Abstract
Sirtuin 1 (SIRT1) is a protein deacetylase, which regulates various physiological activities by deacetylating different protein substrates. An increasing number of studies have revealed critical roles of SIRT1 in different aspects of cancers including metabolism, proliferation, genomic instability, and chemotherapy resistance. Depending on the protein targets in a certain oncogenic context, SIRT1 may play a unique role in each individual blood cancer subtype. Our previous work showed that activation of SIRT1 in primitive leukemia cells of acute myeloid leukemia (AML) and chronic myelogenous leukemia (CML) promotes disease maintenance. On the other hand, an SIRT1 agonist was shown to disrupt maintenance of myelodysplastic syndrome (MDS) stem cells and holds promise as a potential therapeutic approach. Herein, we present a concise summary of the different functions of SIRT1 in hematologic malignancies.
Keywords: Sirtuin 1 (SIRT1), Hematologic malignancy, Stem cell, Drug resistance
1. Introduction
Sirtuins are nicotinamide adenine dinucleotide (NAD)-dependent protein deacetylases, which are highly conserved from yeast to mammalian cells. Seven sirtuins (SIRT1–SIRT7) in mammalian cells exhibit functional significance on aging, diabetes, cardiovascular diseases, and cancers (Chalkiadaki and Guarente, 2015) (Fig. 1). SIRT1, the most extensively studied sirtuin, can deacetylate various histone and non-histone substrates including p53, c-MYC, and FOXO, thereby regulating diverse biological processes such as DNA repair, metabolism, cell cycle, and survival (Brooks and Gu, 2009; Herranz and Serrano, 2010; Chalkiadaki and Guarente, 2015). Earlier studies identified the tumor suppressor p53 as the first non-histone SIRT1 deacetylase target: under stress conditions, such as DNA damage, deacetylation of p53 attenuates its transactivation-dependent apoptosis, thus promoting lung cancer cell survival (Luo et al., 2001; Vaziri et al., 2001). Likewise, E2F1 was also found to be negatively regulated by SIRT1 in the lung cancer cell line (Wang et al., 2006). Therefore, SIRT1 was considered to be an oncogenic protein. However, recent investigations shed a new light on SIRT1 function in stem cell transformation, including its roles in promoting the faithful repair of DNA and inhibiting oncogenic transformation, showing that SIRT1 can serve as a tumor suppressor in some cancers (Chalkiadaki and Guarente, 2015). Here, we highlight the different roles of SIRT1 in several hematologic malignancy subtypes. In each context, we also summarize the possible molecular mechanisms of SIRT1 effects (Fig. 2).
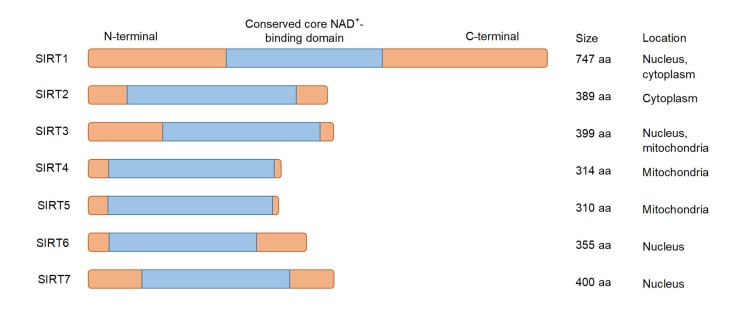
Fig. 1.
Primary structure of seven mammalian sirtuins (SIRTs)
NAD: nicotinamide; aa: amino acids
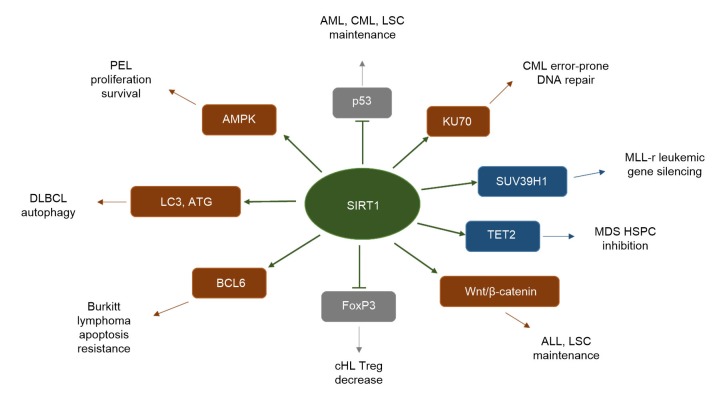
Fig. 2.
Representative targets of SIRT1 in hematologic malignancies
SIRT1 enhances target activity (labeled in brown), promoting hematologic malignancies; SIRT1 decreases target activity (labeled in grey), promoting hematologic malignancies; SIRT1 enhances target activity (labeled in blue), suppressing hematologic malignancies. AML, acute myeloid leukemia; CML, chronic myelogenous leukemia; LSC, leukemia stem cell; MLL-r, mixed-lineage leukemia-rearranged; MDS, myelodysplastic syndrome; HSPC, hematopoietic stem/progenitor cell; ALL, acute lymphoblastic leukemia; cHL, classical Hodgkin lymphoma; DLBCL, diffuse large B-cell lymphoma; PEL, primary effusion lymphoma
2. Role of SIRT1 in acute myeloid leukemia
Acute myeloid leukemia (AML) is a heterogeneous disease characterized by hyperproliferative and immature leukemia blasts expanding in the bone marrow (BM). Leukemia blasts arise from aberrant primitive hematopoietic precursor cells called leukemic stem cells (LSCs). LSCs are a small subset of self-renewing leukemic cells, which are enriched in the CD34+CD38− subset, that persist after conventional therapy and are considered a source of leukemia relapse (Ng et al., 2016). We have found increased SIRT1 protein levels in CD34+CD38− cells of AML BM relative to normal counterparts (Li et al., 2014). In addition, SIRT1 expression was higher in cells from patient specimens with high or intermediate risk compared to those with low risk. Internal tandem duplication in FLT3 (FLT3-ITD) is one of the most frequent mutations in AML and is associated with enhanced relapse rate (Patel et al., 2012). Two independent groups reported higher SIRT1 expression in CD34+ cells from FLT3-ITD+ AML specimens relative to those of FLT3 wild-type AML counterparts (Li et al., 2014; Sasca et al., 2014). Sasca et al. (2014) demonstrated that SIRT1 activity is positively regulated through the FLT3–ATM–DBC1 axis. However, we found that SIRT1 overexpression is related to enhanced expression of the USP22 deubiquitinase (Li et al., 2014). This c-Myc/USP22/SIRT1 post-transcriptional regulatory network in human FLT3-ITD AML LSCs eventually leads to LSC maintenance and drug resistance through downregulation of p53 activity (Li et al., 2014). The two reports converged on p53 and concluded that pharmacological inhibition of SIRT1 can enhance p53 acetylation levels, leading to increased p53 target gene expression, cell growth inhibition, and enhanced sensitivity to tyrosine kinase inhibitor treatment. These results support the idea of an important role of SIRT1 in AML LSCs and suggest SIRT1 inhibition as a potential strategy for precise targeting of AML LSCs. Bradbury et al. (2005) analyzed the role of SIRT1 expression in mononuclear cells from a large cohort of AML specimens and found SIRT1 to be consistently overexpressed in AML samples compared with normal controls.
Another unique upstream regulatory pathway and corresponding function of SIRT1 in AML were identified by Tian et al. (2018). They found that the mRNA level of SIRT1 was negatively correlated with the mRNA level of IRF9, a member of the interferon-regulatory factor (IRF) family. They also showed that IRF9 binds the SIRT1 promoter and represses the transcription of SIRT1, thus activating p53 and inhibiting AML cell growth.
Previous studies have shown that SIRT1 positively regulates autophagy by deacetylating autophagy factors Atg5, Atg7, Atg8, and LC3 (Lee et al., 2008; Huang et al., 2015). As autophagy plays a protective role upon stress, Ou et al. (2014) evaluated connections between SIRT1 activity and reactive oxygen species (ROS)-induced autophagy in murine and human embryonic stem cells (ESCs), and found that SIRT1 promotes PI3K/Beclin 1 and mediates stress-induced autophagy in ESCs. These works revealed an important role of SIRT1 in enhancing autophagy, eventually leading to apoptosis resistance.
Another group focused their attention on epigenetic regulation and found a tumor suppressive role of SIRT1 in mixed-lineage leukemia (MLL)-rearranged leukemia (Chen et al., 2015). Previous studies have shown that leukemia cells driven by MLL-rearrangement are highly sensitive to DOT1L inhibition and H3K79 methylation, which established a heterochromatin-like state around MLL fusion target genes and aberrantly active leukemic gene expression. To determine the exact mechanisms, they conducted a genome-scale RNA interfere (RNAi) screen and found SIRT1 as an antagonist of DOT1L. Further investigation demonstrated that chromatin localization of SIRT1 and the H3K9 methyltransferase SUV39H1 could induce silencing of leukemic genes. Conversely, DOT1L can inhibit this localization and maintain an open chromatin state at MLL fusion target genes, thereby sustaining leukemic gene expression. The results of Chen et al. (2015) indicated that DOT1L inhibition in combination with SIRT1 activation could be a promising strategy in MLL-rearranged leukemia patients.
3. Role of SIRT1 in chronic myeloid leukemia
Chronic myeloid leukemia (CML) is a clonal hematological malignancy resulting from BCR-ABL transformed hematopoietic stem cells (Ren, 2005). There are three clinical phases of CML, progressing from a chronic phase to an accelerated phase and then to a terminal blast crisis. Tyrosine kinase inhibitors (TKIs) target the constitutively activated BCR-ABL kinase, thus leading to longer term remission of CML in the majority of patients, but they do not eliminate LSCs. The relapse that occurs in 50% of patients after stopping treatment with TKIs is likely due to the presence of LSCs (Ross et al., 2013; Rea and Mahon, 2018).
Similar to AML, the expression of SIRT1 is significantly increased in CML LSCs relative to normal counterparts (Li et al., 2012; Yuan et al., 2012). Yuan et al. (2012) showed that BCR-ABL activates SIRT1 through kinase-dependent STAT5 signaling. However, BCR-ABL kinase inhibition or STAT5 knockdown can only partially reduce SIRT1 expression, which suggests that other kinase-independent mechanisms are responsible for increased SIRT1 activity in CML (Yuan et al., 2012). To explore the functional role of SIRT1 in CML, our group inhibited SIRT1 using short hairpin RNA (shRNA) and the small-molecule inhibitor TV-6 and showed that SIRT1 inhibition increased p53 acetylation and activation in CML LSCs, reducing LSC survival and growth (Li et al., 2012). Wang et al. (2015) identified novel LSC markers in the BALB/c mouse model of CML. They demonstrated that the genetic loss of SIRT1 can increase p53 acetylation, depleting CML LSCs.
Previous studies have shown that SIRT1 maintains genome stability and acts as a tumor suppressor (Wang et al., 2008). On the other hand, impaired DNA damage response was also observed in SIRT1-knockout CML cells. Accordingly, Wang et al. (2013) showed that the high expression of SIRT1 in CML can enhance error-prone DNA damage repair and promote mutation acquisition. Similarly, recent work indicated that SIRT1 inhibition impairs non-homologous end joining DNA damage repair by increasing KU70 acetylation in CML (Roth et al., 2016; Zhang et al., 2016). These results reveal a different role of SIRT1 for promoting mutation acquisition, thus leading to drug resistance in CML.
4. Role of SIRT1 in myelodysplastic syndrome
Myelodysplastic syndrome (MDS) is a clonal disease that arises from hematopoietic stem/progenitor cells (HSPCs) harboring genetic alterations. MDS is characterized by morphological dysplasia, ineffective hematopoiesis, and risk of transformation to AML. MDS remains incurable by existing non-transplant treatments (Sperling et al., 2017).
Recently, our lab demonstrated that SIRT1 protein levels were decreased in MDS HSPCs compared to normal counterparts (Sun et al., 2018). Interestingly, SIRT1 mRNA levels were similar in these two cohorts, and thus we next explored post-transcriptional regulation of SIRT1. As a result, microRNA-9 (miR-9) and miR-34a were found to be highly expressed in CD34+ cells and their expression was inversely correlated with SIRT1 protein but not mRNA levels, which suggests that miR-9 and miR-34a downregulate SIRT1 in MDS cells. In this study, we further explored the SIRT1 function and showed that SIRT1 deficiency in MDS HSPCs enhances their growth and self-renewal. To identify the target of SIRT1 in MDS cells, we immunoprecipitated total acetylated lysine proteins and then identified potential targets through mass spectrometry. Through small interfering RNA (siRNA) screening, we identified TET2, an essential enzyme of DNA demethylation, as a possible SIRT1 deacetylation target. Taken together, low SIRT1 levels in MDS HSPCs caused TET2 hyperacetylation and further reduced TET2 catalytic activity, thereby resulting in higher levels of DNA methylation, decreased tumor suppressor gene expression, and enhanced maintenance of MDS HSPCs. Conversely, overexpression or pharmacological activation of SIRT1 enhanced TET2 function and drastically delayed MDS development in vivo. In conclusion, this work uncovered an important tumor suppressive role of SIRT1 in MDS and potentially in other myeloid cancers. Restoring TET2 function through SIRT1 activation seems to be a promising means to target MDS HSPCs. Nevertheless, as mentioned above, SIRT1 activation can inhibit p53 activity and accelerate disease progression in FLT3-ITD+ AML and CML. Although we observed a modest reduction of p53 acetylation levels in SRT1720-treated MDS cells, considering MDS heterogeneity and risk to transform into AML, future trials will require careful consideration of patient selection criteria.
5. Role of SIRT1 in lymphoma
Acute lymphoblastic leukemia/lymphoblastic lymphoma (ALL/LBL) is one of the most common childhood malignancies. Although the cure rates for patients with newly diagnosed ALL/LBL have steadily improved over past decades, little progress has been made in the treatment of the relapsed population (Pui et al., 2004; Bhojwani and Pui, 2013). Similar to what was observed in other myeloid malignancies, the SIRT1 expression level is also elevated in ALL/LBL compared to normal counterparts (Jin et al., 2015; Li et al., 2015). Jin et al. (2015) found that inhibition of SIRT1 by the small molecule inhibitor tenovin-6 can lead to hyperacetylation of p53 and induce cell growth inhibition and apoptosis in ALL cells. Mechanistically, the inhibition of SIRT1/2 activity also decreased Wnt/β-catenin signaling, thus eliminating ALL-initiating cells (Jin et al., 2015). SIRT1 expression levels were also increased in chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL) (Audrito et al., 2011; dal Bo et al., 2015; Bhalla and Gordon, 2016). Audrito et al. (2011) found that nicotinamide can significantly increase miR-34a levels with a concomitant inhibition of SIRT1 in CLL cells, which leads to a block of proliferation and the activation of apoptosis. Another group demonstrated that activation of BCR in CLL cells can upregulate miR-132 levels causing downregulation of SIRT1 and increased acetylation of p53 (dal Bo et al., 2015). Consistently, higher miR-132 levels in CLL/SLL cells were associated with a better clinical course for patients (dal Bo et al., 2015).
Diffuse large B-cell lymphoma (DLBCL) is one of the most common non-Hodgkin lymphomas. A decade ago, researchers observed the correlation between high SIRT1 expression and poor prognosis of DLBCL (Jang et al., 2008). Recently, Kan et al. (2018) revealed a negative association between the SIRT1 single-nucleotide polymorphism (SNP) rs3758391 (an SNP of the SIRT1 gene promoter) and the survival rate of DLBCL patients in the Chinese Han population. Their genotyping results showed that patients with SIRT1 SNP rs3758391 have a better overall survival than those with wild-type (WT) allele, indicating that SIRT1 could serve as a biomarker for DLBCL. Targeting SIRT1 by pharmacological inhibitors like tenovin-6 seems promising. To explore their function in DLBCL, Yuan et al. (2017) treated DLBCL cell lines with tenovin-6 and observed that treatment could inhibit cancer cell proliferation. Interestingly, specific knockdown of SIRT1/2/3 has no effect on DLBCL, suggesting that the inhibitory effects of tenovin-6 are independent of sirtuin. Mechanistically, they found that tenovin-6 can increase the expression level of the autophagy marker LC3B-II in all DLBCL cell lines tested, but the action is not relevant to the SIRT1/p53 axis. As tenovin-6 was used as an SIRT1 inhibitor in these studies, Yuan et al. (2017) unveiled a new mechanism involving autophagy, and thus precautions should be taken in future studies.
There are also examples of SIRT1 regulating other types of lymphomas. Nihal et al. (2014) found increased SIRT1 expression of cutaneous T-cell lymphoma cell lines and tissues relative to normal lymphocytes. SIRT1 inhibition by tenovin-1 resulted in reduced cellular metabolism and proliferation, and increased apoptosis. Similarly, SIRT1 helps to maintain the proliferation and survival of primary effusion lymphoma (PEL) cells in an AMPK-dependent manner (He et al., 2017). Thus, targeting the SIRT1–AMPK axis with tenovin derivatives effectively inhibits the initiation and progression of PEL, and dramatically extends lymphoma xenografted animal survival. Heltweg et al. (2006) identified a compound called cambinol that can inhibit SIRT1/2 in Burkitt lymphoma cells, thus decreasing BCL-6 level and activating checkpoint pathways like p53 concurrently, eventually leading to apoptosis of lymphoma cells.
6. Role of SIRT1 in hematologic malignancy microenvironment
Over the past decades, mounting evidence suggests that immune dysregulation happens in the tumor microenvironment. There is also evidence that SIRT1 participates in immune dysregulation. Previous studies suggest that the presence of cytotoxic TIA-1+ cells with reduced FoxP3+ Treg cells predicts unfavorable outcomes in classical Hodgkin lymphoma (cHL). Briefly, Quesada et al. (2015) focused their attention on the relationship between SIRT1 and the immune microenvironment in cHL. They detected the expression of SIRT1 and FoxP3 from 24 cases and found increased expression of SIRT1 in 21 cases. Further analysis revealed that the patients who suffered a recurrence had a significantly higher ratio of SIRT1 versus FoxP3 than the patients who were in remission. As FoxP3 is one of the deacetylation targets of SIRT1, they hypothesized that SIRT1 deacetylates FoxP3 and reduces T regulatory (FoxP3+) cell function. Thus, SIRT1 inhibition may promote Hodgkin Reed-Sternberg cells to differentiate into CD4+ naive T cells as well as Tregs, contributing to a better clinical outcome. Moreover, Daenthanasanmak et al. (2019) reported that SIRT1 inhibition can attenuate graft versus host disease (GVHD) by promoting Treg cell differentiation and inhibiting interferon-γ production. These observations indicate that SIRT1 can regulate not only tumor cells themselves, but also the microenvironment, affecting clinical outcomes (Table 1).
Table 1.
Role of SIRT1 in hematologic malignancies
| Disease | Upstream | SIRT1 level | Role of SIRT1 |
| AML | IRF9 | Increased | Promote AML development |
| SDF-1α, CXCR4 | Increased | Promote autophagy | |
| FLT3-ITD AML | c-MYC, USP22 | Increased | Promote LSC maintenance |
| ATM, DBC1 | Increased | Inhibit genomic stress-induced apoptosis | |
| MLL-r leukemia | DOT1L | Unknown | Mediate leukemic gene silencing |
| CML | BCR-ABL STAT5 | Increased | Enhance error-prone DNA damage repair |
| HIC1 | Increased | Promote LSC maintenance | |
| MDS | miR-9, miR-34a | Decreased | Disrupt maintenance of MDS stem and progenitor cells |
| ALL/LBL | Unknown | Increased | Promote LSC maintenance |
| CLL/SLL | miR-34a, miR132 | Increased | Promote proliferation and survival |
| PEL | Unknown | Increased | Promote proliferation and survival |
| CTCL | Unknown | Increased | Promote proliferation and survival |
| Burkitt lymphoma | Unknown | Unknown | Promote apoptosis resistance |
| cHL | Unknown | Increased | Decrease Treg |
| BM transplantation | Unknown | Unknown | Promote graft versus host disease |
AML, acute myeloid leukemia; FLT3-ITD, internal tandem duplication in FLT3; MLL-r, mixed-lineage leukemia-rearranged; CML, chronic myelogenous leukemia; MDS, myelodysplastic syndrome; ALL, acute lymphoblastic leukemia; LBL, lymphoblastic lymphoma; CLL, chronic lymphocytic leukemia; SLL, small lymphocytic lymphoma; PEL, primary effusion lymphoma; CTCL, cutaneous T-cell lymphoma; cHL, classical Hodgkin lymphoma; BM, bone marrow; LSC, leukemia stem cell
7. Conclusions
It is now clear that SIRT1 can regulate pro-survival pathways in most of the hematologic malignancies. Moreover, this SIRT1 action can be multifaceted, modulating cell survival, genomic stability, metabolism, and the microenvironment. Though many targets have been reported, the SIRT1 effects can be context-dependent, given that only certain substrates will contribute to the major phenotype. Thus, further evaluation of SIRT1-directed therapy for both preclinical and clinical application is warranted.
Acknowledgments
We thank Marjorie ROBBINS (Beckman Research Institute, City of Hope Medical Center, Duarte, USA) for critical reading of the manuscript.
Footnotes
Project supported by the National Institutes of Health (No. R01HL141336), the Margaret E. Early Medical Research Trust Award, the Stop Cancer Research Career Development Award, V Scholar Award of Cancer Research, and the Gehr Family Center for Leukemia Research, USA
Contributors: Fei-teng HUANG wrote this review. Jie SUN, Lei ZHANG, Xin HE, Ying-hui ZHU, Hao-jie DONG, Han-ying WANG, Lei ZHU, and Jing-ying ZOU participated in the design and editing of the manuscript. Jin-wen HUANG and Ling LI checked and approved the final version.
Compliance with ethics guidelines: Fei-teng HUANG, Jie SUN, Lei ZHANG, Xin HE, Ying-hui ZHU, Hao-jie DONG, Han-ying WANG, Lei ZHU, Jing-ying ZOU, Jin-wen HUANG, and Ling LI declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Audrito V, Vaisitti T, Rossi D, et al. Nicotinamide blocks proliferation and induces apoptosis of chronic lymphocytic leukemia cells through activation of the p53/miR-34a/SIRT1 tumor suppressor network. Cancer Res. 2011;71(13):4473–4483. doi: 10.1158/0008-5472.can-10-4452. [DOI] [PubMed] [Google Scholar]
- 2.Bhalla S, Gordon LI. Functional characterization of NAD dependent deacetylases SIRT1 and SIRT2 in B-cell chronic lymphocytic leukemia (CLL) Cancer Biol Ther. 2016;17(3):300–309. doi: 10.1080/15384047.2016.1139246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhojwani D, Pui CH. Relapsed childhood acute lymphoblastic leukaemia. Lancet Oncol. 2013;14(6):e205–e217. doi: 10.1016/s1470-2045(12)70580-6. [DOI] [PubMed] [Google Scholar]
- 4.Bradbury CA, Khanim FL, Hayden R, et al. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia. 2005;19(10):1751–1759. doi: 10.1038/sj.leu.2403910. [DOI] [PubMed] [Google Scholar]
- 5.Brooks CL, Gu W. How does SIRT1 affect metabolism, senescence and cancer? Nat Rev Cancer. 2009;9(2):123–128. doi: 10.1038/nrc2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalkiadaki A, Guarente L. The multifaceted functions of sirtuins in cancer. Nat Rev Cancer. 2015;15(10):608–624. doi: 10.1038/nrc3985. [DOI] [PubMed] [Google Scholar]
- 7.Chen CW, Koche RP, Sinha AU, et al. DOT1L inhibits SIRT1-mediated epigenetic silencing to maintain leukemic gene expression in MLL-rearranged leukemia. Nat Med. 2015;21(4):335–343. doi: 10.1038/nm.3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daenthanasanmak A, Iamsawat S, Chakraborty P, et al. Targeting Sirt-1 controls GVHD by inhibiting T-cell allo-response and promoting Treg stability in mice. Blood. 2019;133(3):266–279. doi: 10.1182/blood-2018-07-863233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.dal Bo M, D'Agaro T, Gobessi S, et al. The SIRT1/TP53 axis is activated upon B-cell receptor triggering via miR-132 up-regulation in chronic lymphocytic leukemia cells. Oncotarget. 2015;6(22):19102–19117. doi: 10.18632/oncotarget.3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He ML, Tan B, Vasan K, et al. SIRT1 and AMPK pathways are essential for the proliferation and survival of primary effusion lymphoma cells. J Pathol. 2017;242(3):309–321. doi: 10.1002/path.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heltweg B, Gatbonton T, Schuler AD, et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66(8):4368–4377. doi: 10.1158/0008-5472.can-05-3617. [DOI] [PubMed] [Google Scholar]
- 12.Herranz D, Serrano M. SIRT1: recent lessons from mouse models. Nat Rev Cancer. 2010;10(12):819–823. doi: 10.1038/nrc2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang R, Xu YF, Wan W, et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol Cell. 2015;57(3):456–466. doi: 10.1016/j.molcel.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 14.Jang KY, Hwang SH, Kwon KS, et al. SIRT1 expression is associated with poor prognosis of diffuse large B-cell lymphoma. Am J Surg Pathol. 2008;32(10):1523–1531. doi: 10.1097/PAS.0b013e31816b6478. [DOI] [PubMed] [Google Scholar]
- 15.Jin YL, Cao Q, Chen C, et al. Tenovin-6-mediated inhibition of SIRT1/2 induces apoptosis in acute lymphoblastic leukemia (ALL) cells and eliminates ALL stem/progenitor cells. BMC Cancer, 15:226. 2015 doi: 10.1186/s12885-015-1282-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kan YT, Ge P, Wang XY, et al. SIRT1 rs3758391 polymorphism and risk of diffuse large B cell lymphoma in a Chinese population. Cancer Cell Int, 18:163. 2018 doi: 10.1186/s12935-018-0659-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee IH, Cao L, Mostoslavsky R, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA. 2008;105(9):3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, Wang LS, Li L, et al. Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer Cell. 2012;21(2):266–281. doi: 10.1016/j.ccr.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Osdal T, Ho Y, et al. SIRT1 activation by a c-MYC oncogenic network promotes the maintenance and drug resistance of human FLT3-ITD acute myeloid leukemia stem cells. Cell Stem Cell. 2014;15(4):431–446. doi: 10.1016/j.stem.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L, Ye SG, Yang M, et al. SIRT1 downregulation enhances chemosensitivity and survival of adult T-cell leukemia-lymphoma cells by reducing DNA double-strand repair. Oncol Rep. 2015;34(6):2935–2942. doi: 10.3892/or.2015.4287. [DOI] [PubMed] [Google Scholar]
- 21.Luo JY, Nikolaev AY, Imai SI, et al. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107(2):137–148. doi: 10.1016/S0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 22.Ng SWK, Mitchell A, Kennedy JA, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540(7633):433–437. doi: 10.1038/nature20598. [DOI] [PubMed] [Google Scholar]
- 23.Nihal M, Ahmad N, Wood GS. SIRT1 is upregulated in cutaneous T-cell lymphoma, and its inhibition induces growth arrest and apoptosis. Cell Cycle. 2014;13(4):632–640. doi: 10.4161/cc.27523. [DOI] [PubMed] [Google Scholar]
- 24.Ou X, Lee MR, Huang XX, et al. SIRT1 positively regulates autophagy and mitochondria function in embryonic stem cells under oxidative stress. Stem Cells. 2014;32(5):1183–1194. doi: 10.1002/stem.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel JP, Gönen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pui CH, Relling MV, Downing JR. Mechanisms of disease: acute lymphoblastic leukemia. N Engl J Med. 2004;350(15):1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 27.Quesada AE, Assylbekova B, Jabcuga CE, et al. Expression of Sirt1 and FoxP3 in classical Hodgkin lymphoma and tumor infiltrating lymphocytes: implications for immune dysregulation, prognosis and potential therapeutic targeting. Int J Clin Exp Pathol. 2015;8(10):13241–13248. [PMC free article] [PubMed] [Google Scholar]
- 28.Rea D, Mahon FX. How I manage relapse of chronic myeloid leukaemia after stopping tyrosine kinase inhibitor therapy. Br J Haematol. 2018;180(1):24–32. doi: 10.1111/bjh.14973. [DOI] [PubMed] [Google Scholar]
- 29.Ren RB. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer. 2005;5(3):172–183. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 30.Ross DM, Branford S, Seymour JF, et al. Safety and efficacy of imatinib cessation for CML patients with stable undetectable minimal residual disease: results from the TWISTER study. Blood. 2013;122(4):515–522. doi: 10.1182/blood-2013-02-483750. [DOI] [PubMed] [Google Scholar]
- 31.Roth M, Wang ZQ, Chen WY. SIRT1 and LSD1 competitively regulate KU70 functions in DNA repair and mutation acquisition in cancer cells. Oncotarget. 2016;7(31):50195–50214. doi: 10.18632/oncotarget.10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasca D, Hähnel PS, Szybinski J, et al. SIRT1 prevents genotoxic stress-induced p53 activation in acute myeloid leukemia. Blood. 2014;124(1):121–133. doi: 10.1182/blood-2013-11-538819. [DOI] [PubMed] [Google Scholar]
- 33.Sperling AS, Gibson CJ, Ebert BL. The genetics of myelodysplastic syndrome: from clonal haematopoiesis to secondary leukaemia. Nat Rev Cancer. 2017;17(1):5–19. doi: 10.1038/nrc.2016.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun J, He X, Zhu YH, et al. SIRT1 activation disrupts maintenance of myelodysplastic syndrome stem and progenitor cells by restoring TET2 function. Cell Stem Cell. 2018;23(3):355–369e9. doi: 10.1016/j.stem.2018.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian WL, Guo R, Wang F, et al. The IRF9-SIRT1-P53 axis is involved in the growth of human acute myeloid leukemia. Exp Cell Res. 2018;365(2):185–193. doi: 10.1016/j.yexcr.2018.02.036. [DOI] [PubMed] [Google Scholar]
- 36.Vaziri H, Dessain SK, Ng Eaton E, et al. hSIR2SIRT1 functions as an NAD-dependent p53 deacetylase. Cell. 2001;107(2):149–159. doi: 10.1016/S0092-8674(01)00527-X. [DOI] [PubMed] [Google Scholar]
- 37.Wang CG, Chen LH, Hou XH, et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8(9):1025–1031. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- 38.Wang RH, Sengupta K, Li CL, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14(4):312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, Yuan H, Roth M, et al. SIRT1 deacetylase promotes acquisition of genetic mutations for drug resistance in CML cells. Oncogene. 2013;32(5):589–598. doi: 10.1038/onc.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang ZQ, Chen CC, Chen WY. CD150− side population defines leukemia stem cells in a BALB/c mouse model of CML and is depleted by genetic loss of SIRT1. Stem Cells. 2015;33(12):3437–3451. doi: 10.1002/stem.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan HF, Wang ZQ, Li L, et al. Activation of stress response gene SIRT1 by BCR-ABL promotes leukemogenesis. Blood. 2012;119(8):1904–1914. doi: 10.1182/blood-2011-06-361691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan HF, He ML, Cheng F, et al. Tenovin-6 inhibits proliferation and survival of diffuse large B-cell lymphoma cells by blocking autophagy. Oncotarget. 2017;8(9):14912–14924. doi: 10.18632/oncotarget.14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang WJ, Wu HX, Yang M, et al. SIRT1 inhibition impairs non-homologous end joining DNA damage repair by increasing Ku70 acetylation in chronic myeloid leukemia cells. Oncotarget. 2016;7(12):13538–13550. doi: 10.18632/oncotarget.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]