Abstract
Genetically engineered mesenchymal stem cells (MSCs), as non-viral gene delivery platforms, are rapidly evolving in tumor therapy due to their low immunogenicity and natural tumor-homing capacity.
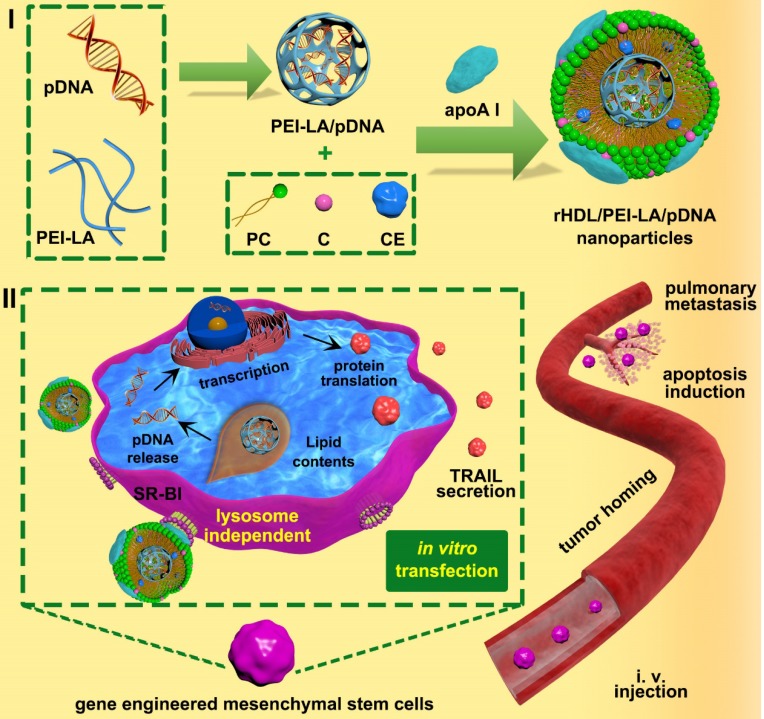
Methods: In this paper, we selected reconstituted high-density lipoprotein (rHDL), a lipoprotein-bioinspired nanovector with specific binding ability to scavenger receptor B type I (SR-BI) expressed on MSCs, as a transfection agent to genetically modify MSCs. pDNA encoding tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) was used as a functional gene to be transfected into the nucleus of MSCs for TRAIL expression. Lauric acid-coupled polyethyleneimine (PEI-LA) as an amphiphilic cationic polymer was synthesized to electrostatically bind to pDNA, and then incorporated into rHDL to form rHDL/PEI-LA/pDNA nanoparticles.
Results: The nanoparticles exhibited homogenous particle size and excellent serum stability in vitro. Meanwhile, this SR-BI-targeted rHDL performed efficient intracellular gene delivery, specific lysosome-independent mechanism of cellular uptake and high transfection of pDNA towards MSCs. Moreover, high TRAIL expression in MSCs was detected after rHDL-mediated transfection. In vitro and in vivo results indicated that genetically engineered MSCs could accurately target to B16F10 cells, thereby producing significant apoptosis-inducing effect on aggressive melanoma.
Conclusion: TRAIL-expressing MSCs engineered by rHDL nanovector was an efficient and hypotoxic method for stem cells-based pulmonary melanoma metastasis-targeting therapy.
Keywords: Mesenchymal stem cells, reconstituted high-density lipoprotein, tumor-targeted therapy, tumor necrosis factor-related apoptosis-inducing ligand, pulmonary melanoma metastasis.
1. Introduction
In recent years, a growing number of drug delivery systems (DDS) have been studied to delivery chemotherapeutics and gene therapeutic agents for tumor therapy 1. However, the immunogenicity and cytotoxicity to normal tissues caused by these DDS are still the main obstacles for antitumor efficacy, which hinder their clinical applications 2. Hence, endogenous DDS including cell-based systems have attracted extensive attention from researchers 3, 4. Mesenchymal stem cells (MSCs) have been used to treat a wide range of diseases as drug delivery platforms, due to its various advantages such as easy accessibility and proliferation in vitro, low immunogenicity in vivo and free of ethical issues 5, 6. More importantly, MSCs can accurately target to inflammatory tissues, so that they are considered as promising candidates for tumor-targeted drug delivery platforms 7, 8.
MSCs can deliver therapeutic cargos as vehicles after genetic modification for tumor-targeting therapy due to its unique tumor-tropism properties 9-11. It should be noted that efficient and safe gene transfection of MSCs is the precondition to achieve antineoplastic effect. However, viral transfection of MSCs may lead to unwanted transformation such as significantly increase the risk of gene mutation or immunogenicity 12, 13. Non-viral gene vectors with better biosafety and easy-handling properties, including cationic polymers and lipoprotein-derived nanocarriers, have become alternative candidates for genetic engineering of MSCs 14, 15.
High-density lipoprotein (HDL), as one of the most essential components of the lipid transport system, plays a critical role in the reverse cholesterol transport from peripheral tissues to liver 16. HDL has been developed as a non-viral vehicle for gene delivery, which is attributed to its specific binding to scavenger receptor class B type I (SR-BI) on the surface of engineered cells 17. Natural HDL is composed of a hydrophilic shell and a hydrophobic core, whose unique shell-core structure provides a broad interior space for hydrophobic drugs 18. HDL can form a SR-BI-based hydrophobic channel on cell membrane when HDL binds to SR-BI, which promotes the efflux and diffusion of lipid contents from HDL into cytoplasm through concentration gradient 19, 20. This unique process can protect gene therapeutic agents from endosome/lysosome degradation. Apolipoprotein A I (apoA I) is a highly helical protein (Mw: 28 kDa) which is the most abundant protein in HDL. It can specifically bind to liver cells or other SR-BI-expressed cells such as MSCs 21. Reconstituted HDL (rHDL), as the synthetic form of natural HDL, is composed of phospholipids (PC), cholesterol (C), cholesteryl esters (CE) and apoA I. rHDL shares the similar physicochemical properties to natural HDL 22 and has advantages of endogenous component, biomimetic structure and endosome/lysosome-independence endocytosis, which make rHDL a successful gene transfection vector for stem cell-based gene therapy 23.
Tumor necrosis factor (TNF)-related apoptosis inducing ligand (TRAIL) is extensively used as a safe and broad-spectrum anticancer protein for tumor therapy, which can specifically bind to death receptor 4 (DR4) or death receptor 5 (DR5) on tumor cells, leading to apoptosis occurrence of tumor cells through extracellular pathway without side effects on normal cells 24, 25.
In this study, we developed a MSCs-based platform, bone marrow-derived MSCs of C57BL/6 mice genetically engineered by rHDL-based nanoparticles to express and secrete TRAIL for metastatic melanoma therapy (shown in Scheme. 1). Based on our understanding, it was a completely innovative strategy to construct MSCs-based platform engineered by an eximious lipoprotein-bioinspired nanovector rHDL in the field of MSCs-mediated cancer therapy. During the construction of rHDL-based gene delivery system, the amphiphilic complexes were assembled by electrostatic binding of plasmid DNA encoding TRAIL protein (pTRAIL) and lauric acid modified PEI 25 K (PEI-LA), followed by loading PEI-LA/pTRAIL complexes into hydrophobic core of rHDL. TRAIL-delivered rHDL nanoparticles could specifically combine with SR-BI expressed on MSCs to form a hydrophobic channel, which facilitated the diffusion of PEI-LA/pTRAIL cargo of rHDL directly into the cytoplasm of MSCs via a lysosome-independent pathway. Then PEI-LA/pTRAIL complexes were dissociated by the negative-charged substances in cytoplasm, followed by TRAIL transfection and expression in MSCs nucleus. These tumor targeting TRAIL-engineered MSCs were eventually administrated to pulmonary melanoma bearing mice to achieve apoptosis induction of tumor cells. We studied the physicochemical properties, cytotoxicity, cellular uptake, intracellular distribution, and transfection efficiency ofrHDL/PEI-LA/pDNA nanoparticles in vitro to establish TRAIL-engineered MSCs platform. Western Blotting Assay and ELISA were used to characterize the expression of TRAIL in MSCs engineered by rHDL, and transwell migration assay was used to determine tumor-targeting of MSCs-TRAIL in vitro. Finally, we validated the therapeutic effect of TRAIL-engineered MSCs anticancer platform for the treatment of aggressive melanoma cell line B16F10 in vitro and in vivo.
2. Methods
Materials
PEI (Mw 25 K, 1.8 K) and lauric acid (LA) were purchased from Sigma-Aldrich (MO, USA). 1-Ethyl-3-(3-dimethyllaminopropyl) carbodiimide hydrochloride (EDCI) and N-hydroxysuccinimide (NHS) were obtained from Aladdin Reagent Database Inc. (Shanghai, China). An expression vector containing open reading frame of the mouse TRAIL (tumor necrosis factor ligand superfamily member 10 [Mus musculus], noted as TNFSF 10 or TRAIL) (pCMV6-Myc-DDK-TNFSF10, 4.944 kb) was purchased from ORIGENE (Beijing, China) The reporter plasmid (pEGFP-C3, 4.7 kb) encoding enhanced green fluorescent protein (GFP) driven by CMV promoter was purchased from Addgene (MA, USA). Both the plasmid was propagated in DH-5α Escherichia coli and purified by Endo-free Plasmid Maxi kit (Omega Bio-Tek, USA) to obtained plasmid coding TRAIL (noted as pTRAIL) and pEGFP-C3, respectively. Soybean phospholipids was purchased from Shanghai A.V.T. Pharmaceutical Co.,Ltd (Shanghai, China); cholesterol, cholesteryl esters were purchased from Sigma-Aldrich (MO, USA). Apolipoprotein A I (apoA I), the major protein component in HDL nanoparticles, was isolated and highly purified from the albumin byproduct in our laboratory according to an established protocol 26. Fetal bovine serum (FBS) and trypsin solution were obtained from Gibco (CA, USA). DMEM-F12 and penicillin-streptomycin was purchased from HyClone (UT, USA). 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) and crystal violet were obtained from KeyGEN BioTECH Corp.,Ltd (Jiangsu, China). Cy5.5-NHS were obtained from Beijing Fanbo Science and Technology Co., Ltd (Beijing, China). YOYO-1, DAPI, LysoTracker Red and DiI were offered by Invitrogen Corporation (CA, USA). Mouse TRAIL ELISA Kit was obtained from Boster Company (Wuhan, China). Annexin V-FITC apoptosis detection kit was purchased from BioVision (CA, USA).
Preparation and characterization of PEI-LA/pDNA complexes
PEI-LA/pDNA binary complexes was prepared by adding a PEI-LA solution dropwise into a pDNA solution at a series of N/P. Then the solution was incubated for 30 min with gentle vortex at room temperature. The encapsulation capacity of PEI-LA/pDNA complexes were assessed by using electrophoresis on a 0.6% agarose gel (containing 5 μL/100 mL of Goldview) at 90 mV for 30 min in 0.5×Tris-Borate-EDTA (TBE) buffer solution. The gel image was visualized under Gel-Pro analyzer (Genegenius, Syngene, UK). Additionally, the particle size and zeta potential of PEI-LA/pDNA complexes were characterized using Zetasizer (Nano-ZS90, Malvern, UK).
Preparation of rHDL/PEI-LA/pDNA nanoparticles
Solvent evaporation method was introduced to prepare lipid contents. rHDL/PEI-LA/pDNA was conducted by incubating the lipid contents with apoA I as described in previous studies with minor modifications 27. Briefly, 1 mg of soybean phospholipids (PC), 0.2 mg of cholesterol (C) and 0.1 mg of cholesteryl esters (CE) were dissolved in 5 mL of organic solvent (ethanol: dichloromethane = 5:2, V/V) and evaporated under vacuum at 37 °C until a thin film was formed. The film was then kept under vacuum for additional 8 h to remove residual organic solvent. After that, a PEI-LA/pDNA complexes solution (the concentration of pDNA was 2 μg/mL) was added to dissolve the thin film for 30 min at 37 °C, followed by sonicated in ice/water bath using a probe-type ultrasonicator (JY 92-2D, Ningbo Scientz Biotechnology Co., Ltd, China) at 120 W for 20 min. The suspension was then filtrated through 0.22 μm filter and incubated with apoA I (0.2 mg in 0.1 mL of PBS) under 300 rpm stirring at 4 °C for 8 h to form the final rHDL/PEI-LA/pDNA nanoparticles. After incubation, free apoA I was eliminated using a Sepharose G-100 column (1.0 × 18 cm) to obtain the final rHDL/PEI-LA/pDNA nanoparticles.
Physicochemical properties of rHDL/PEI-LA/pDNA nanoparticles
The particle size distribution and zeta potential of rHDL/PEI-LA/pDNA nanoparticles were measured by dynamic light scattering (DLS) with a scattering angle 90 º at 25 °C using Zetasizer (Nano-ZS90, Malvern, UK). The morphology of complexes was observed by a transmission electron microscope (TEM, H7650, Hitachi, Japan). The encapsulation efficiency (EE) of pDNA (labeled by YOYO-1 as the direction, noted as pDNAYOYO-1) in rHDL/PEI-LA/pDNA nanoparticles was calculated by the following formulation (1):
| EE% = C/C0 × 100% | (1) |
Where C0 and C were the initial feed concentration of pDNAYOYO-1 and the concentration of pDNAYOYO-1 in the nanoparticles, respectively. The concentration of pDNAYOYO-1 was measured by fluorescence spectrophotometry with excitation at 491 nm and emission at 509 nm.
Serum stability assays of rHDL/PEI-LA/pDNA nanoparticles
Serum stability of rHDL/PEI-LA/pDNA was investigated in the presence of fetal bovine serum (FBS, Gibco). The nanoparticles were mixed with equal volume of FBS for prearranged incubation time (1, 2, 4, 6, 8, 12, 24 h) at 37 °C and the particle size was determined by Zetasizer (Nano-ZS90, Malvern, UK).
Culture of MSCs
C57BL/6 mouse bone marrow derived MSCs were purchased from Cyagen Biosciences Inc. (Suzhou, China). The cells were incubated in DMEM/F12 (HyClone, USA) supplemented with 10% FBS (Gibco, USA), 100 U/mL of penicillin and 100 μg/mL of streptomycin. MSCs were cultured at 37 ± 0.5 °C in a humidified atmosphere containing 95% relative humidity and 5% CO2. Culture medium was changed every other day, and the cells were subcultivated at 80~90% confluence using 0.25% (w/v) trypsin. MSCs within 5 passages were used for this study.
Culture of tumor cells
Mouse B16F10 melanoma cells utilized in this study was purchased from the cell bank of Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI-1640 (HyClone, USA) with 10% (v/v) FBS (Gibco, USA), 100 U/mL of penicillin and 100 μg/mL of streptomycin at 37 ± 0.5 °C under 5% CO2 and 90% relative humidity, and were subcultivated with 0.25% (w/v) trypsin at 80~90% confluence.
Cytotoxicity Assay of rHDL/PEI-LA/pTRAIL Nanoparticles to MSCs in vitro
The cytotoxicity of rHDL/PEI-LA/pTRAIL nanoparticles on MSCs was evaluated using MTT assay. MSCs were seeded at a density of 1 × 104 cells/well into 96-well plates (Costar, USA) and incubated with complete medium for 24 h at 37 °C. Then the medium was replaced with 200 μL of rHDL/PEI-LA/pTRAIL nanoparticles or 200 μL of PEI-LA/pTRAIL complexes suspended in serum-free medium, respectively. The concentration of pTRAIL in each group was set to 2, 4, 6, 8, 10 μg/mL. After 6 h of incubation, the medium was replaced with fresh medium and the cells were cultured for additional 48 h. Subsequently, 20 μL of MTT solution (5 mg/mL) was added into each well for another 4 h incubation at 37 °C. Then the solution in each well was removed, followed by adding 150 μL of DMSO in each well. The absorbance of each well was measured by a multifunctional microplate reader (Synergy2, BioTek, USA) at a wavelength of 570 nm. Cell viability was calculated by the following formulation (2):
| Cell viability (%) = (Asample - Ablank)/(Acontrol - Ablank) × 100% | (2) |
Where Asample represented the absorbance of the cells treated with the corresponding sample; Acontrol and Ablank represented the absorbance of untreated cells and blank culture medium, respectively.
Cellular Uptake of rHDL/PEI-LA/pDNA Nanoparticles to MSCs
The cellular uptake of rHDL/PEI-LA/pDNA nanoparticles was investigated by flow cytometry (FACS Calibur, Becton Dickinson, USA) and confocal laser scanning microscopy (CLSM, SP5, Leica, Germany). pDNA was labeled by YOYO-1 (noted as pDNAYOYO-1) as a guide to track the position of the plasmid. MSCs were seeded in 6-well plates (Costar, USA) at a density of 1 × 105 cells/well until 70~80% confluence. After that, the culture medium was replaced with rHDL/PEI-LA/pDNAYOYO-1 nanoparticles or PEI-LA/pDNAYOYO-1 complexes that were suspended in serum-free medium respectively (the total volume was 1 ml and the concentration of pDNAYOYO-1 was 4 μg/ml). Meanwhile, 4 μg/mL of naked pDNAYOYO-1 was set as control. For competition assay, excessive apoA I (a terminal concentration of 1 mg/mL) was added 2 h prior to rHDL/PEI-LA/pDNAYOYO-1 nanoparticles group. The treated MSCs were rinsed with PBS (pH 7.4) three times after 6 h incubation. The fluorescence intensity in tumor cells was analyzed by flow cytometer with 488 nm excitation, and images of cellular uptake by DAPI stained cells were taken by CLSM.
Intracellular Distribution assay of rHDL/PEI-LA/pDNA Nanoparticles in MSCs
For cellular distribution assay, MSCs were seeded at a density of 1 × 104 cells/dish in 35 mm glass-bottomed dishes and incubated with culture medium containing 10% serum for 24 h. Subsequently, MSCs were treated with PEI-LA/pDNAYOYO-1 complexes or rHDL/PEI-LA/pDNAYOYO-1 nanoparticles for 2 h (the concentration of pDNA was 4 µg/ml). For competition assay, excessive of apoA I (a terminal concentration of 1 mg/mL) was added 2 h prior to rHDL/PEI-LA/pDNAYOYO-1 nanoparticles group. MSCs were stained with 50 nM Lyso-Tracker Red for 1 h prior the 2 h post-administration and then washed three times with PBS to observe the co-localization images using CLSM. In order to track the location of apoA I, Cy5.5-NHS was utilized to decorate apoA I based on the established protocol 28. MSCs were treated with Cy5.5-rHDL/PEI-LA/pDNAYOYO-1 nanoparticles containing 4 µg/mL of pDNA for 6 h followed by washing three times by PBS (pH 7.4). Then DAPI was added to stain the nucleus for 10 min followed by adding of 4% paraformaldehyde for 15min to fix MSCs. Location images of Cy 5.5-apoA I and pDNAYOYO-1 were visualized by CLSM.
Transfection of rHDL/PEI-LA/pEGFP-C3 Nanoparticles to MSCs
MSCs were seeded into a 6-well plate at a density of 1 × 105 cells/well for 24 h. Then rHDL/PEI-LA/pEGFP-C3 nanoparticles, PEI-LA/pEGFP-C3 complexes and Lipofectamine 2000/pEGFP-C3 (containing 4 µg/mL of pDNA per well) were added into transfection medium followed by 6 h incubation. Subsequently, the transfection medium was replaced with fresh complete culture medium and cultured for additional 48 h. Meanwhile, naked pEGFP-C3 with concentration of 4 μg/mL was set as control. For competition assay, excessive of apoA I (a terminal concentration of 1 mg/mL) was added 2 h prior to rHDL/PEI-LA/pEGFP-C3 nanoparticles group. The transfection efficiency defined as the percentage of GFP positive MSCs was quantified by flow cytometry and the expression of GFP in MSCs was observed by CLSM.
Western Blotting Assay of TRAIL Expression in MSCs
The expression of TRAIL protein in MSCs treated with rHDL/PEI-LA/pTRAIL nanoparticles was evaluated by Western Blotting assay. MSCs (1×105 cells/well) were seeded in 6-well plates for 24 h followed by transfection with rHDL/PEI-LA/pTRAIL nanoparticles or PEI-LA/pTRAIL complexes for 6 h (the concentration of pTRAIL was 4 μg/ml). Subsequently, the transfection medium was changed into fresh medium. Meanwhile, naked pTRAIL at a concentration of 4 μg/mL and PBS (pH 7.4) were set as control. For competition assay, excessive of apoA I (a terminal concentration of 1 mg/mL) was added 2 h prior to rHDL/PEI-LA/pTRAIL nanoparticles group. After 5 days of transfection, the treated cells were harvested and processed by standard Western Blotting assay to detect the expression level of TRAIL. The lanes were visualized by Gel-Pro analyzer (Genegenius, Syngene, UK) and normalized to β-actin expression.
ELISA assay of TRAIL Expression in MSCs
TRAIL expression level of MSCs treated with rHDL/PEI-LA/pTRAIL nanoparticles was also evaluated by ELISA. Briefly, MSCs were seeded in 24-well plates (5 × 104 cells/well) for 24 h followed by treating with rHDL/PEI-LA/pTRAIL nanoparticles or PEI-LA/pTRAIL complexes as described in Section Western Blotting Assay of TRAIL Expression in MSCs. After 5 days of culture, the TRAIL concentration in the culture medium supernatant was measured using an ELISA kit according to the manufacturer's instructions.
In vitro Migration Assay of MSCs-TRAIL
The migration capacity of MSCs was determined by a standard transwell migration assay using transwell chambers with 8 μm pore size (Corning Costar, USA). After transfection by rHDL/PEI-LA/pTRAIL nanoparticles or PEI-LA/pTRAIL complexes as described in Section 2.8, the treated MSCs were trypsinized and suspended in 200 μL of culture media to plate in the upper chamber at the density of 2 × 104 cells/mL. The untreated MSCs were set as positive control. Then B16F10 cells with same density were seeded in the lower chamber of transwell plate (the total volume was 500 μL) and the untreated MSCs in the upper chamber and culture medium without B16F10 cells in the lower chamber were acted as negative control. MSCs remaining on the upper face of the filters were removed by cotton swabs after incubated for 24 h at 37 °C, and the migrating MSCs were fixed with 4% paraformaldehyde PBS solution and stained with crystal violet. Cells on the insert membrane were observed by a transmitted-light microscopy (EVOS XL, ThermoFisher Scientific, USA). Results were expressed as the average number of migrating cells for three replicates from five random fields.
In vitro Antitumor Effects of MSCs-TRAIL
To evaluate the antitumor efficiency of TRAIL-expressing MSCs in vitro, MSCs were transfected by PEI-LA/pTRAIL complexes or rHDL/PEI-LA/pTRAIL nanoparticles as described in section Transfection of rHDL/PEI-LA/pEGFP-C3 Nanoparticles to MSCs. Then MSCs were plated in the upper chamber of the transwell plates (0.4 μm pores, Corning Costar, USA) at a density of 4 × 104 cells/well. Meanwhile, B16F10 cells (2 × 104 cells/well) were seeded in the lower chamber. PBS (pH 7.4) and rHDL/PEI-LA/pDNA transfected MSCs were set as controls. After 1, 3 and 5 days of administration, the viability of B16F10 cells in the lower chamber was detected by MTT assay as described in section 2.6. Apoptosis induction of MSCs treated for 5 days was investigated with an Annexin V-FITC/PI apoptosis detection kit according to kit manufacturer's protocol, and was analyzed by flow cytometry.
Animals and pulmonary metastasis model
Female C57BL/6 mice (six week-old) were purchased from Shanghai Laboratory Animal Center (SLAC, Shanghai, China). The mice were treated according to the protocols evaluated and approved by the ethical committee of China Pharmaceutical University. The pulmonary metastasis model of malignant melanoma was established in C57BL/6 mice by intravenous injection of 200 μL of B16F10 cells (2 × 105 cells/mouse) suspended in PBS (pH 7.4).
Tissue distribution of MSCs-TRAIL
C57BL/6 mice bearing B16F10 pulmonary metastasis tumor were randomly divided into 4 groups after the injection of B16F10 cells to tail vein for 8 days. Mice in each group were treated with 200 μL of PBS (pH 7.4), 200μL of untreated MSCs (1× 106 cells), 200 μL of TRAIL-expressing MSCs (1× 106 cells, transfected by PEI-LA/pTRAIL nanoparticles, noted as MSCs-TRAIL (PEI-LA)) and 200 μL of TRAIL-expressing MSCs (1× 106 cells, transfected by rHDL/PEI-LA/pTRAIL nanoparticles, noted as MSCs-TRAIL (rHDL)), respectively. Meanwhile, MSCs were pre-stained by DiI (10 μM) according to manufacturer's protocol. The tumor-bearing lung tissues and other organs in different groups were obtained and imaged using an in vivo imaging system (FX PRO, Kodak, USA) to detect the fluorescence 7 days after administration. Then the tumor-bearing lung tissues were fixed in 4% paraformaldehyde PBS solution, sliced and stained with DAPI. The images were visualized by transmitted-light microscopy (EVOS FL, ThermoFisher Scientific, USA).
In vivo Antitumor Efficacy of MSCs-TRAIL
C57BL/6 mice bearing B16F10 pulmonary metastasis tumor were randomly divided into 4 groups after the injection of B16F10 cells to tail vein for 9 days. Mice in different groups were treated with 200 μL of PBS (pH 7.4), 200 μL of MSCs-pDNA (1× 106 cells, transfected by rHDL/PEI-LA/pDNA nanoparticles), 200 μL of MSCs-TRAIL (PEI-LA) (1× 106 cells) and 200 μL of MSCs-TRAIL (rHDL) (1× 106 cells) by intravenous injection, respectively. All the preparations were suspended in PBS (pH 7.4). Mice in each group were sacrificed at 21 days after establishment of pulmonary metastasis tumor. Lungs were then collected from the mice and the lungs of normal mice were collected as control. The metastatic nodules in lung and lung weight were recorded. Then lungs in each group were fixed in the 4% paraformaldehyde followed by sliced for hematoxylin and eosin (H&E) staining and terminal deoxyribonucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) analysis.
Statistical Analysis
All the data were expressed as mean ± standard deviation (SD) and were calculated using Student's t-test; significant differences between groups were indicated by *P < 0.05, **P < 0.01 and ***P < 0.001, respectively.
3. Results and Discussions
Preparation and Characterization of rHDL/PEI-LA/pDNA Nanoparticles
PEI 25 K, as a “golden standard” for polymeric gene carrier, exhibits higher gene transfection efficiency than other cationic polymer carriers. Moreover, hydrophobic modification of polymers has been frequently studied as an effective way to enhance their gene transfection efficiency because lipid components can significantly alter some physicochemical and biological properties of polymers and enhance their adsorption on the cell membrane 29. Herein, in order to optimize the encapsulation efficiency of rHDL for pDNA, PEI 25 K backbone was hydrophobically modified across amide linkage using LA as a saturated fatty acid by EDCI/NHS mediated chemistry (Figure S1A). Due to the hydrophobic property of LA, it could be more suitable to introduce LA into PEI skeleton to incorporate into the hydrophobic core of rHDL. The chemical structure of PEI-LA was determined by 1H NMR (Figure S1B). The degree of LA grafted on PEI 25 K calculated by proton peak integral was 3.94%. As for FT-IR spectrum (Figure S1C), the peak of PEI-LA for the νNH was observed at 3400~3250 cm-1 while the peak of νC=O was observed at 1647 cm-1, indicating that LA was successfully conjugated to PEI backbone by amide linkage. In addition, the buffering capacity of PEI-LA was shown in Figure S2. The results indicated that LA decoration did not affect the proton buffer capacity of PEI 25 K in PEI-LA.
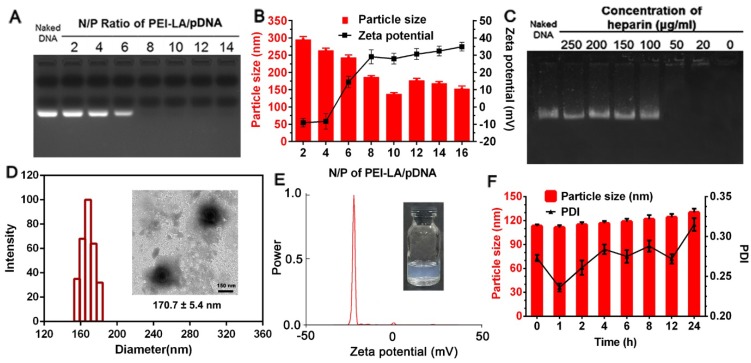
A gene vector with high transfection efficacy and low cytotoxicity is essential for the construction of genetically engineered MSCs to prevent invalid therapy and cellular damage. In our design, PEI 25 K was modified with hydrophobic LA in previous description. However, the relative high positive zeta potential of PEI-LA/pDNA binary complex might still cause MSCs injury after direct transfection. To this point, PEI-LA/pDNA binary complex was loaded into rHDL to conceal this defect, thereby protecting cell membrane of MSCs from direct contact with PEI-LA, which was due to the amphiphilic property of LA-modified PEI 25 K. The physicochemical characterization of PEI-LA/pDNA binary complexes and rHDL/PEI-LA/pDNA nanoparticles were shown in Figure 1. The ability of PEI-LA to condense pDNA was confirmed by gel retardation assay (Figure 1A). When N/P was more than 8, no free pDNA were observed in the lanes, indicating that pDNA were completely encapsulated by PEI-LA. The particle size and zeta potential were also investigated. At N/P of 10, PEI-LA/pDNA exhibited an optimal particle size of 137.4 ± 4.1 nm and positive zeta potential of 27.95 ± 5.12 mV (Figure 1B). The TEM image of PEI-LA/pDNA was showed in Figure S3A. In order to ensure the intracellular DNA release of PEI-LA/pDNA by competing with negatively charged substances, heparin stability assay was conducted at N/P of 10 by gel retardation assay. As shown in Figure 1C, pDNA could be well protected by PEI-LA when the concentration of heparin was less than 100 μg/mL because DNA bands were not reflected in the corresponding lanes. Furthermore, pDNA were released from binary complexes as heparin concentration increased due to the charge competition. This release property could disassemble PEI-LA/pDNA complexes in cytoplasm to achieve DNA transcription and translation.
Figure 1.
(A) Agarose gel electrophoresis retardation assay of PEI-LA/pDNA binary complexes with different N/P ratio. (B) Effect of different N/P ratio on the particle size and zeta potential of PEI-LA/pDNA binary complexes (n = 3). (C) Protection assay of PEI-LA/pDNA binary complexes against different concentration of heparin. (D) Size distribution histogram of rHDL/PEI-LA/pDNA nanoparticles. Inset: TEM image of rHDL/PEI-LA/pDNA nanoparticles. Scar bar: 150 nm. (E) Zeta potential of rHDL/PEI-LA/pDNA nanoparticles. (F) Influence of FBS on the particle size and polydispersity index of rHDL/PEI-LA/pDNA nanoparticles at different time points (n = 3).
Subsequently, PEI-LA/pDNA binary complexes with N/P of 10 were incorporated into the lipid core of rHDL relying on the amphiphilic property of PEI-LA. As shown in Figure 1D and Figure 1E, rHDL/PEI-LA/pDNA nanoparticles with semitransparent appearance (inset of Figure 1E and Figure S3C) had the resulting particle size of 170.7 ± 5.4 nm. The polydispersity index (PDI) of rHDL/PEI-LA/pDNA nanoparticles was 0.192 ± 0.024 and the zeta potential was -24.47 ± 1.88 mV. And the size and morphology with a clear core-shell structure of rHDL/PEI-LA/pDNA nanoparticles were directly revealed by TEM (inset of Figure 1D). As contrast, the results of size, zeta potential and TEM image of rHDL nanoparticles were showed in Figure S3. In addition, the encapsulation efficacy of pDNAYOYO-1 in rHDL/PEI-LA/pDNA nanoparticles determined by the fluorescence spectrophotometry was (84.64 ± 3.82) %, confirming the outstanding pDNA loading capacity of our designed rHDL nanocarrier. The size changes of rHDL/PEI-LA/pDNA nanoparticles were also investigated in the presence of FBS to identify their stability in vitro. As shown in Figure 1F, there was no significant change in particle size (within the increase of 10%) during 6 h incubation with FBS, and PDI was acceptable (less than 0.3), indicating that rHDL/PEI-LA/pDNA nanoparticles could keep stable when incubated with MSCs. Furthermore, rHDL/PEI-LA/pDNA nanoparticles could protect pDNA against DNase I for at least 24 h,as shown in Figure S4.
In vitro Cytotoxicity Study of rHDL/PEI-LA/pTRAIL Nanoparticles
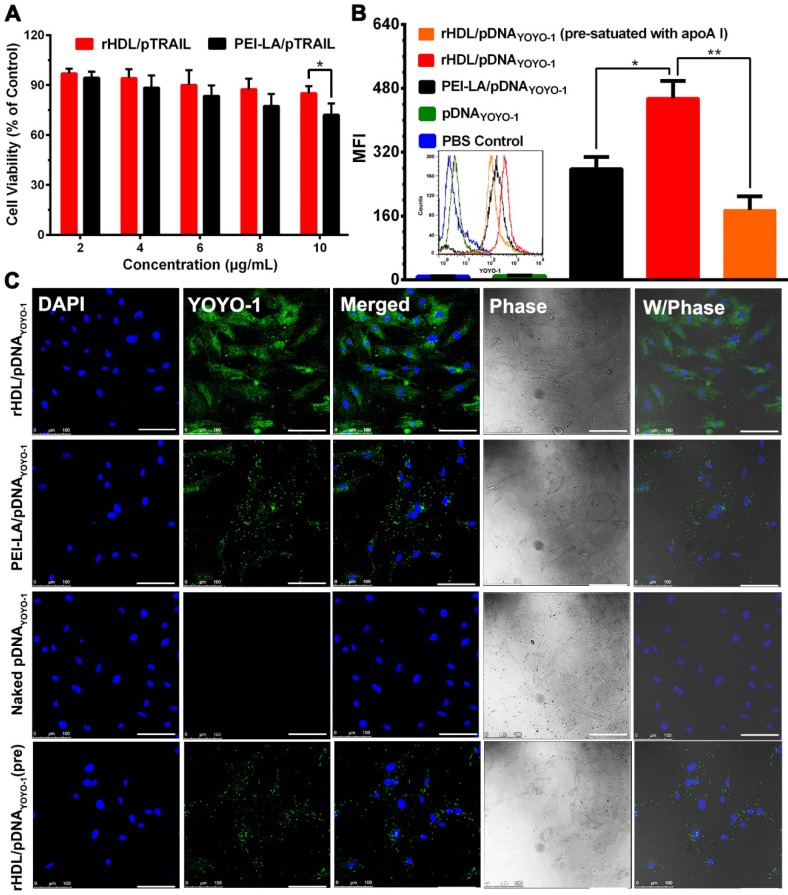
In order to investigate the TRAIL-delivering rHDL we designed on MSCs, the cytotoxicity of PEI-LA/pTRAIL binary complexes and rHDL/PEI-LA/pTRAIL nanoparticles were evaluated by MTT assay. As shown in Figure 2A, rHDL nanoparticles had no significant cytotoxicity to MSCs as pTRAIL concentration changed from 2 μg/mL to 10 μg/mL, indicating that neither rHDL vector nor pTRAIL caused damage to MSCs. In contrast, PEI-LA/pTRAIL showed a little cytotoxicity. PEI-LA/pTRAIL loaded with 10 μg/mL TRAIL plasmid resulted in 72.13% cell survival, which demonstrated that rHDL was an ideal TRAIL-delivered lipoprotein-bioinspired nanovector with superior biocompatibility and could significantly reduce the toxicity of PEI-LA towards MSCs.
Figure 2.
(A) Cytotoxicity of MSCs exposed to PEI-LA/pTRAIL binary complexes and rHDL/PEI-LA/pTRAIL nanoparticles with different concentrations of pTRAIL for 6 h followed by incubation for another 48 h (n = 5). (B) Flow cytometry measurements of YOYO-1 fluorescence intensity in MSCs (n = 3). (C) Representative confocal imaging of rHDL/PEI-LA/pDNAYOYO-1 in MSCs. Naked pDNAYOYO-1, PEI-LA/pDNAYOYO-1 and apoA I pre-treated rHDL/PEI-LA/pDNAYOYO-1 were performed as controls. Scale bar: 100 μm.
Cellular Uptake and Intracellular Distribution of rHDL/PEI-LA/pDNA Nanoparticles
Due to the specific binding between rHDL and SR-BI on cell membrane of MSCs, rHDL was expected to have a tendency to greatly improve the intracellular uptake of its therapeutic gene cargoes. To verify this, pDNA labeled by the fluorescent probe YOYO-1 (named pDNAYOYO-1) was loaded into rHDL nanoparticles and PEI-LA binary complexes, respectively, and the nucleus of MSCs was stained with DAPI followed by characterization with CLSM. As shown in Figure 2B and Figure 2C, the fluorescent intensity of rHDL group was significantly higher than that of PEI-LA group after 6 h incubation with MSCs, both of which were observed by flow cytometry and CLSM. For comparison, naked pDNAYOYO-1 group showed no detectable fluorescent intensity. In addition, rHDL mediated pDNAYOYO-1 intracellular uptake was significantly decreased by 2.6-fold when MSCs were pre-treated with excess free apoA I to competitively saturate SR-BI, indicating that the intracellular uptake of gene-loaded rHDL nanoparticles was mainly mediated by SR-BI.
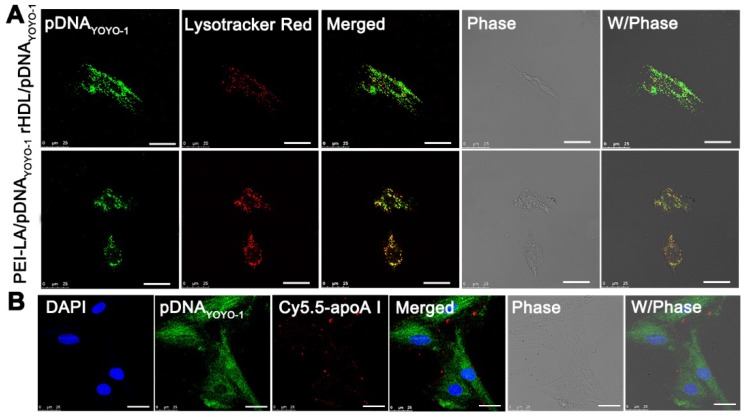
ApoA I as the main apolipoprotein on the surface of rHDL was able to specifically bind to SR-BI on the surface of MSCs, through which the lipid compositions of rHDL could diffuse into cytoplasm, avoiding SR-BI-mediated cytosolic delivery of endosome/lysosome degradation 28. The cellular uptake pathway of rHDL nanoparticles was also investigated by tracing the intracellular co-location of nanocarrier and lysosomes which were stained with Lyso-Tracker Red in MSCs. As shown in Figure 3A, PEI-LA/pDNAYOYO-1 group showed a great amount of yellow fluorescence after 2 h incubation, indicating that pDNAYOYO-1 (green fluorescence) was almost overlapped with lysosome (red fluorescence). In contrast, little yellow fluorescence was observed in rHDL group, demonstrating that the main cellular uptake mechanism of rHDL gene delivery system was mediated by lysosome-independent pathway via non-aqueous channels of SR-BI expressed on MSCs 30.
Figure 3.
(A) Representative images of intracellular trafficking of rHDL/PEI-LA/pDNAYOYO-1 in MSCs at 2 h observed by CLSM. PEI-LA/pDNAYOYO-1 was set as control. Scale bar: 25 μm. (B) Representative images of co-location of Cy5.5-rHDL/pDNAYOYO-1 treated MSCs at 6 h. Scale bar: 25 μm.
To further discuss this presumed intracellular uptake mechanism of rHDL gene delivery system, the co-location of Cy5.5 labeled apoA I and YOYO-1 labeled pDNA were loaded in rHDL followed by observation in MSCs using CLSM. Meanwhile, the nucleus of MSCs was stained with DAPI. As shown in Figure 3B, the apoA ICy5.5 in rHDL/pDNAYOYO-1 group was mainly observed on the cell membrane of MSCs, while pDNAYOYO-1 was distributed in cytoplasm even in nucleus of MSCs after 6 h incubation. As reported in our previous studies, rHDL could selectively delivery the lipid compositions to the cytoplasm of tumor cells via SR-BI based hydrophobic channels 15, 29. Therefore, it was speculated that during cellular internalization of pDNAYOYO-1, apoA I of rHDL could specifically combine with SR-BI expressed on the surface of MSCs and PEI-LA/pDNAYOYO-1 diffused from rHDL nanoparticles to cytoplasm along the concentration gradient. The gene delivery mechanism mediated by this non-aqueous “channel” of SR-BI on MSCs was similar to HDL mediated cholesteryl esters transport. This lipid cargo-selective cellular uptake mechanism of rHDL could protect therapeutic gene from various enzymes and acidic environment of lysosome.
In vitro Transfection Mediated by rHDL Nanoparticles
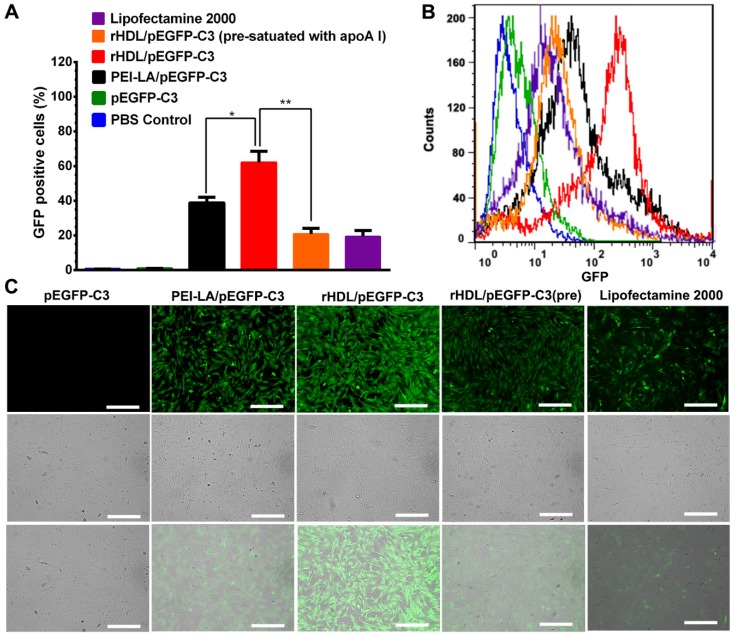
The rHDL with unique lysosome-independent mechanism of cellular uptake exhibited high gene transfection efficiency in MCF-7 cells compared with cationic vectors 17. Herein, enhanced green fluorescent protein plasmid (pEGFP-C3) was selected to be encapsulated in rHDL to evaluate the transfection efficiency of rHDL in MSCs in vitro. The results were presented by the fluorescence positive rate in total cells determined by flow cytometry. Meanwhile, the fluorescence intensity was also observed by CLSM. As shown in Figure 4A and Figure 4B, MSCs treated with rHDL/PEI-LA/pEGFP-C3 exhibited the highest ratio of GFP positive cells as 62.16%, while PEI-LA/pEGFP-C3 group showed only 38.91% of GFP positive cells. As comparison, few fluorescence signal was detected in the naked pEGFP-C3 group, and the lipofectamine 2000/pEGFP-C3 group as positive control showed low level of transfection with 19.44% of GFP positive cells. Furthermore, in the competitive suppression group, the ratio of GFP positive MSCs dropped to 20.89% due to SR-BI pre-satuation of apoA I. Similar results were also observed in CLSM images that rHDL/PEI-LA/pEGFP-C3 group exhibited higher green fluorescence intensity compared with PEI-LA/pEGFP-C3 group and apoA I pre-treated group (shown in Figure 4C). The above results confirmed that rHDL significantly enhanced the gene transfection efficacy in MSCs due to rHDL-mediated SR-BI and lysosome-independent mechanism of cellular uptake for gene cargoes of rHDL.
Figure 4.
(A) Transfection efficiency in MSCs defined as the percentage of GFP positive cells detected by flow cytometry (n = 3). (B) Flow cytometry analysis of GFP positive MSCs. (C) Representative image of in vitro GFP expression in MSCs obtained by CLSM. Scale bar: 250 μm.
TRAIL Expression in MSCs
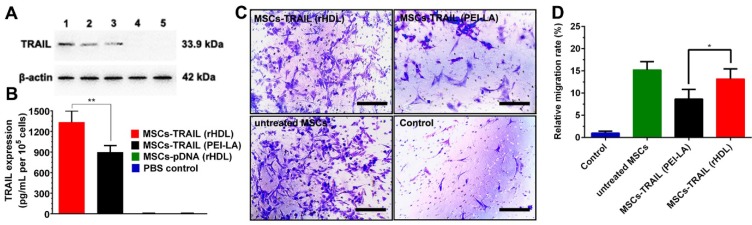
The expression of TRAIL in MSCs transduced by rHDL/PEI-LA/pTRAIL nanoparticles was evaluated using Western Blotting assay and ELISA assay. As shown in Figure 5A, the clear protein band of rHDL/PEI-LA/pTRAIL nanoparticles group indicated that TRAIL was successfully expressed in MSCs. As comparison, naked pTRAIL group and PBS control group did not express TRAIL. Meanwhile, MSCs treated with PEI-LA/pTRAIL showed less TRAIL expression compared with rHDL group, and apoA I pre-incubated rHDL group also showed low expression of TRAIL protein, indicating that pre-saturation of SR-BI resulted in a decrease in cellular uptake of rHDL-based nanoparticles. Furthermore, the expression level of TRAIL secreted by MSCs was also measured by ELISA (shown in Figure 5B). MSCs transfected by rHDL/PEI-LA/pTRAIL nanoparticles exhibited a relatively high protein expression after transfection for 5 days (1332.4 pg/mL per 105 cells), while PEI-LA/pTRAIL group showed less protein expression level (896.45 pg/mL per 105 cells). Results above demonstrated that TRAIL could be efficiently delivered and transfected into SR-BI-expressed MSCs mediated by rHDL nanoparticles owing to rHDL-mediated SR-BI targeting and lysosome-independent intracellular gene delivery, causing high expression of TRAIL protein in MSCs, which laid foundation for the MSCs-based tumor therapy platform.
Figure 5.
(A) Western blotting analysis of TRAIL expression in MSCs at 5 days post-transfection. Lane 1: rHDL/PEI-LA/pTRAIL; Lane 2: PEI-LA/pTRAIL; Lane 3: apoA I pre-treated rHDL/PEI-LA/pTRAIL; Lane 4: naked pTRAIL; Lane 5: PBS control. (B) The secretion of TRAIL in culture media of different formulation transfected MSCs tested by mouse TRAIL ELISA kit at 5 days (n = 3). (C) Representative photomicrographs of in vitro migration of different formulations transfected MSCs in response to B16F10 cells using Transwell plates (8 μm pores) for 24 h. (D) Quantitative migration results of different formulations transfected MSCs from five random fields.
In vitro Migration Assay of MSCs-TRAIL
In order to verify whether rHDL-mediated TRAIL transfection influenced the tumor- homing ability of MSCs, standardized MSCs migration assay towards B16F10 cells in vitro was performed in this study. As shown in Figure 5C and Figure 5D, MSCs-TRAIL treated with rHDL/PEI-LA/pTRAIL nanoparticles exhibited relatively high MSCs migration rate (13.19 times compared with control group), and the non-treated MSCs group exhibited equal level of migration rate that was 15.23 times of the control. In contrast, MSCs migration rate of PEI-LA/pTRAIL transfected group was just 8.67 times compared with control group. Results above indicated that both rHDL-based gene delivery system and TRAIL protein expression had no obvious side effect on the migration capacity of MSCs. Therefore, the drug delivery platform based on rHDL-engineered MSCs, with good biocompatibility and efficient tumor-targeting, would have a great potential for further active targeting tumor therapy.
In vitro Antitumor Efficacy of MSCs-TRAIL
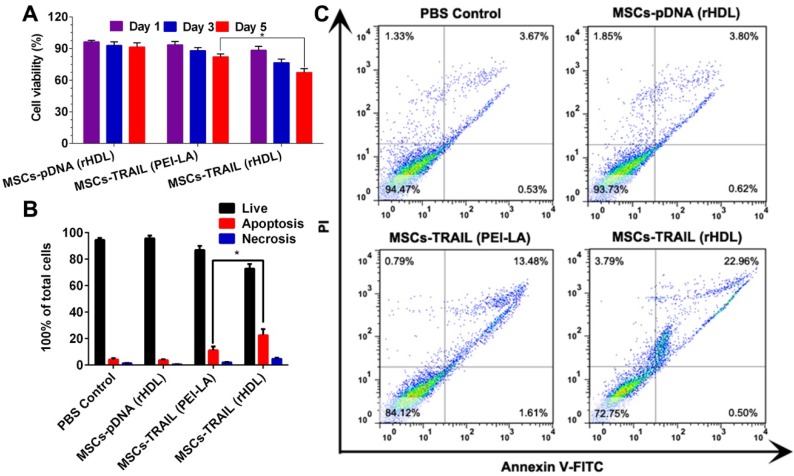
We tested the viability of B16F10 cells by cytotoxicity assay in vitro after incubated with TRAIL-expressing MSCs. Transwell plates (the pore size of semiporous membranes was 0.4 μm) were used to separate MSCs and B16F10 cells in this experiment. MTT results shown in Figure 6A exhibited a time-dependent inhibition of B16F10 cell growth by TRAIL-expressing MSCs. In addition, the viability of B16F10 cells declined to < 70% for 5 days after administration with TRAIL-expressing MSCs transfected with rHDL.
Figure 6.
(A) Cytotoxicity of B16F10 cells exposed to different formulations transfected MSCs for 1, 3 and 5 days (n = 5). (B) Statistical analysis of apoptotic and necrotic cells of B16F10 cells at 5 days induced by different formulations transfected MSCs (n = 3). (C) Cell apoptosis of B16F10 cells at 5 days induced by different formulations transfected MSCs using flow cytometry analysis.
The apoptosis induction of B16F10 cells after incubated with TRAIL-expressing MSCs was investigated by flow cytometry to quantify the apoptotic and necrotic cells. As shown in Figure 6B and Figure 6C, TRAIL-expressing MSCs (transfected by rHDL/PEI-LA/pTRAIL nanoparticles) caused (22.54 ± 4.65)% apoptosis of B16F10 cells, while TRAIL-expressing MSCs transfected by PEI-LA only induced (11.14 ± 2.83)% apoptosis. The decreased cell apoptosis induction indicated that PEI-LA might be harmful to MSCs thereby reducing the TRAIL transfection efficacy and influencing B16F10 cell apoptosis. In contrast, rHDL showed reliable safety and effective transfection on MSCs, resulting in significant apoptosis induction to B16F10 cells. Results above proved the importance of rHDL encapsulation on pTRAIL transfection. This TRAIL-expressing MSCs transfected by rHDL as a bioinspired nonviral vector could significantly inhibit the proliferation of B16F10 cells, thereby showing great potential in anticancer agent delivery system.
In vivo Migration of MSCs-TRAIL
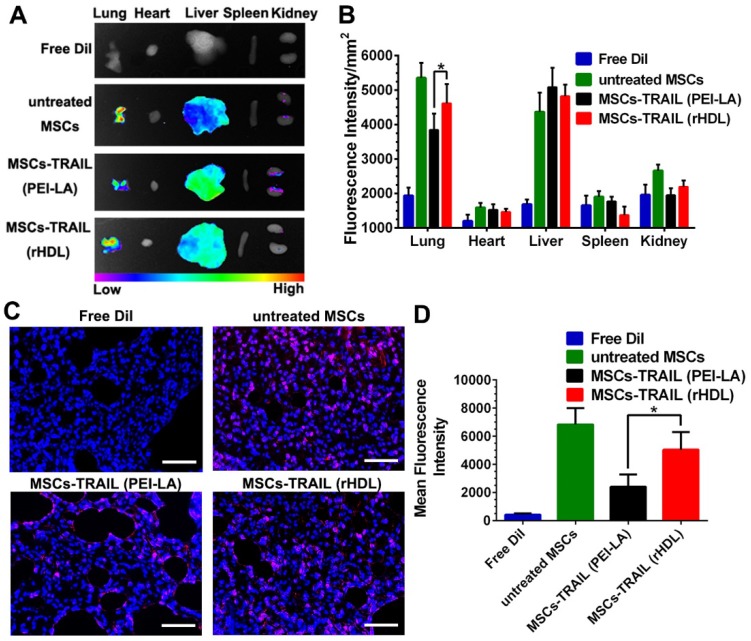
Compared with conventional nanosystems, MSCs owned the advantage of natural tumor homing ability. Therefore, we studied the tumor targeting effect of gene-engineered MSCs in vivo. In order to verify the tumor homing ability of TRAIL-expressing MSCs in lung metastasis model, MSCs were labeled with cell membrane fluorescent probe DiI to trace their own behavior in vivo before administration. Meanwhile, free DiI suspended in PBS was set as control. The in vivo biodistribution of DiI-labeled TRAIL-expressing MSCs at day 7 post-injection was shown in Figure 7A. The fluorescence of rHDL transfected TRAIL-expressing MSCs could still be detected at B16F10 pulmonary melanoma metastasis as well as natural MSCs group. Moreover, the fluorescence intensity of these two groups were significantly higher than that of PEI-LA transfected TRAIL-expressing MSCs group, as shown in Figure 7B. The images of lung tumor tissue sections in each group were shown in Figure 7C and their quantifications were shown in Figure 7D. Untreated MSCs group and MSCs-TRAIL (rHDL) group showed strong fluorescence signal of DiI during metastasis, while MSCs-TRAIL (PEI-LA) group exhibited less fluorescence signal. The results demonstrated that rHDL mediated TRAIL transfection did not reduce the tumor homing effect of MSCs, whereas PEI-LA impaired biological function of MSCs resulting in their less efficient target to pulmonary melanoma metastasis. Moreover, the TRAIL-loading MSCs possessed the tropism capacity for pulmonary melanoma metastasis in vivo.
Figure 7.
(A-B) Accumulation of DiI-MSCs in pulmonary melanoma metastasis and major organs detected by ex vivo imaging at day 7 post-injection of different formulations (n = 5). (C) Representative photomicrographs of in vivo migration of MSCs in lung metastasis slides observed under fluorescence microscope (Scale bar: 50 μm). (D) Quantification of in vivo migration of MSCs in lung metastasis slides observed under fluorescence microscope (n = 5, *P < 0.05).
In vivo Antitumor Efficacy of MSCs-TRAIL
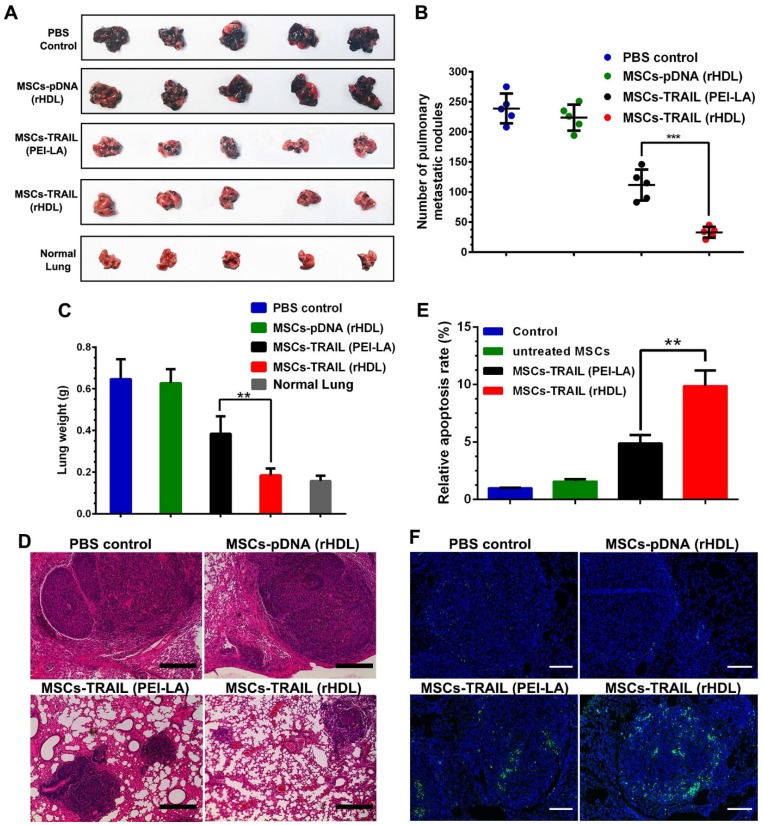
To investigate the antitumor effects of rHDL transfected TRAIL-secreting MSCs, B16F10 pulmonary melanoma metastasis model was established by intravenous injection of B16F10 cells into C57BL/6 mice. MSCs transfected by different formulations were administered on day 9 after tumor implantation. After inoculation of B16F10 tumor cells for 21 days, the treated animals were sacrificed to collected lungs. Meanwhile, the metastatic nodules on lungs were counted by visual inspection. As shown in Figure 8A-C, compared with the control group, the growth of pulmonary metastasis tumor was significantly inhibited by rHDL transfected TRAIL-expressing MSCs according to the number of metastatic nodules and lung weight. Meanwhile, images of tumor tissue section stained by H&E showed the smallest tumor nests after administration of rHDL transfected TRAIL-expressing MSCs, which further indicated their antitumor activity in vivo (shown in Figure 8D). Moreover, significant apoptosis in TUNEL slides was detected in MSCs-TRAIL (rHDL) group (shown in Figure 8E and Figure 8F). The safety test results of MSCs-TRAIL (rHDL) demonstrated that rHDL transfected TRAIL-expressing MSCs showed superior safety in vivo as shown in Figure S5. Based on the results, the tumor-targeting TRAIL-delivering MSCs platform exhibited obvious antitumor effect on B16F10 pulmonary melanoma metastasis and showed satisfactory safety for in vivo application.
Figure 8.
(A-C) Inhibition of pulmonary metastasis of B16F10 cells after treatment with different formulations and the lungs of normal mice were set as control (n = 5, **P < 0.01, ***P < 0.001). (D) H&E stained sections from each group (Scale bar: 200 μm). (E) Quantification of TUNEL in tumor sections after treatment with different formulations (n = 5, **P < 0.01). (F) TUNEL in tumor sections after treatment with different formulations (Scale bar: 100 μm).
4. Conclusions
In this paper, we constructed a bioinspired rHDL as a new member of non-viral gene delivery vectors for genetic engineering of MSCs. Gene-loaded rHDL nanoparticles exhibited special binding capacity to SR-BI expressed on MSCs surface, and used an endosome/lysosome-independent cellular uptake pathway to protect gene cargo from degradation. Moreover, rHDL-based nanoparticles also exhibited high transfection efficiency and low cytotoxicity. The rHDL-mediated TRAIL-engineered MSCs owned a promising targeting ability and effective treatment for pulmonary melanoma metastasis both in vitro and in vivo. Therefore, this gene-delivered rHDL engineering MSCs-based platform represented a step forward in the field of MSCs genetic engineering and would have important application potential in MSCs-based cancer therapy.
Supplementary Material
Supplementary figures and methods.
Scheme 1.
Schematic illustration of (I) construction of biomimetic rHDL-based gene delivery system, (II) construction of MSCs-based gene delivery system for tumor targeted therapy to pulmonary melanoma metastasis.
Acknowledgments
This study was financially supported by the National Major Scientific and Technological Special Project for "Significant New Drugs Development" (No. 2016ZX09101031), the National Natural Science Foundation of China (Nos. 31872756 and 81102398), the Jiangsu Six Talent Peaks Program (No. JY-079), and the “Double First-Class” Innovation Team Project of China Pharmaceutical University (Nos. CPU2018GY23 and CPU2018GY26).
Abbreviations
- ApoA I
apolipoprotein A I
- C
cholesterol
- CE
cholesteryl esters
- CLSM
confocal laser scanning microscopy
- DDS
drug delivery systems
- DLS
dynamic light scattering
- DMSO
dimethylsulfoxide
- DR4
death receptor 4
- DR5
death receptor 5
- EDCI
1-Ethyl-3-(3-dimethyllaminopropyl) carbodiimide hydrochloride
- EE
encapsulation efficiency
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- HDL
High-density lipoprotein
- H&E
hematoxylin and eosin
- LA
lauric acid
- MSCs
mesenchymal stem cells
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
- NHS
N-hydroxysuccinimide
- PC
phospholipids
- PDI
polydispersity index
- PEI
polyethyleneimine
- PEI-LA
Lauric acid-coupled polyethyleneimine
- pTRAIL
plasmid DNA encoding TRAIL protein
- rHDL
reconstituted high-density lipoprotein
- SD
standard deviation
- SR-BI
scavenger receptor B type I
- TBE
Tris-Borate-EDTA
- TEM
transmission electron microscope
- TRAIL
tumor necrosis factor- related apoptosis-inducing ligand
- TNF
tumor necrosis factor
References
- 1.Wicki A, Witzigmann D, Balasubramanian V. et al. Nanomedicine in cancer therapy: challenges, opportunities, and clinical applications. J Control Release. 2015;200:138–57. doi: 10.1016/j.jconrel.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 2.Blanco E, Shen H, Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat Biotechnol. 2015;33:941–51. doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeo RWY, Lai RC, Zhang B. et al. Mesenchymal stem cell: an efficient mass producer of exosomes for drug delivery. Adv Drug Deliv Rev. 2013;65:336–41. doi: 10.1016/j.addr.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Xue JW, Zhao ZK, Zhang L. et al. Neutrophil-mediated anticancer drug delivery for suppression of postoperative malignant glioma recurrence. Nat Nanotechnol. 2017;12:692–700. doi: 10.1038/nnano.2017.54. [DOI] [PubMed] [Google Scholar]
- 5.Gao Z, Zhang L, Hu J. et al. Mesenchymal stem cells: a potential targeted-delivery vehicle for anti-cancer drug, loaded nanoparticles. Nanomedicine. 2013;9:174–84. doi: 10.1016/j.nano.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Kucerova L, Durinikova E, Toro L. et al. Targeted antitumor therapy mediated by prodrug-activating mesenchymal stromal cells. Cancer Lett. 2017;408:1–9. doi: 10.1016/j.canlet.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 7.Rhee KJ, Lee JI, Eom YW. Mesenchymal stem cell-mediated effects of tumor support or suppression. Int J Mol Sci. 2015;16:30015–33. doi: 10.3390/ijms161226215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lourenco S, Teixeira VH, Kalber T. et al. Macrophage migration inhibitory factor-CXCR4 is the dominant chemotactic axis in human mesenchymal stem cell recruitment to tumors. J Immunol. 2015;194:3463–74. doi: 10.4049/jimmunol.1402097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin PT, Shah S, Pasquale NJ. et al. Stem cell-based gene therapy activated using magnetic hyperthermia to enhance the treatment of cancer. Biomaterials. 2016;81:46–57. doi: 10.1016/j.biomaterials.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mangraviti A, Tzeng SY, Gullotti D. et al. Non-virally engineered human adipose mesenchymal stem cells produce BMP4, target brain tumors, and extend survival. Biomaterials. 2016;100:53–66. doi: 10.1016/j.biomaterials.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JS, Yi SW, Kim HJ. et al. Sunflower-type nanogels carrying a quantum dot nanoprobe for both superior gene delivery efficacy and tracing of human mesenchymal stem cells. Biomaterials. 2016;77:14–25. doi: 10.1016/j.biomaterials.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Dewey RA, Morrissey G, Cowsill CM, Chronic brain inflammation and persistent herpes simplex virus 1 thymidine kinase expression in survivors of syngeneic glioma treated by adenovirus-mediated gene therapy: Implications for clinical trials. Nat Med; 1999. p. 5. 1256-63. [DOI] [PubMed] [Google Scholar]
- 13.Stohlman SA, Hinton DR. Viral induced demyelination. Brain Pathol. 2001;11:92–106. doi: 10.1111/j.1750-3639.2001.tb00384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Rorke S, Keeney M, Pandit A. Non-viral polyplexes: Scaffold mediated delivery for gene therapy. Prog Polym Sci. 2010;35:441–458. [Google Scholar]
- 15.Shahzad MMK, Mangala LS, Han HD. et al. Targeted delivery of small interfering RNA using reconstituted high-density lipoprotein nanoparticles. Neoplasia. 2011;13:309–19. doi: 10.1593/neo.101372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenson RS, Brewer HB, Davidson WS. et al. Cholesterol efflux and atheroprotection advancing the concept of reverse cholesterol transport. Circulation. 2012;125:1905–19. doi: 10.1161/CIRCULATIONAHA.111.066589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W, Chen KR, Su YJ. et al. Lysosome-independent intracellular drug/gene codelivery by lipoprotein-derived nanovector for synergistic apoptosis-inducing cancer-targeted therapy. Biomacromolecules. 2018;19:438–48. doi: 10.1021/acs.biomac.7b01549. [DOI] [PubMed] [Google Scholar]
- 18.Lacko AG, Sabnis NA, Nagarajan B. et al. HDL as a drug and nucleic acid delivery vehicle. Front Pharmacol. 2015;6:247–53. doi: 10.3389/fphar.2015.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang ZH, Cao WG, Jin HL. et al. Biomimetic nanocarrier for direct cytosolic drug delivery. Angew Chem Int Ed Engl. 2009;48:9171–5. doi: 10.1002/anie.200903112. [DOI] [PubMed] [Google Scholar]
- 20.Rodrigueza WV, Thuahnai ST, Temel RE. et al. Mechanism of scavenger receptor class B type I-mediated selective uptake of cholesteryl esters from high density lipoprotein to adrenal cells. J Biol Chem. 1999;274:20344–50. doi: 10.1074/jbc.274.29.20344. [DOI] [PubMed] [Google Scholar]
- 21.Xu JF, Qian JY, Xie XX. et al. High density lipoprotein cholesterol promotes the proliferation of bone-derived mesenchymal stem cells via binding scavenger receptor-B type I and activation of PI3K/Akt, MAPK/ERK1/2 pathways. Mol Cell Biochem. 2012;371:55–64. doi: 10.1007/s11010-012-1422-8. [DOI] [PubMed] [Google Scholar]
- 22.Simonsen JB. Evaluation of reconstituted high-density lipoprotein (rHDL) as a drug delivery platform - a detailed survey of rHDL particles ranging from biophysical properties to clinical implications. Nanomedicine. 2016;12:2161–79. doi: 10.1016/j.nano.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Ng KK, Lovell JF, Zheng G. Lipoprotein-inspired nanoparticles for cancer theranostics. Acc Chem Res. 2011;44:1105–13. doi: 10.1021/ar200017e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemke J, von Karstedt S, Zinngrebe J. et al. Getting TRAIL back on track for cancer therapy. Cell Death Differ. 2014;21:1350–64. doi: 10.1038/cdd.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Miguel D, Lemke J, Anel A. et al. Onto better TRAILs for cancer treatment. Cell Death Differ. 2016;23:733–47. doi: 10.1038/cdd.2015.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lerch PG, Fortsch V, Hodler G. et al. Production and characterization of a reconstituted high density lipoprotein for therapeutic applications. Vox Sang. 1996;71:155–64. doi: 10.1046/j.1423-0410.1996.7130155.x. [DOI] [PubMed] [Google Scholar]
- 27.Wang RN, Zhao ZQ Han Y. et al. Natural particulates inspired specific-targeted codelivery of sirna and paclitaxel for collaborative antitumor therapy. Mol Pharm. 2017;14:2999–3012. doi: 10.1021/acs.molpharmaceut.7b00192. [DOI] [PubMed] [Google Scholar]
- 28.Ding Y, Wang YZ, Zhou JP. et al. Direct cytosolic siRNA delivery by reconstituted high density lipoprotein for target-specific therapy of tumor angiogenesis. Biomaterials. 2014;35:7214–27. doi: 10.1016/j.biomaterials.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 29.Liu ZH, Zhang ZY, Zhou CR. et al. Hydrophobic modifications of cationic polymers for gene delivery. Prog Polym Sci. 2010;35:1144–62. [Google Scholar]
- 30.Ding Y, Wang W, Feng MQ. et al. A biomimetic nanovector-mediated targeted cholesterol-conjugated siRNA delivery for tumor gene therapy. Biomaterials. 2012;33:8893–905. doi: 10.1016/j.biomaterials.2012.08.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and methods.