Abstract
Clinical trials and studies are increasingly using systems, such as REDCap, to capture data in electronic form. However these tools are not designed to mimic the capture of clinical information for regular clinical care and lack support for sharing the data effectively. In this paper we describe the implementation of a transformation engine, FHIRCap, that allows defining rules to map REDCap forms into FHIR resources. To assess the feasibility of the system, a case study with one of the Australian Genomics clinical demonstration projects was done. The case study showed that the transformation language is flexible enough to handle most of the data being captured in REDCap for a typical clinical trial. A number of design issues in the forms were identified and a series of recommendations were provided to enable a more accurate transformation. These results show that it is possible to transform most data in existing REDCap projects to FHIR resources without having to modify the forms. This is significant because it demonstrates that most data in existing clinical trials and studies can be made available in a standardised manner.
Introduction
In recent years, clinical trials and studies have increasingly started using electronic systems to capture data required to conduct a range of analysis, such as the effectiveness of a new treatment or its economic value. However, even though these tools allow creating electronic forms easily, they are not designed to capture clinical data, impose few constraints on what should be captured and also have limited data sharing capabilities.
One of the most popular tools currently used to capture research data is REDCap, a web application created at Vanderbilt University1. In the Australian Genomics (AG), REDCap has been selected to capture patient data in most Flagship projects. AG is an alliance of 31 organisations undertaking translational research for the implementation of genomics into clinical care. AG has developed minimum data sets for each of the clinical demonstration projects representing the minimal clinical data that should be captured to support the implementation of a genomics-based care model. This data represents a patient phenotype. However, data is not captured in a standardised manner, making it hard to share between projects and organisations.
In this project, we overcome this limitation by implementing FHIRCap, a rules-based transformation engine that allows exporting data in REDCap as FHIR resources. The result of the transformation is stored in a centralised FHIR repository. This paper describes the methods used to develop the system, a case study that was done with one of the AG Flagship projects to evaluate the feasibility of the approach and our conclusions.
1. Related Work
To our knowledge, no one has developed a system that allows transforming REDCap forms into FHIR resources. However, there have been attempts to transform standards, such as the Operational Data Model (ODM), into FHIR resources and also to transform REDCap data into other formats such as the Clinical Data Interchange Standards Consortium (CDISC) Study Data Tabulation Model (SDTM) format.
Doods et al. transformed the CDISC ODM 1.3.2 standard into a FHIR Questionnaire resource2. They established a mapping between the ODM elements and attributes and their FHIR resource counterparts. The transformation was implemented in Java, but the reverse transformation, that is, FHIR to ODM, was not implemented because the structure of FHIR Questionnaires was found to be more flexible. This work is related to ours because it is possible to export REDCap projects in ODM format. However, the FHIR Questionnaire resources provide limited interoperability and limited search capabilities. In order to make any clinical data searchable in a standardised way, it needs to be transformed into FHIR clinical resources, such as Observations and Conditions. This cannot be done automatically and this is the gap that FHIRCap attempts to address.
Yamamoto et al. developed a method to transform REDCap data into the CDISC SDTM format3. The method requires annotating REDCap forms with mapping annotations in the Field Annotation field (a flexible-use field available in REDCap since version 6.5). The application uses these annotations to generate mapping templates that are then completed manually and later used to generate the output data. This application uses an approach similar to ours but the target format is different and also simpler, and therefore does not require the implementation of a transformation language. Also, our application does not require modifying the REDCap forms.
Leroux et al. describe a mapping between ODM and FHIR resources and implement it as a semi-automatic process4. This work predates FHIRCap and helped inform the requirements of the REDCap to FHIR transformation language.
2. Methods
FHIRCap is the core component of a larger system, which was conceived as a mechanism to collect disparate pheno-type data into a standardised, centralised repository. Figure 1 shows a high level view of the system’s architecture. The entire system is made up of four components: one or more REDCap systems, the FHIRCap transformation engine, the CSIRO FHIR server and Ontoserver, the CSIRO FHIR terminology server. Any REDCap system can be registered with FHIRCap by providing its URL and an API token provided by the REDCap administrator.
Figure 1:
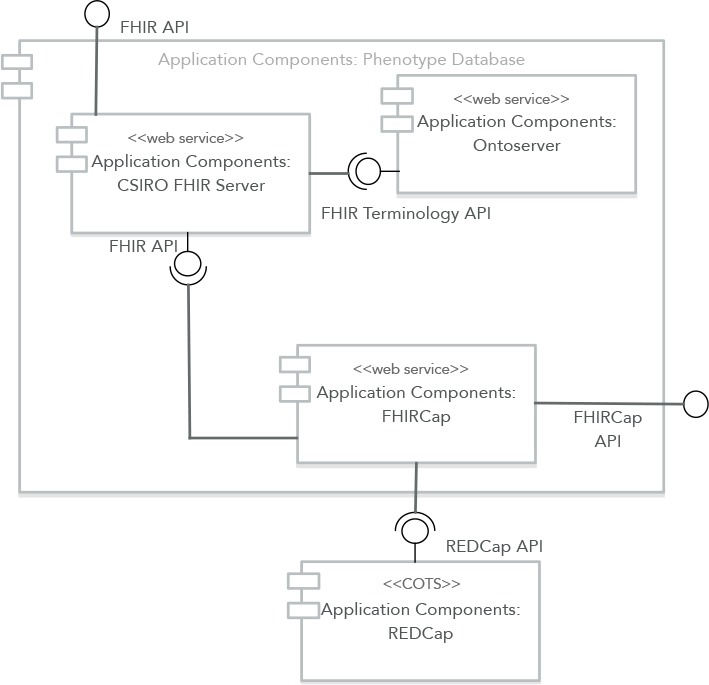
High-level architecture of the Phenotype Database.
FHIRCap is implemented as a Spring Boot micro-service and exposes a REST API that can be used to register REDCap projects, define transformation rules and mappings to standardised code systems, and upload the resulting FHIR resources to a centralised FHIR server. The CSIRO FHIR server is a modified version of the HAPIJPA open source FHIR server1 that adds the capability of plugging in an external FHIR terminology server to handle all terminology-related requests. The reason for this modification is that Ontoserver2, the final component in the application, is a specialised FHIR terminology server that provides more functionality and better performance than the default HAPI implementation.
The three key challenges in the implementation of the system are the transformation language, dealing with free text and synchronising the state of all the systems involved. A case study with one of the AG flagships was conducted to validate the feasibility of the proposed approach.
2.1. Transformation Language
The design of the transformation language was guided by two main goals: the need for an easy mechanism to express how a collection of REDCap forms should be represented as FHIR resources, which enables interoperability at the information model level, and the need for a mechanism to map custom codes defined in REDCap into standardised code systems such as SNOMED CT and HPO, which enables semantic interoperability.
The grammar was designed to be declarative, so the order of the rules does not affect the outcome of the transformation. The rules take the form
[condition]:
[resource_type]<[resource_id]> →
([attribute] = [value]) *
where the condition determines if the rule runs and the resource indicates the type and id of a FHIR resource to create, along with which attributes to set. Part of the grammar is generated automatically from the FHIR metadata, which allows changing the generated parser easily whenever the FHIR specification changes.
Consider the example REDCap field in Figure 2. This is a dropdown box used to select a primary tumour site. There are two main issues with this type of field: the codes used to represent the sites are not standardised, which hinders interoperability, and the number of choices is limited, which means that some sites might not be present and will end up in the ‘other’ category.
Figure 2:
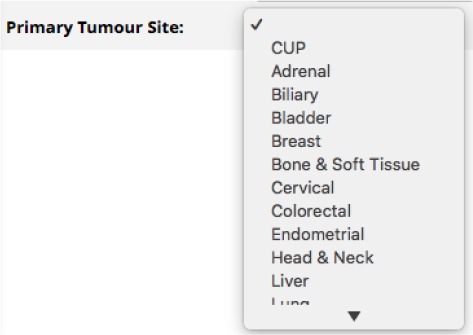
Simple REDCap drop-down ield example.
Using FHIRCap, a FHIR Condition resource can be created for each tumour site collected using this form. The rule
VALUE(ts) != “27” :
Condition<c1> →
bodySite[0] = CONCEPTJSELECTED(ts ),
code = CONCEPT.VALUE (
http ://snomed. info/sct 1108369006
);
states that if this form element (whose id is not shown in the figure but is ‘ts’) has a value other than 27 (which is the code for ‘Other’), then create or retrieve a Condition resource with id c1 and assign the first element of the bodySite attribute (which has multiplicity 0..*) to the concept that has been assigned to the selected code (more on this later) and set the Condition’s code to the SNOMED CT concept Neoplasm (108369006).
Note that FHIRCap is able to help overcome the first limitation by transforming the REDCap data into FHIR resources, which are interoperable, but cannot do anything about the second limitation, the limited number of choices available. REDCap has an option that allows populating text fields with values from a biomedical ontology. When this option is selected the field becomes an autocomplete widget. We are currently developing a component that provides similar functionality but using a FHIR terminology server to search for concepts. Using a FHIR terminology server has several advantages, such as the ability to use value sets to constrain the search.
The ‘CONCEPT_SELECTED’ statement instructs FHIRCap to look for a user-defined mapping between the custom REDCap code and a standardised equivalent. Once the rules are written, the system generates a list of required mappings that the user can retrieve as a spreadsheet or through a web interface. If the user does not enter a mapping, a default FHIR code system is created.
The CONDITION section of a rule determines if the rule runs. In this example, the rule is always run unless the value of the dropdown box is ‘Other’. There are many other conditions that can be used to trigger a rule. Table 1 shows a summary of all the available conditions.
Table 1:
Summary of rule conditions.
| Condition | Description |
|---|---|
| TRUE | Rule always runs. |
| FALSE | Rule never runs. |
| NOT condition | Rule runs if condition is false. |
| condition AND condition | Only runs if both conditions are true. |
| condition OR condition | Runs if either condition is true. |
| NOTNULL(id) | Rule runs if the REDCap field is populated. |
| VALUE(id) op vaue | Runs depending on the evaluation of the REDCap field against the value based on the operator, op which can be =, ≠, <, >, ≤ or ≥. |
| RESOURCES EXIST(id1, id2, ...) | Rule runs if all the referenced resources exist. |
The example rule also shows how values are assigned to the target resource attributes. In this case, the body site is assigned using the CONCEPT-SELECTED clause and the code is assigned using the CONCEPT VALUE clause. Table 2 shows a summary of all the available expressions used to assign values to resource attributes.
Table 2:
Summary of expressions used to assign resource attributes.
| Condition | Description |
|---|---|
| TRUE | True literal. |
| FALSE | False literal. |
| STRING_VALUE | A string literal. |
| NUMERIC_VALUE | A number literal. |
| REFERENCE | A reference to a FHIR resource defined in the rules. |
| CONCEPT | The concept mapped to a REDCap field. |
| CONCEPT_VALUE (code literal) | A concept literal. This has the form ‘system | code’. |
| CONCEPT_SELECTED(id) | The user-mapped concept for the selected value of a REDCap field. |
| Only applies to dropdown and radio button fields. | |
| SYSTEM(id) | The system part of a user-mapped concept. |
| CODE (id) | The code part of a user-mapped concept. |
| VALUE(id) | The value of a REDCap field. Works according to the type of field:
|
| LABEL(id) | The label of a REDCap field. |
| LABEL SELECTED(id) | The label of the selected option. Only applies to dropdown and radio button fields. |
2.2. Free Text Handling
REDCap fields with coded values, such as drop-down boxes, radio buttons and check boxes, can be mapped statically to standardised code systems. However, there are cases where free text fields are used to capture data that should have been coded, for example, when there is an ‘other’ category in the list, and the user is given the option of entering the name of the missing option.
FHIRCap deals with these cases by creating codeable concepts that contain just the free text that was entered by the user. Future work will explore adding an NLP module that can partially automate the assignment of codes to some of these entries. Note that this is not a trivial task and manual intervention is likely to be required. Also, if the text represents more than one element, for example, a list of differential diagnoses written in a single diagnosis field, it may be necessary to create more than one target resource to accurately represent what is captured in the source field.
2.3. Synchronisation Mechanism
One of the main challenges in the design of the system is keeping everything synchronised. Changes can originate from the following sources:
Changes in REDCap data. This is the most frequent source of changes. It happens when a REDCap user adds a new patient or modifies existing data, for example, to add information collected in a follow up visit. The transformation needs to be run again and some of the generated FHIR resources might need to be updated and any new resources added to the FHIR server.
Changes in REDCap metadata. This should not happen very often, especially if the REDCap project is in production. It is possible, however, that forms change at any time and this will impact the generated resources.
Changes in the transformation rules. This should not happen very often. However, if the transformation rules change, then the resulting FHIR resources might also change and might need to be updated.
Changes in the code mappings. This shouldn’t happen very often. However, if some of the mappings to standardised code systems change, because an incorrect mapping was done or because a code becomes inactive, for example, then the resulting FHIR resources might change and might need to be updated.
FHIRCap implements two mechanisms to deal with changes that originate in REDCap. The first one exposes an endpoint where REDCap can notify FHIRCap when data has changed, using the Data Entry Trigger plugin. Once a notification is received, FHIRCap will query the REDCap API and update the local information with any changes. This mechanism requires the REDCap administrator to configure the FHIRCap endpoint, so an alternative mechanism is also provided that doesn’t require any additional configuration in REDCap. This second mechanism regularly calls the REDCap API and checks for changes. The frequency of these checks can be configured. The disadvantage of this approach is that there will more, potentially unnecessary calls to the REDCap API. Also, the frequency needs to be set to a value that will not create excessive load on the REDCap server but will maintain the FHIR server up to date frequently enough.
Whenever the metadata in REDCap changes, there is the possibility that some of the transformation rules become invalid if, for example, a REDCap field that is referenced in the rules is deleted. The system was designed with this in mind, so invalid transformation rules are not rejected by the server but rather stored along with any errors, so the author can later edit them and fix any issues.
The changes that originate in FHIRCap, such as a change in the transformation rules, only affect the generated FHIR resources. FHIRCap also needs to synchronise the FHIR resources that are stored in the central FHIR server. Even though a whole new bundle of resources could be generated and uploaded each time something changes, this would cause the technical version of the unchanged resources to increase unnecessarily. Therefore, FHIRCap tags the generated resources so these can be retrieved and compared with the newly generated resources whenever something changes. This allows submitting only a delta with the resources that have changed to the central FHIR server.
2.4. Case Study
In order to validate the feasibility of the approach, a case study with the Somatic Cancer Flagship project was done. This project was chosen because it was, at the time, the most advanced in terms of patient recruitment, with data for 57 patients already in the system. Because data had already been captured, one of the restrictions was that the REDCap forms could not be modified.
A centralised REDCap instance is used in AG to store data for all its projects. This instance contains a single REDCap project that includes forms used to collect common data, such as study identifiers, demographics and personal contact information, and forms used to collect patient data specific to each Flagship project. A drop down box in one of the common forms is used to indicate to which Flagship project the patient belongs to and branching logic is used to display only the forms applicable to that project. This means, however, that huge rows of data with mostly empty values are created for every patient.
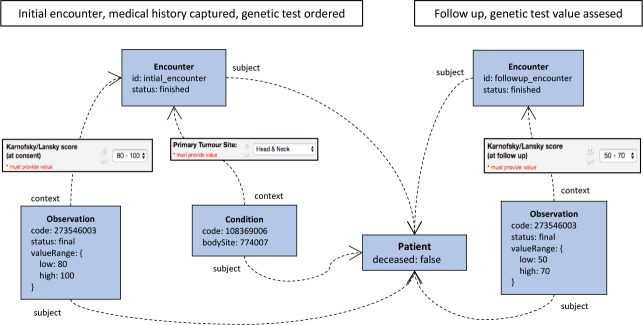
The patient-specific form for the Somatic Cancer Flagship is large, with 292 variables defined in REDCap. The form is designed to capture information in two encounters with the patient, an initial one where medical history is captured and a genetic test is ordered (at this stage the patient has already been recruited) and a follow up encounter where the value of the genetic test is assessed.
3. Results
3.1. Transformation Rules
The case study required a collection of 155 rules to produce the desired FHIR model. These rules were initially authored by an expert in FHIR and then iteratively validated by several clinicians in the flagship. The rules used in the Somatic Cancer Flagship transformation include the following information:
Study identifiers
Patient demographics
Personal contact information
Data entered at recruitment and after consent
Data entered after result review (3 months after reporting)
The following information was not included:
Data entered when the genomic report is received, because this can be captured more accurately from the curated variants
Figure 3 shows a high-level view of the transformation. Most of the patient’s phenotype is modelled using Conditions and Observations.
Figure 3:
High-level view of the REDCap to FHIR transformation for the Somatic Cancer Flagship.
The case study demonstrated that the proposed rules language is flexible enough to transform most data available in REDCap into a FHIR representation. However, if the REDCap forms are poorly designed, which is possible given the degree of flexibility offered by the REDCap form designer, some information might be impossible to extract. In Section 3.3, several recommendations about REDCap form authoring are given based on the findings from the case study.
3.2. Custom Code Mapping
Most custom REDCap codes were mapped to SNOMED CT and some codes related to previous tests were mapped to Logical Observation Identifiers Names and Codes (LOINC), the Human Genome Organisation Gene Nomenclature Committee (HGNC) and Sequence Variant Nomenclature codes.
3.3. REDCap Form Design Recommendations
The case study highlighted some patterns used by the form authors that made some subsets difficult to transform into FHIR resources. These patterns are illustrated through examples, the difficulties explained and some more suitable alternatives proposed.
3.3.1. Multiple data items captured in a single form element
When multiple data items are captured using a single form element, it can be hard to separate them during the transformation. Figure 4 shows an example where a single REDCap field is used to capture the onset of cancer for multiple relatives. Even though a format is suggested (e.g. mother- 60, sister-40 for the onset age) this is not enforced. This makes extracting this information very hard, especially when the suggested format is not used.
Figure 4:
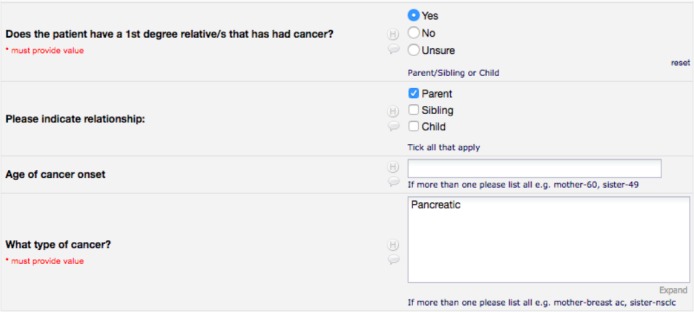
Fields in the Somatic Cancer REDCap form used to capture information about cancer in relatives.
This section of the form could be modelled as shown in Figure 5. Having a group of fields per relative makes extraction much easier and eliminates any potential ambiguities.
Figure 5:
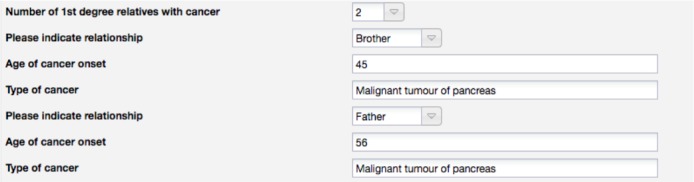
Proposed redesign of the fields used to capture cancer in relatives in the Somatic Cancer REDCap form.
3.3.2. Ambiguous form fields
Some fields are defined in a way that their interpretation is ambiguous. For example, Figure 6 shows the REDCap field used to capture the current or most recent treatment at time of consent. The problem with this design is that it is impossible to determine if the treatment is currently active or not, i.e., the treatment can be either active (current) or the most recent but not currently active, but it is impossible to determine this from the form. This can be fixed by simply adding a checkbox to indicate if the treatment is currently active or not.
Figure 6:

REDCap field used to capture the most recent treatment at consent.
3.3.3. Multiple elements are not grouped together
Figure 7 shows the forms elements used to capture a cancer diagnosis. In this case, the user can select the type of cancer diagnosis. If “Newly diagnosed metastatic” or “Progressive Metastatic or Metastatic Relapse” is selected as the diagnosis type, then an additional input for the date of the metastasis diagnosis is shown. This date can be different from the date of the primary diagnosis, which suggests that these are two diagnostic reports, one for the initial diagnosis and a second one for the diagnosis of the metastasis. However, the additional data associated to these diagnostic reports, such as the tissue sources, is captured in a single field, making it hard to determine how to associate it to each report.
Figure 7:

REDCap fields used to capture clinical history of cancer.
Figure 8 shows the proposed redesign of these form elements (only a subset of the grouped fields is shown for brevity). Using this approach, it is easy to determine which tissue samples belong to each diagnostic report.
Figure 8:
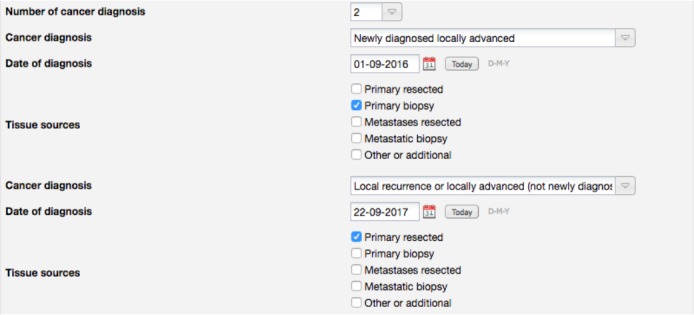
Proposed redesign of the REDCap fields used to capture clinical history of cancer.
3.3.4. Free text
Free text fields can be used to capture different types of data in REDCap. Figure 9 shows an acceptable use of a free text field. In this case, it is used to capture a numeric value that represents an estimate of the level of smoking of the patient. The field is set to the correct type in REDCap, which validates that the user enters a numeric value.
Figure 9:

REDCap field used to capture an estimate of pack years for smokers.
In other cases, free text fields are used to allow the user to add a value that is not present in a set of predefined choices. Figure 10 shows a free text field that can be used to enter a tumour site if it is not available in the drop-down choices. In this case, FHIRCap provides a very simple mechanism that assigns a generic code to the free text and allows replacing that code with an existing code from a terminology such as SNOMED CT. However, this mapping has to be done manually for each new value that is entered and therefore generates additional maintenance work. The system will be able to detect if the same concept has been entered before and assign the correct mapping in those cases.
Figure 10:
REDCap fields used to capture the patient’s primary tumour site.
As mentioned previously, we are currently developing a component that provides auto-complete style searching capabilities for REDCap fields backed by a FHIR-based terminology server. We recommend using this component when it is released. In the meantime, we recommend using the biomedical ontology searching capability already available in REDCap.
This recommendation applies only to fields that could be coded. Other free text fields might be needed, especially in a research context where some concepts may still not be available in an ontology or where more detailed/nuanced information is required. In addition to the clinical resources produced by the transformation rules, FHIRCap generates a set of Questionnaire FHIR resources that represent the REDCap forms, so all the free text entered by the users will be available.
4. Discussion
Having high quality clinical data from disparate sources available in a standardised manner is important when doing data analysis. In practice, however, data is still being collected in non-standard ways. This is usually manageable in the context of a single study but becomes problematic when data from multiple studies across different organisations is required.
Out tool provides an effective mechanism to extract data from REDCap into a standardised FHIR representation, with custom codes mapped to standard terminologies such as SNOMED CT. Even though there is significant amount of effort involved in writing and verifying the correctness of the rules, the work only needs to be done once for every project.
The case study provided valuable insight into the way REDCap forms are currently being authored and helped produce a set of recommendations around form authoring that aim to facilitate the transformation into a FHIR model. The list of recommendations is by no means comprehensive, but it is useful as a guide to understanding common anti-patterns found in REDCap forms.
The coded FHIR representation has many advantages over the original data stored in REDCap. In addition to standardising data from heterogeneous sources, an interesting feature provided by the FHIR standard, when the resources are coded with a terminology such as SNOMED CT, is semantic searching. This allows using search operators such as :below to search for resources that have a code subsumed by the specified search code, and therefore enables queries such as “give me all the patients with a condition that is any kind of kidney lesion”. This type of search is not possible with the original REDCap data.
One limitation of this work is that the case study was done with a single Flagship project. Even though the REDCap form used to capture data was complex and the rules language was successfully used to extract most data, there could be additional functionality that might be important to support that was not present in this form.
Another limitation is that the FHIRCap rules language does not support operators such as addition or subtraction. This means that only static values and data available in the REDCap form can be used to populate the target FHIR resources. A decision was made not to support these operators, at least in the initial version, mostly because they are already available in REDCap, so in most cases it should be possible to create a field in REDCap with the desired computation. In our case study, however, this is problematic, because in the Somatic Cancer REDCap form, the date for the follow up encounter is not captured directly and even though we know that it is supposed to happen roughly three months later, we cannot tell FHIRCap to add three months to the initial encounter’s date. This could be easily fixed by adding a calculated field in REDCap, but unfortunately, for this case study, it was not desirable to modify the forms.
Finally, it is important to note that FHIRCap is a generic platform that allows creating any FHIR resource, but does not provide any guidance around which resources to use in a specific domain. This is not in the scope of this work but it is necessary to achieve interoperability.
5. Conclusion
In this paper we described a system that enables exporting data captured in REDCap as FHIR resources and mapping custom-coded values to standardised codes from terminologies such as SNOMED CT and LOINC. This approach does not require modifications to the REDCap forms and can be used to integrate multiple REDCap systems into a single FHIR repository.
A case study with one of the Australian Genomics Flagship projects showed that most data can be extracted using the rules language but also highlighted that certain patterns used to author the forms can make data hard to extract automatically. Some recommendations around form authoring were given in order to facilitate the extraction process. Also, the case study showed that most of the clinical concepts in the forms were available in standardised terminologies.
Even though authoring the transformation rules is a manual process that takes some effort, it only needs to be done once and has many advantages, such as allowing users to continue working on a platform they already know while enabling a centralised repository based on the FHIR standard that can then be queried in a standard way.
6 Acknowledgements
Australian Genomics is supported by the National Health and Medical Research Council (GNT1113531). The authors would like to thank Donna Truran for mapping the custom codes in REDCap to standard clinical terminologies and the Somatic Cancer Flagship for sharing their data and reviewing the transformation.
Footnotes
References
- 1.Paul A. Harris, Robert Taylor, Robert Thielke, Jonathon Payne, Nathaniel Gonzalez, Jose G. Conde. Research electronic data capture (redcap) - a metadata-driven methodology and workflow process for providing translational research informatics support. Journal of Biomedical Informatics. 2009;42(2):377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doods J, Neuhaus P, Dugas M. Converting odm metadata to fhir questionnaire resources. Studies in health technology and informatics. 2016;228:456. [PubMed] [Google Scholar]
- 3.Keiichi Yamamoto, Keiko Ota, Ippei Akiya, Ayumi Shintani. A pragmatic method for transforming clinical research data from the research electronic data capture redcap to clinical data interchange standards consortium (cdisc) study data tabulation model (sdtm): Development and evaluation of redcap2sdtm. Journal of Biomedical Informatics. 2017;70:65–76. doi: 10.1016/j.jbi.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Hugo Leroux, Alejandro Metke-Jimenez, Michael J. Lawley. Towards achieving semantic interoperability of clinical study data with fhir. Journal of Biomedical Semantics. 2017 Sep;8(1):41. doi: 10.1186/s13326-017-0148-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender D, Sartipi K. In Proceedings of the 26th IEEE International Symposium on Computer-Based Medical Systems. 2013. Jun, Hl7 fhir: An agile and restful approach to healthcare information exchange; pp. 326–331. [Google Scholar]
- 6.Joshua D. Franklin, Alicia Guidry, James F. Brinkley. Journal of Biomedical Informatics. Vol. 44. AMIA Joint Summits on Translational Science 2011; 2011. A partnership approach for electronic data capture in small-scale clinical trials; pp. S103–S108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Julie A. McMurry, Sebastian Kohler, Nicole L, Washington James P. Balhoff, Charles Borromeo, Matthew Brush, Seth Carbon, Tom Conlin, Nathan Dunn, Mark Engelstad, Erin Foster, Jean-Philippe Gourdine, Julius O.B, Ja-cobsen Daniel Keith, Bryan Laraway, Jeremy Nguyen Xuan, Kent Shefchek, Nicole A. Vasilevsky, Zhou Yuan, Suzanna E. Lewis, Harry Hochheiser, Tudor Groza, Damian Smedley, Peter N. Robinson, Christopher J. Mungall, Melissa A. Haendel. Navigating the phenotype frontier: The monarch initiative. Genetics. 2016;203(4):1491–1495. doi: 10.1534/genetics.116.188870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamed Hassanzadeh, Anthony Nguyen, Bevan Koopman. In Proceedings of the Australasian Language Technology Association Workshop 2016. 2016. Evaluation of medical concept annotation systems on clinical records; pp. 15–24. [Google Scholar]