Abstract
Despite the recent developments in commercial Question Answering (QA) systems, medical QA remains a challenging task. In this paper, we study the factors behind the complexity of consumer health questions and potential improvement tracks. In particular, we study the impact of information source quality and question conciseness through three experiments. First, an evaluation of a QA method based on a Question-Answer collection created from trusted NIH resources, which outperformed the best results of the medical LiveQA challenge with an average score of 0.711. Then, an evaluation of the same approach using paraphrases and summaries of the test questions, which achieved an average score of 1.125. Our results provide an empirical evidence supporting the key role of summarization and reliable information sources in building efficient CHQA systems. The latter finding on restricting information sources is particularly intriguing as it contradicts the popular tendency ofrelying on big data for medical QA.
1. Introduction
The integration of artificial intelligence into daily life has increased the importance of Question Answering (QA). QA was one of the earliest NLP tasks. The first QA systems, Baseball (1961) and Lunar (1973), were domain specific, both exploiting knowledge bases written manually by domain experts. Several other QA systems were proposed in the literature for closed and open domains4,7,9,12 as well as in the context of international challenges1,16. Recent years have seen advances in the introduction of large QA collections8,18 and in the methods applied to QA through different variations of deep neural networks and natural logic3,9.
From a medical perspective, the application of these methods to consumer heath question answering (CHQA) is not straightforward. For instance, the lack of large training datasets as well as task-specific requirements constrain the range of relevant methods. In addition, the goal of CHQA is not only to ease the search of medical information but also to overcome the use of popular search engines which can mislead the users with harmful information. CHQA has attracted several efforts focusing on the construction of new resources14,17 and the development of relevant QA methods such as question focus/topic identification11,15, question entailment recognition6, and answering consumer health questions using existing questions and answers13,20.
Recently, we organized the first CHQA task in the LiveQA track5 at the Text Retrieval Conference1 TREC 2017. The best participating system in the medical task achieved an average score of 0.637 on a 0-3 scale, a performance well below the average score of 1.139 when the same deep learning approach was applied to open-domain questions.
In this paper, we build upon the data from LiveQA medical task and study the factors behind the complexity of consumer health questions and potential improvement tracks. We developed a medical QA system and evaluated the retrieved answers to study the impact of information source restriction and question conciseness through three experiments using (i) a Question-Answer collection created from trusted NIH resources, (ii) paraphrases of the test questions and (iii) summaries of the questions. Our results show that paraphrasing or summarizing the questions alleviate some of the linguistic challenges in answering consumer health questions. They also show that restricting information sources according to reliability improves the overall efficiency of CHQA, which suggests that well selected information sources can outperform larger datasets.
The paper is organized as follows. In Section 2, we summarize the methods and results of LiveQA participants and share insights related to the CHQA task. In Section 3, we describe our medical QA system based on finding similar answered questions from trusted resources and present our three experiments. Section 4 describes the evaluation methodology and the obtained results. Finally, we discuss the results and give some insights for future research in Section 5.
2. Consumer Health QA Competition
The medical task5 at TREC 2017 LiveQA track focused on automatically retrieving answers to medical questions received by the U.S. National Library of Medicine (NLM). The NLM2 receives more than 100,000 requests a year, including over 10,000 consumer health questions. The medical task at TREC 2017 LiveQA was organized in the scope of the CHQA project3 at the NLM.
2.1. Test Dataset
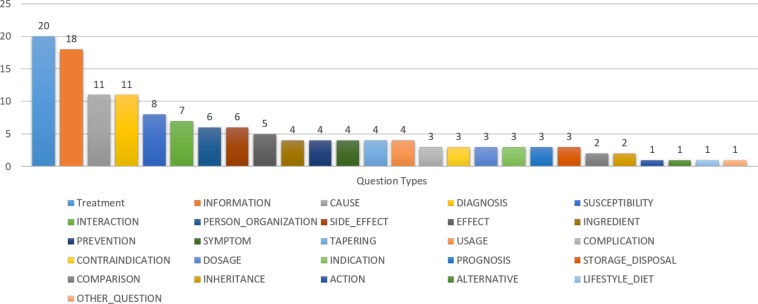
To evaluate the performance and effectiveness of the participating QA systems on real consumer health questions, we provided a test set of 104 NLM questions. Figure 1 presents an example from the test dataset with the associated annotations and reference answers. All LiveQA’17 medical training and test datasets are publicly available4. The test dataset covers different medical entities and a wide variety of question types such as Treatment or Inheritance of a Medical Problem, and Dosage or Tapering of a Drug. Figure 2 presents the question types covered by the test dataset.
Figure 1:
A question from LiveQA’17 test dataset and the associated annotations and reference answers.
Figure 2:
Questions types covered by the medical test questions of LiveQA 2017.
2.2. Current Approaches to QA and their Limitations
The aim of running the medical task at LiveQA was to develop techniques for answering complex questions such as consumer health questions, as well as to identify relevant answer sources that can comply with the sensitivity of medical information retrieval.
The CMU-OAQA system19 achieved the best performance of 0.637 on the medical task by using an attentional encoder-decoder model for paraphrase identification and answer ranking. The Quora question-similarity dataset was used for training. The PRNA system10 achieved the second best performance in the medical task with 0.49 average score using Wikipedia as the first answer source and Yahoo and Google searches as secondary answer sources. Each medical question was decomposed into several subquestions. To extract the answer from the selected text passage, a bi-directional attention model trained on the SQUAD dataset was used.
Deep neural network models have been pushing the limits of performance achieved in QA related tasks using large training datasets. The results obtained by CMU-OAQA and PRNA showed that large open-domain datasets were beneficial for the medical domain. However, the best system (CMU-OAQA) relying on the same training data obtained a score of 1.139 on the LiveQA open-domain task.
While this gap in performance can be explained in part by the discrepancies between the medical test questions and the open-domain questions, it also highlights the need for larger medical datasets to support deep learning approaches in dealing with the linguistic complexity of consumer health questions and the challenge of finding correct and complete answers. Another technique was used by ECNU-ICA team2 based on learning question similarity via two long short-term memory (LSTM) networks applied to obtain the semantic representations of the questions. To construct a collection of similar question pairs, they searched community question answering sites such as Yahoo! and Answers.com. The ECNU-ICA system achieved the best performance of 1.895 in the open-domain task but an average score of only 0.402 in the medical task. As the ECNU-ICA approach also relied on a neural network for question matching, this result shows that training attention-based decoder-encoder networks on the Quora dataset generalized better to the medical domain than training LSTMs on similar questions from Yahoo! and Answers.com.
The CMU-LiveMedQA team21 designed a specific system for the medical task. Using only the provided training datasets and the assumption that each question contains only one focus, the CMU-LiveMedQA system obtained an average score of 0.353. They used a convolutional neural network (CNN) model to classify a question into a restricted set of 10 question types and crawled “relevant” online web pages to find the answers. However, the results were lower than those achieved by the systems relying on finding similar answered questions. These results support the relevance of similar question matching for the end-to-end QA task as a new way of approaching QA instead of the classical QA approaches based on Question Analysis and Answer Retrieval.
The above analysis suggested we need to answer two questions: (i) to what extent does the question surface form affect finding similar questions, and (ii) do the available reliable sources of answers to consumer health questions contain answers to all test questions. To answer these questions, we developed a medical QA system based on retrieving answered questions from a collection of trusted question-answer pairs. We present the developed QA system in the following section.
3. Methods
3.1. Baseline QA System
We built a medical QA system based on a combination of two information retrieval models retrieving similar questions that already have correct answers. We used a collection of 47,527 medical question-answer pairs extracted from 12 websites from the National Institutes of Health (NIH)5 including MedlinePlus, Genetics Home Reference (GHR) and National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). The question-type taxonomy includes 16 question types about Diseases (e.g. Treatment, Prevention, Susceptibility), 20 question types about Drugs (e.g. Interactions, Contraindication, Usage). One generic question type (Information) is associated with all possible topics/focus including Procedures and Medical exams.
We translate the QA task to retrieving N relevant questions mqj, j ∈ [1, N], to the submitted consumer health question chq and returning their existing responses to answer the new chq. We used the Terrier search engine6 to retrieve similar questions. In order to improve the performance of question retrieval, we indexed each question with the synonyms of its focus and the trigger words corresponding to its question type. The focus synonyms and the question type were extracted automatically from the Question-Answer collection. The triggers of each question type were defined in our taxonomy of 36 question types. For instance, the list of trigger words associated with the question type “Treatment” includes: relieve, manage, cure, remedy and therapy.
As result fusion has shown improved performance in different tracks in TREC, we merge the results of two IR models TF-IDF (Term Frequency - Inverse Document Frequency model) and In_expB2 (Inverse Expected Document Frequency model). Let be the set of questions retrieved by the first IR model V and be the set of questions retrieved by the second IR model W, we merge both sets by summing the scores of each question mqj in both lists.
3.2. Compared Methods
To study the difficulty of understanding consumer health questions and to examine the resources used to find relevant answers, we conducted three experiments. In the first experiment (M1-TQs), we use the LiveQA test questions and the baseline QA system to retrieve relevant answers from the medical QA collection. In the second experiment (M2-PAR), we use paraphrases of the test questions created by NIST7 assessors for the official evaluation of LiveQA participating systems. This experiment aims to study the difficulty of the original questions by evaluating the impact of reformulating the questions on the QA process. In a third experiment (M3-SUM), we create short summaries of the consumer health questions to study the effect of conciseness.
The summaries were created by a medical doctor and expert in QA to ensure the correctness and the adequacy of the short questions to the QA task. Figure 3 presents an example of a LiveQA test question, the associated NIST paraphrase, and the NLM summary.
Figure 3:
Example of a LiveQA’17 test question and the associated NIST paraphrase and NLM summary.
The manual process of creating these paraphrases and summaries is crucial to the relevance of our study. It avoids conflating the effects of automated summarization and paraphrase generation, and allows us to analyze the results and find insights that would be biased otherwise.
The following section describes the evaluation methodology used to compare these three QA methods and the obtained results on LiveQA medical test questions.
4. Evaluation Methodology and Results
In this section, we describe the evaluation interface, the evaluation metrics and the results of the compared methods.
4.1. Evaluation Methodology
The official results of the LiveQA track relied on one assessor per question. Similarly, in our experiments, a medical doctor evaluated manually the answers returned by the three IR-based methods, using the same LiveQA’17 reference answers as guidance. The reference answers were also used by NIST assessors to judge the answers retrieved by the participating systems.
For a relevant comparison, we used the same judgment scores as the LiveQA track: ”Correct and Complete Answer” (4), ”Correct but Incomplete” (3), ”Incorrect but Related” (2) and ”Incorrect” (1). Figure 4 presents the evaluation interface used to evaluate the answers retrieved by each method.
To have an idea about the inter-annotator agreement (IAA) in such tasks, a second assessor (a medical librarian) evaluated the answers of the first method (M1-TQs). The IAA is based on F1 score computed by considering one of the assessors as reference. First, we computed the True Positives (TP) and False Positives (FP) over all ratings and the Precision and F1 score. As there are no negative labels (only true or false positives for each category), Recall is 100%. We then computed a partial IAA by grouping the ”Correct and Complete Answer” and ”Correct but Incomplete” ratings (as Correct), and the ”Incorrect but Related” and ”Incorrect” ratings (as Incorrect). In this evaluation, the strict IAA is 89.38%. The partial agreement on distinguishing the Correct and Incorrect answers is 94.81 %.
We computed LiveQA measures to evaluate the first retrieved answer for each question:
avgScore(0-3): the average score over all questions, transferring 1-4 level grades to 0-3 scores. This is the main score used to rank LiveQA runs.
succ@i+: the number of questions with score i or above (i∈{2..4}) divided by the total number of questions.
prec@i+: the number of questions with score i or above (i∈{2..4}) divided by number of questions answered by the system.
Additionally, we used Mean Average Precision (MAP) and Mean Reciprocal Rank (MRR) which are commonly used in QA to evaluate the top-10 answers for each question. We consider answers rated as ”Correct and Complete Answer” or ”Correct but Incomplete” as correct answers, as the test questions contain multiple subquestions while each answer in our QA collection can cover only one subquestion.
MAP is the mean of the Average Precision (AvgP) scores over all questions.
(1)
Q is the number of questions. AvgPi is the AvgP of the ith question.
K is the number of correct answers. rankn is the rank of nth correct answer.
MRR is the average of the reciprocal ranks for each question. The reciprocal rank of a question is the multiplicative inverse of the rank of the first correct answer.
(2)
Q is the number of questions, ranki is the rank of the first correct answer for the ith question.
4.2. Evaluation Results
We compare the results of the three methods M1-TQs, M2-PAR and M3-SUM presented in Section 3.2. Table 1 presents the LiveQA measures AvgScore, Success and Precision, used to evaluate the first retieved answer for each test question. The M1-TQs method achieves 0.711 average score using the original questions and outperforms the best results achieved in the medical challenge at LiveQA’17. The M3-SUM method achieves the best performance of 1.125 average score, getting closer to the performance achieved in open domain. Table 2 presents the results of MAP@10 and MRR@10 used to evaluate the top-10 answers. The compared methods M1-TQs, M2-PAR and M3-SUM achieve 0.28, 0.3 and 0.38 MAP@10, and allow to correctly answer 51, 56 and 64 questions, respectively.
Table 1:
LiveQA Measures: Average Score (main score), Success@i+ and Precision@i+ on LiveQA’17 Test Data. Evaluation of the first retrieved answer for each question.
| Measures | M1-TQs | M2-PAR | M3-SUM | LiveQA’17 Best Results | LiveQA’17 Median |
| avgScore(0-3) | 0.711 | 0.913 | 1.125 | 0.637 | 0.431 |
| succ@2+ | 0.442 | 0.567 | 0.663 | 0.392 | 0.245 |
| succ@3+ | 0.192 | 0.240 | 0.317 | 0.265 | 0.142 |
| succ@4+ | 0.077 | 0.106 | 0.144 | 0.098 | 0.059 |
| prec@2+ | 0.46 | 0.567 | 0.663 | 0.404 | 0.331 |
| prec@3+ | 0.2 | 0.240 | 0.317 | 0.273 | 0.178 |
| prec@4+ | 0.08 | 0.106 | 0.144 | 0.101 | 0.077 |
Table 2:
Common Measures: MAP and MRR on LiveQA’17 Test Questions. Evaluation of top 10 answers.
| Measures | M1-TQs | M2-PAR | M3-SUM |
| Correctly answered questions | 51 | 56 | 64 |
| MAP@10 | 0.282 | 0.308 | 0.380 |
| MRR@10 | 0.281 | 0.334 | 0.417 |
5. Discussion
5.1. Relevance of Similar-Question Retrieval from Trusted Resources
The first part of our results (M1-TQs method) shows that relying on similar-question retrieval to answer consumer health questions is a viable QA strategy, which outperformed the best systems participating in the medical task at TREC 2017 LiveQA. The fact that our system used only a collection of 47,527 QA pairs from NIH resources to find the answers also supports the feasibility, and to some extent the efficiency, of relying on trusted answer sources for CHQA despite the limited size of such collections.
We also looked closely at the resources used manually to provide reference answers for LiveQA test questions. The most frequent resources are MedlinePlus Encyclopedia, DailyMed, Mayoclinic, MedlinePlus, GHR, CDC, Pubmed, PMC, and other NIH websites such as NHLBI and NIDDK. Other trusted resources were also used to find reference answers such as aafp.org, cancer.org, nature.com, umm.edu, and who.int.
Private websites had answers for 6% of the test questions. For instance, the ConsumerLab website was useful to answer a question about the ingredients of a Drug (COENZYME Q10). Similarly, the eHealthMe website was used to answer a test question asking about interactions between two drugs (Phentermine and Dicyclomine) when no information was found in DailyMed. eHealthMe provides healthcare big data analysis and private research and studies including self-reported adverse drug effects by patients. But the question remains on the extent to which such big data and other private websites could be used to automatically answer medical questions if information is otherwise unavailable. Unlike medical professionals, patients do not necessarily have the knowledge and tools to validate such information. An alternative approach could be to put limitations on CHQA systems in terms of the questions that can be answered (e.g. ”What is my diagnosis for such symptoms”) and build classifiers to detect such questions and warn the users about the dangers of looking for their answers online.
More generally, medical QA systems should follow some strict guidelines regarding the goal and background knowledge and resources of each system in order to protect the consumers from misleading or harmful information. Such guidelines could be based on the source of the information such as health and medical information websites sponsored by the U.S. government, not-for-profit health or medical organizations, and university medical centers, or on conventions such as the code of conduct of the HON Foundation issued (HONcode) which addresses the reliability and usefulness of medical information on the Internet.
Our experiments show that limiting the number of answer sources with such guidelines is not only feasible, but it could also enhance the performance of the QA system from an information retrieval perspective.
5.2. Complexity of Consumer Health Question Answering
The M3-SUM method achieves 1.125 average score, getting closer to the performance achieved in open domain. This substantial improvement through the summarization of the questions points out the difficulty of the automatic analysis of the original questions.
Both experiments using paraphrases of the original questions (M2-PAR and M3-SUM) show that the obtained improvement in answer retrieval is important. The best results were obtained with the concise summaries but not with the NIST paraphrases which attempted to keep as much information as possible from the user questions. Retrieving answers with NIST paraphrases yielded a 28.41% increase in AvgScore and 9.22% increase in MAP@10.
Using summaries provided overall better results than paraphrases as the questions are shorter and more to the point, achieving a 58.22% increase in LiveQA AvgScore and 34.75% increase in MAP@10. These results highlight the fact that the complexity of consumer health questions is only partially related to the vocabulary, and that a substantial area of improvement is reformulating and summarizing consumer health questions.
For further analysis, we describe below three examples of questions and their retrieved answers, starting with the easiest question, correctly answered by all methods (all top-10 answers are correct).
Example 1:
-
–
Question: Beckwith-Wieddeman Syndrome. I would like to request further knowledge on this specific disorder.
-
–
Paraphrase: I want information on Beckwith-Wieddeman Syndrome.
-
–
Summary: Where can I find information on Beckwith-Wieddeman Syndrome?
The QA system provided 10 correct answers to this question. The same results were obtained using the paraphrases. This is a simple question as there is only one focus (Beckwith-Wieddeman Syndrome). Also, there is no specific question type, therefore all question types retrieved by the QA system are relevant (Information, Treatment, Genetic changes, Causes, Prevention, Symptoms, Complications, Susceptibility, Frequency and Inheritance). Moreover, many resources provide relevant information about this medical problem such as A.D.A.M. Medical Encyclopedia8 (including more than 4,000 physician-reviewed and physician-updated articles) and GHR9.
Example 2:
-
–
Question: SUBJECT: Gluten information. Re:NDC# 0115-0672-50 Zolmitriptan tabkets 5mg. I have celiac disease & need to know ifthese contain gluten, Thank you!
-
–
Paraphrase: Do 5 mg. Zolmitriptan tabkets contain gluten?
-
–
Summary: Do Zolmitriptan 5mg tablets contain gluten?
The first method M1-TQs provided 10 incorrect answers to this question. All top-10 retrieved answers were about celiac disease. The paraphrase and summary do not contain ”celiac disease”, which eliminated all wrong answers and provided correct answers. This points out the importance of Paraphrasing and also Focus Recognition in QA systems.
Example 3:
-
–
Question: calcitonin salmon nasal spray. I picked up a bottle of above but noted it had NOT been refrigerated for at least the 3 days since Rx was filled. Box and literature state ”refrigerate until opened.” Pharmacist insisted it was ok ”for 30 days” although I said that meant after opening. Cost is $54.08 plus need to know if it will be as effective as should be. Thank you.
-
–
Paraphrase: Will an unopened, unrefrigerated calcitonin salmon nasal spray be as effective as ifit had been refrigerated? The directions say it needs to be refrigerated.
-
–
Summary:How long can unopened calcitonin salmon nasal spray be left unrefrigerated?
The first method did not find any answer to this question. However a correct answer was found when using the paraphrase and the summary. The answer was found in the MedlinePlus article related to ”Calcitonin Salmon Nasal Spray” and in the section ”What should I know about storage and disposal of this medication?”. But this answer was ranked lower than all other answers extracted from the same page10 such as ”How should this medicine be used?” and ”What special precautions should I follow?”. The relation between ”duration/validity of an unopened and unrefrigerated medication ” and ”storage and disposal of the medication” is needed to improve the ranking of this answer.
Defining semantic and inference relations between the question types can lead to more relevant answers. For instance, if answers are not found for questions about the susceptibility to a condition, the consumer could be redirected to answers about the prevention or causes of the same condition. A similar reasoning can be applied to medical entities. For instance, for a question like ”Is dementia genetically passed down or could anyone get it”, both the broader answers about dementia in general and answers for specific types (e.g. Alzheimer’s) are relevant. The same for a question about ”Trisomy 7 causes”, general information on causes of duplication is relevant, if there is no specific information. Hence, formalizing the semantics of background medical knowledge and automated inference are potential tracks of improvement in question analysis and answer retrieval.
6. Conclusion
We studied the difficulty of understanding consumer questions by comparing the answers of the original questions with the answers of the paraphrased and summarized interpretations. We also studied the effect of relying on restricted and trusted information sources for CHQA. Our experiments and results on data from the medical task at LiveQA 2017 showed that the QA approach based on retrieving similar answered questions from such resources outperforms the best official results of the challenge, which relied on much larger document collections. Our results also showed that paraphrasing and summarizing the questions leads to a substantial improvement in the QA performance according to standard metrics. Future areas of study include the design of relevant techniques for automatic question reformulation and summarization, as well as inference methods relying on background medical knowledge.
Figure 4:
Evaluation interface developed and used to evaluate the retrieved answers for each medical question.
Acknowledgements
This work was supported by the intramural research program at the U.S. National Library of Medicine, National Institutes of Health. We thank Sonya Shooshan for her help with the manual evaluation.
Footnotes
The U.S. National Institute of Standards and Technology www.nist.gov
ghr.nlm.nih.gov
References
- [1].Eugene Agichtein, David Carmel, Dan Pelleg, Yuval Pinter, Donna Harman. Overview of the TREC 2015 liveqa track. In Proceedings of The Twenty-Fourth Text REtrieval Conference, TREC 2015, Gaithersburg, Maryland, USA, November 17-20, 2015.2015. [Google Scholar]
- [2].Weijie An, Qin Chen, Wei Tao, Jiacheng Zhang, Jianfeng Yu, Yan Yang, Qinmin Hu, Liang He, Bo Li. Ecnu at 2017 liveqa track: Learning question similarity with adapted long short-term memory networks. In Proceedings of The Twenty-Sixth Text REtrieval Conference, TREC 2017, Gaithersburg, Maryland, USA.2017. [Google Scholar]
- [3].Gabor Angeli, Neha Nayak, Christopher D. Manning. Combining natural logic and shallow reasoning for question answering. Long Papers; In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics, ACL 2016; Berlin, Germany: 2016 2016. Aug 7-12, [Google Scholar]
- [4].Sofia J. Athenikos and Hyoil Han. Biomedical question answering: A survey. Comput. Methods Prog. Biomed. 2010 Jul;99(1):1–24. doi: 10.1016/j.cmpb.2009.10.003. [DOI] [PubMed] [Google Scholar]
- [5].Asma Ben Abacha, Eugene Agichtein, Yuval Pinter, Dina Demner-Fushman. Overview of the medical question answering task at trec 2017 liveqa. In TREC 2017. 2017.
- [6].Asma Ben Abacha, Dina Demner-Fushman. In AMIA 2016, American Medical Informatics Association Annual Symposium. Chicago, IL, USA: 2016. Nov, Recognizing question entailment for medical question answering. [PMC free article] [PubMed] [Google Scholar]
- [7].Asma Ben Abacha, Pierre Zweigenbaum. MEANS: A medical question-answering system combining NLP techniques and semantic web technologies. Information Processing and Management Journal. 2015;51(5):570–594. [Google Scholar]
- [8].Antoine Bordes, Nicolas Usunier, Sumit Chopra, Jason Weston. Large-scale simple question answering with memory networks. CoRR, abs/1506.02075. 2015.
- [9].Zihang Dai, Lei Li, Wei Xu. CFO: conditional focused neural question answering with large-scale knowledge bases. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics, ACL 2016 August 7-12 2016 Berlin, Germany Volume 1 Long Papers.2016. [Google Scholar]
- [10].Vivek Datla, Tilak Arora, Joey Liu, Viraj Adduru, Sadid A. Hasan, Kathy Lee, Ashequl Qadir, Yuan Ling, Aaditya Prakash, Oladimeji Farri. Prna at the TREC 2017 liveqa track. In Proceedings of The Twenty-Sixth Text REtrieval Conference, TREC 2017, Gaithersburg, Maryland, USA.2017. [Google Scholar]
- [11].Haihong Guo, Xu Na, Li Hou, Jiao Li. Classifying Chinese Questions Related to Health Care Posted by Consumers Via the Internet. JMIR medical informatics. 2017;19(6) doi: 10.2196/jmir.7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Konrad Hoffner, Sebastian Walter, Edgard Marx, Ricardo Usbeck, Jens Lehmann, Axel-Cyrille Ngonga Ngomo. Survey on challenges of question answering in the semantic web. Semantic Web. 2017;8(6):895–920. [Google Scholar]
- [13].Jiwoon Jeon, Croft W. Bruce, Joon Ho Lee. Finding similar questions in large question and answer archives. In Proceedings of the 2005 ACM CIKM International Conference on Information and Knowledge Management; Bremen, Germany: 2005 2005. November October. pp. 84–90. [Google Scholar]
- [14].Halil Kilicoglu, Asma Ben Abacha, Yassine Mrabet, Sonya E. Shooshan, Laritza Rodriguez, Kate Masterton, Dina Demner-Fushman. Semantic annotation of consumer health questions. BMC Bioinformatics. 2018;19(1):34:1–34:28. doi: 10.1186/s12859-018-2045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yassine Mrabet, Halil Kilicoglu, Kirk Roberts, Dina Demner-Fushman. In AMIA 2016, American Medical Informatics Association Annual Symposium. Chicago, IL, USA: 2016. Nov, Combining open-domain and biomedical knowledge for topic recognition in consumer health questions. [PMC free article] [PubMed] [Google Scholar]
- [16].Anastasios Nentidis, Konstantinos Bougiatiotis, Anastasia Krithara, Georgios Paliouras, Ioannis A. Kakadi-aris. In BioNLP 2017, Vancouver. Canada: 2017 2017. Aug 4, Results of the fifth edition of the bioasq challenge; pp. 48–57. [Google Scholar]
- [17].João R, Palotti M, Guido Zuccon Jimmy, Pavel Pecina, Mihai Lupu, Lorraine Goeuriot, Liadh Kelly, Allan Hanbury. CLEF 2017 task overview: The IR task at the ehealth evaluation lab - evaluating retrieval methods for consumer health search. In Working Notes of CLEF 2017 - Conference and Labs of the Evaluation Forum; Dublin, Ireland: 2017 2017. Sep 11-14, [Google Scholar]
- [18].Pranav Rajpurkar, Jian Zhang, Konstantin Lopyrev, Percy Liang. Squad: 100,000+questions for machine comprehension of text. CoRR,abs/1606.05250. 2016.
- [19].Di Wang, Eric Nyberg. Cmu oaqa at trec 2017 liveqa: A neural dual entailment approach for question paraphrase identification. In Proceedings of The Twenty-Sixth Text REtrieval Conference, TREC 2017; Gaithersburg Maryland, USA; 2017. [Google Scholar]
- [20].Papis Wongchaisuwat, Diego Klabjan, Siddhartha Reddy Jonnalagadda. A Semi-Supervised Learning Approach to Enhance Health Care Community-Based Question Answering: A Case Study in Alcoholism. JMIR medical informatics. 2016;4(3) doi: 10.2196/medinform.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yuan Yang, Jingcheng Yu, Ye Hu, Xiaoyao Xu, Eric Nyberg. Cmu livemedqa at trec 2017 liveqa: A consumer health question answering system. In Proceedings of The Twenty-Sixth Text REtrieval Conference, TREC 2017; Gaithersburg, Maryland, USA; 2017. [Google Scholar]