Abstract
Drug-resistant tuberculosis (TB) remains a public health threat to the United States and worldwide control of TB. Rapid and reliable drug susceptibility testing (DST) is essential for aiding clinicians in selecting an optimal treatment regimen for TB patients and to prevent ongoing transmission. Growth-based DST results for culture-confirmed cases are routinely reported to the U.S. Centers for Disease Control and Prevention through the National TB Surveillance System (NTSS). However, the NTSS currently lacks the capacity and functionality to accept laboratory results from advanced molecular methods that detect mutations associated with drug resistance. The objective of this study is to design and implement novel comprehensive data exchange formats that utilize the Health Level Seven (HL7) version 2.5.1 messaging hierarchy to capture, store, and monitor molecular DST data, thereby, improving the quality of data, specifications and exchange formats within the NTSS as well as ensuring full reporting of drug-resistant TB.
Introduction
Drug-resistant tuberculosis (TB) is a communicable disease that remains a public health threat to the United States and worldwide control of TB1, 2. Rapid and reliable drug susceptibility testing (DST) is essential for aiding clinicians in selecting an optimal treatment regimen for TB patients and to prevent ongoing transmission. DST is crucial for detecting primary or emerging resistance and for monitoring the incidence of drug-resistant TB cases.
Molecular DST methods provide a rapid and robust diagnosis of drug-resistant TB case detection with credible sensitivity and specificity3. For some drugs, molecular DST methods are able to detect genetic mutations associated with lower levels of phenotypic resistance that may be missed by certain growth-based DST methods2, 4. The ability to identify these genetic mutations, therefore, may be clinically “useful in optimizing treatment by switching the more potent drug regimens: an advantage over conventional DST that reports only the results from the critical concentration tested”2 (p. 3).
Growth-based DST results are routinely collected with the Report of Verified Case of Tuberculosis (RCVT) form and reported to the U.S. Centers for Disease Control and Prevention (CDC) through the National TB Surveillance System (NTSS)5, 6. However, the NTSS currently lacks the capacity and functionality to accept electronic laboratory test results from the advanced rapid and robust molecular methods3, 7-10 that detect genetic mutations associated with drug resistance. This limitation prohibits the comprehensive capture of data for drug-resistant cases and impedes accurate reporting11. The objective of this study is to design and implement novel comprehensive data exchange formats that utilize the Health Level Seven (HL7) version 2.5.1 messaging hierarchy to capture, store, and monitor molecular DST data, thereby, improving the quality of data, specifications and exchange formats within the NTSS as well as ensuring full reporting of drug-resistant TB through the revised RVCT to be implemented in 2020.
Methods
The design setting utilized a congruence or similarity of standardization protocols that involved many published implementation guides by HL7 International and the use of reference terminology standards.
Use of Published Implementation Guides
Designing the Division of Tuberculosis Elimination (DTBE) message hierarchy utilized two published reference documents. These included HL7 Version 2.5.1 Implementation Guide: Electronic Laboratory Reporting to Public Health, Release 1 (US Realm) and HL7 Version 2.5.1 Implementation Guide: Electronic Laboratory Reporting to Public Health, Release 2 (US Realm)12, 13. Both guides contain necessary specifications and constraints for reporting laboratory test results to appropriate federal, state, local and territorial health agencies. In particular, these two guides address messaging content and formats related to the transmission of reportable laboratory result messages or ELR using HL7 v2.5.1 ORUˆR01 Unsolicited Observation Message standard12, 13.
In order to support data representation of molecular DST, additional implementation guides, published by the HL7 Clinical Genomics Work Group, were used and adopted for the purpose of this study. The implementation guides are HL7 Version 2 Implementation Guide: Clinical Genomics; Fully LOINC-Qualified Genetic Variation Model, Release 2 (U.S. Realm), and HL7 Version 2 Implementation Guide: Clinical Genomics Coded Reporting – “Lite”, Release 1 (1st DSTU Ballot) – US Realm, Standard for Trial Use, July 201614, 15. These implementation guides are modeled after established laboratory reporting standards and detail how to structure genetic test results into electronic health records (EHRs) utilizing HL7 v2.5.1 messaging standard specifications14, 15. For example, the HL7 Version 2 Implementation Guide: Clinical Genomics; Fully LOINC-Qualified Genetic Variation Model, Release 2 (U.S. Realm) specifically “covers the reporting of genetic test results for sequencing and genotyping based tests where identified DNA variants are located within a gene”14 (p. 2). In the context of molecular DST data representation, the interest is on identified DNA variants located within the TB genome.
Use of Standard Vocabulary and Value Sets
The study also used a set of recommended standard vocabulary and value sets including but not limited to Logical Observation Identifiers Names and Codes (LOINC) and Systematized Nomenclature of Medicine - Clinical Terms (SNOMED CT). Whereas the LOINC codes and associated long common names were used for identifying resulted laboratory tests, the SNOMED CT concept identifiers and associated preferred terms were used for reporting specimen types and microbiology related test results. Use of these recommended standard vocabulary and value sets not only simplifies the complexity of the message components of the molecular genetic data, but also enhances the quality of segments and components of the TB case notification messages generated by the variety of molecular DST platforms14.
Identifying drug resistance-associated genetic mutations in isolates of Mycobacterium tuberculosis complex (MTBC) required the use of a reference terminology known as RxNorm for antituberculosis drug names. The RxNorm system, initially developed by the U.S. National Library of Medicine in November 2004, is a single, multipurpose standard terminology for representing medications or clinical drug components16. More specifically, RxNorm is a standardized nomenclature for clinical drugs that enables storage, retrieval, analysis, and interoperability of clinical data16-19.
Results
The Role of Electronic Laboratory Reporting (ELR) in Public Health Surveillance
Electronic Laboratory Reporting (ELR) plays an integral role in improved communicable disease surveillance by representing and transmitting exchangeable nationally notifiable laboratory data to public health agencies. Participating in ELR allows incoming laboratory data to be translated, processed, and routed to appropriate public health recipients (e.g., Local Health Departments and State Programs) for rapid and immediate public health action20. ELR is, therefore, critical for an effective public health response to nationally notifiable diseases and potential bioterrorism agents21, 22.
Wurtz and Cameron21 defined ELR as the “direct, automated messaging of reportable disease laboratory information from clinical laboratory information management systems (LIMS) directly to the appropriate public health jurisdiction’s communicable disease reporting system” (p. 1639). Specifically, ELR relies solely on the electronic exchange of laboratory data with participating public health agencies. ELR has also been demonstrated to improve the completeness and timeliness of communicable disease surveillance and reduce manual data entry errors21, 23-25.
The Health Information Technology for Economic and Clinical Health (HITECH) Act authorized the Centers for Medicare and Medicaid Services (CMS) to incentivize the nationwide implementation and adoption of ELR for communicable disease surveillance through a program commonly known as Meaningful Use (MU)23, 25, 26. The stages 2 and 3 of the MU criteria provide incentive payments to eligible professionals, eligible hospitals, and critical access hospitals who adopt and successfully demonstrate electronic submission of laboratory data for reportable disease cases to public health departments25. These MU requirements, coupled with incentive payments, have directed many federal and state public health agencies and standards development organizations (SDOs) to create standard specifications for secure systems for electronic data transmission to not only support the MU requirements, but also improve the quality of the electronic exchange of laboratory data with external public health agencies22, 23, 26. As a case in point, CDC has accelerated the use and adoption of ELR by advancing standards for messaging, vocabulary, and data formats; and by conducting an extensive outreach campaign to state and local public health departments22, 25, 27. Moreover, HL7 International, which is an ANSI-accredited SDO, has developed and published many implementation guides for ELR involving the use of HL7 v2 messaging standard.
Designing the DTBE ORUˆRO1 Message Hierarchy
Laboratory information is usually reported through the HL7 Observation Report – Unsolicited (ORU) trigger event R01 (represented as ORUˆR01) message to public health agencies. The ELR message is a constrained ORUˆR01 message for transmitting reportable laboratory results and observations from the testing source to public health agencies12, 13. The ORUˆR01 message is the only acceptable message format or standard to follow for creating ELR messages for MU in order to support electronic data exchange of laboratory results of reportable diseases20, 28, 29.
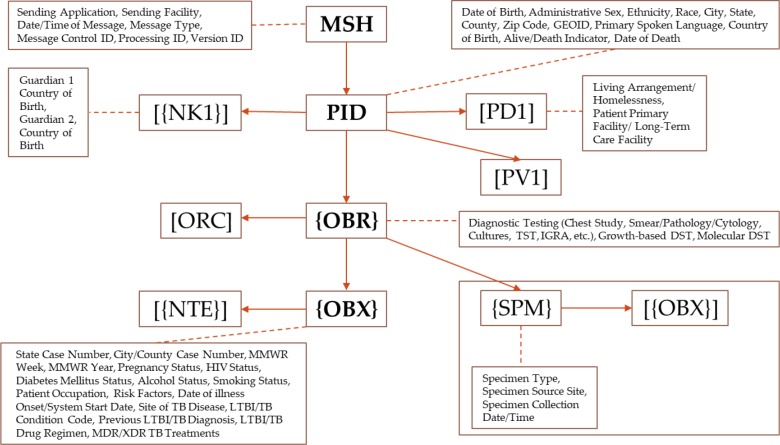
The ORUˆR01 message structure or hierarchy for the HL7 v2.3.1 ELR standard, released in 2005, utilized Message Header (MSH), Patient Identification (PID), Next of Kin (NK1), Common Order (ORC), Observation Request (OBR), Observation Result (OBX), and Notes and Comments (NTE) segments. The release of HL7 v2.5.1 ELR standard in 2010, however, expanded the optionality to include segments such as Software (SFT) and laboratory specimens (SPM). This expansion not only addressed requests from state public health departments and clinical laboratories, but also resolved a conflict with the eXtensible Markup Language (XML) implementations of the HL7 standard12. The Venn diagram (Figure 1) depicts the key differences in supported segments between the HL7 v2.3.1 ELR message and the HL7 v2.5.1 ELR message. Segments displayed without brackets are required (e.g., PID). Segments enclosed in square [] brackets are optional (e.g., [ORC]). Segments enclosed in curly {} brackets are required and may repeat (e.g., {OBR}). Segments enclosed in both square [] and curly {} brackets are optional, but if included these segments may repeat (e.g., [{SPM}])12, 20.
Figure 1.
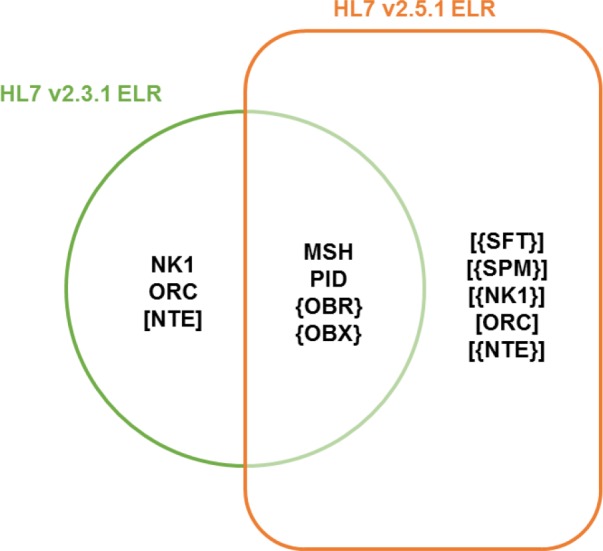
Supported segments between the HL7 v2.3.1 ELR message and the HL7 v2.5.1 ELR message.
Using the HL7 v2.5.1 ELR message specifications as the foundation, the DTBE ORUˆR01 message hierarchy (Figure 2) was developed to support comprehensive data representation, capture, storage and monitoring of all proposed RCVT 2020 data elements that include molecular DST data. Whereas the Additional Demographic (PD1) segment contains demographic information that is likely to change about the patient, the Patient Visit (PV1) segment is used to communicate information about the patient’s visit to institutions and healthcare facilities30.
Figure 2.
DTBE ORUˆR01 Message Hierarchy with key RVCT 2020 Data Elements.
Identification of Molecular DST Data Elements
Molecular DST methods are gaining popularity in TB diagnostics because they offer robust and rapid detection of genetic mutations associated with resistance to antituberculosis drugs1, 31-35. Another contributing factor to their popularity is the rapidly changing laboratory methodology for TB diagnostics and sensitivity testing.
Lin and Desmond3 categorized molecular DST into two types: Probe-based and Sequence-based methods. Whereas Probe-based methods report the presence or absence of mutations in the gene, Sequence-based methods report the identity of specific mutations in the gene1, 3. Examples of sequence-based methods include pyrosequencing, Sanger sequencing, next generation sequencing and whole genome sequencing. Probe-based or non-sequencing methods include Cepheid GeneXpert® MTB/RIF, Hain MTBDRplus and MTBDRsl, and INNO-LiPA RIF3. It is recommended that detected genetic mutations from non-sequencing methods (e.g., Cepheid GeneXpert®) is confirmed with sequence-based methods3, 36. The CDC’s Molecular Detection of Drug Resistance (MDDR) service utilizes the combination of Sanger sequencing and pyrosequencing1, 3, 10, 31. For the purposes of this study, the focus or interest was primarily on two key questions: 1) How should we capture and store Rifampin resistant results from Cepheid GeneXpert® MTB/RIF? and 2) How should we capture and store specific genetic mutations detected using sequence- based assays (e.g., CDC’s MDDR Service)?
Data Representation of Rifampin Resistant Results from Cepheid GeneXpert® MTB/RIF
The Cepheid GeneXpert® MTB/RIF has U.S. Food and Drug Administration (FDA) market authorization for use with raw sputum or sputum sediments37. The Cepheid GeneXpert® MTB/RIF assay utilizes a self-contained, disposable cartridge to provide rapid and robust detection of mutations associated with RIF resistance3, 38. Essentially, the Cepheid GeneXpert® MTB/RIF test is a cartridge-based fully automated nucleic acid amplification test (NAAT) for simultaneous detection of MTBC and mutations associated with rifampin resistance1, 39.
Hence, in the context of the Cepheid GeneXpert® data representation, there are two resulted laboratory reports to be considered: (a) identification of MTBC via NAAT method; and (b) detection of mutation in the Rifampin resistance (rpoB) gene. At its simplest, the majority of coded results for GeneXpert® will fall into three categories: MTBC identification with NAA probe detection (i.e., 38379-4ˆMycobacterium tuberculosis complex DNA [Presence] in Unspecified specimen by NAA with probe detectionˆLN), MTBC detected (i.e., 260373001ˆDetectedˆSCT) or not detected (i.e., 260415000ˆNot detectedˆSCT) as well as whether a mutation in the Rifampin resistance (rpoB) gene has been detected or not been detected. Table 1 depicts the key molecular DST data elements and their associated vocabulary and minimum data representations in HL7 v2.5.1 ELR. The convenience and automation of the Cepheid GeneXpert® assay has the potential to provide rapid access to patient laboratory results38.
Table 1.
Key Cepheid GeneXpert® molecular DST data elements and their associated minimum data representations in HL7 v2.5.1 ELR.
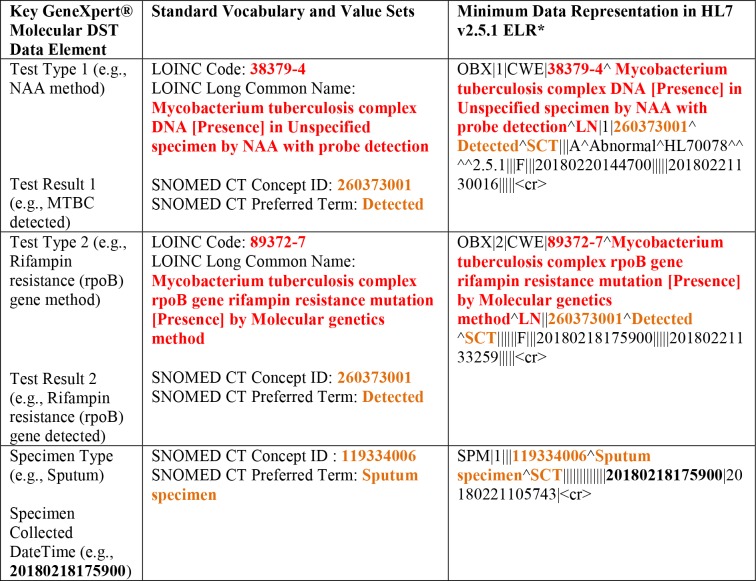
|
See Illustration A for full representation of supported segments.
Data Representation of Specific Genetic Mutations Detected from CDC’s MDDR Service
Since September 2009, the Mycobacteriology Laboratory Branch of the DTBE at CDC has offered Clinical Laboratory Improvement Amendments (CLIA)-compliant molecular diagnostics reference service known as Molecular Detection of Drug Resistance (MDDR)31, 37. The CDC’s MDDR service uses conventional DNA sequencing (Sanger sequencing) for the identification of genetic mutations associated with multidrug-resistant (MDR) and extensively drug-resistant (XDR) TB33, 37, 40. In June 2012, however, the service was expanded by incorporating pyrosequencing into the testing algorithm to more efficiently perform the service and serve as an initial screen for MDR TB31, 32, 40. If mutations are detected by pyrosequencing, then the full panel of Sanger sequencing assay is performed.
The CDC’s MDDR service examines DNA from isolates or NAAT positive sediments at specific targets to determine if mutations known to be associated with resistance are present. Growth-based DST is performed concurrently33, 37. The data representation of molecular DST from the CDC’s MDDR service, therefore, focuses on capturing and storing data elements relating to antituberculosis drug names, TB gene names, amino acid changes, indels, nucleic acid changes, specimen types, test types, and results indicating if genetic mutation is present or not at each relevant target (Table 2). Even though this study used the CDC’s MDDR service as an example, the minimum data representation in HL7 v2.5.1 ELR would be the same for any clinical or public health laboratories performing sequencing-based assays.
Table 2.
Key CDC’s MDDR Service molecular DST data elements and their associated minimum data representations in HL7 v2.5.1 ELR.
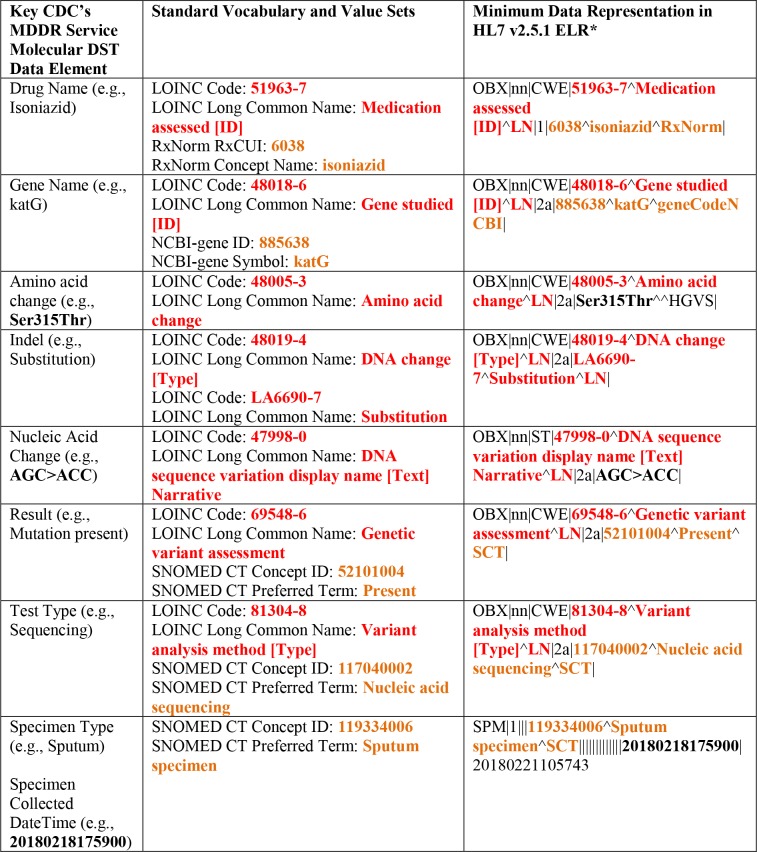
|
See Illustration B for full representation of supported segments.
Discussion
Molecular DST data are challenging and difficult to standardize for ELR. The majority of complicated genetic and molecular DST results are currently reported by clinical and public health laboratories in pure narrative text formats (e.g., Word or PDF format) with no computer accessible coding and processing of the results15, 24. The overarching goal of this study is to make it easier to structure molecular DST results with standardized vocabulary, thereby, enabling the delivery of structured data that could be used in clinical decision support systems and medical record queries to facilitate evidence-based medicine and translational research. Early implementation and adoption of the developed data exchange formats in this study could be relatively simple for clinical and public health laboratories that already use HL7 v2.5.1 ELR15.
The data exchange formats, therefore, would (a) optimize early detection and reporting of drug-resistant TB cases; (b) provide a national standardized protocol for public health and clinical laboratories to use reference terminologies such as LOINC, SNOMED CT, and RxNorm to report molecular DST data to state public health departments that would use the same to report to CDC; (c) provide a high-quality data-driven decision-making process for TB public health administrators; (d) generate high-quality datasets to enhance reporting or analyses of TB surveillance data and drug resistance; and (e) ensure that critical or important clinical information is not hidden in the notes and comments segment of the HL7 v2.5.1 ELR.
The study supports structural and semantic interoperability as it not only uses HL7 v2.5.1 ELR message as the preferred data format for the molecular DST data, but also uses LOINC, RxNorm, and SNOMED CT as the preferred terminologies, respectively, for laboratory tests, antituberculosis drugs, and specimen types. The study also provides a comprehensive informatics platform to continuously, systematically, and seamlessly collect and monitor molecular DST data to support epidemiological studies of drug-resistant TB cases for public health surveillance. Appropriate use of the developed data exchange formats will support accurate, timely, coherent and consistent data representations for public health laboratories that identify reportable or nationally notifiable conditions to support comprehensive public health surveillance.
Beyond the flexibility of implementing the data exchange formats, it is important to understand that molecular DST methods have several caveats and limitations that differ depending on the drug3, 37. One of the key limitations is that molecular DST is not available for all antituberculosis drugs. Not all mutations confer resistance and not all mutations associated with resistance are known. More importantly, there exist some discordance between molecular and growth- based DST results3, 32, 33, 37. Also, sensitivity and specificity for molecular DST are less than 100%3. These limitations inherently affect the reliability of the data exchange formats and might subsequently affect the results of epidemiological studies.
Conclusion
The implementation of the data exchange formats is still progressing at DTBE. There is also a plan to pilot molecular genetic data exchange in the NTSS using California Department of Public Health and New York State Department of Health as external partners. In conclusion, this study demonstrates that it is possible to apply standardized protocols to enhance data specifications and exchange formats within the NTSS, thereby streamlining the seamless exchange of drug-resistant TB incident cases in an integrated public health environment supporting TB surveillance, informatics, and translational studies. Moreover, the methodology applied in this study could be replicated across other disease surveillance systems that seek to exchange molecular genetic data and maximize data quality of information exchanges.
Disclaimer
The views expressed are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention (CDC) or the United States Government. References in this manuscript to any specific commercial products, process, service, manufacturer, or company does not constitute its endorsement or recommendation by the U.S. Government or CDC.
Acknowledgments
This work was supported in part by an appointment to the Public Health Informatics Fellowship Program at the Centers for Disease Control and Prevention (CDC). There have been contributions to this work by the Association of Public Health Laboratories (APHL) and California Department of Public Health (CDPH).
Illustration A: Sample Supported Segments for Cepheid GeneXpert®
OBR|1|||89371-9ˆMTB complex DNA and rpoB RIF resistance mutation panel [Presence] - Isolate or SpecimenˆLN|||20180218175900|||||||||||||||20180220144700|||F|<cr>
OBX|1|CWE|38379-4ˆ Mycobacterium tuberculosis complex DNA [Presence] in Unspecified specimen by NAA with probe detectionˆLN|1|260373001ˆDetectedˆSCT|||AˆAbnormalˆHL70078ˆˆˆˆ2.5.1|
||F|||20180220144700|||||20180221130016|||||<cr>
OBX|2|CWE|89372-7ˆMycobacterium tuberculosis complex rpoB gene rifampin resistance mutation [Presence] by Molecular genetics methodˆLN||260373001ˆDetectedˆSCT||||||F|
||20180218175900|||||20180221133259|||||<cr>
SPM|1|||119334006ˆSputum specimenˆSCT|||||||||||||20180218175900|20180221105743|<cr>
Illustration B: Sample Supported Segments for CDC’s MDDR Service
OBR|1|||81247-9ˆMaster HL7 genetic variant reporting panelˆLN|||20160729|||||||||||||||20160805|||F|
OBX|1|CWE|51963-7ˆMedication assessed [ID]ˆLN|1.a|9384ˆRifampinˆRxNorm|
OBX|2|CWE|51963-7ˆMedication assessed [ID]ˆLN|1.b|6038ˆisoniazidˆRxNorm|
OBX|3|CNE|48018-6ˆGene(s) assessedˆLN|1.a|888164ˆrpoBˆgeneCodeNCBI|
OBX|4|CNE|48018-6ˆGene(s) assessedˆLN|1.b|885638ˆkatGˆgeneCodeNCBI|
OBX|5|CWE|48018-6ˆGene studied [ID]ˆLN|2a|888164ˆrpoBˆgeneCodeNCBI|
OBX|6|CWE|48005-3ˆAmino acid changeˆLN|2a|Ser531LeuˆˆHGVS|
OBX|7|CWE|48019-4ˆDNA change [Type]ˆLN|2a|LA6690-7ˆSubstitutionˆLN |
OBX|8|ST|47998-0ˆDNA sequence variation display name [Text] NarrativeˆLN|2a|TCG>TTG|
OBX|9|CWE|69548-6ˆGenetic variant assessmentˆLN|2a|52101004ˆPresentˆSCT|
OBX|10|CWE|81304-8ˆVariant analysis method [Type]ˆLN|2a|117040002ˆNucleic acid sequencingˆSCT|
OBX|11|CWE|48018-6ˆGene studied [ID]ˆLN|2b|885638ˆkatGˆgeneCodeNCBI|
OBX|12|CWE|48005-3ˆAmino acid changeˆLN|2b|Ser315ThrˆˆHGVS|
OBX|13|CWE|48019-4ˆDNA change [Type]ˆLN|2b|LA6690-7ˆSubstitutionˆLN|
OBX|14|ST|47998-0ˆDNA sequence variation display name [Text] NarrativeˆLN|2b|AGC>ACC|
OBX|15|CWE|69548-6ˆGenetic variant assessmentˆLN|2b|52101004ˆPresentˆSCT|
OBX|16|CWE|81304-8ˆVariant analysis method [Type]ˆLN|2b|117040002ˆNucleic acid sequencingˆSCT|
SPM|1|3000654137||258589002ˆLymph node sampleˆSCT|||||||||||||20160729|
Figures & Table
References
- 1.Curry International Tuberculosis Center and California Department of Public Health. Drug-Resistant Tuberculosis: A Survival Guide for Clinicians. (3rd ed) 2016 [Google Scholar]
- 2.Barnard M, Parsons L, Miotto P, Cirillo D, Feldmann K, Gutierrez C, Somoskovi A. Molecular Detection of Drug-Resistant Tuberculosis By Line Probe Assay - Laboratory Manual for Resource-Limited Settings. 2012 [Google Scholar]
- 3.Lin SY, Desmond EP. Molecular diagnosis of tuberculosis and drug resistance. Clin Lab Med. 2014;34(2):297–314. doi: 10.1016/j.cll.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48(Suppl 1(suppl_1)):5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 5.Stewart RJ, Tsang CA, Pratt RH, Price SF, Langer AJ. Tuberculosis - United States, 2017. MMWR Morb Mortal Wkly Rep. 2018;67(11):317–23. doi: 10.15585/mmwr.mm6711a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonney W, Price SF, Miramontes R. Improving the Quality of Data Exchange Formats in the U.S. National Tuberculosis Surveillance System. Online J Public Health Inform. 2018;10(1):e48. [Google Scholar]
- 7.Davis JL, Kawamura LM, Chaisson LH, Grinsdale J, Benhammou J, Ho C, et al. Impact of GeneXpert MTB/RIF on patients and tuberculosis programs in a low-burden setting. a hypothetical trial. Am J Respir Crit Care Med. 2014;189(12):1551–9. doi: 10.1164/rccm.201311-1974OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helb D, Jones M, Story E, et al. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol. 2010;48(1):229–37. doi: 10.1128/JCM.01463-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hillemann D, Rusch-Gerdes S, Boehme C, Richter E. Rapid molecular detection of extrapulmonary tuberculosis by the automated GeneXpert MTB/RIF system. J Clin Microbiol. 2011;49(4):1202–5. doi: 10.1128/JCM.02268-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin SY, Rodwell TC, Victor TC, Rider EC, Pham L, Catanzaro A, et al. Pyrosequencing for rapid detection of extensively drug-resistant Mycobacterium tuberculosis in clinical isolates and clinical specimens. J Clin Microbiol. 2014;52(2):475–82. doi: 10.1128/JCM.01821-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shak EB, France AM, Cowan L, Starks AM, Grant J. Representativeness of Tuberculosis Genotyping Surveillance in the United States, 2009–2010. Public Health Rep. 2015;130(6):596–601. doi: 10.1177/003335491513000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.HL7 International. HL7 Version 2.5.1 Implementation Guide: Electronic Laboratory Reporting to Public Health, Release 1 (US Realm) 2010 [cited June 11, 2018] Available from: http://www.hl7.org/implement/standards/product_brief.cfm?product_id=98.
- 13.HL7 International. HL7 Version 2.5.1 Implementation Guide: Electronic Laboratory Reporting to Public Health, Release 2 (US Realm) 2014 [cited June 11, 2018] Available from: http://www.hl7.org/implement/standards/product_brief.cfm?product_id=329.
- 14.HL7 International. HL7 Version 2 Implementation Guide: Clinical Genomics; Fully LOINC-Qualified Genetic Variation Model (US Realm) 2013 [cited June 11, 2018] Available from: http://www.hl7.org/implement/standards/product_brief.cfm?product_id=23.
- 15.HL7 International. HL7 Version 2 Implementation Guide: Clinical Genomics Coded Reporting - “Lite,” Release 1 (1st DSTU Ballot) (US Realm) 2016 [cited June 11, 2018] Available from: http://www.hl7.org/Special/committees/clingenomics/index.cfm.
- 16.Nelson SJ, Zeng K, Kilbourne J, Powell T, Moore R. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc. 2011;18(4):441–8. doi: 10.1136/amiajnl-2011-000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenbloom ST, Miller RA, Johnson KB, Elkin PL, Brown SH. Interface terminologies: facilitating direct entry of clinical data into electronic health record systems. J Am Med Inform Assoc. 2006;13(3):277–88. doi: 10.1197/jamia.M1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fung KW, McDonald C, Bray BE. RxTerms - a drug interface terminology derived from RxNorm. AMIA Annu Symp Proc. 2008;2008:227–31. [PMC free article] [PubMed] [Google Scholar]
- 19.U.S. National Library of Medicine. RxNorm Technical Documentation 2018 [updated June 22, 2018; cited June 27, 2018] Available from: https://www.nlm.nih.gov/research/umls/rxnorm/docs/2018/rxnorm_doco_full_2018-1.html#s1_0.
- 20.Oregon Public Health Division. Oregon Electronic Laboratory Reporting Implementation Guide 2016 [updated September 13, 2016; cited May 22, 2018] Available from: https://www.oregon.gov/oha/PH/DISEASESCONDITIONS/COMMUNICABLEDISEASE/REPORTINGCOMMUNICABLEDISEASE/ELECTRONICLABREPORTING/Documents/OR%20ELR%20HL7%20251%20Implementation%20Guide.pdf.
- 21.Wurtz R, Cameron BJ. Electronic laboratory reporting for the infectious diseases physician and clinical microbiologist. Clin Infect Dis. 2005;40(11):1638–43. doi: 10.1086/429904. [DOI] [PubMed] [Google Scholar]
- 22.Abisa M. Meaningful Use and Electronic Laboratory Reporting: Challenges Health Information Technology Vendors Face in Kentucky. Online J Public Health Inform. 2017;9(3):e196. doi: 10.5210/ojphi.v9i3.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson MG, Williams J, Lee A, Bradley KK. Completeness and timeliness of electronic vs. conventional laboratory reporting for communicable disease surveillance--Oklahoma, 2011. Public Health Rep. 2014;129(3):261–6. doi: 10.1177/003335491412900308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Overhage JM, Grannis S, McDonald CJ. A comparison of the completeness and timeliness of automated electronic laboratory reporting and spontaneous reporting of notifiable conditions. Am J Public Health. 2008;98(2):344–50. doi: 10.2105/AJPH.2006.092700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.U.S. Centers for Disease Control and Prevention (CDC) Electronic Laboratory Reporting (ELR) 2016 [updated May 19, 2016; cited 2018 June 7, 2018] Available from: http://www.cdc.gov/ehrmeaningfuluse/elr.html.
- 26.Dixon BE, Gibson PJ, Grannis SJ. Estimating increased electronic laboratory reporting volumes for meaningful use: implications for the public health workforce. Online J Public Health Inform. 2014;5(3):225. doi: 10.5210/ojphi.v5i3.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen TQ, Thorpe L, Makki HA, Mostashari F. Benefits and barriers to electronic laboratory results reporting for notifiable diseases: the New York City Department of Health and Mental Hygiene experience. Am J Public Health. 2007;97(Suppl 1(Suppl 1)):S142–5. doi: 10.2105/AJPH.2006.098996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Indiana State of Public Health. Electronic Laboratory Reporting (ELR): HL7 version 2.5.1 Message Structure Reference Guide Version 1.0 2017 [updated March 28, 2017; cited June 11, 2018] Available from: https://eportal.isdh.in.gov/MeaningfulUse/Documents/ISDH_HL7251_ELR_Message_Structure_Reference_Guide_Version_1-0.pdf.
- 29.Florida Department of Health. Florida Department of Health HL7 2.5.1 Specifications for ELR Implementation Guide 2015 [updated October 9, 2015; cited 2018 May 22, 2018] Available from: http://www.floridahealth.gov/diseases-and-conditions/disease-reporting-and-management/_documents/florida-meainingful-use/elr-implementation-guide.pdf.
- 30.HL7 International. HL7 Messaging Standard Version 2.5.1: An application protocol for electronic data exchange in healthcare environments 2007 [cited May 2, 2018] Available from: https://www.hl7.org/implement/standards/product_brief.cfm?product_id=144.
- 31.Campbell PJ, Morlock GP, Sikes RD, Dalton TL, Metchock B, Starks AM, et al. Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2011;55(5):2032–41. doi: 10.1128/AAC.01550-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yakrus MA, Driscoll J, Lentz AJ, Sikes D, Hartline D, Metchock B, et al. Concordance between molecular and phenotypic testing of Mycobacterium tuberculosis complex isolates for resistance to rifampin and isoniazid in the United States. J Clin Microbiol. 2014;52(6):1932–7. doi: 10.1128/JCM.00417-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yakrus MA, Driscoll J, McAlister A, Sikes D, Hartline D, Metchock B, et al. Molecular and Growth-Based Drug Susceptibility Testing of Mycobacterium tuberculosis Complex for Ethambutol Resistance in the United States. Tuberc Res Treat. 2016;2016:3404860. doi: 10.1155/2016/3404860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yakrus MA, Metchock B, Starks AM. Evaluation of a U.S. public health laboratory service for the molecular detection of drug resistant tuberculosis. Tuberc Res Treat. 2015;2015:701786. doi: 10.1155/2015/701786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Association of Public Health Laboratories (APHL) Molecular Biology 101 n.d [cited May 12, 2018] Available from: https://www.aphl.org/programs/infectious_disease/tuberculosis/TBCore/10_Molecular-Biology-101.pdf.
- 36.Div of Tuberculosis Elimination, National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention, CDC. Availability of an Assay for Detecting Mycobacterium tuberculosis, Including Rifampin-Resistant Strains, and Considerations for Its Use — United States, 2013. MMWR Morb Mortal Wkly Rep. 2013;62(41):821–4. [PMC free article] [PubMed] [Google Scholar]
- 37.Metchock B. New Tools in the Diagnosis of TB: Principles, Indications and Limitations (Understanding TB Laboratory Results and Challenges of Reporting Results) 2014 [cited 2018 June 6, 2018] Available from: http://166.67.66.226/tb/documents/2_MetchockVA2014.pdf.
- 38.Association of Public Health Laboratories (APHL) Laboratory Considerations for Use of Cepheid GeneXpert MTB/RIF Assay 2013 [cited May 12, 2018] Available from: https://www.aphl.org/AboutAPHL/publications/Documents/ID_2013Nov_Cepheid-Xpert-Fact-Sheet.pdf.
- 39.World Health Organization (WHO) Frequently Asked Questions on Xpert MTB/RIF assay n.d [cited June 12, 2018] Available from: http://www.who.int/tb/laboratory/xpert_faqs.pdf.
- 40.Driscoll J, Lentz A, Sikes D, Hartline D, Metchock B. The first month of a new diagnostic service for molecular detection of MDR and XDR tuberculosis. A93 New Strategies for Diagnosing Latent and Active Tuberculosis: American Thoracic Society. 2010:A2259–A. [Google Scholar]