Abstract
The stem/progenitor cell has long been regarded as a central cell type in development, homeostasis, and regeneration, largely owing to its robust self-renewal and multilineage differentiation abilities. The balance between self-renewal and stem/progenitor cell differentiation requires the coordinated regulation of cell cycle progression and cell fate determination. Extensive studies have demonstrated that cell cycle states determine cell fates, because cells in different cell cycle states are characterized by distinct molecular features and functional outputs. Recent advances in high-resolution epigenome profiling, single-cell transcriptomics, and cell cycle reporter systems have provided novel insights into the cell cycle regulation of cell fate determination. Here, we review recent advances in cell cycle-dependent cell fate determination and functional heterogeneity, and the application of cell cycle manipulation for cell fate conversion. These findings will provide insight into our understanding of cell cycle regulation of cell fate determination in this field, and may facilitate its potential application in translational medicine.
Keywords: Cell cycle, Cell fate, Heterogeneity, Fate conversion, Stem/progenitor cell
1. Introduction
Cell proliferation is one of the fundamental biological activities that take place during development, homeostasis, and regeneration (Matson and Cook, 2017). The sequence of stages during the cell proliferation process is generally termed the cell cycle, and is divided into a synthesis phase (S) and a mitotic segregation phase (M), with two intervenient gap phases (G1 and G2) preceding the S and M phases (Orford and Scadden, 2008). The progression of the G1 phase is tightly regulated by a “start point” (in yeast) or a “restriction point” (in mammals) (Johnson and Skotheim, 2013), which separates the G1 phase into early and late phases. A combination of intrinsic factors, such as the rate of protein synthesis, and extrinsic factors, such as mitogenic signals (growth factors, etc.), determines whether cells enter the cell cycle by passing through the “restriction point” (Zetterberg et al., 1995). However, the absence of these essential factors results in cells exiting the cell cycle and entering a well-defined quiescent state known as the G0 phase. Cells already entering the cell cycle are further controlled by three “checkpoints”: the G1/S, G2/M, and mitotic spindle checkpoints (Hartwell and Weinert, 1989; Pietenpol and Stewart, 2002; Barnum and O'Connell, 2014). At the molecular level, there is a growing body of evidence that cell cycle progression in eukaryotes is driven by an evolutionarily conserved central mechanism comprising cyclin-dependent kinases (CDKs), which are serine/threonine protein kinases that promote DNA synthesis and chromosome segregation by phosphorylating key targets. However, the catalytic activity of CDKs may be negatively regulated by CDK inhibitors (CKIs) (Lim and Kaldis, 2013; Barnum and O'Connell, 2014). According to their evolutionary origins, structures, and CDK specificities, two CKI gene families have been defined: the INK4 (inhibitors of CDK4/CDK6) family and the Cip/Kip family. The INK4 family includes p15INK4b, p16INK4a, p18INK4c, and p19INK4d; and the Cip/Kip family includes p21Waf1/Cip1 (p21, encoded by cdkn1a), p27Kip1 (p27, encoded by cdkn1b), and p57Kip2 (p57, encoded by cdkn1c) (Harper et al., 1993; Polyak et al., 1994; Toyoshima and Hunter, 1994; Lee et al., 1995; Matsuoka et al., 1995; Sherr and Roberts, 1999; Besson et al., 2008). The correct cooperation between cyclin, CDK, and CKI is critical to ensure the ordered progression through an intact cell cycle.
Fate determination of stem/progenitor cells is strictly orchestrated by extrinsic signals and intrinsic regulators. Emerging evidence suggests that changes in cell cycle states affect the fate determination of stem/progenitor cells (Roccio et al., 2013; Lauridsen et al., 2018; Lu et al., 2018). For example, previous studies on mammalian embryonic stem cells (ESCs) have demonstrated that cells in the G1 phase exhibit increased differentiation properties (Maimets et al., 2008; Koledova et al., 2010; Sela et al., 2012), and cells in distinct stages of the G1 phase generate different germ layer cells (Dalton, 2013; Pauklin and Vallier, 2013) (Fig. 1). These findings reveal that stem/progenitor cells stimulated by extrinsic signals remodel their cell cycle states—for example by changing the phase duration or arresting cycling progression—to activate intrinsically instructive signaling pathways (Lange et al., 2009; Salomoni and Calegari, 2010; Calder et al., 2013; Dalton, 2015; Mende et al., 2015).
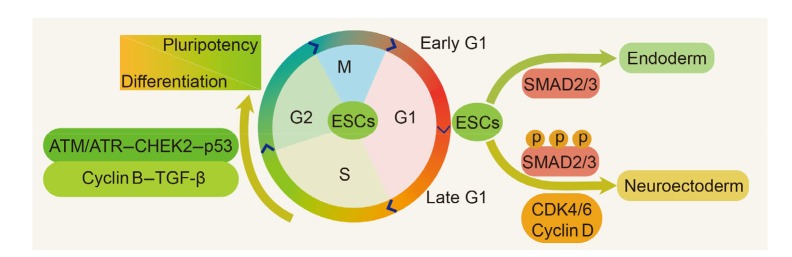
Fig. 1.
Cell cycle-dependent fate determination in embryonic stem cells
In the early G1 phase, the TGF-β-SMAD2/3 pathway promotes the differentiation of ESCs into endodermal cells by activating endodermal genes, whereas the cyclin D-CDK4/6 complex governs the fate conversion of ESC-derived progenies from endoderm cells to neuroectoderm cells by phosphorylating SMAD2/3 in the late G1 phase. Once the cell cycle of ESCs is arrested in the S and G2 phases, DNA damage checkpoint factors ATM/ATR are activated, and subsequently stimulate p53 and cyclin B. Activated cyclin B enhances TGF-β/activin/nodal signaling, which can trigger the selective preference of pluripotency in ESCs. ESCs: embryonic stem cells; TGF-β: transforming growth factor-β; CDK: cyclin-dependent kinase; ATM: ataxia telangiectasia mutated; ATR: ATM and Rad3-related; CHEK2: checkpoint kinase 2
In the present review, we discuss recent advances in genomics profiling techniques and cell cycle reporter systems, with a focus on the cell cycle regulation of cell fate determination. First, we introduce the molecular mechanisms of cell cycle-dependent fate determination. We then describe the cell cycle regulation of cellular functional heterogeneity. Finally, we propose that cell cycle-dependent fate conversion may serve as a potential strategy for the treatment of cells with malignant transformations.
2. Molecular mechanisms of cell cycle-dependent fate determination
Given that cell fate determination is intrinsically associated with cell cycle regulation (Dalton, 2015; Vallier, 2015), it is essential to fully understand the molecular features of cell cycle dynamics. To investigate cell cycle dynamics, chemical drugs have been widely employed to synchronize the cell cycle progression of various cells; however, such drugs can have side effects on cell differentiation (Pauklin and Vallier, 2013). With the help of spatial-temporal observation systems for cell cycle progression (Sakaue-Sawano et al., 2008; Coronado et al., 2013; Zerjatke et al., 2017), it is feasible to separate cells of interest into subpopulations with different cell cycle states, and to characterize their native properties. Furthermore, with the prevalent application of single-cell transcriptomics, there is emerging evidence that the cell cycle states of individual cells contribute to gene expression variation, and cell cycle-related genes also have an impact on cell type/subtype identification (Buettner et al., 2015; McDavid et al., 2016; Skinner et al., 2016; Sun et al., 2017; Lauridsen et al., 2018). Moreover, several studies have also shown that developmental cells are characterized by different cell cycle states (Su et al., 2018; Lu et al., 2019). Overall, the revolution in technologies related to the cell cycle has boosted our understanding of cell cycle dynamics at the molecular level.
2.1. G1 phase-dependent cell fate determination
G1 is crucial for cell fate determination, as reflected by the fact that stem/progenitor cells are particularly susceptible to differentiation signals in that phase. For instance, by employing the fluorescent ubiquitination-based cell cycle indicator (FUCCI) system to monitor the cell cycle states of human ESCs (hESCs), it has been revealed that ESCs respond to differentiation signals primarily occurring during a narrow window of the early G1 phase, whereas these cells remain insensitive during other cell cycle phases. Upon stimulation by SMAD2/3 transcription factors (TFs), hESCs differentiate into endodermal cells. In contrast, hESCs are converted into neuroectodermal cells following overexpression of the cyclin D-CDK4/6 complex. Mechanistically, the cyclin D-CDK4/6 complex inhibits the transcriptional activity of SMAD2/3 during the late G1 phase, thereby ensuring the fate conversion of hESC-derived progenies from endodermal to neuroectodermal cells (Pauklin and Vallier, 2013). This result demonstrates that stem cells initiate fate determination via activation of cell cycle-regulated instructive factors in the G1 phase (Dalton, 2013, 2015). Moreover, accumulating evidence suggests that a transient high expression of TFs, such as GATA6 and SOX17, in response to differentiation signals also occurs in the G1 phase in hESCs, and this transcriptional regulation is a major contributor to heterogeneity in those cells (Singh et al., 2013).
Furthermore, the transition from the M phase to the next G1 phase is associated with a dynamic change in the epigenetic landscape, involving such factors as chromosomal architecture (Thomson et al., 2004; Dalton, 2015), histone modification (Singh et al., 2013, 2015; Gonzales et al., 2015), and DNA methylation (Singh et al., 2013; Ma et al., 2015). Specifically, the epigenetic modification of 5-hydroxymethylcytosine (5hmC) peaks in the G1 phase and subsequently declines in the S phase. The 5-methylcytosine (5mC)/5hmC ratio during cell cycle progression may dictate active transcription in the G1 phase (Singh et al., 2013). Notably, the cell cycle-dynamics of chromosomal organization have been profiled at single-cell resolution using high-resolution chromosome conformation capture techniques (Nagano et al., 2013). It has been proposed that cell cycle progression makes a major contribution to chromosomal dynamics, and together with the accompanying gene regulatory network may be a prerequisite for cell fate determination (Nagano et al., 2017) (Fig. 2). Taken together, these findings demonstrate that the G1 phase serves as a special window that enables the genetic/epigenetic regulation of cell fate-related genes to initiate the process of cell fate determination.
Fig. 2.
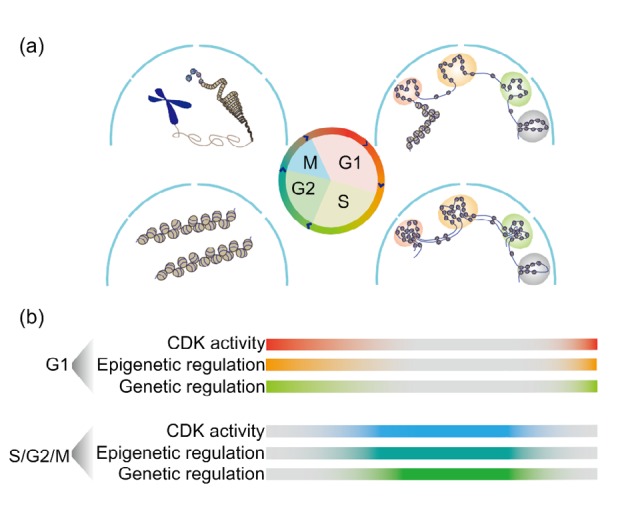
Cell cycle dynamics of molecular regulatory mechanisms
(a) A schematic model showing the dynamics of chromosomal architecture during the cell cycle. (b) The potential mechanisms of cell cycle-dependent fate determination. Cell cycle-specific machinery, cooperating with epigenetic and genetic regulators, can directly orchestrate the cell fate determination of stem/progenitor cells. CDK: cyclin-dependent kinase
2.2. G1 phase-independent cell fate determination
During cell differentiation, stem/progenitor cells experience various biological events, such as DNA damage, chromatin remodeling, and checkpoint activation, which lead to the downregulation of signaling pathways associated with pluripotency and the upregulation of differentiation-signaling pathways (Singh et al., 2013; Akdemir et al., 2014; Gonzales et al., 2015). In addition to the role of the G1 phase in regulating stem/progenitor cell fate determination, the regulatory mechanisms of the S and G2 phases in such cell fate determination have also been gradually decoded. Systematic genomics studies have greatly advanced our knowledge of the regulatory network involved in hESC differentiation (Chia et al., 2010). High-throughput RNA interference (RNAi) screening combined with small-molecule inhibitor treatment has revealed that the S and G2 phases have an intrinsic propensity to rapidly attenuate pluripotency in hESCs. Particularly when progression of the hESC S and G2 phases is perturbed, the DNA damage checkpoint factors ataxia telangiectasia mutated (ATM)/ATM and Rad3-related (ATR) stimulate the activity of p53/cyclin B, and subsequently enhance transforming growth factor-β (TGF-β)/activin/nodal signaling, which can trigger a selective preference for pluripotency (Betschinger et al., 2013; Gonzales et al., 2015) (Fig. 1). Taken together, these studies demonstrate that stem/progenitor cells in the G1 phase respond sensitively to differentiation signals, and subsequently lose their pluripotency in the S and G2 phases, indicating that stem/progenitor cells initiate cell fate determination in the G1 phase while committing to a specified fate in the S and G2 phases (Vallier, 2015).
Dynamic changes to epigenetic modification, such as chromatin remodeling, also occur in the S and M phases (Fig. 2), and may play a role in cell fate determination. Two essential cell cycle events occur in the S and M phases, and result in chromatin remodeling: first, new DNA synthesized in the S phase is assembled with newly synthesized histones to re-establish chromatin and the corresponding epigenetic modifications; second, the loose chromatin is condensed into chromosomes in the M phase, numerous chromatin-remodeling complexes and transcriptional complexes dissociate from the chromosome, and the nuclear envelope ultimately decomposes (Ma et al., 2015). Thus, histone acetylation, nucleosome remodeling, and widespread DNA demethylation, which take place during the S and M phases, contribute to the tightly regulated processes of cell fate determination (Singh et al., 2013; Gonzales et al., 2015).
3. Cell cycle regulation of functional heterogeneity
Although the study of molecular mechanisms can help elucidate the interplay between cell cycle regulators and cell fate determinants, it is necessary to link molecular mechanisms to cellular functions in order to understand the contribution made by the cellular state to cell heterogeneity during development and homeostasis (Lein et al., 2017; Haas et al., 2018). For example, it has been demonstrated that quiescent and cycling stem/progenitor cells are distinguished by their metabolic states, morphologies, and differentiation outputs (Ohnuma and Harris, 2003; Li and Clevers, 2010; Lugert et al., 2010; Salazar-Roa and Malumbres, 2017; Haas et al., 2018). In particular, quiescent mammalian hematopoietic stem cells (HSCs) have more robust long-term reconstitution ability in blood system-impaired recipients than cycling HSCs (Haug et al., 2008; Li and Clevers, 2010; Lauridsen et al., 2018). This ability is associated with the low reactive oxygen species production, low molecular synthesis rate, and inactive metabolic state of HSCs (Ito and Suda, 2014; Walter et al., 2015; Qian et al., 2016). Moreover, a recent study of Drosophila neural stem cells (NSCs) revealed that quiescent stem cells can be arrested in the G2 phase, as well as in the G0 phase, which is well known (Cheung and Rando, 2013). Once stimulated by insulin signals, quiescent NSCs in the G2 phase re-enter the cell cycle before quiescent NSCs in the G0 phase. Functionally, this unexpected phenomenon may ensure the orderly development of neurons into appropriate neural circuits (Otsuki and Brand, 2018). These results demonstrate that heterogeneous cell cycle states lead to differences in cellular function (Pauklin and Vallier, 2013; Gonzales et al., 2015; Vallier, 2015; Gruenheit et al., 2018). Cell cycle speed is also associated with differentiation outputs of mouse HSCs and progenitor cells (Lu et al., 2018; Upadhaya et al., 2018). These studies indicate that disparate cell cycle speeds may also serve as an essential determinant of cellular functional output. Taken together, heterogenous cell cycle states—such as quiescence versus cycling, G0 versus G2, and early G1 versus late G1—and disparate cell cycle speeds contribute to the functional heterogeneity of adult stem/progenitor cells.
4. Cell fate conversion by manipulating cell cycle
Cell fate conversion through defined factors provides a novel strategy for tissue/organ regeneration and disease treatment (Lis et al., 2017; Liu et al., 2017). During development, TFs play a crucial role in the generation and maintenance of cells with distinct fates (Gurdon, 2016). Several researchers have accomplished direct fate conversion by manipulating the master TFs in hematopoietic and nervous systems (Xie et al., 2004; Vierbuchen et al., 2010; Zhang et al., 2018). As mentioned above, cell cycle regulators also participate in fate determination by remodeling cell cycle states or cooperating with TFs. Studies on pre-artery/artery specification have revealed that the reduced expression of G1 phase-specific genes resulting from changes in blood flow is required for the expression of artery-generating genes. Specifically, the Notch-GJA4-CKI and chicken ovalbumin upstream promoter transcription factor 2 (COUP-TF 2) signaling pathways are involved in the cycle regulation of pre-artery/artery specification (Fang et al., 2017; Su et al., 2018). Arresting the cell cycle in the G1 phase combined with p53 suppression can markedly improve the efficiency of fate conversion from human fibroblasts to dopaminergic neurons when mediated by TFs and microRNA. Mechanistically, highly efficient fate conversion requires the overexpression of epigenetic regulator Tet1, and cell cycle-mediated remodeling of cellular states is essential for overcoming the fate conversion barrier (Jiang et al., 2015). Fate conversion by manipulating the cell cycle has now been accomplished in several cell types, indicating its potential application in translational medicine and disease modeling.
5. Concluding remarks
The development of high-resolution epigenome profiling techniques, single-cell transcriptomics, and cell cycle reporter systems has resulted in a better understanding of the relationships between molecular features, cell cycle progression, and fate determination (Fig. 3). At the molecular level, when cyclin-CDK/CKI complexes are coordinated with master TFs, they are capable of determining the fate of a cell. Specifically, cell fate determination occurs in the early G1 phase, and the initiation of cell differentiation takes place in the S and G2 phases. When responding to differentiation signals, stem/progenitor cells execute cell fate determination primarily in the early G1 phase owing to transcriptional restart and chromatin reorganization. Subsequently, stem/progenitor cells lose their pluripotency and commit to a specified fate in the S and G2 phases, when cells establish new chromatin/epigenetic modifications and undergo dramatic changes to their nuclear architectures. Furthermore, stem/progenitor cells in various cell cycle states—such as quiescent cells versus cycling cells, G0 quiescent cells versus G2 quiescent cells, and early G1 cells versus late G1 cells—are distinguished by their morphologies, metabolic states, self-renewal abilities, and differentiation outputs. Cell cycle speed and phase duration can also contribute to the functional heterogeneity of stem/progenitor cells. Although the cell cycle serves as an essential fate determinant, caution is required when analyzing cell cycle information from transcriptomics data (Sun et al., 2017).
Fig. 3.
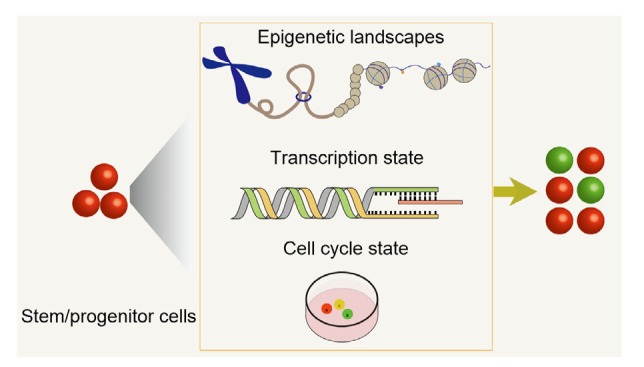
Three deterministic factors of stem/progenitor cell fates
Cell fate determination is attributed to epigenetic landscapes, transcription states, and the cell cycle states of stem/progenitor cells. Epigenomic analysis techniques, such as single-cell Hi-C (high-resolution chromosome conformation capture), can capture the chromosomal conformation of individual cells during the cell cycle. Transcriptomic analyses can reveal the transcription state during the cell cycle. Cell cycle reporter systems can dynamically trace cell cycle progression by live imaging
Currently, studies on cell cycle-dependent cell fate determination are based on cell cycle perturbation. Therefore, the regulatory mechanisms underlying normal cell cycle progression in cell fate determination remain largely unknown. Furthermore, there is no comprehensive framework for the integrative analysis of the cell cycle dynamics of chromosomal architecture, epigenetic modification, or gene expression during cell fate determination (Nagano et al., 2017). The establishment of a comprehensive framework would enable the elucidation of the molecular mechanisms underlying cell cycle-dependent fate determination on a genome-wide scale. Furthermore, using cell cycle reporter systems, it is now feasible to dynamically trace cell cycle progression. It would be helpful if future studies focused on the real-time visualization of the cell cycle dynamics of various fate-determined regulators at single-cell or subcellular resolution. However, it is still unclear whether the mechanisms identified in ESCs in vitro are conserved in the cell fate determination of adult stem/progenitor cells in vivo. Importantly, the direct targeting of cell cycle regulators by small molecules or by genetic perturbation has produced favorable results with regard to the suppression of pathologic neuron degeneration and tumorigenesis. However, these experiments were performed in vitro in mammalian tumor cell lines, or in vivo using genetically engineered disease models (Kar et al., 2006; Chatterjee et al., 2016; Biswas et al., 2017; Tomás-Loba et al., 2019). Therefore, the translation of basic research into clinical treatment remains a critical challenge. Addressing the questions mentioned above will promote our understanding of cell cycle regulation in cell fate determination, and may facilitate its application in translational medicine.
Glossary
ESC (embryonic stem cell): a totipotent stem cell derived from the inner cell mass of a blastocyst that can differentiate into three germ layers and produce any type of cell within the embryo.
HSC (hematopoietic stem cell): a multipotent stem cell residing in adult bone marrow that can yield all mature blood cells, including myeloid, lymphoid, and erythroid cells.
NSC (neural stem cell): a multipotent stem cell residing in the hippocampal dentate gyrus of the adult brain that can differentiate into neurons, astrocytes, and oligodendrocytes.
FUCCI (fluorescent ubiquitination-based cell cycle indicator): a reporter system that can be used to monitor the phase transition of the cell cycle in living cells. The fluorescent proteins are fused with cell cycle regulators Cdt1 and geminin, which are destabilized by APC/C (anaphase-promoting complex or cyclosome)-and SCFSkp2 (Skp1/Cul1/F-box protein)-mediated ubiquitination, enabling the accurate visualization of living cells in various phases of the cell cycle.
Acknowledgments
The study was also supported by the State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing, China.
Footnotes
Project supported by the Ministry of Science and Technology of China (No. 2016YFA0100500), the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDA16010207), and the National Natural Science Foundation of China (Nos. 31425016, 81530004, and 31830061)
Contributors: Su-wei GAO and Feng LIU conceived the idea for the review and wrote the paper. Both authors read and approved the final manuscript.
Compliance with ethics guidelines: Su-wei GAO and Feng LIU declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by either of the authors.
References
- 1.Akdemir KC, Jain AK, Allton K, et al. Genome-wide profiling reveals stimulus-specific functions of p53 during differentiation and DNA damage of human embryonic stem cells. Nucleic Acids Res. 2014;42(1):205–223. doi: 10.1093/nar/gkt866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnum KJ, O'Connell MJ. Cell cycle regulation by checkpoints. In: Noguchi E Gadaleta MC., editor. Cell Cycle Control: Methods in Molecular Biology (Methods and Protocols), Vol. 1170. Humana Press, New York; 2014. pp. 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Dev Cell. 2008;14(2):159–169. doi: 10.1016/j.devcel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Betschinger J, Nichols J, Dietmann S, et al. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell. 2013;153(2):335–347. doi: 10.1016/j.cell.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas SC, Sanphui P, Chatterjee N, et al. Cdc25A phosphatase: a key cell cycle protein that regulates neuron death in disease and development. Cell Death Dis. 2017;8(3):e2692. doi: 10.1038/cddis.2017.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buettner F, Natarajan KN, Casale FP, et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33(2):155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 7.Calder A, Roth-Albin I, Bhatia S, et al. Lengthened G1 phase indicates differentiation status in human embryonic stem cells. Stem Cells Dev. 2013;22(2):279–295. doi: 10.1089/scd.2012.0168. [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee N, Sanphui P, Kemeny S, et al. Role and regulation of Cdc25A phosphatase in neuron death induced by NGF deprivation or β-amyloid. Cell Death Discov, 2:16083. 2016 doi: 10.1038/cddiscovery.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung TH, Rando TA. Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol. 2013;14(6):329–340. doi: 10.1038/nrm3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chia NY, Chan YS, Feng B, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468(7321):316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- 11.Coronado D, Godet M, Bourillot PY, et al. A short G1 phase is an intrinsic determinant of naïve embryonic stem cell pluripotency. Stem Cell Res. 2013;10(1):118–131. doi: 10.1016/j.scr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Dalton S. G1 compartmentalization and cell fate coordination. Cell. 2013;155(1):13–14. doi: 10.1016/j.cell.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Dalton S. Linking the cell cycle to cell fate decisions. Trends Cell Biol. 2015;25(10):592–600. doi: 10.1016/j.tcb.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang JS, Coon BG, Gillis N, et al. Shear-induced Notch-Cx37-p27 axis arrests endothelial cell cycle to enable arterial specification. Nat Commun, 8:2149. 2017 doi: 10.1038/s41467-017-01742-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzales KA, Liang HQ, Lim YS, et al. Deterministic restriction on pluripotent state dissolution by cell-cycle pathways. Cell. 2015;162(3):564–579. doi: 10.1016/j.cell.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Gruenheit N, Parkinson K, Brimson CA, et al. Cell cycle heterogeneity can generate robust cell type proportioning. Dev Cell. 2018;47(4):494–508e4. doi: 10.1016/j.devcel.2018.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurdon JB. Cell fate determination by transcription factors. Curr Top Dev Biol. 2016;116:445–454. doi: 10.1016/bs.ctdb.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Haas S, Trumpp A, Milsom MD. Causes and consequences of hematopoietic stem cell heterogeneity. Cell Stem Cell. 2018;22(5):627–638. doi: 10.1016/j.stem.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Harper JW, Adami GR, Wei N, et al. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75(4):805–816. doi: 10.1016/0092-8674(93)90499-G. [DOI] [PubMed] [Google Scholar]
- 20.Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246(4930):629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 21.Haug JS, He XC, Grindley JC, et al. N-cadherin expression level distinguishes reserved versus primed states of hematopoietic stem cells. Cell Stem Cell. 2008;2(4):367–379. doi: 10.1016/j.stem.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 22.Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol. 2014;15(4):243–256. doi: 10.1038/nrm3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang HB, Xu ZM, Zhong P, et al. Cell cycle and p53 gate the direct conversion of human fibroblasts to dopaminergic neurons. Nat Commun, 6:10100. 2015 doi: 10.1038/ncomms10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson A, Skotheim JM. Start and the restriction point. Curr Opin Cell Biol. 2013;25(6):717–723. doi: 10.1016/j.ceb.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kar S, Wang MF, Yao W, et al. PM-20, a novel inhibitor of Cdc25A, induces extracellular signal-regulated kinase 1/2 phosphorylation and inhibits hepatocellular carcinoma growth in vitro and in vivo . Mol Cancer Ther. 2006;5(6):1511–1519. doi: 10.1158/1535-7163.Mct-05-0485. [DOI] [PubMed] [Google Scholar]
- 26.Koledova Z, Kafkova LR, Calabkova L, et al. Cdk2 inhibition prolongs G1 phase progression in mouse embryonic stem cells. Stem Cells Dev. 2010;19(2):181–194. doi: 10.1089/scd.2009.0065. [DOI] [PubMed] [Google Scholar]
- 27.Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5(3):320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 28.Lauridsen FKB, Jensen TL, Rapin N, et al. Differences in cell cycle status underlie transcriptional heterogeneity in the HSC compartment. Cell Rep. 2018;24(3):766–780. doi: 10.1016/j.celrep.2018.06.057. [DOI] [PubMed] [Google Scholar]
- 29.Lee MH, Reynisdóttir I, Massagué J. Cloning of p57KIP2, a cyclin-dependent kinase inhibitor with unique domain structure and tissue distribution. Genes Dev. 1995;9(6):639–649. doi: 10.1101/gad.9.6.639. [DOI] [PubMed] [Google Scholar]
- 30.Lein E, Borm LE, Linnarsson S. The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science. 2017;358(6359):64–69. doi: 10.1126/science.aan6827. [DOI] [PubMed] [Google Scholar]
- 31.Li LH, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327(5965):542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140(15):3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- 33.Lis R, Karrasch CC, Poulos MG, et al. Conversion of adult endothelium to immunocompetent haematopoietic stem cells. Nature. 2017;545(7655):439–445. doi: 10.1038/nature22326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu ZQ, Wang L, Welch JD, et al. Single-cell transcriptomics reconstructs fate conversion from fibroblast to cardiomyocyte. Nature. 2017;551(7678):100–104. doi: 10.1038/nature24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu CJ, Fan XY, Guo YF, et al. Single-cell analyses identify distinct and intermediate states of zebrafish pancreatic islet development. J Mol Cell Biol, mjy064. 2019 doi: 10.1093/jmcb/mjy064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu YC, Sanada C, Xavier-Ferrucio J, et al. The molecular signature of megakaryocyte-erythroid progenitors reveals a role for the cell cycle in fate specification. Cell Rep. 2018;25(8):2083–2093e4. doi: 10.1016/j.celrep.2018.10.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lugert S, Basak O, Knuckles P, et al. Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell. 2010;6(5):445–456. doi: 10.1016/j.stem.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 38.Ma YQ, Kanakousaki K, Buttitta L. How the cell cycle impacts chromatin architecture and influences cell fate. Front Genet, 6:19. 2015 doi: 10.3389/fgene.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maimets T, Neganova I, Armstrong L, et al. Activation of p53 by nutlin leads to rapid differentiation of human embryonic stem cells. Oncogene. 2008;27(40):5277–5287. doi: 10.1038/onc.2008.166. [DOI] [PubMed] [Google Scholar]
- 40.Matson JP, Cook JG. Cell cycle proliferation decisions: the impact of single cell analyses. FEBS J. 2017;284(3):362–375. doi: 10.1111/febs.13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsuoka S, Edwards MC, Bai C, et al. p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev. 1995;9(6):650–662. doi: 10.1101/gad.9.6.650. [DOI] [PubMed] [Google Scholar]
- 42.McDavid A, Finak G, Gottardo R. The contribution of cell cycle to heterogeneity in single-cell RNA-seq data. Nat Biotechnol. 2016;34(6):591–593. doi: 10.1038/nbt.3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mende N, Kuchen EE, Lesche M, et al. CCND1-CDK4-mediated cell cycle progression provides a competitive advantage for human hematopoietic stem cells in vivo. J Exp Med. 2015;212(8):1171–1183. doi: 10.1084/jem.20150308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagano T, Lubling Y, Stevens TJ, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502(7469):59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagano T, Lubling Y, Várnai C, et al. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature. 2017;547(7661):61–67. doi: 10.1038/nature23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohnuma S, Harris WA. Neurogenesis and the cell cycle. Neuron. 2003;40(2):199–208. doi: 10.1016/S0896-6273(03)00632-9. [DOI] [PubMed] [Google Scholar]
- 47.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9(2):115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 48.Otsuki L, Brand AH. Cell cycle heterogeneity directs the timing of neural stem cell activation from quiescence. Science. 2018;360(6384):99–102. doi: 10.1126/science.aan8795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pauklin S, Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155(1):135–147. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pietenpol JA, Stewart ZA. Cell cycle checkpoint signaling: cell cycle arrest versus apoptosis. Toxicology. 2002;181-182:475–481. doi: 10.1016/S0300-483X(02)00460-2. [DOI] [PubMed] [Google Scholar]
- 51.Polyak K, Lee MH, Erdjument-Bromage H, et al. Cloning of p27kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell. 1994;78(1):59–66. doi: 10.1016/0092-8674(94)90572-X. [DOI] [PubMed] [Google Scholar]
- 52.Qian PX, He XC, Paulson A, et al. The Dlk1-Gtl2 locus preserves LT-HSC function by inhibiting the PI3K-mTOR pathway to restrict mitochondrial metabolism. Cell Stem Cell. 2016;18(2):214–228. doi: 10.1016/j.stem.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roccio M, Schmitter D, Knobloch M, et al. Predicting stem cell fate changes by differential cell cycle progression patterns. Development. 2013;140(2):459–470. doi: 10.1242/dev.086215. [DOI] [PubMed] [Google Scholar]
- 54.Sakaue-Sawano A, Kurokawa H, Morimura T, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132(3):487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 55.Salazar-Roa M, Malumbres M. Fueling the cell division cycle. Trends Cell Biol. 2017;27(1):69–81. doi: 10.1016/j.tcb.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 56.Salomoni P, Calegari F. Cell cycle control of mammalian neural stem cells: putting a speed limit on G1. Trends Cell Biol. 2010;20(5):233–243. doi: 10.1016/j.tcb.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 57.Sela Y, Molotski N, Golan S, et al. Human embryonic stem cells exhibit increased propensity to differentiate during the G1 phase prior to phosphorylation of retinoblastoma protein. Stem Cells. 2012;30(6):1097–1108. doi: 10.1002/stem.1078. [DOI] [PubMed] [Google Scholar]
- 58.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13(12):1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 59.Singh AM, Chappell J, Trost R, et al. Cell-cycle control of developmentally regulated transcription factors accounts for heterogeneity in human pluripotent cells. Stem Cell Rep. 2013;1(6):532–544. doi: 10.1016/j.stemcr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh AM, Sun YH, Li L, et al. Cell-cycle control of bivalent epigenetic domains regulates the exit from pluripotency. Stem Cell Rep. 2015;5(3):323–336. doi: 10.1016/j.stemcr.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Skinner SO, Xu H, Nagarkar-Jaiswal S, et al. Single-cell analysis of transcription kinetics across the cell cycle. eLife, 5:e12175. 2016 doi: 10.7554/eLife.12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su TY, Stanley G, Sinha R, et al. Single-cell analysis of early progenitor cells that build coronary arteries. Nature. 2018;559(7714):356–362. doi: 10.1038/s41586-018-0288-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun N, Yu XM, Li F, et al. Inference of differentiation time for single cell transcriptomes using cell population reference data. Nat Commun. 2017;8(1):1856. doi: 10.1038/s41467-017-01860-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomson I, Gilchrist S, Bickmore WA, et al. The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr Biol. 2004;14(2):166–172. doi: 10.1016/j.cub.2003.12.024. [DOI] [PubMed] [Google Scholar]
- 65.Tomás-Loba A, Manieri E, González-Terán B, et al. p38γ is essential for cell cycle progression and liver tumorigenesis. Nature. 2019;568(7753):557–560. doi: 10.1038/s41586-019-1112-8. [DOI] [PubMed] [Google Scholar]
- 66.Toyoshima H, Hunter T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell. 1994;78(1):67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 67.Upadhaya S, Sawai CM, Papalexi E, et al. Kinetics of adult hematopoietic stem cell differentiation in vivo. J Exp Med. 2018;215(11):2815–2832. doi: 10.1084/jem.20180136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vallier L. Cell cycle rules pluripotency. Cell Stem Cell. 2015;17(2):131–132. doi: 10.1016/j.stem.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 69.Vierbuchen T, Ostermeier A, Pang ZP, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463(7284):1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walter D, Lier A, Geiselhart A, et al. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015;520(7548):549–552. doi: 10.1038/nature14131. [DOI] [PubMed] [Google Scholar]
- 71.Xie HF, Ye M, Feng R, et al. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117(5):663–676. doi: 10.1016/S0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 72.Zerjatke T, Gak IA, Kirova D, et al. Quantitative cell cycle analysis based on an endogenous all-in-one reporter for cell tracking and classification. Cell Rep. 2017;19(9):1953–1966. doi: 10.1016/j.celrep.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zetterberg A, Larsson O, Wiman KG. What is the restriction point? Curr Opin Cell Biol. 1995;7(6):835–842. doi: 10.1016/0955-0674(95)80067-0. [DOI] [PubMed] [Google Scholar]
- 74.Zhang MY, Dong Y, Hu FX, et al. Transcription factor Hoxb5 reprograms B cells into functional T lymphocytes. Nat Immunol. 2018;19(3):279–290. doi: 10.1038/s41590-018-0046-x. [DOI] [PMC free article] [PubMed] [Google Scholar]