Abstract
Normalization maps clinical terms in medical notes to standardized medical vocabularies. In order to capture semantic similarity between different surface expressions of the same clinical concept, we develop a hybrid normalization system that incorporates a deep learning model to complement the traditional dictionary lookup ap- proach. We evaluate our system against the ShARe/CLEF 2013 challenge data in which 30% of the mentions have no concept mapping. When evaluating against the mentions which may be normalized to existing concepts, our hybrid system achieves 90.6% accuracy, obtaining a statistically significant improvement of 2.6% over a strong edit-distance and dictionary lookup combined baseline. Our analysis of semantic similarity between concepts and mentions reveals existing inconsistencies in ShARe/CLEF data, as well as problematic ambiguities in the UMLS. Our results suggest the potential of the proposed deep learning approach to further improve the performance of normalization by utilizing semantic similarity.
Introduction
The information about findings, symptoms, diseases, diagnoses, and medications recorded in the medical notes is invaluable for clinical applications. The information extracted from medical notes has been used in clinical decision- making1–3, mortality prediction4, 5, adverse drug effect analysis6, 7 among others. A lot of work has focused on named entity recognition (NER) for clinical notes8–12 which identifies text spans (“mentions”) of the clinically-relevant con- cepts. However, without linking such information to standardized medical vocabularies, the proliferation of similar- sounding acronyms, and conversely, the presence of synonyms that can be used interchangeably to refer to the same clinical concept, coupled with misspellings, reduce the usefulness of this information for generalization. For example, one may use “heart attack”, “MI”, and “myocardial infarction” to refer to the same general concept, and if different words are used to express this concept in patient records, we would fail to identify similarity between such patients, unless we can correctly identify the underlying concept in each case. In this paper, we focus specifically on the entity normalization task. As distinct from NER, the normalization task involves mapping individual mentions of clinically relevant entities to the entries in the standardized medical vocabulary.
There are very few annotated datasets available for the task of clinical entity normalization. One such dataset was provided by task 1 in ShARe/CLEF eHealth 2013 challenge* which defined two subtasks: (a) named entity recogni-tion, and (b) normalization of disorders. In this research, we focus on the normalization subtask, defined as mapping a given mention to a concept unique identifier (CUI) from the Unified Medical Language System (UMLS)13. Following ShARe/CLEF 2013, SemEval-2014 Task 7† and SemEval-2015 Task 14‡ included additional annotated medical notes for the same task. Subtask (a), the named entity recognition, has been well explored in the Clinical NLP research community, with a number of NER systems, including MetaMap8, cTAKES9, CLAMP10, CliNER11, and RapTAT12, available. Although such well-known tools as MetaMap, cTAKES and CLAMP contain normalization modules, they are based on morphological variation. Such approaches have obvious drawbacks, as previously pointed out by Kate14 and elsewhere, specifically, (1) the systems based on morphological variation may not capture the semantic similar- ity between medical mentions and concepts, and (2) TF-IDF15 representation, commonly used in the normalization modules, may be inappropriate for short medical terms.
Amongst the top-ranked teams in subtask (b) of ShARe/CLEF 2013, DNorm16 learned a normalization weight matrix using pairwise ranking17 over TF-IDF representation and UTHealth18 normalized clinical terms based on cosine sim- ilarity over TF-IDF representation. In SemEval 2014 and 2015, UTH-CCB19, 20 used a similar TF-IDF representation and cosine similarity method for normalization. ezDI21, 22 adopted dictionary lookup over lexical variants generated by Lexical Variants Generation§, with Levenshtein edit distance23 used to normalize the mentions that fail to be nor-malized by exact match. UWM24, 25 learned normalization rules automatically based on edit distance at character level. Following UWM, D’Souza and Ng26 proposed a sieve-based approach which includes a series of lexical trans- formations combined with exact match and rule-based partial match. In summary, most of the existing systems use either a vector representation, typically TF-IDF or lexical transformations, in combination with similarity ranking for the normalization task. Because of the characteristics of the dataset, dictionary lookup and its variants may produce high performance. However, such normalization methods are challenged by the remaining mentions which can not be resolved in this way and require semantic analysis14.
Li et al.27 recently applied convolutional neural network (CNN) to rank the candidate concepts in the normalization task. Their rule-based system first generates candidate concepts which are morphologically similar to a given mention. Then, CNN-based ranking is used to find the optimal normalization from the candidate concepts. However, this ap- proach does not generalize to the cases when a concept is semantically similar but morphologically different. Inspired by the recent success of Bidirectional Long Short-Term Memory (Bi-LSTM) models28 in sequence modeling tasks, and specifically, in NER29, 30, we apply a LSTM-based model to the normalization task.
In this work, we propose a hybrid method which combines the traditional dictionary lookup with a deep learning approach to the normalization task and investigate its capability for matching concept mentions to the standardized clinical concepts at semantic level. We represent concept mentions and their context using dense lexical embeddings that capture semantic similarity between words as input to a deep learning model. Concept mentions and their context are processed by three parallel Bi-LSTM layers in a recurrent neural network model, with threshold-based concept assignment. The model is trained on a combination of labeled data and concept synonyms provided by the UMLS, ensuring wider coverage and better generalization. Evaluated on the normalized mentions from the ShARe/CLEF dataset, our deep learning hybrid system achieves 90.6% accuracy, obtaining a statistically significant (0.05 alpha) improvement of 2.6% over the accuracy of 88.0% for the edit distance and dictionary lookup combined.
Dataset
ShARe/CLEF eHealth 2013
The publicly available ShARe/CLEF 2013 dataset contains de-identified clinical free-text notes from the MIMIC II31 database. The dataset contains 199 notes in the training set and 99 notes in the test set. A disorder mention is defined as any span of text which can be mapped to a CUI in the Systematized Nomenclature of Medicine – Clinical Terms (SNOMED CT)¶ terminology and which belongs to one of the 11 UMLS semantic types including “Congenital Ab-normality”, “Acquired Abnormality”, “Injury or Poisoning”, “Pathologic Function”, “Disease or Syndrome”, “Mental or Behavioral Dysfunction”, “Cell or Molecular Dysfunction”, “Experimental Model of Disease”, “Anatomical Ab- normality”, “Neoplastic Process”, and “Signs and Symptoms”. If no appropriate CUI can be found for a disorder mention, it is assigned to the “CUI-less” category. Due to limited access to the expanded dataset from SemEval 2014 Task 7 and SemEval 2015 Task 14, the development and evaluation of our work is based on ShARe/CLEF dataset.
UMLS and SNOMED CT
In order to minimize the variance caused by different UMLS releases, we used sources in English included in the active UMLS 2013 AA release which was when the ShARe/CLEF data was released. Following the specification of the task, we retrieved CUIs belonging to one of the 11 semantic types that occur in SNOMED CT. We also collected the concept synonyms for each retrieved CUI from all UMLS sources, and normalized them to lowercase. UMLS provides a list of concept synonyms for each CUI. We describe how these are used in our normalization approach in the methods section.
Medical Information Mart for Intensive Care III (MIMIC III)
MIMIC III32 database was used to train word2vec33 word embeddings. This data were collected between 2001 and 2012 at Boston’s Beth Israel Deaconess Medical Center. The database contains over 2 million notes from 40K patients. The following pre-processing steps were applied to the notes: (1) using regular expressions to replace 64 Protected Health Information (PHI) patterns with 56 unique tokens; (2) removing punctuation; (3) replacing floating numbers with a unique placeholder token; (4) lowercasing.
Methods
We created two baseline systems as comparison to our hybrid deep learning approach. The first baseline is an exact- match system. The second baseline is the edit-distance system which is the top-ranked team in the SemEval 2014 and 2015 clinical tasks. Our hybrid normalization system utilizing deep learning approach is built on the state-of-the-art edit-distance system. We describe the details of two baseline systems and the architecture of our hybrid normalization method which combines the exact match and edit distance in the following sections.
Exact Match Baseline
The exact-match method contains two components: (1) exact match against the mentions in the training set, and (2) exact match against the UMLS concept synonyms mentioned in the dataset section above. We consider a mention to be an exact match if it occurs in the same typographical form in one of the above sources.
If a mention can be matched exactly to a unique CUI, it is normalized to that CUI. Otherwise, the mention is considered ambiguous and is assigned to the CUI-less category. For example, the mention “lung cancer” is treated as CUI-less because there are two CUIs (C0684249 and C0242379) bound to the concept synonym “lung cancer”. One common approach to resolving ambiguity in such case is to randomly assign the first CUI to the mention. However, this approach produced worse accuracy on the development dataset, therefore we do not use it. If no exact match is found, we also treat the mention as CUI-less.
Edit Distance Baseline
Ghiasvand and Kate24, 25 and Kate14 implemented automatically learned edit-distance method to the normalization task and were top-ranked teams in SemEval 2014 and 2015 clinical task. We followed their approach to build a similar exact-match baseline and used their open-source edit-distance system for comparison as well as a component in our hybrid method. We use the open-source implementation of the edit-distance baseline by Kate14 with the default settings, but we omit the abbreviation disambiguation step. If multiple full forms exist for a given abbreviation, their strategy is to arbitrarily pick the first full form in the dictionary. We found that this arbitrary disambiguation strategy did not have a substantial impact on the relative performance improvement obtained by our hybrid system.
Deep Learning Model
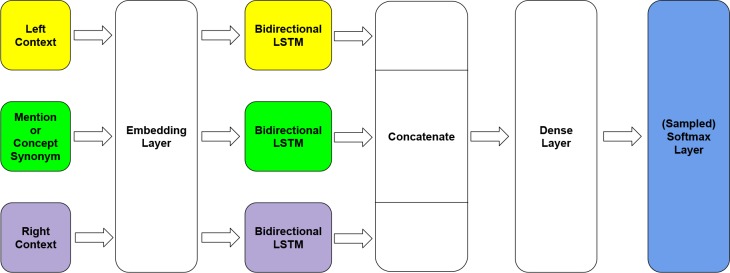
Our deep learning model contains four layers: (1) an embedding layer that converts input tokens into word embeddings, (2) three Bi-LSTMs which are used to process the concept mention, the left context, and the right context, (3) a dense layer of size 2048 neurons, and (4) a softmax output layer. The model architecture is illustrated in Figure 1. Occurrence contexts are extracted with a window size +/- 10 tokens. The outputs from the Bi-LSTMs are concatenated and fed into the dense layer. Because the number of possible classes (CUI assignments) is large, we use a sampled softmax loss|| during training and a regular softmax in prediction. Dropout34 with 20% probability is applied to avoid model overfitting.
Figure 1.
Architecture of the deep learning model.
The text spans for the concept mention and for the left and right contexts are represented as sequences of word2vec word vectors. We used Gensim** to train 200-dimensional word embeddings over all MIMIC III notes, using skip-gram in order to consider word order. We used hierarchical softmax and the frequency cutoff 3. The embeddings were tuned continuously during model training.
Importantly, we expand our training data by creating additional training instances using UMLS concept synonyms. Specifically, for the all valid disease CUIs, the retrieved concept synonyms are used at the training stage as mentions with zero left and right context, with the corresponding CUI as a label. We exclude from training the instances which contain out of vocabulary words for our word2vec model, i.e. words that occurred less than three times in MIMIC. We also exclude concept synonyms that map to multiple CUIs.
According to the released data statistics, 30% of mentions in the ShARe/CLEF data are not mapped to any UMLS CUI, because no appropriate concept could be found. These mentions are assigned to the “CUI-less” category, which serves as a catch-all label for a variety of different concepts. Although we treat the normalization task as a classification problem, we do not assign a mention to a CUI-less category when it has the highest probability, but instead use a threshold determined on the validation data. That is, the CUI-less label is assigned whenever the probability of the most likely CUI is below that threshold. The output of the model is a probability distribution over all 64798 CUIs, including the CUI-less category. We divide the training set into 72% training (dev-train), 8% validation (dev-val), and 20% development test (dev-test), and use dev-val for threshold selection.
Hybrid Method
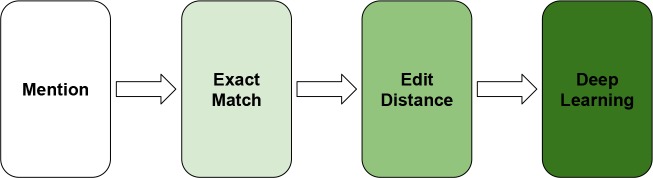
Since the ShARe/CLEF data contains solely the mentions of disorders, the relatively unique and precise surface lex- ical expressions make the dictionary lookup a very strong baseline. We therefore propose a hybrid approach which combines exact-match module, edit-distance module, and a deep learning model as illustrated in Figure 2 . Our hybrid method normalizes concept mentions using a sequential pipeline. If a given concept mention is not associated with a CUI at a given stage in the pipeline, the mention is passed on to the next stage. We compared the performance of two hybrid configurations: (1) exact match + edit distance (EM + ED), and (2) exact match + edit distance + deep learning (EM + ED + DL).
Figure 2.
Architecture of the hybrid deep learning model.
Results and Discussion
Normalization Task Evaluation
In the ShARe/CLEF 2013 challenge, the tasks of NER and normalization were evaluated together, and only the com- bined score for these tasks was reported for participating systems. Therefore, we cannot directly compare our normal- ization system to the participating systems scores reported in the ShARe/CLEF 2013 challenge paper. Instead, we use the open-source implementation from Kate14, which reports high scores on SemEval 2014 and 2015 challenge data, as one of our baselines.
Following prior work14, we evaluate our system under two settings: on the full dataset, including the CUI-less men- tions, and only on the mentions which could be normalized to existing UMLS concepts. Table 1 shows system performance for the first setting, when the CUI-less category is included in evaluation. Table 2 shows the results of evaluation with the CUI-less category excluded, i.e. only on the mentions mapped to existing CUIs. We also report F- measure, precision, and recall following prior work14. The reported weighted metrics†† consider label imbalance by weighting label metrics based the number of true instances for each label.
Table 1.
Evaluation of Exact Match (EM), Exact Match + Edit Distance (EM + ED), and Exact Match + Edit Distance + Deep Learning (EM + ED + DL) models against all mentions including the CUI-less category.
| EM | EM+ED | EM+ED+DL | ||
|---|---|---|---|---|
| Accuracy | 87.4 | 89.8 | 89.6 | |
| Micro | F-measure | 87.9 | 90.7 | 91.2 |
| Precision | 88.5 | 91.6 | 92.9 | |
| Recall | 87.4 | 89.8 | 89.6 | |
| Macro | F-measure | 71.5 | 77.8 | 80.4 |
| Precision | 74.5 | 79.9 | 81.5 | |
| Recall | 70.7 | 77.7 | 81.1 | |
| Weighted | F-measure | 85.4 | 88.9 | 89.8 |
| Precision | 85.6 | 89.4 | 91.0 | |
| Recall | 87.4 | 89.8 | 89.6 | |
Table 2.
Evaluation against mentions normalized to existing UMLS concepts.
| EM | EM+ED | EM+ED+DL | ||
|---|---|---|---|---|
| Accuracy | 83.4 | 88.0 | 90.6 | |
| Micro | F-measure | 90.5 | 92.9 | 94.1 |
| Precision | 98.9 | 98.4 | 97.9 | |
| Recall | 83.4 | 88.0 | 90.6 | |
| Macro | F-measure | 72.1 | 78.5 | 81.4 |
| Precision | 75.5 | 81.0 | 83.3 | |
| Recall | 70.7 | 77.7 | 81.1 | |
| Weighted | F-measure | 86.0 | 89.7 | 91.5 |
| Precision | 90.9 | 93.2 | 93.8 | |
| Recall | 83.4 | 88.0 | 90.6 | |
Under the setting in which the CUI-less mentions are included, the EM + ED and EM + ED + DL configurations show comparable performance. However, when they are excluded from evaluation, the hybrid method with the deep learning component performs better on most of the metrics.
Semantic Similarity between Concepts and Concept Mentions
In order to understand the information captured by the deep learning model, we further examined the CUIs selected by the model on the dev-test data. Some examples of the concepts identified by the model for the mentions labeled as CUI-less in the annotated data are shown in Table 3. While not all examples selected by the model are equally reasonable, it is nonetheless clear that in many cases the model is able to capture semantic similarity between concepts and concept mentions, and ultimately provide a performance boost over the methods relying on dictionary lookup using lexical transformations. However, this also suggests that at least some of the potential improvement that could be provided by the deep learning model is lost due to underspecified labelling choices made during annotation. The latter may also be an artifact of the annotation methodology. For example, if the UMLS search interface is used to identify possible concept mappings for “suicidal ideation”, a human annotator would not be able to associate it with C0522178 “Thoughts of self harm”, since it will not be listed.
Table 3.
UMLS concepts predicted by the deep learning model for selected CUI-less mentions.
| CUI Annotation | CUI Prediction | Mention | Concept Synonym |
|---|---|---|---|
| CUI-less | C0522178 | suicidal ideation | Thoughts of self harm |
| CUI-less | C0023380 | unarousable | Lethargy |
| CUI-less | C0575090 | poor balance | Impairment of balance/ Problem with balance |
| CUI-less | C0013404 | trouble breathing | Difficulty breathing |
| CUI-less | C0234246 | Abdomen rebound | Rebound tenderness |
| CUI-less | C0576690 | Position sense impaired | Impaired body position sense |
| CUI-less | C0333133 | mucus plugging event | Mucus cast / Mucus plug |
| CUI-less | C0238997 | dullness to percussion | Chest dull to percussion |
Another source of discrepancies between the gold standard annotation and possible semantically matching concepts is the presence of multiple similar concepts in the UMLS Metathesaurus itself. For example, the mention “depres- sion” may be appropriately normalized to either C0011581 “Depressive disorder” or C0011570 “Mental Depression”. The same synonym can often be given for multiple CUIs, making them essentially equivalent. Thus, “chronic renal insufficiency” may be associated with either C0022661 “Kidney Failure, Chronic” or C0403447 “Chronic Kidney Insufficiency” both of which are listed with the synonym, “chronic renal insufficiency”.
Some of such inconsistencies have been or are being handled in the later UMLS releases, so in the future, one can expect improved performance of the models that rely on this information. For example, in the 2011 UMLS release used for the ShARe/CLEF data annotation, “infectious source” could be associated with either C0021311 “Infection” or with C0009450 “Communicable disease” and these two concepts were merged in later UMLS releases.
Note that in this work, we have not attempted to address the disambiguation of abbreviations and acronyms, which we believe should be treated as a separate task. Effectively, this means that our system represents them in the same way as regular tokens, that is, using dense word2vec embeddings. However, the context representation used by the deep learning model is nonetheless able to identify the appropriate concepts for some of the acronyms and abbreviations that have multiple disambiguations. For example, our deep learning model was able to correctly normalize “Breast CA” to C0006142 “Malignant neoplasm of breast”, “pelvic fx” to C0149531 “Fracture of pelvis”, and “Schizoaffective D/O” to C0036337 “Schizoaffective Disorder”, and “C. difficile” to C0343386 “Clostridium difficile infection”. This suggests that it may be possible to include the disambiguation of acronyms and abbreviations in our hybrid deep learning system as a separate component in the future.
Conclusion
In this work, we have proposed a novel hybrid method for linking clinically relevant entity mentions in free-text medical notes to standardized medical vocabularies. This method combines traditional dictionary lookup with a deep learning model, showing an improvement over existing state-of-the-art and the potential of the deep learning model to better capture semantic similarity between concepts and concept mentions, compared to a strong edit distance and dictionary lookup-based baseline. We believe that the two main factors contributed to the relatively modest improvement the deep learning model was able to achieve: (1) the presence of the semantically diverse catch-all CUI-less category in the data, for which no consistent representation could be learned, and (2) the relatively small training dataset size, which is a known impediment to improved performance by deep learning techniques. The qualitative examination of the CUIs predicted by the deep learning model also highlighted existing inconsistencies and ambiguities in the ShARe/CLEF data, as well as in the UMLS, which will need to be addressed for normalization tasks in the future.
Acknowledgment
This work was supported in part by a research grant from Philips HealthCare.
Footnotes
ShARe/CLEF eHealth 2013. https://sites.google.com/site/shareclefehealth/
SemEval-2014 Task 7: Analysis of Clinical Text. http://alt.qcri.org/semeval2014/task7/
SemEval-2015 Task 14: Analysis of Clinical Text. http://alt.qcri.org/semeval2015/task14/
Lexical Variants Generation. https://lexsrv3.nlm.nih.gov/LexSysGroup/Projects/lvg/2014/docs/userDoc/tools/lvg.html
SNOMED CT. https://www.snomed.org/snomed-ct
Sampled Softmax Loss. https://www.tensorflow.org/api docs/python/tf/nn/sampled softmax loss
Gensim. https://radimrehurek.com/gensim/
Scikit-learn Weighted Metrics. http://scikit-learn.org/stable/modules/generated/sklearn.metrics.f1 score.html
Figures & Table
References
- 1.Shao Y, Mohanty AF, Ahmed A, Weir CR, Bray BE, Shah RU, et al. Identification and use of frailty indicators from text to examine associations with clinical outcomes among patients with heart failure; AMIA Annual Symposium Proceedings; American Medical Informatics Association; 2016. p. 1110. [PMC free article] [PubMed] [Google Scholar]
- 2.Topaz M, Lai K, Dowding D, Lei VJ, Zisberg A, Bowles KH, et al. Automated identification of wound information in clinical notes of patients with heart diseases: developing and validating a natural language processing application. International journal of nursing studies. 2016;64:25–31. doi: 10.1016/j.ijnurstu.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Zhang P, Wang F, Hu J, Sorrentino R. Towards personalized medicine: leveraging patient similarity and drug similarity analytics. AMIA Summits on Translational Science Proceedings. 2014:132. [PMC free article] [PubMed] [Google Scholar]
- 4.Jo Y, Loghmanpour N, Rosé CP. Time series analysis of nursing notes for mortality prediction via a state transition topic model; Proceedings of the 24th ACM international on conference on information and knowledge management; ACM; 2015. pp. 1171–1180. [Google Scholar]
- 5.Luo YF, Rumshisky A. AMIA Annual Symposium Proceedings. American Medical Informatics Association; 2016. Interpretable topic features for post-ICU mortality prediction; p. 827. [PMC free article] [PubMed] [Google Scholar]
- 6.LePendu P, Liu Y, Iyer S, Udell MR, Shah NH. Analyzing patterns of drug use in clinical notes for patient safety. AMIA Summits on Translational Science Proceedings. 2012:63. [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Salmasian H, Vilar S, Chase H, Friedman C, Wei Y. A method for controlling complex confounding effects in the detection of adverse drug reactions using electronic health records. Journal of the American Medical Informatics Association. 2014;21(2):308–314. doi: 10.1136/amiajnl-2013-001718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aronson AR. Proceedings of the AMIA Symposium. American Medical Informatics Association; 2001. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program; p. 17. [PMC free article] [PubMed] [Google Scholar]
- 9.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association. 2010;17(5):507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soysal E, Wang J, Jiang M, Wu Y, Pakhomov S, Liu H, et al. CLAMP–a toolkit for efficiently building customized clinical natural language processing pipelines. Journal of the American Medical Informatics Association. 2017 doi: 10.1093/jamia/ocx132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boag W, Wacome K, Naumann T, Rumshisky A. CliNER: a lightweight tool for clinical named entity recognition. AMIA Joint Summits on Clinical Research Informatics (poster) 2015 [Google Scholar]
- 12.Gobbel GT, Reeves R, Jayaramaraja S, Giuse D, Speroff T, Brown SH, et al. Development and evaluation of Rap-TAT: a machine learning system for concept mapping of phrases from medical narratives. Journal of biomedical informatics. 2014;48:54–65. doi: 10.1016/j.jbi.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 13.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic acids research. 2004;32(suppl 1):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kate RJ. Normalizing clinical terms using learned edit distance patterns. Journal of the American Medical Informatics Association. 2016;23(2):380–386. doi: 10.1093/jamia/ocv108. [DOI] [PubMed] [Google Scholar]
- 15.Salton G, McGill MJ. Introduction to modern information retrieval. New York, NY, USA: McGraw-Hill, Inc.; 1986. [Google Scholar]
- 16.Leaman R, Khare R, Lu Z. NCBI at 2013 ShARe/CLEF eHealth shared task: disorder normalization in clinical notes with DNorm; Proceedings of the Conference and Labs of the Evaluation Forum; 2013. [Google Scholar]
- 17.Bai B, Weston J, Grangier D, Collobert R, Sadamasa K, Qi Y, et al. Learning to rank with (a lot of) word features. Information retrieval. 2010;13(3):291–314. [Google Scholar]
- 18.Tang B, Wu Y, Jiang M, Denny JC, Xu H. Recognizing and encoding disorder concepts in clinical text using machine learning and vector space; Proceedings of the Conference and Labs of the Evaluation Forum; 2013. [Google Scholar]
- 19.Zhang Y, Wang J, Tang B, Wu Y, Jiang M, Chen Y, et al. UTH CCB: a report for SemEval 2014–task 7 analysis of clinical text. Proceedings of the 8th International Workshop on Semantic Evaluation (SemEval 2014) 2014:802–806. [Google Scholar]
- 20.Xu J, Zhang Y, Wang J, Wu Y, Jiang M, Soysal E, et al. UTH-CCB: the participation of the SemEval 2015 challenge–Task 14. Proceedings of the 9th International Workshop on Semantic Evaluation (SemEval 2015) 2015:311–314. [Google Scholar]
- 21.Pathak P, Patel P, Panchal V, Choudhary N, Patel A, Joshi G. ezDI: a hybrid CRF and SVM based Model for detecting and encoding disorder mentions in clinical notes. Proceedings of the 8th International Workshop on Semantic Evaluation (SemEval 2014) 2014:278–283. [Google Scholar]
- 22.Pathak P, Patel P, Panchal V, Soni S, Dani K, Patel A, et al. ezDI: a supervised NLP system for clinical narrative analysis. Proceedings of the 9th International Workshop on Semantic Evaluation (SemEval 2015) 2015:412–416. [Google Scholar]
- 23.Levenshtein VI. Binary codes capable of correcting deletions, insertions, and reversals. Soviet physics doklady. 1966;vol. 10:707–710. [Google Scholar]
- 24.Ghiasvand O, Kate R. UWM: disorder mention extraction from clinical text using CRFs and normalization using learned edit distance patterns. Proceedings of the 8th International Workshop on Semantic Evaluation (SemEval 2014) 2014:828–832. [Google Scholar]
- 25.Ghiasvand O, Kate R. UWM: a simple baseline method for identifying attributes of disease and disorder mentions in clinical text. Proceedings of the 9th International Workshop on Semantic Evaluation (SemEval 2015) 2015:385–388. [Google Scholar]
- 26.D'souza J, Ng V. Sieve-based entity linking for the biomedical domain; Proceedings of the 53rd Annual Meeting of the Association for Computational Linguistics and the 7th International Joint Conference on Natural Language Processing; 2015. pp. 297–302. [Google Scholar]
- 27.Li H, Chen Q, Tang B, Wang X, Xu H, Wang B, et al. CNN-based ranking for biomedical entity normalization. BMC bioinformatics. 2017;18(11):385. doi: 10.1186/s12859-017-1805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hochreiter S, Schmidhuber J. Long short-term memory. Neural computation. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 29.Chiu JPC, Nichols E. Named entity recognition with bidirectional LSTM-CNNs. Transactions of the Association for Computational Linguistics. 2016:4. [Google Scholar]
- 30.Lample G, Ballesteros M, Subramanian S, Kawakami K, Dyer C. Neural architectures for named entity recognition; Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies; Association for Computational Linguistics; 2016. pp. 260–270. [Google Scholar]
- 31.Saeed M, Villarroel M, Reisner AT, Clifford G, Lehman LW, Moody G, et al. Multiparameter intelligent monitoring in intensive care II (MIMIC-II): a public-access intensive care unit database. Critical care medicine. 2011;39(5):952. doi: 10.1097/CCM.0b013e31820a92c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson AEW, Pollard TJ, Shen L, LwH Lehman, M Feng, M Ghassemi, et al. MIMIC-III, a freely accessible critical care database. Scientific Data. 2016 May;:3. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. Advances in neural information processing systems. 2013:3111–3119. [Google Scholar]
- 34.Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. The Journal of Machine Learning Research. 2014;15(1):1929–1958. [Google Scholar]