Abstract
Aluminum (Al) is the most abundant metal element in the earth’s crust. On acid soils, at pH 5.5 or lower, part of insoluble Al-containing minerals become solubilized into soil solution, with resultant highly toxic effects on plant growth and development. Nevertheless, some plants have developed Al-tolerance mechanisms that enable them to counteract this Al toxicity. One such well-documented mechanism is the Al-induced secretion of organic acid anions, including citrate, malate, and oxalate, from plant roots. Once secreted, these anions chelate external Al ions, thus protecting the secreting plant from Al toxicity. Genes encoding the citrate and malate transporters responsible for secretion have been identified and characterized, and accumulating evidence indicates that regulation of the expression of these transporter genes is critical for plant Al tolerance. In this review, we outline the recent history of research into plant Al-tolerance mechanisms, with special emphasis on the physiology of Al-induced secretion of organic acid anions from plant roots. In particular, we summarize the identification of genes encoding organic acid transporters and review current understanding of genes regulating organic acid secretion. We also discuss the possible signaling pathways regulating the expression of organic acid transporter genes.
Keywords: Acid soil, Aluminum (Al) toxicity, Expression regulation, Organic acid anion, Transporter
1. Introduction
Because it is the most abundant metal element in the earth’s crust, aluminum (Al) is ubiquitous in soils. However, it is not taken as an essential element by living organisms, possibly because of its stronger metal activity than comparable metal elements, such as manganese, zinc, iron, copper, and molybdenum. Furthermore, Al, when present in ionic form, and especially in the Al3+ form, is highly toxic to living organisms (Rengel, 2004).
Worldwide, close to 50% of potential arable land is acidic, with Al being solubilized into soil solution in cationic form (von Uexküll and Mutert, 1995). The resultant Al toxicity is now widely acknowledged to be one of the most serious factors limiting global plant growth and productivity, sparking recent efforts to understand plant Al toxicity and the Al-tolerance mechanisms of plants grown in acid soils. Actually, the realization that Al toxicity is related to the adverse effects of acid soils can be traced back more than 100 years (Hartwell and Pember, 1918). Reports concerning the “crestamento” (burning or toasting) symptoms displayed by crops grown on certain soils emerged in Brazil, and by 1942, these symptoms were finally attributed to Al toxicity in low-pH soil conditions (Matzenbacher, 1988). These early reports were the forerunners of our modern understanding of Al toxicity in acid soils and of how plants use tolerance mechanisms to overcome that toxicity.
In this review, we provide a historical perspective on research into plant Al toxicity and Al-tolerance mechanisms. We particularly focus on the importance of Al-induced root organic acid (OA) anion secretion for Al tolerance, and provide insights into the identification of genes encoding OA anion transporters. We also outline the mechanisms via which the expression of these genes is regulated, describing both the work of our own research group and that of others.
2. Plant Al toxicity
The primary visible symptom of Al toxicity is the rapid inhibition of root growth, and this index has therefore been widely used to measure Al resistance. Whilst a number of possible mechanisms have been proposed to explain Al-induced root elongation inhibition, the exact mechanism remains controversial. Because Al is such an active metal, it may simultaneously target sites at multiple different cellular and molecular levels, with these combined effects collectively contributing to the arrest of root elongation. There are a number of review papers summarizing understanding of plant Al toxicity mechanisms (Kochian, 1995; Matsumoto, 2000; Barceló and Poschenrieder, 2002; Zheng and Yang, 2005; Ma, 2007; Singh et al., 2017).
Recently, a counterintuitive explanation for the Al-induced inhibition of root elongation has been proposed, based on screens for mutant suppressors of the Al hypersensitivity of the Arabidopsis als3-1 mutant (Rounds and Larsen, 2008; Nezames et al., 2012; Sjogren et al., 2015; Sjogren and Larsen, 2017). According to this explanation, Al-induced root elongation inhibition is due to active triggering of cell cycle arrest, terminal differentiation of the root tip, and loss of the root quiescent center, all processes associated with loss of DNA integrity. Four genes, ATAXIA TELANGIECTASIA AND RAD3 RELATED (ATR), ALUMINUM TOLERANCE2 (ALT2), SUPPRESSOR OF GAMMA RESPONSE1 (SOG1), and SENSITIVE TO UV 2 (SUV2), were isolated based on the ability of loss-of-function mutations in these genes to reverse the als3-1 phenotype. ATR is considered to be a master eukaryotic cell-cycle checkpoint component, which detects and responds to persistent single-stranded DNA. The other three genes appear to be involved in an ATR-dependent DNA damage response. However, because a chronic exposure regime (lasting several days) was used in these studies, whilst Al rapidly (within hours) inhibits root elongation, it is possible that these effects are the consequence rather than the cause of Al toxicity. The likelihood of this latter possibility has recently been strengthened by a study demonstrating that ATR is involved in root growth inhibition due to internal Al toxicity, but not in inhibition due to external Al toxicity (ZhangY et al., 2018). Interestingly, another recent study provided evidence that the SOG1 loss-of-function mutant displays increased sensitivity to Al when higher Al concentrations are applied in the growth medium (Chen et al., 2019). Considering the importance of the apoplast in the expression of Al toxicity (Horst et al., 2010), it is likely that other mechanisms directly related to cell elongation processes will in the future be implicated in the rapid inhibition of root elongation in response to Al stress.
3. Plant Al-tolerance mechanisms
As early as the 1920s, genetic differences in Al toxicity or tolerance were identified in different Brazilian cereal varieties. However, the reasons underlying these differences remained unclear until the 1990s. Taylor (1991) proposed two potential strategies by which plants might cope with Al toxicity. The first strategy involves the exclusion of Al from the root apex (external exclusion), whilst the second strategy involves mechanisms of tolerance to Al once it has entered plant cells (internal tolerance). Possible mechanisms involved in external exclusion include the secretion of Al chelators, increases in rhizosphere pH, secretion of mucilage, immobilization of Al by the cell wall and Al efflux. In contrast, possible mechanisms involved in internal tolerance include complexation, compartmentalization, and sequestration of internal Al. Among these several strategies and mechanisms, the one best documented is the mechanism of Al exclusion by the secretion of OA anions from roots (Fig. 1). From the 1990s onwards, various studies demonstrated convincingly that some plants can indeed resist Al toxicity by releasing OA anions from their roots. To date, there is a plethora of reports demonstrating that root secretion of OA anions is responsible for genotypic differences in plant Al toxicity responses (Ryan et al., 2001; Kochian et al., 2004, 2015).
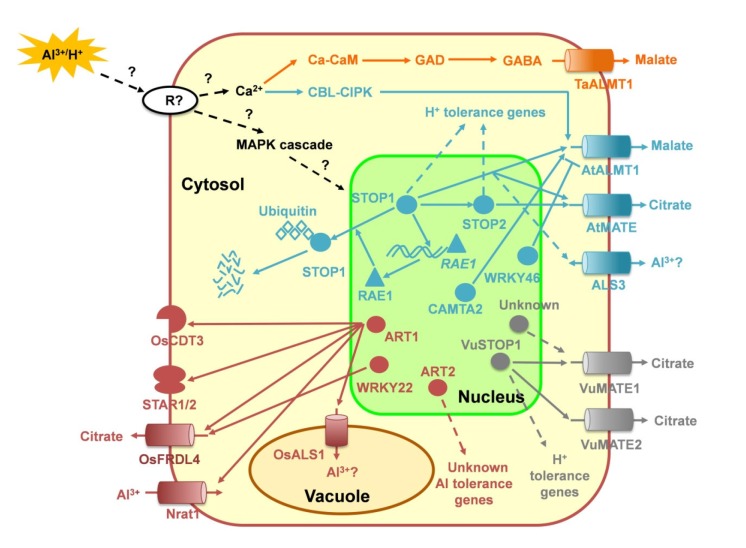
Fig. 1.
Model illustrating putative aluminum (Al3+)/proton (H+)-mediated signal transduction and transcriptional regulation pathways in several plant species
The model is based primarily on experimental evidence from the literatures for a number of plant species (blue, gray, brown, and orange colors represent pathways in Arabidopsis, rice bean, rice, and wheat, respectively). Al3+/H+ activates unknown receptors (R) firstly. The increase in cytosolic Ca2+ leads to the activation of calmodulin (CaM), which binds to glutamate decarboxylase (GAD), converting it from the inactive to the active form. Glutamate is then converted to γ-aminobutyric acid (GABA), which is already known to be involved in regulating Triticum aestivum TaALMT1 activity. On the other hand, calcineurin B-like protein (CBL)-CBL-interacting protein kinase (CIPK) network is involved in the regulation of expression of AtALMT1. In addition, mitogen-activated protein kinase (MAPK) cascade pathways are implicated in regulating the expression of organic acid (OA) anion transporter genes or transcription factors mediated by OA secretion through unidentified pathways. In Arabidopsis thaliana, SENSITIVE TO PROTON RHIZOTOXICITY1 (STOP1) regulate Al-tolerance genes including AtALMT1, AtMATE, and ALS3, and several H+ tolerance genes, suggesting that STOP1 is a core component controlling Al and H+ tolerance in Arabidopsis. STOP2 regulated by STOP1 confers H+ tolerance by regulating the expression of several H+ tolerance genes, but not for Al tolerance because of only a very limited ability to regulate AtALMT1 and ALS3 expression (Kobayashi et al., 2014). Additional transcription factors such as CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR2 (CAMTA2) also regulates AtALMT1 expression, while AtWRKY46 negatively regulates AtALMT1, thus contributing to Al tolerance. In Oryza sativa, ART1 regulates the expression of at least 31 downstream genes involved in Al resistance, most of which have been shown to be involved in internal Al detoxification (including Nrat1, encoding a natural resistance-associated macrophage protein specific for Al3+; OsALS1, encoding a tonoplast-localized half-size ABC transporter; and CDT3, encoding a plasmamembrane-localized cysteine-rich peptide) or external detoxification (including STAR1/STAR2, encoding an ABC transporter; and OsFRDL4, encoding an Al-induced MATE transporter). Additional transcription factors such as WRKY22 also regulate OsFRDL4 expression, while ART2 is involved in Al tolerance by regulating unknown Al-tolerance genes, which is independent of the ART1-regulated pathway. In Vigna umbellata, VuSTOP1 regulates VuMATE1 and VuMATE2 expression in different ways. VuSTOP1 predominantly regulates VuMATE2 expression by interacting with an ART1-like GGGAGG cis-acting element, but regulates VuMATE1 expression by binding to promoter sequences that are not critical for the Al-inducible expression of VuMATE1, thus suggesting that there are other unknown transcription factors activating the expression of VuMATE1
4. Physiology of plant Al-induced OA anion secretion
The first report of plant Al-induced OA anion secretion was from Kitagawa (1986), who investigated the possible role of Al-induced malate secretion in causing genotypic differences in wheat Al responses. Later, Miyasaka et al. (1991) reported that Al-resistant snapbean cultivar (Phaseolus vulgaris) secreted considerably more citrate from their roots than did Al-sensitive cultivar. However, the most convincing evidence that OA anion secretion plays a role in Al tolerance came from studies of a pair of near isogenic wheat (Triticum aestivum) lines, ET3 and ES3. These lines differ in Al tolerance, and it was considered likely that allelic variation at a single gene locus controlled these differences (Delhaize et al., 1993a, 1993b). These studies enabled two critical conclusions to be drawn. First, Al-induced malate secretion occurs precisely in the several millimeters length apical region of the root, which is in many cases the region that incurs the most severe Al toxicity. Second, the onset of malate secretion is triggered rapidly by Al, indicating that the “biochemical machinery” for secretion was already present in the absence of Al, and that Al simply activates it. These findings greatly stimulated further studies on Al-induced OA anion secretion from the roots of several different plant species. For example, Al was shown to induce oxalate secretion from taro (Colocasia esculenta) (Ma and Miyasaka, 1998), buckwheat (Fagopyrum esculentum Moench) (Zheng et al., 1998), and spinach (Spinacia oleracea) (Yang et al., 2005), and citrate secretion from soybean (Glycine max) (Yang et al., 2001), rice bean (Vigna umbellata) (Yang et al., 2006a), and Cassia tora (Ma et al., 1997; Yang et al., 2006b). In some cases, plants secrete two different OA anions in response to Al. For example, maize (Zea mays) and rye (Secale cereale) roots secret both citrate and malate (Pellet et al., 1995; Li et al., 2000), while amaranth (Amaranthus hypochondriacus) roots simultaneously secret both oxalate and citrate in response to Al stress (Fan et al., 2016). Up to now, citrate, oxalate, and malate have been identified as being the major OA anions secreted by plant roots exposed to Al stress. Although all three of these anions are able to chelate Al, thus making the Al non-toxic to plant cells, their chelating abilities differ, following the order citrate>oxalate>malate (Li et al., 2009). Exactly why different plant species secrete different forms of OA anions is unclear, although Ryan and Delhaize (2010) have proposed convergent evolution of plant Al resistance as a possible explanation.
Based on the timing of Al-induced OA anion secretion from different plant species, two secretion patterns have been proposed (Ma, 2000). In pattern I, Al stimulates the secretion of OA anions from plant roots almost immediately upon exposure. In contrast, in pattern II, there is a lag phase between the onset of Al stress and the onset of secretion. It was also proposed that the underlying basis of these two different patterns could be attributed to the “biochemical machinery” being either already present or needing to be induced de novo. Taking advantage of the broad-spectrum protein translation inhibitor cycloheximide, we provided experimental evidence that de novo protein biosynthesis is required for Al-induced citrate secretion from C. tora (a pattern II plant), whereas de novo protein biosynthesis is not involved in Al-induced oxalate secretion from buckwheat (a pattern I plant) (Yang et al., 2006b). More recently, we found that Al-induced citrate secretion from rice bean root apices consists of two phases. In the early phase, only a little citrate is secreted in response to Al, whilst, after longer exposure, a large amount of citrate is secreted in a second phase. We further demonstrated that two different citrate transporters belonging to the same transporter protein family are specifically implicated in these two distinct phases of citrate secretion (Liu et al., 2018).
5. Transporters responsible for the secretion of OA anions
Because cytoplasmic pH is approximately neutral, cytoplasmic OAs exist mainly in anionic form. Prior to the cloning of genes encoding OA anion permeable transporters, it was assumed that anion channels or transporters are responsible for OA efflux. Electrophysiological studies with maize confirmed that Al activates inward Cl− current (presumably anion efflux) (Piñeros and Kochian, 2001), and Al-induced anion channel is permeable to both malate and citrate (Kollmeier et al., 2001). However, by employing a suppression subtraction hybridization approach, a new gene, ALMT1 (ALUMINUM-ACTIVATED MALATE TRANSPORTER1) was identified in wheat isogenic lines ET8 and ES8 (Sasaki et al., 2004). The encoded ALMT1 belongs to a previously unidentified protein family, and heterologous expression in Xenopus oocytes showed that it exhibits malate but not citrate transport activity. Furthermore, ALMT1 expression is constitutive in the root apices of the Al-resistant ET8 line, consistent with previous proposals that the “biochemical machinery” is pre-existing. When ALMT1 is introduced into barley (Hordeum vulgare), the resultant transgenic barley exhibits improved Al resistance and increased malate secretion, suggesting that ALMT1 is a bona fide plant Al-resistant gene (Delhaize et al., 2004). Soon after the initial discovery of AMLT1, Arabidopsis AtALMT1, the first of what are now known to be 14 Arabidopsis ALMT family members, was functionally characterized (Hoekenga et al., 2006). Subsequently, orthologous genes have been identified in rape (Brassica napus) (Ligaba et al., 2006), rye (Collins et al., 2008), soybean (Liang et al., 2013), Medicago sativa (Chen Q et al., 2013), and cabbage (Brassica oleracea) (Zhang L et al., 2018). It is now known that the physiological roles of ALMTs are relevant not only to Al resistance but also to stomata movement, anion homeostasis, fruit quality, seed development, and response to pathogen attack (Sharma et al., 2016).
In fact, citrate is considerably more effective as an Al chelator than is malate (Li et al., 2009). Accordingly, much work went into the molecular identification of the gene encoding the plant Al-activated citrate transporter. In 2007, two independent research groups reported, at more or less the same time, the identification of HvAACT1 (aluminum-activated citrate transporter 1) in barley (Furukawa et al., 2007), and of SbMATE (multidrug and toxic compound extrusion) in sorghum (Sorghum bicolor) (Magalhaes et al., 2007), both via map-based cloning. The proteins encoded by both of these genes are responsible for Al-induced citrate secretion. Both HvAACT1 and SbMATE are members of the MATE protein family, one of the largest of plant transporter protein families. MATE transporters are capable of transporting a broad range of molecular substrates, including OA anions, hormones, and secondary metabolites (Takanashi et al., 2014). In fact, prior to the characterization of HvAACT1 and SbMATE, a functional Arabidopsis homolog, FERRIC REDUCTASE DEFECTIVE3 (FRD3), had been shown to be involved in the translocation of iron from roots to shoots by mediating the secretion of citrate into the xylem (Durrett et al., 2007). To date, citrate permeable MATE transporters have been identified in Arabidopsis (Liu et al., 2009), wheat (Ryan et al., 2009), maize (Maron et al., 2010), rye (Yokosho et al., 2010), rice (Oryza sativa) (Yokosho et al., 2011), and our group has identified them in rice bean (Yang XY et al., 2011; Liu et al., 2018).
The mechanism of Al stress-induced plant root oxalate secretion remains unknown, although such secretion has been observed in taro (Ma and Miyasaka, 1998), buckwheat (Zheng et al., 1998), tea (Camellia sinensis) (Morita et al., 2011), spinach (Yang et al., 2005), tomato (Lycopersicon esculentum) (Yang JL et al., 2011), and Polygonum species (You et al., 2005). It seems that Al-induced oxalate secretion is common to plant species abundant in oxalate (Yang et al., 2008). Recently, we found that a gene encoding formate dehydrogenase, which is involved in formate catabolism, is involved not only in the Al resistance but also in the low-pH tolerance of rice bean (Lou et al., 2016a). Later again, we further found that the oxalate acetylation degradation pathway is implicated in formate production in Al stress conditions and showed that a gene encoding acyl activating enzyme3 is involved in rice bean oxalate acetylation (Lou et al., 2016b). Unlike citrate and malate, which are primary metabolites of the tricarboxylic acid (TCA) cycle, oxalate is regarded as an end-product of secondary metabolism. We demonstrated that oxalate accumulation is detrimental to cellular functions and needs to be tightly controlled.
6. Regulation of Al-induced OA anion secretion
The identification of genes encoding Al-induced OA anion transporters enabled investigation of the transcript-level regulation of these genes in response to Al stress. These studies identified some general expression pattern characteristics of these transporter genes. First, their expression is higher in Al-resistant genotypes than in Al-sensitive genotypes. Second, they are predominantly expressed in root apices. Third, in most cases, their expression can be further induced by Al stress. These observations suggest that regulation of the expression of genes encoding transporter proteins enables adaptation to Al toxicity conditions, although post-transcriptional regulation is also likely involved.
6.1. Modulation of transcription factors
The first discovered transcription factor regulating Al tolerance was identified by mutant screening and map-based gene cloning. The sensitive to proton rhizotoxicity1 (stop1) Arabidopsis mutant displays hypersensitivity to low pH (Iuchi et al., 2007), and is also Al-sensitive, because AtALMT1 expression is abolished. STOP1 encodes a C2H2 zinc-finger transcription factor (STOP1), whose expression is induced only by extremely high Al concentrations or very low pH (Iuchi et al., 2007). Subsequent studies revealed that two additional Al-tolerance genes, AtMATE and ALS3 are regulated by STOP1 (Liu et al., 2009; Sawaki et al., 2009). AtALMT1 and AtMATE mediate the secretion of malate and citrate, respectively, while ALS3 encodes a half-type ATP-binding cassette transporter, which participates in intracellular Al redistribution (Larsen et al., 2005). Additionally, comparative microarray analysis revealed that STOP1 regulates the expression of other genes regulating proton and Al tolerance (Sawaki et al., 2009). Obviously, STOP1 is a core component controlling Al and proton tolerance in Arabidopsis (Fig. 1). Among the genes regulated by STOP1, STOP2 is a unique paralog of STOP1, having a shorter C-terminus than STOP1. When introduced into stop1 mutant, STOP2 is able to rescue proton tolerance by regulating the expression of several proton-tolerance genes. However, STOP2 is not able to rescue Al tolerance of stop1 mutant because of only a very limited ability of STOP2 to regulate AtALMT1 and ALS3 expression. Therefore, STOP1 might control Al tolerance and proton tolerance differentially via control of its downstream transcription factors when receiving signals from different stressors. Alternatively, the transcriptional activation of major Al-tolerance genes (e.g., AtALMT1 and ALS3) by STOP1 requires additional mechanisms (e.g., co-activators or post-translational mechanisms) which are themselves sensitive to STOP1 protein structure. Recently, an F-box protein-encoding gene regulating AtALMT1 expression1 (RAE1) was shown to regulate STOP1 protein abundance. The RAE1 protein interacts with STOP1 and hence promotes STOP1 ubiquitination and subsequent STOP1 degradation in the 26S proteasome (Zhang et al., 2019). Because STOP1 also promotes RAE1 transcription via direct binding to the RAE1 promoter, STOP1 and RAE1 together form a negative feedback loop that regulates STOP1 accumulation (Zhang et al., 2019). Whilst these observations further illustrate that post-translational modification is required for maintenance of STOP1 homeostasis in Al tolerance, the mechanism via which Al prevents RAE1-mediated degradation of STOP1 remains unknown.
Amongst small-grain cereals, rice is the most Al-tolerance crop, displaying an Al tolerance that is 6–10 times higher than that of other major cereals, such as maize, wheat, barley, and sorghum (Famoso et al., 2011). ART1, a gene encoding ART1, a rice STOP1 ortholog, was cloned following molecular analysis of mutants obtained in screens for Al sensitivity (Fig. 1). However, ART1 expression, unlike that of STOP1, is not Al-induced. Microarray analysis revealed that ART1 regulates the expression of at least 31 downstream genes involved in Al resistance, most of which have been shown to be involved in internal Al detoxification (including Nrat1, encoding a natural resistance-associated macrophage protein specific for Al3+; OsALS1, encoding a tonoplast-localized half-size ABC transporter; and CDT3, encoding a plasmamembrane-localized cysteine-rich peptide) (Xia et al., 2010, 2013; Huang et al., 2012) or external detoxification (including STAR1/STAR2, encoding an ABC transporter; and OsFRDL4, encoding an Al-induced MATE transporter) (Huang et al., 2009; Yokosho et al., 2011). Although Al-activated citrate secretion plays only a minor role in rice Al tolerance, OsFRDL4-mediated secretion of citrate from root cells contributes to overall Al tolerance (Yokosho et al., 2011). Recently, we found that OsFRDL4 is regulated by binding of transcription factor OsWRKY22 to W-box elements in the OsFRDL4 promoter, indicating that OsWRKY22 is an additional essential transcription factor that functions together with ART1 in the control of Al-induced OsFRDL4 expression and citrate secretion (Li et al., 2018). Additionally, there are seven STOP1 homologs in rice (Huang et al., 2009), although the functions of these genes, with the exception of ART1, are little understood. Nevertheless, one of these ART1 homologs, ART2, was shown to regulate rice Al tolerance independently of the ART1-regulated pathway. ART2 expression is induced by Al but is not involved in activation of genes regulated by ART1. Moreover, the contribution of ART2 to Al tolerance is smaller than that of ART1, suggesting that ART1 and ART2 regulate different Al-tolerance pathways, and that ATR2 plays a supplementary role in rice Al tolerance (Che et al., 2018). Overall, these various results suggest that rice Al tolerance is a complex quantitative trait controlled by multiple genes.
The identification and characterization of ART1/STOP1 indicate that STOP1-like proteins are likely to regulate the expression of multiple Al-tolerance genes in a relatively wide range of plant species. At the time of writing, functional STOP1-like gene orthologs have been identified in a range of plant species, including wheat (Garcia-Oliveira et al., 2013), tobacco (Nicotiana tabacum), Populus nigra, Lotus japonicas, Physcomitrella patens (Ohyama et al., 2013), Eucalyptus (Sawaki et al., 2014), rice bean (Fan et al., 2015), pigeonpea (Cajanus cajan) (Daspute et al., 2018), tea (Zhao et al., 2018), soybean (Wu et al., 2018), sweet sorghum (Huang et al., 2018), and cotton (Gossypium hirsutum) (Kundu et al., 2019). Whilst these STOP1-like proteins are functionally conserved with respect to proton tolerance, their ability to rescue Al tolerance in the Arabidopsis stop1 mutant differs. Despite having conserved C2H2 zinc finger domains, the N-and C-termini of these STOP-1 like proteins are highly variable. Further studies are needed to determine the structure–function relationships of these STOP1-like proteins with respect to their differing roles in regulating the expression of Al-tolerance genes, and especially that of OA transporter genes. Furthermore, the pattern of expression of the STOP1-like genes in response to Al stress differs in different plant species. We showed that in rice bean, the mRNA levels of three STOP1/ART1-like genes, including the previously identified VuSTOP1, are increased by exposure to Al (Fan et al., 2019). In addition, a constitutively expressed STOP1/ART1-like gene was found (Fan et al., 2019). Study of the functions of the rice bean STOP1/ART1-like proteins is expected to further reveal the role of STOP1/ART1-like proteins in the mechanism of Al tolerance.
It is also obvious that the mechanism underlying STOP1-like regulation of the expression of OA transporter genes is complicated, because several other transcription factors (both activators and suppressors) are involved, and because variation in cis-acting elements exists among different species (Fig. 1). Tokizawa et al. (2015) used computation of overrepresented octamers to characterize eight transcription factor-interacting cis-acting elements in the AtALMT1 promoter. In addition to STOP1 and CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR2 (CAMTA2) binding regions, some further unknown regions suppressed the expression of AtALMT1. For example, the AtWRKY46, AtALMT1 repressor, was shown in our laboratory to bind the AtALMT1 promoter independently of the above identified eight transcription factor-binding regions (Ding et al., 2013). Similarly, Tsutsui et al. (2011) identified a consensus GGN(T/g/a/C)V(C/A/g)S(C/G) sequence as the core binding element of rice ART1. Of the 31 promoters regulated by ART1, 29 contain this element. However, because of the small base number and relatively large sequence variation of this consensus, the chance of accidental occurrence in any gene promoter is very high, thus greatly limiting the utilization of this cis element. We also found that two MATE-type transporter genes are responsible for citrate secretion from rice bean root apices in response to Al stress. However, VuSTOP1 regulates VuMATE1 and VuMATE2 expression in different ways (Liu et al., 2018). VuSTOP1 predominantly regulates VuMATE2 expression by interacting with an ART1-like GGGAGG cis-acting element, but regulates VuMATE1 expression by binding to promoter sequences that are not critical for the Al-inducible expression of VuMATE1, thus suggesting that there are other unknown transcription factors activating the expression of VuMATE1 (Fan et al., 2015; Liu et al., 2018). Sequence analysis of STOP1-binding region in Arabidopsis AtALMT1 promoter also showed that the binding range of STOP1 far exceeds the length of five bases (Tokizawa et al., 2015). These various results have revealed the complexity of STOP1-regulated expression of OA transporter genes, and their associated pleiotropic functions.
6.2. Regulation of transposable elements
Accumulating evidence has suggested the involvement of transposable elements (TEs) or transposons in Al tolerance in plants. TEs are able to change their position in the genome, i.e. “transpose,” which confers them ability to regulate gene expression. To date, TE insertions have been reported to be involved in regulating Al-resistant genes in wheat (Sasaki et al., 2006; Ryan et al., 2010; Tovkach et al., 2013), sorghum (Magalhaes et al., 2007; Melo et al., 2013), barley (Fujii et al., 2012; Kashino-Fujii et al., 2018), Holcus lanatus (Chen ZC et al., 2013) and rice (Yokosho et al., 2016). Very recently, Pereira and Ryan (2019) have summarized the role of TEs in the evolution of Al tolerance in plants. Thus, in this review, we will only introduce briefly to this topic.
In barley, root tips of some Al-resistant cultivars constitutively expressed HvAACT1 that facilitates citrate secretion from barley root apices in response to Al stress. Fujii et al. (2012) identified a 1023-bp CACTA-like transposon inserted into the 5' untranslated region (5'-UTR) approximately 4.8 kb upstream of the HvAACT1 start codon in Al-resistant barley cultivars, and found that this insertion regulates HvAACT1 expression at root tip. Moreover, an independent multiretrotransposon-like (MRL) sequence insertion (at least 15.3 kb in length) affecting barley Al resistance was detected 6.6 kb upstream of HvAACT1 (Kashino-Fujii et al., 2018). However, the MRL insertion was not directly related to the expression levels of HvAACT1. Kashino-Fujii et al. (2018) further found that the regulation of DNA methylation of this MRL insertion influenced the expression of HvAACT1. It is interesting for the future to clarify the DNA methylation mechanisms in barley in response to Al stress.
The malate secretion mediated by TaALMT1 and the citrate secretion mediated by TaMATE1 play important roles in detoxifying Al in wheat, with Al-activated malate secretion being the major contributor to wheat Al resistance. Whilst the TaALMT1 amino acid sequence encoded by TaALMT1 is conserved both in Al-tolerant and sensitive cultivars, TaALMT1 is substantially polymorphic in the promoter region (Sasaki et al., 2006; Ryan et al., 2010). A comparative analysis of the promoter sequences of 69 different accessions identified both double and triple tandem repeats of the promoters of Al-resistant TaALMT1 alleles, resulting in enhanced TaALMT1 expression (Sasaki et al., 2006). In contrast, the TaMATE1B gene (located on chromosome 4B) confers wheat Al resistance by mediating citrate secretion from root apices. TaMATE1B possesses an 11.1-kb TE (contains a 3.9-kb Sukkula-like TE) insertion 25 bp upstream of the TaMATE1B start codon, which increased TaMATE1B expression (Tovkach et al., 2013). TaALMT1 and TaMATE1B are both constitutively and highly expressed in Al-resistant cultivars, and some cultivars from Brazil (e.g., cv. Carazinho, cv. IAC5-Maringá, and cv. Toropi) combining the superior TaALMT1 and TaMATE1B alleles, exhibit strong Al resistance (Pereira et al., 2015; Aguilera et al., 2016). In contrast, expression of SbMATE in sorghum is induced by Al. Polymorphism analysis of SbMATE allele promoters revealed the existence of 1 to 5 Tourist-like miniature inverted repeats (MITEs) insertions, with the relative abundance of Al-induced SbMATE mRNAs being related to the number of MITEs (Magalhaes et al., 2007). Additional trans-acting elements may also affect SbMATE expression because the contribution of AltSB locus (the major Al-tolerance locus in sorghum) to Al resistance relies on the genetic backgrounds where AltSB resides (Melo et al., 2013). Using expression-quantitative trait locus (QTL) mapping and expression genome-wide association mapping, Melo et al. (2019) further demonstrated that SbMATE expression is related to transcriptional interplay between cis and trans elements, and identified a WRKY and a zinc finger-DHHC transcription factors as being involved in the trans-activation of SbMATE.
In rice, regulation of OsFRDL4 by ART1 and OsWRKY22 contributes to citrate secretion and resultant Al detoxification (Yokosho et al., 2011; Li et al., 2018). Some of the phenotypic variation for Al resistance in japonica rice lines can be explained by increased OsFRDL4 expression linked with a 1213-bp solo long terminal repeat (LTR) (which contains nine ART1-binding cis-acting elements) inserted 615 bp upstream of the OsFRDL4 transcription start site (TSS). However, this insertion does not alter the spatial expression of OsFRDL4 or the cellular localization of OsFRDL4 (Yokosho et al., 2011, 2016). These results indicate that the LTR insertion in the OsFRDL4 promoter region in japonica subspecies is responsible for an increased OsFRDL4 expression level, due to the increased number of ART1-binding cis-acting elements. Similarly, Chen ZC et al. (2013) reported that H. lanatus plants grown in acidic soils secrete more malate than those grown in neutral soils. The reason is that there are more ART1-binding cis-acting elements in the promoter of the Al-resistant HlALMT allele, thus increasing the expression of HlALMT1.
6.3. Gene copy number
The differences between the Al resistances of different rye genotypes are related to the relative amounts of malate and citrate secreted by those genotypes (Li et al., 2000). Collins et al. (2008) showed that both copy number and expression level of the rye ScALMT1 gene, carried on chromosome 7RS, differed between Al-resistant and -sensitive genotypes of rye. Whilst the Al-resistant haplotype (M39A-1–6) contained five copies of ScALMT1, of which two were induced by Al treatment, the Al-sensitive haplotype (M77A-1) had but two copies of ScALMT1, of which only a single one was induced by Al (Collins et al., 2008). At present, the relationship between allele sequence difference and the different expression properties of these different rye ScALMT1 genes is not clear. In maize, the extent of Al-induction of expression of ZmMATE1, a gene encoding a citrate transporter, is greater in Al-resistant lines than in Al-sensitive lines. This difference in expression levels is not ascribed to polymorphisms in either gene or cis-regulatory regions (Maron et al., 2013). Instead, the number of ZmMATE1 copies is associated with gene expression, such that Al-sensitive lines (e.g., L53) have a single copy, whilst Al-resistant lines (e.g., Al237) have three copies (Maron et al., 2013).
6.4. Reversible protein phosphorylation
Although significant advances have been made in the cloning and identification of genes encoding OA transporters, the nature of the signal transduction pathways regulating the expression of those genes remains unclear. Reversible protein phosphorylation is one potential mechanism of regulation. For example, Osawa and Matsumoto (2001) first confirmed that the protein kinase inhibitors K252a or staurosporine inhibit wheat root tip Al-induced malate secretion. Later studies showed that the Al-inducible expression of the Arabidopsis AtALMT1 gene is significantly decreased by pretreatment with K252a or staurosporine. Whilst malate secretion is inhibited by K252a or staurosporine following Al pretreatment, the expression of AtALMT1 is not reduced (with transcription being already completed), indicating that the regulation of Al-induced malate secretion in Arabidopsis works via effects on AtALMT1 function at both transcriptional and post-translational levels, the latter via phosphorylation (Kobayashi et al., 2007). Similarly, we also found protein phosphorylation to be involved both in transcriptional activation of VuMATE1 expression and post-transcriptional regulation of VuMATE1 protein activity in rice bean (Liu et al., 2013). Further evidence provided by Ligaba et al. (2009) showed that the activity of wheat TaALMT1 is associated with phosphorylation by protein kinase C. When the serine (amino acid residue 384) of TaALMT1 was converted to alanine, TaALMt1 activity was inhibited and the mutant protein displayed insensitivity to protein kinase inhibitors or activators. In contrast to the above, Kobayashi et al. (2007) reported that the protein phosphatase inhibitor calyculin A solely inhibited Arabidopsis Al-induced AtALMT1 transcript level increases, but did not exert effects at the post-translational level, suggesting that protein regulatory dephosphorylation may occur only at the transcriptional level for this particular gene. These various results indicate that reversible protein phosphorylation is involved in the regulation of Al-induced OA secretion. However, which particular phosphorylation/dephosphorylation enzymes are involved in these processes remains to be clarified.
7. Perspective
Al-induced secretion of OA anions is one of the most important mechanisms via which plants cope with Al toxicity. Whilst great progress has been made in understanding of the physiology of Al-induced OA secretion, and with the identification of the relevant transporter genes and transcription factors involved in the regulation of expression of those genes (particularly for citrate and malate), the genes encoding oxalate transporters have yet to be identified in plants. Because cytoplasmic oxalate content needs to be precisely controlled (Lou et al., 2016b; Chen et al., 2017), it is possible that the genes responsible for oxalate transport are functionally redundant, making it difficult to clone them via mutant screening methods. Large-scale metabolomic analysis of oxalate content from a panel of cultivars or species may provide an alternative approach to the cloning of these genes. In addition, understanding of the signaling pathways enabling regulation of the expression of these transporter genes remains elusive. When suffering from Al stress, plant cells trigger the expression of specific genes to combat that stress, and a signal transduction cascade is presumably responsible for this differential gene regulation (Fig. 1). Indeed, it is already well known that mitogen-activated protein kinase (MAPK) pathways play crucial roles in signal transmission following initial sensing of the Al stress signal. These MAPK pathways typically consist of three sequentially activated protein kinases: MAPK kinase kinase (MAPKKK), MAPK kinase (MAPKK), and MAPK. Thus, revealing the possible role of the MAPK pathways in regulating the expression of OA transporter genes may prove to be profitable topic for exploration in the near future.
In addition to MAPK pathways, Ca2+ often acts as a second messenger in plant signal transduction pathways. There are two possible ways of transducing the Ca2+ signal, firstly via calcium-calmodulin (Ca-CaM) and alternatively via the calcineurin B-like protein (CBL)-CBL-interacting protein kinase (CIPK) network. Calcium and CaM are already well known to act as second messengers of external stimuli, and indeed, Al stress triggers changes in cytosolic Ca2+ concentrations (Zheng and Yang, 2005; Kochian et al., 2015). The increase in cytosolic Ca2+ leads to the activation of CaM, which binds to glutamate decarboxylase (GAD), converting it from the inactive to the active form. Glutamate is then converted to γ-aminobutyric acid (GABA), which is already known to be involved in regulating ALMT activity (Ramesh et al., 2015). In addition to CaM, the CBL-CIPK network is involved in regulating responses to diverse plant abiotic stresses. Recent experiments have shown that CBL1 is involved in the regulation of expression of AtALMT1 but not of AtMATE, suggesting that the CBL-CIPK network is involved in the regulation of expression of genes encoding OA transporters (Ligaba-Osena et al., 2017). Whether additional members of the CBL-CIPK network are involved in the regulation of expression of OA transporter genes merits future investigation.
Finally, the mechanism via which plants sense Al ions and transduce the Al signal to activate various downstream Al-resistant strategies is a major fundamental frontier issue in plant science. Currently, STOP1 is considered to be the central regulator in Al signal transduction as well as in Al resistance. It was for a long time unknown how STOP1 is responsive to Al stress, but very recently Zhang et al. (2019) discovered an F-box protein, RAE1, that mediates the ubiquitination and degradation of STOP1. Nevertheless, it remains unclear how Al inhibits the degradation of STOP1, and this open question is now amongst the most important in plant Al stress research. Thus, future work will focus on the identification of key genes responsible for Al-dependent accumulation of STOP1, using genetic and biochemical approaches. For example, a mutant library could be constructed in the search for suppressors of Al-induced accumulation of STOP1, and the post-translational modification of STOP1 protein should also be considered. In addition, since Al is not an essential element for plant growth, it is reasonable to speculate that the perception of Al might operate outside of the cell, and that the potential Al receptor or sensor might thus be an outward-facing protein located in the plasma membrane. Following this supposition, known functional membrane protein classes such as receptor-like proteins, receptor-like kinases or G proteins, and protein classes that are usually involved in perception of external signals and subsequent signal transduction, might be good candidates for early Al-sensing components.
Acknowledgments
Thanks are given to Prof. Nicholas P. HARBERD from the University of Oxford (UK) for polishing the English.
Footnotes
Project supported by the National Natural Science Foundation of China (Nos. 31572193, 31760615, and 31760584), 111 Project (No. B14027), and the Changjiang Scholars Program of China
Contributors: Jian-li YANG and Wei FAN wrote and edited the manuscript. Shao-jian ZHENG designed the structure of the paper and edited the manuscript. All authors read and approved the final manuscript.
Compliance with ethics guidelines: Jian-li YANG, Wei FAN, and Shao-jian ZHENG declare that they have no conflict of interests.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Aguilera JG, Minozzo JAD, Barichello D, et al. Alleles of organic acid transporter genes are highly correlated with wheat resistance to acidic soil in field conditions. Theor Appl Genet. 2016;129(7):1317–1331. doi: 10.1007/s00122-016-2705-3. [DOI] [PubMed] [Google Scholar]
- 2.Barceló J, Poschenrieder C. Fast root growth responses, root exudates, and internal detoxification as clues to the mechanisms of aluminium toxicity and resistance: a review. Environ Exp Bot. 2002;48(1):75–92. doi: 10.1016/S0098-8472(02)00013-8. [DOI] [Google Scholar]
- 3.Che J, Tsutsui T, Yokosho K, et al. Functional characterization of an aluminum (Al)-inducible transcription factor, ART2, revealed a different pathway for Al tolerance in rice. New Phytol. 2018;220(1):209–218. doi: 10.1111/nph.15252. [DOI] [PubMed] [Google Scholar]
- 4.Chen PY, Sjogren CA, Larsen PB, et al. A multi-level response to DNA damage induced by aluminium. Plant J, in press. 2019 doi: 10.1111/tpj.14231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Q, Wu KH, Wang P, et al. Overexpression of MsALMT1, from the aluminum-sensitive Medicago sativa, enhances malate exudation and aluminum resistance in tobacco. Plant Mol Biol Rep. 2013;31(3):769–774. doi: 10.1007/s11105-012-0543-2. [DOI] [Google Scholar]
- 6.Chen WW, Fan W, Lou HQ, et al. Regulating cytoplasmic oxalate homeostasis by Acyl activating enzyme3 is critical for plant Al tolerance. Plant Signal Behav. 2017;12(1):e1276688. doi: 10.1080/15592324.2016.1276688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen ZC, Yokosho K, Kashino M, et al. Adaptation to acidic soil is achieved by increased numbers of cis-acting elements regulating ALMT1 expression in Holcus lanatus . Plant J. 2013;76(1):10–23. doi: 10.1111/tpj.12266. [DOI] [PubMed] [Google Scholar]
- 8.Collins NC, Shirley NJ, Saeed M, et al. An ALMT1 gene cluster controlling aluminum tolerance at the Alt4 locus of rye (Secale cereale L.) Genetics. 2008;179(1):669–682. doi: 10.1534/genetics.107.083451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daspute AA, Kobayashi Y, Panda SK, et al. Characterization of CcSTOP1; a C2H2-type transcription factor regulates Al tolerance gene in pigeonpea. Planta. 2018;247(1):201–214. doi: 10.1007/s00425-017-2777-6. [DOI] [PubMed] [Google Scholar]
- 10.Delhaize E, Craig S, Beaton CD, et al. Aluminum tolerance in wheat (Triticum aestivum L.): I. uptake and distribution of aluminum in root apices. Plant Physiol. 1993;103(3):685–693. doi: 10.1104/pp.103.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delhaize E, Ryan PR, Randall PJ. Aluminum tolerance in wheat (Triticum aestivum L.): II. aluminum-stimulated excretion of malic acid from root apices. Plant Physiol. 1993;103(3):695–702. doi: 10.1104/pp.103.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delhaize E, Ryan PR, Hebb DM, et al. Engineering high-level aluminum tolerance in barley with the ALMT1 gene. Proc Natl Acad Sci USA. 2004;101(42):15249–15254. doi: 10.1073/pnas.0406258101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding ZJ, Yan JY, Xu XY, et al. WRKY46 functions as a transcriptional repressor of ALMT1, regulating aluminum-induced malate secretion in Arabidopsis. Plant J. 2013;76(5):825–835. doi: 10.1111/tpj.12337. [DOI] [PubMed] [Google Scholar]
- 14.Durrett TP, Gassmann W, Rogers EE. The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol. 2007;144(1):197–205. doi: 10.1104/pp.107.097162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Famoso AN, Zhao KY, Clark RT, et al. Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet. 2011;7(8):e1002221. doi: 10.1371/journal.pgen.1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan W, Lou HQ, Gong YL, et al. Characterization of an inducible C2H2-type zinc finger transcription factor VuSTOP1 in rice bean (Vigna umbellata) reveals differential regulation between low pH and aluminum tolerance mechanisms. New Phytol. 2015;208(2):456–468. doi: 10.1111/nph.13456. [DOI] [PubMed] [Google Scholar]
- 17.Fan W, Xu JM, Lou HQ, et al. Physiological and molecular analysis of aluminium-induced organic acid anion secretion from grain amaranth (Amaranthus hypochondriacus L.) roots. Int J Mol Sci. 2016;17(5):608. doi: 10.3390/ijms17050608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan W, Xu JM, Wu P, et al. Alleviation by abscisic acid of Al toxicity in rice bean is not associated with citrate efflux but depends on ABI5-mediated signal transduction pathways. J Integr Plant Biol. 2019;61(2):140–154. doi: 10.1111/jipb.12695. [DOI] [PubMed] [Google Scholar]
- 19.Fujii M, Yokosho K, Yamaji N, et al. Acquisition of aluminium tolerance by modification of a single gene in barley. Nat Commun, 3:713. 2012 doi: 10.1038/ncomms1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furukawa J, Yamaji N, Wang H, et al. An aluminum-activated citrate transporter in barley. Plant Cell Physiol. 2007;48(8):1081–1091. doi: 10.1093/pcp/pcm091. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Oliveira AL, Benito C, Prieto P, et al. Molecular characterization of TaSTOP1 homoeologues and their response to aluminium and proton (H+) toxicity in bread wheat (Triticum aestivum L.) BMC Plant Biol, 13:134. 2013 doi: 10.1186/1471-2229-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartwell BL, Pember FR. The presence of aluminum as a reason for the difference in the effects of so-called acid soil on barley and rye. Soil Sci. 1918;6(4):259–280. doi: 10.1097/00010694-191810000-00001. [DOI] [Google Scholar]
- 23.Hoekenga OA, Maron LG, Piñeros MA, et al. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc Natl Acad Sci USA. 2006;103(25):9738–9743. doi: 10.1073/pnas.0602868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horst WJ, Wang YX, Eticha D. The role of the root apoplast in aluminium-induced inhibition of root elongation and in aluminium resistance of plants: a review. Ann Bot. 2010;106(1):185–197. doi: 10.1093/aob/mcq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang CF, Yamaji N, Mitani N, et al. A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell. 2009;21(2):655–667. doi: 10.1105/tpc.108.064543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang CF, Yamaji N, Chen ZC, et al. A tonoplast-localized half-size ABC transporter is required for internal detoxification of aluminum in rice. Plant J. 2012;69(5):857–867. doi: 10.1111/j.1365-313X.2011.04837.x. [DOI] [PubMed] [Google Scholar]
- 27.Huang S, Gao J, You JF, et al. Identification of STOP1-like proteins associated with aluminum tolerance in sweet sorghum (Sorghum bicolor L.) Front Plant Sci, 9:258. 2018 doi: 10.3389/fpls.2018.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iuchi S, Koyama H, Iuchi A, et al. Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc Natl Acad Sci USA. 2007;104(23):9900–9905. doi: 10.1073/pnas.0700117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kashino-Fujii M, Yokosho K, Yamaji N, et al. Retrotransposon insertion and DNA methylation regulate aluminum tolerance in European barley accessions. Plant Physiol. 2018;178(2):716–727. doi: 10.1104/pp.18.00651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitagawa T. Genotypic variations in Al resistance in wheat and organic acid secretion. Jpn J Soil Sci Plant Nutr. 1986;57:352–358. [Google Scholar]
- 31.Kobayashi Y, Hoekenga OA, Itoh H, et al. Characterization of AtALMT1 expression in aluminum-inducible malate release and its role for rhizotoxic stress tolerance in Arabidopsis. Plant Physiol. 2007;145(3):843–852. doi: 10.1104/pp.107.102335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobayashi Y, Ohyama Y, Kobayashi Y, et al. STOP2 activates transcription of several genes for Al-and low pH-tolerance that are regulated by STOP1 in Arabidopsis . Mol Plant. 2014;7(2):311–322. doi: 10.1093/mp/sst116. [DOI] [PubMed] [Google Scholar]
- 33.Kochian LV. Cellular mechanisms of aluminum toxicity and resistance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:237–260. doi: 10.1146/annurev.pp.46.060195.001321. [DOI] [Google Scholar]
- 34.Kochian LV, Hoekenga OA, Piñeros MA. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu Rev Plant Biol. 2004;55:459–493. doi: 10.1146/annurev.arplant.55.031903.141655. [DOI] [PubMed] [Google Scholar]
- 35.Kochian LV, Piñeros MA, Liu JP, et al. Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annu Rev Plant Biol. 2015;66:571–598. doi: 10.1146/annurev-arplant-043014-114822. [DOI] [PubMed] [Google Scholar]
- 36.Kollmeier M, Dietrich P, Bauer CS, et al. Aluminum activates a citrate-permeable anion channel in the aluminum-sensitive zone of the maize root apex. A comparison between an aluminum-sensitive and an aluminum-resistant cultivar. Plant Physiol. 2001;126(1):397–410. doi: 10.1104/pp.126.1.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kundu A, Das S, Basu S, et al. GhSTOP1, a C2H2 type zinc finger transcription factor is essential for Aluminum and proton stress tolerance and lateral root initiation in cotton. Plant Biol. 2019;21(1):35–44. doi: 10.1111/plb.12895. [DOI] [PubMed] [Google Scholar]
- 38.Larsen PB, Geisler MJB, Jones CA, et al. ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J. 2005;41(3):353–363. doi: 10.1111/j.1365-313X.2004.02306.x. [DOI] [PubMed] [Google Scholar]
- 39.Li GZ, Wang ZQ, Yokosho K, et al. Transcription factor WRKY22 promotes aluminum tolerance via activation of OsFRDL4 expression and enhancement of citrate secretion in rice (Oryza sativa) New Phytol. 2018;219(1):149–162. doi: 10.1111/nph.15143. [DOI] [PubMed] [Google Scholar]
- 40.Li XF, Ma JF, Matsumoto H. Pattern of aluminum-induced secretion of organic acids differs between rye and wheat. Plant Physiol. 2000;123(4):1537–1544. doi: 10.1104/pp.123.4.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li YY, Zhang YJ, Zhou Y, et al. Protecting cell walls from binding aluminum by organic acids contributes to aluminum resistance. J Integr Plant Biol. 2009;51(6):574–580. doi: 10.1111/j.1744-7909.2009.00825.x. [DOI] [PubMed] [Google Scholar]
- 42.Liang CY, Piñeros MA, Tian J, et al. Low pH, aluminum, and phosphorus coordinately regulate malate exudation through GmALMT1 to improve soybean adaptation to acid soils. Plant Physiol. 2013;161(3):1347–1361. doi: 10.1104/pp.112.208934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ligaba A, Katsuhara M, Ryan PR, et al. The BnALMT1 and BnALMT2 genes from rape encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells. Plant Physiol. 2006;142(3):1294–1303. doi: 10.1104/pp.106.085233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ligaba A, Kochian L, Piñeros M. Phosphorylation at S384 regulates the activity of the TaALMT1 malate transporter that underlies aluminum resistance in wheat. Plant J. 2009;60(3):411–423. doi: 10.1111/j.1365-313X.2009.03964.x. [DOI] [PubMed] [Google Scholar]
- 45.Ligaba-Osena A, Fei ZJ, Liu JP, et al. Loss-of-function mutation of the calcium sensor CBL1 increases aluminum sensitivity in Arabidopsis . New Phytol. 2017;214(2):830–841. doi: 10.1111/nph.14420. [DOI] [PubMed] [Google Scholar]
- 46.Liu JP, Magalhaes JV, Shaff J, et al. Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 2009;57(3):389–399. doi: 10.1111/j.1365-313X.2008.03696.x. [DOI] [PubMed] [Google Scholar]
- 47.Liu MY, Chen WW, Xu JM, et al. The role of VuMATE1 expression in aluminium-inducible citrate secretion in rice bean (Vigna umbellata) roots. J Exp Bot. 2013;64(7):1795–1804. doi: 10.1093/jxb/ert039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu MY, Lou HQ, Chen WW, et al. Two citrate transporters coordinately regulate citrate secretion from rice bean root tip under aluminum stress. Plant Cell Environ. 2018;41(4):809–822. doi: 10.1111/pce.13150. [DOI] [PubMed] [Google Scholar]
- 49.Lou HQ, Gong YL, Fan W, et al. A formate dehydrogenase confers tolerance to aluminum and low pH. Plant Physiol. 2016;171(1):294–305. doi: 10.1104/pp.16.01105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lou HQ, Fan W, Xu JM, et al. An oxalyl-CoA synthetase is involved in oxalate degradation and aluminum tolerance. Plant Physiol. 2016;172(3):1679–1690. doi: 10.1104/pp.16.01106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma JF. Role of organic acids in detoxification of aluminum in higher plants. Plant Cell Physiol. 2000;41(4):383–390. doi: 10.1093/pcp/41.4.383. [DOI] [PubMed] [Google Scholar]
- 52.Ma JF. Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int Rev Cytol. 2007;264:225–252. doi: 10.1016/S0074-7696(07)64005-4. [DOI] [PubMed] [Google Scholar]
- 53.Ma JF, Zheng SJ, Matsumoto H. Specific secretion of citric acid induced by Al stress in Cassia tora L. Plant Cell Physiol. 1997;38(9):1019–1025. doi: 10.1093/oxfordjournals.pcp.a029266. [DOI] [Google Scholar]
- 54.Ma Z, Miyasaka SC. Oxalate exudation by taro in response to Al. Plant Physiol. 1998;118(3):861–865. doi: 10.1104/pp.118.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Magalhaes JV, Liu JP, Guimarães CT, et al. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet. 2007;39(9):1156–1161. doi: 10.1038/ng2074. [DOI] [PubMed] [Google Scholar]
- 56.Maron LG, Piñeros MA, Guimarães CT, et al. Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. Plant J. 2010;61(5):728–740. doi: 10.1111/j.1365-313X.2009.04103.x. [DOI] [PubMed] [Google Scholar]
- 57.Maron LG, Guimarães CT, Kirst M, et al. Aluminum tolerance in maize is associated with higher MATE1 gene copy number. Proc Natl Acad Sci USA. 2013;110(13):5241–5246. doi: 10.1073/pnas.1220766110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsumoto H. Cell biology of aluminum toxicity and tolerance in higher plants. Int Rev Cytol. 2000;200:1–46. doi: 10.1016/S0074-7696(00)00001-2. [DOI] [PubMed] [Google Scholar]
- 59.Matzenbacher RG. Advances made in developing wheats with better aluminum toxicity tolerance in Brazil. In: Klatt AR , editor. Wheat Production Constraints in Tropical Environments. CIMMYT, Mexico; 1988. pp. 285–304. [Google Scholar]
- 60.Melo JO, Lana UGP, Piñeros MA, et al. Incomplete transfer of accessory loci influencing SbMATE expression underlies genetic background effects for aluminum tolerance in sorghum. Plant J. 2013;73(2):276–288. doi: 10.1111/tpj.12029. [DOI] [PubMed] [Google Scholar]
- 61.Melo JO, Martins LGC, Barros BA, et al. Repeat variants for the SbMATE transporter protect sorghum roots from aluminum toxicity by transcriptional interplay in cis and trans . Proc Natl Acad Sci USA. 2019;116(1):313–318. doi: 10.1073/pnas.1808400115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miyasaka SC, Buta JG, Howell RK, et al. Mechanism of aluminum tolerance in snapbeans: root exudation of citric acid. Plant Physiol. 1991;96(3):737–743. doi: 10.1104/pp.96.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morita A, Yanagisawa O, Maeda S, et al. Tea plant (Camellia sinensis L.) roots secrete oxalic acid and caffeine into medium containing aluminum. Soil Sci Plant Nutr. 2011;57(6):796–802. doi: 10.1080/00380768.2011.629176. [DOI] [Google Scholar]
- 64.Nezames CD, Sjogren CA, Barajas JF, et al. The Arabidopsis cell cycle checkpoint regulators TANMEI/ALT2 and ATR mediate the active process of aluminum-dependent root growth inhibition. Plant Cell. 2012;24(2):608–621. doi: 10.1105/tpc.112.095596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohyama Y, Ito H, Kobayashi Y, et al. Characterization of AtSTOP1 orthologous genes in tobacco and other plant species. Plant Physiol. 2013;162(4):1937–1946. doi: 10.1104/pp.113.218958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Osawa H, Matsumoto H. Possible involvement of protein phosphorylation in aluminum-responsive malate efflux from wheat root apex. Plant Physiol. 2001;126(1):411–420. doi: 10.1104/pp.126.1.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pellet DM, Grunes DL, Kochian LV. Organic acid exudation as an aluminum-tolerance mechanism in maize (Zea mays L.) Planta. 1995;196(4):788–795. doi: 10.1007/BF01106775. [DOI] [Google Scholar]
- 68.Pereira JF, Ryan PR. The role of transposable elements in the evolution of aluminium resistance in plants. J Exp Bot. 2019;70(1):41–54. doi: 10.1093/jxb/ery357. [DOI] [PubMed] [Google Scholar]
- 69.Pereira JF, Barichello D, Ferreira JR, et al. TaALMT1 and TaMATE1B allelic variability in a collection of Brazilian wheat and its association with root growth on acidic soil. Mol Breeding. 2015;35(8):169. doi: 10.1007/s11032-015-0363-9. [DOI] [Google Scholar]
- 70.Piñeros MA, Kochian LV. A patch-clamp study on the physiology of aluminum toxicity and aluminum tolerance in maize. Identification and characterization of Al3+-induced anion channels. Plant Physiol. 2001;125(1):292–305. doi: 10.1104/pp.125.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramesh SA, Tyerman SD, Xu B, et al. GABA signalling modulates plant growth by directly regulating the activity of plant-specific anion transporters. Nat Commun, 6:7879. 2015 doi: 10.1038/ncomms8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rengel Z. Aluminium cycling in the soil-plant-animal-human continuum. Biometals. 2004;17(6):669–689. doi: 10.1007/s10534-004-1201-4. [DOI] [PubMed] [Google Scholar]
- 73.Rounds MA, Larsen PB. Aluminum-dependent root-growth inhibition in Arabidopsis results from AtATR-regulated cell-cycle arrest. Curr Biol. 2008;18(19):1495–1500. doi: 10.1016/j.cub.2008.08.050. [DOI] [PubMed] [Google Scholar]
- 74.Ryan PR, Delhaize E. The convergent evolution of aluminium resistance in plants exploits a convenient currency. Funct Plant Biol. 2010;37(4):275–284. doi: 10.1071/FP09261. [DOI] [Google Scholar]
- 75.Ryan PR, Delhaize E, Jones DL. Function and mechanism of organic anion exudation from plant roots. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:527–560. doi: 10.1146/annurev.arplant.52.1.527. [DOI] [PubMed] [Google Scholar]
- 76.Ryan PR, Raman H, Gupta S, et al. A second mechanism for aluminum resistance in wheat relies on the constitutive efflux of citrate from roots. Plant Physiol. 2009;149(1):340–351. doi: 10.1104/pp.108.129155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ryan PR, Raman H, Gupta S, et al. The multiple origins of aluminium resistance in hexaploid wheat include Aegilops tauschii and more recent cis mutations to TaALMT1 . Plant J. 2010;64(3):446–455. doi: 10.1111/j.1365-313X.2010.04338.x. [DOI] [PubMed] [Google Scholar]
- 78.Sasaki T, Yamamoto Y, Ezaki B, et al. A wheat gene encoding an aluminum-activated malate transporter. Plant J. 2004;37(5):645–653. doi: 10.1111/j.1365-313X.2003.01991.x. [DOI] [PubMed] [Google Scholar]
- 79.Sasaki T, Ryan PR, Delhaize E, et al. Sequence upstream of the wheat (Triticum aestivum L.) ALMT1 gene and its relationship to aluminum resistance. Plant Cell Physiol. 2006;47(10):1343–1354. doi: 10.1093/pcp/pcl002. [DOI] [PubMed] [Google Scholar]
- 80.Sawaki Y, Iuchi S, Kobayashi Y, et al. STOP1 regulates multiple genes that protect Arabidopsis from proton and aluminum toxicities. Plant Physiol. 2009;150(1):281–294. doi: 10.1104/pp.108.134700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sawaki Y, Kobayashia Y, Kihara-Doi T, et al. Identification of a STOP1-like protein in Eucalyptus that regulates transcription of Al tolerance genes. Plant Sci. 2014;223:8–15. doi: 10.1016/j.plantsci.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 82.Sharma T, Dreyer I, Kochian L, et al. The ALMT family of organic acid transporters in plants and their involvement in detoxification and nutrient security. Front Plant Sci, 7:1488. 2016 doi: 10.3389/fpls.2016.01488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Singh S, Tripathi DK, Singh S, et al. Toxicity of aluminium on various levels of plant cells and organism: a review. Environ Exp Bot. 2017;137:177–193. doi: 10.1016/j.envexpbot.2017.01.005. [DOI] [Google Scholar]
- 84.Sjogren CA, Larsen PB. SUV2, which encodes an ATR-related cell cycle checkpoint and putative plant ATRIP, is required for aluminium-dependent root growth inhibition in Arabidopsis. Plant Cell Environ. 2017;40(9):1849–1860. doi: 10.1111/pce.12992. [DOI] [PubMed] [Google Scholar]
- 85.Sjogren CA, Bolaris SC, Larsen PB. Aluminum-dependent terminal differentiation of the Arabidopsis root tip is mediated through an ATR-, ALT2-, and SOG1-regulated transcriptional response. Plant Cell. 2015;27(9):2501–2515. doi: 10.1105/tpc.15.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takanashi K, Shitan N, Yazaki K. The multidrug and toxic compound extrusion (MATE) family in plants. Plant Biotechnol. 2014;31(5):417–430. doi: 10.5511/plantbiotechnology.14.0904a. [DOI] [Google Scholar]
- 87.Taylor GJ. Current views of the aluminum stress response; the physiological basis of tolerance. Curr Top Plant Biochem Physiol. 1991;10:57–93. [Google Scholar]
- 88.Tokizawa M, Kobayashi Y, Saito T, et al. SENSITIVE TO PROTON RHIZOTOXICITY1, CALMODULIN BINDING TRANSCRIPTION ACTIVATOR2, and other transcription factors are involved in ALUMINUM-ACTIVATED MALATE TRANSPORTER1 expression. Plant Physiol. 2015;167(3):991–1003. doi: 10.1104/pp.114.256552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tovkach A, Ryan PR, Richardson AE, et al. Transposon-mediated alteration of TaMATE1B expression in wheat confers constitutive citrate efflux from root apices. Plant Physiol. 2013;161(2):880–892. doi: 10.1104/pp.112.207142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tsutsui T, Yamaji N, Ma JF. Identification of a cis-acting element of ART1, a C2H2-type zinc-finger transcription factor for aluminum tolerance in rice. Plant Physiol. 2011;156(2):925–931. doi: 10.1104/pp.111.175802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.von Uexküll HR, Mutert E. Global extent, development and economic impact of acid soils. Plant soil. 1995;171(1):1–15. doi: 10.1007/BF00009558. [DOI] [Google Scholar]
- 92.Wu WW, Lin Y, Chen QQ, et al. Functional conservation and divergence of soybean GmSTOP1 members in proton and aluminum tolerance. Front Plant Sci, 9:570. 2018 doi: 10.3389/fpls.2018.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xia JX, Yamaji N, Kasai T, et al. Plasma membrane-localized transporter for aluminum in rice. Proc Natl Acad Sci USA. 2010;107(43):18381–18385. doi: 10.1073/pnas.1004949107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xia JX, Yamaji N, Ma JF. A plasma membrane-localized small peptide is involved in rice aluminum tolerance. Plant J. 2013;76(2):345–355. doi: 10.1111/tpj.12296. [DOI] [PubMed] [Google Scholar]
- 95.Yang JL, Zheng SJ, He YF, et al. Aluminium resistance requires resistance to acid stress: a case study with spinach that exudes oxalate rapidly when exposed to Al stress. J Exp Bot. 2005;56(414):1197–1203. doi: 10.1093/jxb/eri113. [DOI] [PubMed] [Google Scholar]
- 96.Yang JL, Zhang L, Li YY, et al. Citrate transporters play a critical role in aluminium-stimulated citrate efflux in rice bean (Vigna umbellata) roots. Ann Bot. 2006;97(4):579–584. doi: 10.1093/aob/mcl005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang JL, Zheng SJ, He YF, et al. Comparative studies on the effect of a protein-synthesis inhibitor on aluminium-induced secretion of organic acids from Fagopyrum esculentum Moench and Cassia tora L. roots. Plant Cell Environ. 2006;29(2):240–246. doi: 10.1111/j.1365-3040.2005.01416.x. [DOI] [PubMed] [Google Scholar]
- 98.Yang JL, Zhang L, Zheng SJ. Aluminum-activated oxalate secretion does not associate with internal content among some oxalate accumulators. J Integr Plant Biol. 2008;50(9):1103–1107. doi: 10.1111/j.1744-7909.2008.00687.x. [DOI] [PubMed] [Google Scholar]
- 99.Yang JL, Zhu XF, Peng YX, et al. Aluminum regulates oxalate secretion and plasma membrane H+-ATPase activity independently in tomato roots. Planta. 2011;234(2):281–291. doi: 10.1007/s00425-011-1402-3. [DOI] [PubMed] [Google Scholar]
- 100.Yang XY, Yang JL, Zhou Y, et al. A de novo synthesis citrate transporter, Vigna umbellata multidrug and toxic compound extrusion, implicates in Al-activated citrate efflux in rice bean (Vigna umbellata) root apex. Plant Cell Environ. 2011;34(12):2138–2148. doi: 10.1111/j.1365-3040.2011.02410.x. [DOI] [PubMed] [Google Scholar]
- 101.Yang ZM, Nian H, Sivaguru M, et al. Characterization of aluminium-induced citrate secretion in aluminium-tolerant soybean (Glycine max) plants. Physiol Plantarum. 2001;113(1):64–71. doi: 10.1034/j.1399-3054.2001.1130109.x. [DOI] [Google Scholar]
- 102.Yokosho K, Yamaji N, Ma JF. Isolation and characterisation of two MATE genes in rye. Funct Plant Biol. 2010;37(4):296–303. doi: 10.1071/FP09265. [DOI] [Google Scholar]
- 103.Yokosho K, Yamaji N, Ma JF. An Al-inducible MATE gene is involved in external detoxification of Al in rice. Plant J. 2011;68(6):1061–1069. doi: 10.1111/j.1365-313X.2011.04757.x. [DOI] [PubMed] [Google Scholar]
- 104.Yokosho K, Yamaji N, Fujii-Kashino M, et al. Retrotransposon-mediated aluminum tolerance through enhanced expression of the citrate transporter OsFRDL4. Plant Physiol. 2016;172(4):2327–2336. doi: 10.1104/pp.16.01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.You JF, He YF, Yang JL, et al. A comparison of aluminum resistance among Polygonum species originating on strongly acidic and neutral soils. Plant Soil. 2005;276(1-2):143–151. doi: 10.1007/s11104-005-3786-y. [DOI] [Google Scholar]
- 106.Zhang L, Wu XX, Wang JF, et al. BoALMT1, an Al-induced malate transporter in cabbage, enhances aluminum tolerance in Arabidopsis thaliana . Front Plant Sci, 8:2156. 2018 doi: 10.3389/fpls.2017.02156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang Y, Guo JL, Chen M, et al. The cell cycle checkpoint regulator ATR is required for internal aluminum toxicity-mediated root growth inhibition in Arabidopsis . Front Plant Sci, 9:118. 2018 doi: 10.3389/fpls.2018.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang Y, Zhang J, Guo JL, et al. 2019. [Google Scholar]
- 109.Zhao H, Huang W, Zhang YG, et al. Natural variation of CsSTOP1 in tea plant (Camellia sinensis) related to aluminum tolerance. Plant Soil. 2018;431(1-2):71–87. doi: 10.1007/s11104-018-3746-y. [DOI] [Google Scholar]
- 110.Zheng SJ, Yang JL. Target sites of aluminum phytotoxicity. Biol Plant. 2005;49(3):321–331. doi: 10.1007/s10535-005-0001-1. [DOI] [Google Scholar]
- 111.Zheng SJ, Ma JF, Matsumoto H. High aluminum resistance in buckwheat. I. Al-induced specific secretion of oxalic acid from root tips. Plant Physiol. 1998;117(3):745–751. doi: 10.1104/pp.117.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]