Abstract
Diabetic Kidney Disease (DKD) is a critical and morbid complication of diabetes and the leading cause of chronic kidney disease in the developed world. Electronic medical records (EMRs) hold promise for supporting clinical decision-making with its nationwide adoption as well as rich information characterizing patients’ health care experience. However, few retrospective studies have fully utilized the EMR data to model DKD risk. This study examines the effectiveness of an unbiased data driven approach in identifying potential DKD patients in 6 months prior to onset by utilizing EMR on a broader spectrum. Meanwhile, we evaluate how different levels of data granularity of Medications and Diagnoses observations would affect prediction performance and knowledge discovery. The experimental results suggest that different data granularity may not necessarily influence the prediction accuracy, but it would dramatically change the internal structure of the predictive models.
Keywords: DKD; Predictive Modeling; Data Representation; EMR ontology, Gradient Boosting Machine
Introduction
Diabetic kidney disease (DKD) is one of the most frequent and dangerous complications of Diabetes Mellitus (DM), affecting about 20% to 40% of patients with type 1 or type 2 DM. DKD is a major cause of morbidity and mortality in DM patients and single most common cause of end-stage renal disease (ESRD)1. Thus, it is extremely important to develop predictive models for the early identification of patients at risk for developing DKD and implement appropriate interventions.
In order to build an accurate predictive model, choosing appropriate candidate predictors is critical. The widely adopted approaches for DKD risk stratification are mainly hypothesis-driven, which combine expert opinions with systematic literature review of prognostic factors that are already known to be associated with the target outcome2-5. However, based on findings from previous studies, clinical intuition may not be suitable for identifying candidate predictors, owing to the fact that this type of selection is subjective and can miss out the potential unknown predictors6. An unbiased data-driven approach is to utilize all available data to build the model and let the algorithm identify the top ranked predictors. Electronic Medical Record (EMR) has become a primary data resource for such approach. To facilitate the reuse of EMR data for research, the National Institutes of Health (NIH) funded the Informatics for Integrating Biology and the Bedside (i2b2) National Center for Biomedical Computing to provide an open-source clinical data informatics framework7. Since i2b2’s first release in 2007, over 240 scholarly articles have been published using data derived from i2b2-based repositories.
However, the built-in hierarchical representation of clinical knowledge in the i2b2-based repositories has been under- exploited. Granularity at which clinical features are represented can make a big difference in predictive modeling and knowledge discovery. For example, “250” is the general ICD9 code assigned for “Diabetes Mellitus”, which can be specified as “250.1” for “Diabetes with ketoacidosis” and further specified as “250.10” for “Diabetes with ketoacidosis, type II or unspecified type, not stated as uncontrolled”, depicting an increasing richness of diagnostic detail. In addition, the specificity can be extended from a different perspective by differentiating the types or sources of diagnosis, for example, primary or non-primary diagnosis. Another example, “Furosemide” is a generic drug name for a type of “Loop Diuretics”, which can be generally grouped as “Diuretics” and further as “Cardiovascular Medications”, while “Furosemide” can also be sub-classified into different form, strength and dosage, as well as associated with clinical activities, i.e. inpatient or outpatient orders. In practice, an arbitrary decision is usually made on which level of abstraction should be used for predictive model development, or simply mapping to one of many external ontologies with minimal manual work without much discussion9,10.
The contribution of this study is two-fold. First, we attempted to leverage the full breadth of the i2b2-structured clinical data in the purpose of improving overall DKD prediction accuracy and discovering potentially new discriminating factors over a more comprehensive diabetic population. Second, we investigated how well the knowledge, in relation to DKD prediction, got retained with respect to representations at different granularity. More specifically, when we kept the features at their leaf levels, we were able to view each feature with their finest granularity (e.g., dosage information for medications or diagnosis with very specific details). However, we might misrepresent a “complete” feature by a bunch of independent “partial” features and risk diffusing the real predictive power of that “complete” feature. This discussion tied into the contrast between feature selection and feature extraction methods11, as the former is designed to eliminate redundant features while the latter is to combine correlated features. Instead of resorting to more complex feature extraction techniques, we took advantage of the hierarchical ontology for “Medications” and “Diagnoses” data built in i2b2, which provided a convenient feature extraction solution by rolling up the features to higher level along the ontological trees.
Methods
Diabetes Definition
We adopted the SUPREME-DM definition of diabetes in this study rather than simply relying on diagnosis codes. Diabetes was defined based on a) use of glucose-lowering medications (insulin or oral hypoglycemic medications); or b) level of hemoglobin A1C of 6.5% or greater, random glucose of 200 mg/dL or greater, or fasting glucose of 126 mg/dL on at least two different dates within two years; or c) any two type 1 and type 2 DM diagnoses been given on two different days within 2 years; or d) any two distinct types of events among a),b),or c); e) excluding any gestational diabetes (temporary glucose raise during pregnancy)12.
DKD Definition
Diabetic Kidney Disease (DKD) was defined as diabetes with the presence of microalbuminuria (or even proteinuria), impaired glomerular filtration rate (GFR), or both13,14. More specifically, microalbuminuria was defined as ratio of urine albumin to creatinine (ACR) being 30 mg/g or greater (similarly, proteinuria was defined as ratio of urine protein to creatinine being 30 mg/g or greater) 13,14. Impaired GFR was defined as the estimated GFR (eGFR), an age, gender, race adjusted serum creatinine concentration, being less than 60 mL/min/1.73m2. Since impaired GFR is also a manifestation of acute kidney injury (AKI), which may not necessarily indicate an immediate transition to chronic kidney disease, any low eGFR encounter that was concurrent with an AKI session was excluded, where AKI session was identified by diagnosis codes ICD9:584 or ICD10:N17.
Study Cohort
A retrospective cohort of 35,779 DM patients, who had at least one valid eGFR or ACR record, was eligible for this study. We excluded all the patients who had any kidney disease manifestation (e.g. chronic kidney disease diagnosis, low eGFR, or microalbuminuria) prior to DM onset. The case group included all DKD patients with their DKD onset time defined as the first time of their abnormal eGFR or ACR. The control group was defined as DM patients whose eGFRs had been all above or equal to 60 mL/min/1.73m2 and had never had microalbuminuria, with the endpoint defined as the last time of their normal eGFR or ACR. In the final cohort, we collected 20,718 patients and 7,834 (38.6%) were DKD patients.
HERON Data
At the University of Kansas Medical Center (KUMC), we have established an i2b2-based Healthcare Enterprise Repository for Ontological Narration (HERON) that integrated data from disparate information systems with available data types including patient demographics, medication, laboratory results, diagnoses data, etc. and continuously evolved to better serve the needs of researchers15. Clinical observations, or facts, in HERON can be roughly classified into 11 data types based on the source system (Table 1), which are well linked at patient and encounter level16. Each data type is a mix of categorical and numerical data elements. For data types such as laboratory tests and vital signs, the values are more crucial in reflecting a patient’s status rather than the mere presence of the facts. As a result, we created an indicator variable equal to 1 for the presence of a categorical feature and 0 otherwise, while kept the original values for all the numerical features. As aforementioned, HERON provides an additional attribute indicating either the type/source of a fact (e.g. primary diagnosis (@Primary) or non-primary diagnosis (@Non-Primary)) or different aspects of the same fact (e.g. in-, or out- patient medications prescribed (@InPatient, @OutPatient), or prescribed dosage (@Dose|mg)) which was used to further decompose a bulk feature into granular but more meaningful pieces.
Table 1.
Root Data Types in HERON
| Data Type | Descriptions | No. of Features1 | Patients Frequency (%) |
|---|---|---|---|
| ALERTS | Includes drug interaction, dose warnings, drug interactions, medication administration warnings, and best practice alerts | 3804 | 15733 (75.9%) |
| DEMOGRAPHICS | Basic demographics such as age, gender, race, and etc., as well as their reachability, and some geographical information | 123 | 20718 (100.0%) |
| DIAGNOSES | Mostly organized using ICD9 and ICD10 hierarchies but also includes Intelligent Medical Objects interface terms that are mapped to ICD9 and ICD10 codes | 47711 | 19712 (95.1%) |
| HISTORY | Contains family, social (i.e. smoking), and surgical history | 806 | 16458 (79.4%) |
| LABORATORY TESTS | Results of a variety of laboratory tests, including cardiology labs. Note that the actual lab values are used in modeling, if available | 5335 | 15753 (76.0%) |
| MEDICATIONS | Includes dispensing, administration, prescriptions, as well as home medication reconciliation at KUH. | 28315 | 11525 (55.6%) |
| PROCEDURES | Includes CPT professional services and inpatient ICD9 billing procedure codes | 2548 | 18842 (90.9%) |
| ORDERS | Includes physician orders for non-medications such as culture and imaging orders | 3223 | 19070 (92.0%) |
| REPORTS | Includes structured elements from physician notes, such as progress notes and operative notes | 3090 | 15567 (75.1%) |
| VIZIENT | (formerly UHC) Includes both billing classifications such as Diagnostic Related Groups (DRG), comorbidities, discharge placement, LOS, and national quality metrics. | 1538 | 3897 (18.8%) |
| VISIT DETAILS | Includes visit types, vital signs collected at the visit, discharge disposition and clinical services providing care. | 1127 | 20687 (99.8%) |
This is not all distinct concepts from the entire HERON system, but only the total number of distinct features that had ever been recorded for at least one patient in the study cohort.
For each patient, we extracted their most recent values for all the available features from the 11 types of data at least 6 months prior to their DKD onset time or endpoint. Initially, a total of 96,605 distinct features were available for our study cohort with more than half of them coming from Medications and Diagnoses. Note that serum creatinine and albumin were removed from the candidate feature list even though they had been shown to be predictive in other prospective studies17, because they were collinearly related to the two labs (eGFR and ACR) which we used to define DKD.
Data Representation
High dimensionality is a key challenge in this data set, which, in particular, stemmed from the two data types: Medications and Diagnoses (Table 1). The HERON hierarchical ontology provides a plausible solution for seeking possible lower-dimensional representation of Medications and Diagnoses data such that sufficient information on the original data of these two types can still be preserved. Medication concepts are carefully mapped from names in our Epic EMR system to Semantic Clinical Drug Form (SCDF) or Semantic Clinical Brand Form (SCBF) and grouped under Veterans Administration (VA) class defined by National Drug File Reference Terminology (NDF-RT). Diagnosis codes can be grouped by levels of specificity implied by the number of digits in ICD codes and be further grouped by broader disease groups. Table 2 and Table 3 demonstrate two examples of different representations of Medications and Diagnoses data, respectively.
Table 2.
Example of hierarchical ontology for a medication concept in HERON
| Representation Type | Representation Value |
|---|---|
| RX_RAW | Canagliflozin 100 MG PO Tab@Dose|mg = 300 MG |
| RX_CONCEPT | Canagliflozin 100 MG PO Tab |
| RX_CLASS_LEV (SCDF or SCBF) | Canagliflozin Oral Tablet |
| RX_CLASS_LEV3 (VA_LEV3) | [HS502] Oral Hypoglycemic Agents, Oral |
| RX_CLASS_LEV2 (VA_LEV2) | [HS500] Blood Glucose Regulation Agents |
| RX_CLASS_LEV1 (VA_LEV1) | [HS000] Hormones/Synthetics/Modifiers |
Table 3.
Example of hierarchical ontology for a diagnosis concept in HERON
| Representation Type | Representation Value |
|---|---|
| DX_RAW | 250.13@Primary |
| DX_CONEPT | 250.13 Diabetes with Ketoacidosis, Type I, Uncontrolled |
| DX_CLASS_LEV4 | 250.1 Diabetes with Ketoacidosis |
| DX_CLASS_LEV3 | 250 Diabetes Mellitus |
| DX_CLASS_LEV2 | 249-259.99 Diseases of Other Endocrine Glands |
| DX_CLASS_LEV1 | 240-279.99 Endocrine, Nutritional and Metabolic Diseases, and Immunity Disorders |
Experimental Methodology
Considering the multi-way correlation, or multi-collinearity, which has always been an issue in EMR-based learning, we adopted a decision-tree-based ensemble method, Gradient Boosting Machine (GBM), as the base learner for building predictive models throughout the experiment. GBM is a family of powerful machine-learning techniques that have shown considerable successes in a wide range of practical applications. GBM has been known for its prediction accuracy, performance consistency, and ability to learn non-linear relations or correlations in many practical applications18-22. It is an ensemble learning technique, which combines a large number of weak and simple learners to obtain a stronger ensemble prediction. We chose GBM as our base learner not only for its robustness against high- dimensionality and collinearity, but also because it embeds a feature selection scheme within the process of model development23. In addition, the adopted GBM algorithm used a default strategy to handle missing values: instead of requiring extra imputation, the algorithm always accounted for a missing value split at each tree node within the ensemble24.
We used the area under the receiver operating character curve (AUC), sensitivity and specificity as the consensus metrics for comparing predictive performance. To evaluate the importance of a feature or “gain”, the averaged importance was taken across all boosted trees, while each tree-specific importance was calculated as the cumulative improvement of AUC attributed to splitting by that feature weighted by the number of observations the node is responsible for, which was then normalized to a percentage. The higher the “gain” was, the more relative contribution the feature had made in separating DKD patients from non-DKD ones. “Rank” was based on the “gain” in a descending order. Cumulative “gain” of features from the same type was used to measure the importance of that data type.
To control for overfitting, we randomly partitioned the study cohort into training and testing sets as 70/30, where GBM model was built on the training set and AUC calculated on the testing set for comparison. As a GBM model performance is highly related to hyper-parameters such as learning rate, number of trees, and depth of trees, we tuned the hyper-parameters within the training set using 10-fold cross validation.
Results
Baseline Model
Starting from all eligible clinical observations available for the training cohort and using their raw values (RX_RAW, DX_RAW), the baseline GBM model was first built based on 96,605 distinct features. The baseline model achieved an AUC of 0.8594 with a 95% confidence interval of (0.8488, 0.8681), with contributions from each data type listed in Table 4. “Number of Features” counts the number of distinct features selected by the baseline model of each data type, while “Best rank” (best rank of features of the same type) and “Median rank” (median rank of features of the same type), as well as “Gain” implied how each data type contributed to the model. Overall, the model selected 2,524 features from the 11 root data types, which received positive “Gain”, or had been evoked by at least one of the decision trees. It is worth noting that Medications and Diagnoses, being the most high-dimensional feature spaces, were both contributing less than 10% with their features typically ranked lower than the other data types in terms of the “Median rank”, which were 1712.5 and 1632 respectively. This finding motivated us to further investigate if the loss of predictive power from Medications and Diagnoses features was a result of “curse of cardinality”.
Table 4.
Contribution distribution among data types using raw input (ordered in decreasing order by “gain”)
| Data Type | Number of Features | Gain | Best rank | Median rank |
|---|---|---|---|---|
| LABORATORY TESTS | 314 | 28.73% | 7 | 670 |
| VISIT DETAILS | 212 | 22.27% | 1 | 884 |
| DEMOGRAPHICS | 47 | 13.61% | 2 | 292 |
| ALERTS | 114 | 10.10% | 3 | 1188.5 |
| DIAGNOSES (DX) | 576 | 7.15% | 39 | 1712.5 |
| PROCORDERS | 249 | 6.36% | 8 | 1227 |
| REPORTS | 353 | 4.36% | 180 | 1291 |
| MEDICATIONS (RX) | 200 | 2.18% | 152 | 1632 |
| PROCEDURES | 151 | 2.11% | 179 | 1351 |
| HISTORY | 77 | 1.98% | 41 | 946 |
| UHC | 63 | 1.14% | 92 | 1344 |
Data Representation
A typical depth of ontology for a particular medication concept or a diagnosis concept is five, as demonstrated in Tables 2 and 3. By adding up “modifiers”, there was an exhaustive list of 36 (= 6×6) possible combinations of different representations for Medications or Diagnoses data. Overall out-of-sample AUC and the significance of its improvement25 over the baseline model, as well as the optimal sensitivity (Sens) and specificity (Spec) are reported in Table 5.
Table 5.
AUC comparisons for different Medication and Diagnoses data representations
| AUC / Sens / Spec | DX RAW | DX CONCEPT | DX CLASS_LEV4 | DX CLASS_LEV3 | DX CLASS_LEV2 | DX CLASS_LEV1 |
|---|---|---|---|---|---|---|
| RX RAW | 0.8594 / 0.7635 / 0.7797 | 0.8590 / 0.7471 / 0.7922 | 0.8607 / 0.7472 / 0.7946 | 0.8597 / 0.7538 / 0.7802 | 0.8592 / 0.7581 / 0.7798 | 0.8573 / 0.7829 / 0.7678 |
| RX CONCEPT | 0.8589 / 0.7537 / 0.7857 | 0.8601 / 0.7655 / 0.7741 | 0.8599 / 0.7488 / 0.7924 | 0.8595 / 0.7629 / 0.7777 | 0.8592 / 0.7617 / 0.7751 | 0.8578 / 0.7575 / 0.7498 |
| RX CLASS_LEV4 | 0.8582 / 0.7641 / 0.7747 | 0.8624** / 0.7646 / 0.7786 | 0.8617* / 0.7730 / 0.7694 | 0.8610 / 0.7771 / 0.7640 | 0.8598 / 0.7701 / 0.7676 | 0.8573 / 0.7871 / 0.7600 |
| RX CLASS_LEV3 | 0.8599 / 0.7646 / 0.7724 | 0.8616* / 0.7642 / 0.7754 | 0.8590 / 0.7604 / 0.7800 | 0.8605 0.7479 / 0.7895 | 0.8604 / 0.7577 / 0.7725 | 0.8584 / 0.7751 / 0.7747 |
| RX CLASS_LEV2 | 0.8588 / 0.7502 / 0.7877 | 0.8608 / 0.7637 / 0.7812 | 0.8595 / 0.7634 / 0.7759 | 0.8614 / 0.7555 / 0.7853 | 0.8605 / 0.7626 / 0.7745 | 0.8573 / 0.7246 / 0.7732 |
| RX CLASS_LEV1 | 0.8578 0.7705 / 0.7668 | 0.8589 / 0.7582 / 0.7792 | 0.8588 / 0.7605 / 0.7766 | 0.8589 / 0.7732 / 0.7656 | 0.8591 / 0.7635 / 0.7695 | 0.8568 / 0.7455 / 0.7542 |
suggest significant AUC increase over baseline model (DX RAW and RX RAW), where * indicates weak significance with p-value between 0.01 and 0.05 and ** indicates strong significance with p-value less than 0.01.
The raw values (DX_RAW, RX_RAW) were used in building the baseline model, which was used as the reference for evaluating if any AUC change was significant. Besides a strong significant increase of AUC to 0.8624 (p-value < 0.01) identified at (DX_CONCEPT, RX_CLASS_LEV4). The prediction performance was not affected by changes of data granularity, nor affected with certain visible pattern.
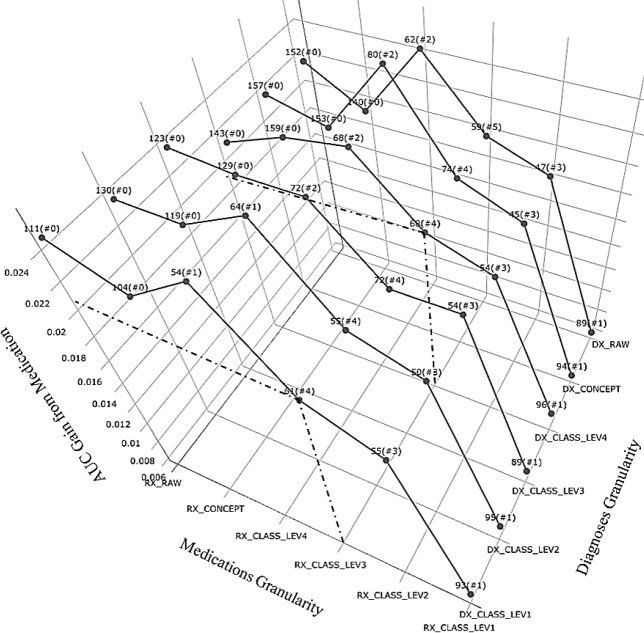
Figure 1 and Figure 2 both depicted how data representation variation changed the internal structure of the ensemble model, or feature importance ranking. In Figure 1, it shows that even though RX_RAW always contributed to AUC improvement relatively the most, it only pushed the medication features to better rankings when we rolled the concepts up to MED_CLASS_LEV4, MED_CLASS_LEV3 or MED_CLASS_LEV2, but not necessarily higher. It is worth noting that Figure 1 also suggested that such trend persisted across different Diagnosis granularities.
Figure 1.
AUC Gain from Medication (with best rank, i.e. # features ranked among top 100, marked)
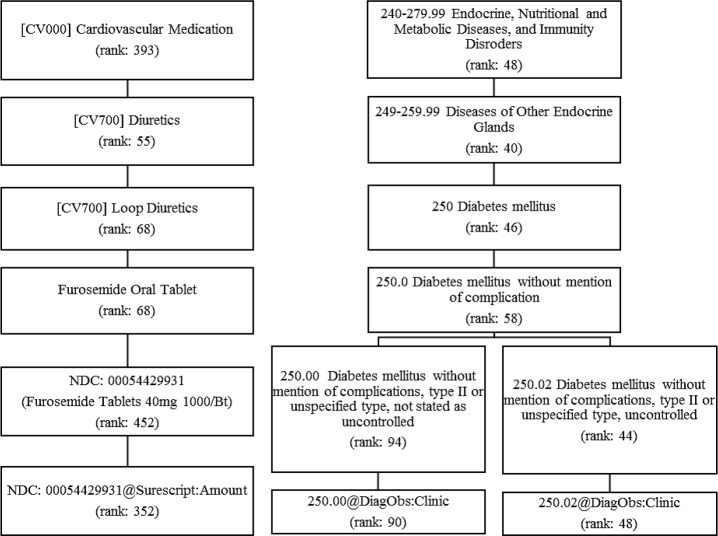
Figure 2.
Examples of important Medications and Diagnoses features with various levels of granularity
To take a closer look at the important medication features, we picked out the examples by fixing Medications at RX_CLASS_LEV3 since the most numbers of medication features made to the top 100 importance list at this level. Two types of Diuretics, Loop Diuretics and Potassium Sparing/Combinations Diuretics, Insulin and Calcium Channel Blockers were identified as part of top drivers of DKD risk, which could be potential medication signals being ignored when we broke them down into granular terms. For better illustration, we highlighted an example of medication feature in Figure 2. When the “Furosemide Oral Tablet”, a generic drug name of Loop Diuretics got subdivided into finer pieces as “Furosemide Tablets 40mg 1000/bottle”, or when the class “Diuretics” got grouped to more coarse-grained granularity as “Cardiovascular Medications”, the “rank” would suddenly drop either way.
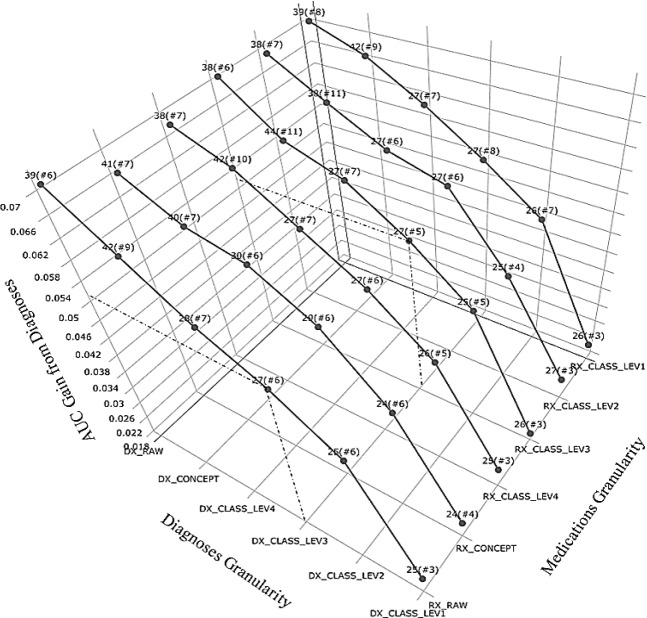
Data granularity affected Diagnoses features in a bit more notable way as shown in Figure 3. On one hand, it consistently presented a negatively monotonic relationship between importance of Diagnoses features and diagnosis granularity. It indicated that even though there existed some dominantly predictive diagnosis groups, having them decomposed into finer features that carry more specific information could collectively strengthen the impact of Diagnoses.
Figure 3.
AUC Gain from Diagnoses (with best rank, i.e. # features ranked among top 100, marked)
On the other hand, the top rankings of Diagnoses features stayed relatively stable around above 50 and the number of top 100 ranked features remained around 7 when data being grouped above level DX_CONCEPT and below DX_CLASS_LEV1. As displayed in Figure 2, the predictability of DM2 diagnoses was preserved across different levels of granularity, for which the ranks did not change considerably until reaching the level of DX_CONCEPT. When DM diagnosis of 250.0 broke down into 250.02 and 250.00, the feature importance ranking got stratified further into a much higher ranking for one (250.02, DM2 stated as uncontrolled) and a lower for the other (250.00, DM2 not stated as uncontrolled).
Discussion
It is worth noting that the HERON medication hierarchy is derived by a method of knowledge mapping across multiple resources such as Epic and RxNorm tables, which, to some extent, can be perceived as a feature extraction process guided by an expert. According to our experimental results, it seemed that how Medication features helped describe the DKD phenomena could be quite sensitive to how those features were represented or abstracted. Grouping the medication concepts into broader classes would potentially enhance their signals for predicting DKD. For example, two types of Diuretics (loop-acting and potassium-sparing diuretics), which had been discovered to be associated with adverse renal outcomes26, got recognized by our model better (Figure 3) when they were grouped at higher level in spite of losing some drug details.
In contrast, the feature importance of Diagnoses appeared to be more robust against data representation than Medications. It could be accredited to Diagnoses features being stably predictive by their nature or was caused by practices that physicians or billers usually follow when they recorded the diagnosis codes. Take diabetes diagnosis as an example, among a total of 10,712 patients in HERON who had ever been assigned a diagnosis code within the group of “249-259.99”, 98% of them were “250”, among which 85% were further defaulted to “250.0” or even “250.00”. As a result, for DKD prediction, it may help purify the signals from Diagnoses by separating the carefully recorded diagnoses from the others with more granular information.
While not the focus of this paper, the baseline model picked out some interesting features that may further our discussion on data granularity. For example, the most important feature that came from “Visit Details” was the “Superscript Encounter” indicator, which was recorded as 1 if a medication-related activity (e.g. fill or refill) occurred at pharmacy for an outpatient and 0 otherwise. Our baseline model suggested a protective effect of “Superscript Encounter”, that is, having at least 1 “Surescript Encounter” would decrease the risk of getting DKD, which was in line with the notion that “Surescript Encounter” carried some information about whether an outpatient was compliant with his/her prescriptions or not. It would make better practical sense or even achieve better prediction performance if we could associate such compliance indicator with a particular drug or drug class.
Limitations
The data granularity discussion was only done on Medications and Diagnoses in this study, which could be extended for examining other categorical facts, in particular those with higher prediction impact such as Alerts or Procedures, where such justifiable hierarchical ontology could be available in EMR. For example, the procedure of “Endoscopy Procedures on the Heart and Pericardium (CPT:1006197)” is a type of “Surgical Procedures on the Heart and Pericardium (CPT:1006057)”, which is under the umbrella of “Surgical Procedures on the Cardiovascular system (CPT:1006056)”.
When it comes to numerical observations like drug dosages, strength and amount, we adopted a simple approach of dropping the actual values but using the mere exposure of that drug, which could be handled with better complexity resembling the “morphine milligram equivalent (MME)” system27. However, it may not be generalizable to other numerical observations.
We only followed i2b2 ontology to uncover the influence of different levels of data granularity on predicting DKD. It is also possible to compare with other data abstraction methods like Clinical Classification Software (CCS) for collapsing diagnoses28, Generic Product Identifier (GPI) for classifying drugs from their primary therapeutic use down to the unique interchangeable product regardless of manufacturer or package size29, or even algorithm-based feature extraction techniques such as latent factor analysis30. In addition, it is also worthwhile to extend this data granularity work to other machine learning algorithms to check consistency.
Conclusion
Our experiments have shown great promises on improving DKD risk predictions by fully exploiting the diversity of the i2b2-based EMR database. We have also identified the utility of the hierarchical structure, built within i2b2, as a valuable but under-utilized resource for representing expert knowledge and facilitating interpretable data abstraction. Moreover, we have shown how the model specification, which was directly reflected by feature importance ranking, can be significantly affected by data abstraction at different levels of granularity and further impact what knowledge could be learned from EMR data. Our findings have potential implications for a number of studies based on EMR data by raising the attention on the role of data representation in knowledge discovery.
Acknowledgement
This research was partially supported by the Major Research Plan of the National Natural Science Foundation of China (Key Program, Grant No. 91746204), the Science and Technology Development in Guangdong Province (Major Projects of Advanced and Key Techniques Innovation, Grant No.2017B030308008), and Guangdong Engineering Technology Research Center for Big Data Precision Healthcare (Grant No.603141789047). The dataset used for analysis described in this study was obtained from KUMC’s HERON clinical data repository, which was supported by institutional funding and by the KUMC CTSA grant UL1TR002366 from NCRR/NIH.
Figures & Table
Reference
- 1.de Boer IH, Rue TC, Hall YN, Heagerty PJ, Weiss NS, Himmelfarb J. Temporal trends in the prevalence of diabetic kidney disease in the United States. JAMA. 2011 Jun 22;305(24):2532–2539. doi: 10.1001/jama.2011.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zoppini G, Targher G, Chonchol M, et al. Predictors of estimated GFR decline in patients with type 2 diabetes and preserved kidney function. Clinical journal of the American Society of Nephrology: CJASN. 2012 Mar;7(3):401–408. doi: 10.2215/CJN.07650711. [DOI] [PubMed] [Google Scholar]
- 3.Ueda H, Ishimura E, Shoji T, et al. Factors affecting progression of renal failure in patients with type 2 diabetes. Diabetes care. 2003 May;26(5):1530–1534. doi: 10.2337/diacare.26.5.1530. [DOI] [PubMed] [Google Scholar]
- 4.Rossing K, Christensen PK, Hovind P, Tarnow L, Rossing P, Parving HH. Progression of nephropathy in type 2 diabetic patients. Kidney international. 2004 Oct;66(4):1596–1605. doi: 10.1111/j.1523-1755.2004.00925.x. [DOI] [PubMed] [Google Scholar]
- 5.Yokoyama H, Kanno S, Takahashi S, et al. Determinants of decline in glomerular filtration rate in nonproteinuric subjects with or without diabetes and hypertension. Clinical journal of the American Society of Nephrology: CJASN. 2009 Sep;4(9):1432–1440. doi: 10.2215/CJN.06511208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Randolph AG, Guyatt GH, Calvin JE, Doig G, Richardson WS. Understanding articles describing clinical prediction tools. Crit. Care Med. 1998;26:1603–12. doi: 10.1097/00003246-199809000-00036. PMID: 9751601. [DOI] [PubMed] [Google Scholar]
- 7.Murphy SN, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) JAMIA. 2010 Mar-Apr;17(2):124–30. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waitman LR, Warren JJ, Manos EL, Connolly DW. Expressing observations from electronic medical record flowsheets in an i2b2 based clinical data epository to support research and quality improvement. AMIA Annu Symp Proc. 2011;2011:1454–63. Epub 2011 Oct 22. PubMed PMID: 22195209; PubMed Central PMCID: PMC3243191. [PMC free article] [PubMed] [Google Scholar]
- 9.Stuart JN, Kelly Z, John K, Tammy P, Robin M. Normalized names for clinical drugs: RxNorm at 6 years. AMIA. 2011 Jul-Aug;18(4):441–448. doi: 10.1136/amiajnl-2011-000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajkomar A, Oren E, Dean J. Scalable and accurate deep learning with electronic health records. NPJ Digital Medicine, Nature. 2018 May 18;2018 doi: 10.1038/s41746-018-0029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Addison JFD, Wermter S, Arevian GZ. A comparison of feature extraction and selection techniques; Int. Conf. Artificial Neural Networks (ICANN) Proc; 2003. pp. 212–215. [Google Scholar]
- 12.Nichols GA, et al. Construction of a Multisite DataLink Using Electronic Health Records for the Identification, Surveillance, Prevention, and Management of Diabetes Mellitus: The SUPREME-DM Project. Preventing Chronic Disease. 2012;9:110311. doi: 10.5888/pcd9.110311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.KDOQI. KDOQI clinical practice guidelines and clinical practice recommendations for diabetes and chronic kidney disease. Am J Kidney Dis. 2007;49(2(suppl 2)):S12–S154. doi: 10.1053/j.ajkd.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 14.American Diabetes Association. Standards of medical care in diabetes—2010. Diabetes Care. 2010;33(suppl 1):S11–S61. doi: 10.2337/dc10-S011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waitman LR, Warren JJ, Manos EL, Connolly DW. Expressing observations from electronic medical record flowsheets in an i2b2 based clinical data epository to support research and quality improvement. AMIA Annu Symp Proc. 2011;2011:1454–63. Epub 2011 Oct 22. PubMed PMID: 22195209; PubMed Central PMCID: PMC3243191. [PMC free article] [PubMed] [Google Scholar]
- 16.Damle R, Alavi K. The University HealthSystem Consortium Clinical Database: An Emerging Resource in Colorectal Surgery Research. Sem. in Colon Rectal Surg., Big Data and Colorectal Surgery. 2016 Jun;27(2):92–95. [Google Scholar]
- 17.Jardine MJ, et al. Prediction of Kidney-Related Outcomes in Patients With Type 2 Diabetes. AJKD. 2012 Nov;60(5):770–778. doi: 10.1053/j.ajkd.2012.04.025. [DOI] [PubMed] [Google Scholar]
- 18.Hutchinson RA, Liu LP, Dietterich TG. Incorporating boosted regression trees into ecological latent variable models. AAAI. 2011:1343–1348. [Google Scholar]
- 19.Johnson R, Zhang T. Learning Nonlinear Functions Using Regularized Greedy Forest. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2014 May;36(5):942–954. doi: 10.1109/TPAMI.2013.159. [DOI] [PubMed] [Google Scholar]
- 20.Kevin He, et al. Component-wise gradient boosting and false discovery control in survival analysis with high- dimensional covariates. Bioinformatics. 2016 Jan 1;32(1):50–57. doi: 10.1093/bioinformatics/btv517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torlay L, Perrone-Bertolotti M, Thomas E, Baciu M. Machine Learning XGBoost Analysis of language networks to classify patients with epilepsy. Brain Informatics. 2017 Apr;4(3):159–169. doi: 10.1007/s40708-017-0065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalousis A, Prados J, Hilario M. Stability of feature selection algorithms: a study on high-dimensional spaces. Knowl Inf Syst. 2007 May;12(1):95–116. [Google Scholar]
- 23.Friedman JH. Greedy boosting approximation: a gradient boosting machine. Ann. Stat. 2001 Oct;29(5):1189–1232. [Google Scholar]
- 24.Chen TQ, Guestrin C. XGBoost: A Scalable Tree Boosting System; Proceedings of the 22nd acm SIGKDD international conference; 2016. pp. 785–794. [Google Scholar]
- 25.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 26.Khan YH, Sarriff A, Adnan AS, Khan AH, Mallhi TH. Chronic Kidney Disease, Fluid Overload and Diuretics: A Complicated Triangle 2016. PLoS ONE. 11(7):e0159335. doi: 10.1371/journal.pone.0159335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Volkow ND. AT ML. Opioid abuse in chronic pain—misconceptions and mitigation strategies. N Engl J Med. 2016;374(13):1253–63. doi: 10.1056/NEJMra1507771. [DOI] [PubMed] [Google Scholar]
- 28.Dey S, Simon G, Westra B, Steinbach M, Kumar V. Mining Interpreatble and Predictive Diagnosis Codes from Multi-source Electronic Health Records; SIAM International Conference on Data Mining; 2014. pp. 1055–1063. [Google Scholar]
- 29.Miller SC, Pedro G, Vincent M. Washington, DC: Office of The Assistant Secretary for Planning and Evaluation, U.S. Department of Health & Human Services; 2000. Mar, Appendix A. Data and Variables Used for Hospice in Nursing Facility Analyses“. Outcomes and Utilization for Hospice and Non-Hospice Nursing Facility Decedents (Report) Drug Data. [Google Scholar]
- 30.Russell TW, Ross L. Evaluating a proposed modification of the Guttman rule for determining the number of factors in an exploratory factor analysis. Psychological Test and Assessment Modeling. 2014;56:104–123. [Google Scholar]