00482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birman-Deych E, Waterman AD, Yan Y, Nilasena DS, Radford MJ, Gage BF. Accuracy of ICD-9-CM codes for identifying cardiovascular and stroke risk factors. Med Care. 2005:480–485. doi: 10.1097/01.mlr.0000160417.39497.a9. [DOI] [PubMed] [Google Scholar]
- 22.Bodenreider O, McCray AT. Exploring semantic groups through visual approaches. JBI. 2003;36(6):414–432. doi: 10.1016/j.jbi.2003.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Walji MF. TURF: Toward a unified framework of EHR usability. JBI. 2011;44(6):1056–1067. doi: 10.1016/j.jbi.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Fan R-E, Chang K-W, Hsieh C-J, Wang X-R, Lin C-J. LIBLINEAR: A library for large linear classification. J Mach Learn Res. 2008;9(Aug):1871–1874. [Google Scholar]
- 25.Schein AI, Ungar LH. Active learning for logistic regression: an evaluation. Mach Learn. 2007;68(3):235–265. [Google Scholar]
- 26.Arora S, Nyberg E, Rosé CP. NAACL HLT 2009 Workshop on Active Learning for Natural Language Processing. Association for Computational Linguistics; 2009. Estimating annotation cost for active learning in a multi-annotator environment; pp. 18–26. [Google Scholar]
- 27.Seung HS, Opper M, Sompolinsky H. Query by committee. The Fifth Annual Workshop on Computational Learning Theory. 1992:287–294. [Google Scholar]
- 28.Settles B, Craven M. An analysis of active learning strategies for sequence labeling tasks. EMNLP. 2008:1070–1079. [Google Scholar]