Abstract
In 2013 we established a cryoelectron microscopy (cryo-EM) technique called microcrystal electron diffraction (MicroED). Since that time, data collection and analysis schemes have been fine-tuned, and structures for over 40 different proteins, oligopeptides and organic molecules and been determined. Here, we review the MicroED technique and place it in context with other structure determination methods. We showcase example structures solved by MicroED and provide practical advice to perspective users.
Cryo-EM encompasses a variety of techniques using a transmission electron microscope (TEM) to determine the structures of frozen-hydrated beam-sensitive samples (Fig. 1). Cryo-EM techniques can make use of two modes of data collection: those which use images of biological samples – purified biomolecules or whole cells - and crystallographic approaches based on diffraction. In single particle cryo-EM, images of individual biomolecules are collected, followed by image processing to generate 3D reconstructions of these biomolecules1. In electron cryo-tomography, images of biomolecules within their native cellular environment are collected2; however, while important biological insights can be gained from tomography, the resolution in this method is significantly lower than that of all other cryo-EM methods.
Figure 1: Methods within the field of Cryo-EM.
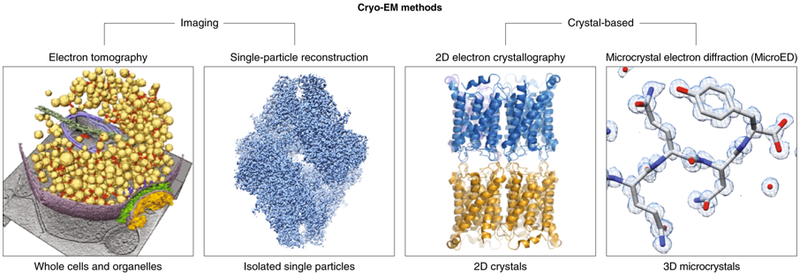
The diverse imaging-based or diffraction-based techniques within cryo-electron microscopy can provide structural information from a wide range of samples. Images shown for the different techniques are the synaptosome98 (electron tomography), 2.2 Å reconstruction of beta-galactosidase99 (single particle reconstruction), 1.9 Å structure of aquaporin-010 (2D-electron crystallography), and 1.0 Å structure of the NNQQNY Sup35 prion fragment49 (MicroED).
The crystallographic cryo-EM techniques of 2D electron crystallography3, 4 and microcrystal electron diffraction (MicroED)5, 6, both use crystalline arrays of material. However, because crystals 2D in electron crystallography and 3D in MicroED, the data collection and processing are substantially different between the two techniques rendering them distinct branches of cryo-EM modalities. For example, a key difference is that in 2D electron crystallography, the orientation of the 2D array can be determined from a single diffraction pattern because the one crystallographic axis is always parallel to the beam in every crystal. In contrast, it is rare for any crystallographic axis to be parallel to the beam in 3D crystals. Therefore with MicroED a wedge of data in reciprocal space must be collected7, 8 rather than a single diffraction pattern and this, in turn, is processed differently than the data for 2D crystals. 2D electron crystallography has a long and storied history; many of the first high-resolution cryo-EM structures were determined from 2D crystals9-15. Thin 3D protein crystals have also been investigated by electron diffraction for several decades16-21. Despite these early observations that 3D protein microcrystals can produce high-resolution diffraction data, the use of 3D microcrystals for structure determination by cryo-EM was not achieved until 2013 with the development of MicroED7.
MicroED takes advantage of highly sensitive modern cryo-EM detectors to determine protein structures from nanocrystals only ~10 layers thick, as we demonstrated with the first complete high-resolution electron diffraction structure of lysozyme7. The initial “still-diffraction” MicroED data collection procedure7 facilitated the correct indexing, data processing and structure determination of lysozyme to 2.9 Å resolution using specialized software22. For this, the crystal was tilted at defined angles within the cryo-TEM while diffraction was recorded.
In early 2014, we improved MicroED data collection by introducing “continuous rotation” where the crystal is continuously rotated within the electron beam as data are recorded on a high-speed detector as a movie8 (Fig. 2). Since continuous rotation is analogous to the rotation method used in X-ray crystallography, data collected by this method can be processed using well established software that was developed for X-ray crystallography. Continuous rotation therefore improved the quality of the raw data by increasing sampling of reciprocal space, reducing dynamic scattering (Box 1), improving data processing, all of which ultimately combined to yield improved final structures. The structure of lysozyme determined by continuous rotation was initially reported at 2.5 Å with significantly improved data processing statistics8. With further data processing improvements23, 24 this structure was improved to 1.8 Å resolution (Fig. 3).
Figure 2: MicroED overview.
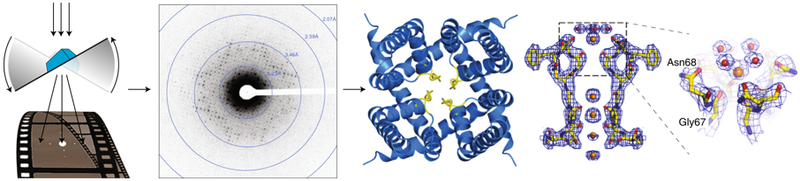
MicroED data are collected as movies while the stage of the cryo-EM is continuously rotated. This produces a series of high-resolution diffraction patterns that can be processed to produce high-resolution structures directly from microcrystals. Here the structure of the non-selective ion channel NaK is illustrated100.
Box 1. About Dynamical Scattering and crystal thickness.
The effects of dynamic scattering, also known as multiple scattering, have been a topic of much investigation and debate in all areas of electron diffraction. Structure determination efforts generally assume that electrons are diffracted by the crystal a single time, known as kinematic scattering; however, as crystals become thicker, the probability that electrons are diffracted multiple times within the crystal increases until the intensities of the reflections have been so severely redistributed that the diffraction patterns can no longer be reliably used for structure interpretation. This is known as dynamical scattering. Despite early reports in 1975 that were performed on thin 3D crystals of catalase demonstrating that kinematic scattering could be assumed for protein crystals in electron diffraction17, it was alternatively commonly assumed that dynamic scattering would be much too severe for protein crystal diffraction data, essentially halting progress in the field for decades. Moreover, simulations indicated that when crystals thicker than only ~50–100 nm are used, dynamical scattering would be so severe that the underlying relationship between the structure factors and the recorded intensity would be completely lost94. The initial proof of concept MicroED publication in 2013 confirmed that multiple scattering did not preclude structure determination from thin 3D crystals, which in this case were approximately 200nm thick lysozyme crystals. By analyzing the systematic absences of the lysozyme diffraction data, it was determined that data derived from still diffraction patterns contained approximately 5% error that was attributed to dynamical effects7.
Precession electron diffraction is a method used in materials science to reduce the effect of dynamic scattering by pivoting the electron beam using the coils of the TEM during exposure95. In this approach, dynamic scattering is diminished because as the beam is rotated, the secondary scattering is averaged, leading to an ensemble of near kinematical recorded diffraction intensities96. Precession electron diffraction has been successfully used to determine several structures in materials science from inorganic samples that are radiation hardy through the ADT and RED methods97. Continuous rotation MicroED is similar to precession except rather than the beam being rotated, the crystal is rotated in the beam during exposure. With continuous rotation MicroED we demonstrated a significant reduction in the error associated with multiple scattering in lysozyme (estimated at 2.5% for the same lysozyme crystals that without continuous rotation showed a 5% error) and an improvement in the final integrated data obtained by MicroED8.
The most convincing evidence for the negligible effects of dynamic scattering on continuous rotation MicroED data came from the ab initio structure determination of the yeast prion protein Sup3549. These crystals were estimated to be 300–400nm thick and produced diffraction data which extended to 1 Å. This level of resolution allowed the phasing of the data by direct methods. Because direct methods only use the measured intensities, the successful ab initio phasing and structure determination of these samples demonstrated that dynamic scattering represents a minor component of the error observed in MicroED data. Consistent with these results, Friedel pair analysis of the data indicated no detectable difference between the reflection pairs, strongly demonstrating the validity of the kinematic scattering assumption for continuous rotation MicroED. Taken together, these results, and those from several other structures that followed, clearly show that highly accurate data can be collected by continuous rotation MicroED from crystals with a range of sub-micron thicknesses.
Figure 3: Improvements in MicroED data quality.
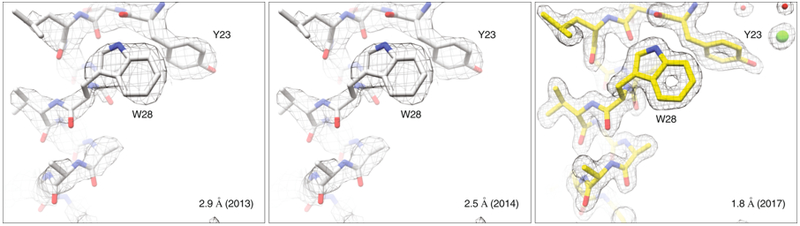
Continual development of MicroED has led to steady improvements in the quality of structures obtained. This can be seen by the increases in resolution that are possible from similar lysosome microcrystals. Density maps (2Fo-Fc) in gray are contoured at 1.5σ for all structures shown.
Continuous-rotation data collection represents the standard data collection approach by which all MicroED structures (except for the very first lysozyme structure from 2013) have been determined to date. Thus far, approximately 40 protein, peptide and small molecule structures have been determined by MicroED since 2013; several of these are novel structures which resisted other methods (Fig 4, Table 1 and described in detail below). Centers around the globe are adopting the technology and several structures have been published by others in the past couple of years25, 26.
Figure 4: Examples of novel structures determined by MicroED.
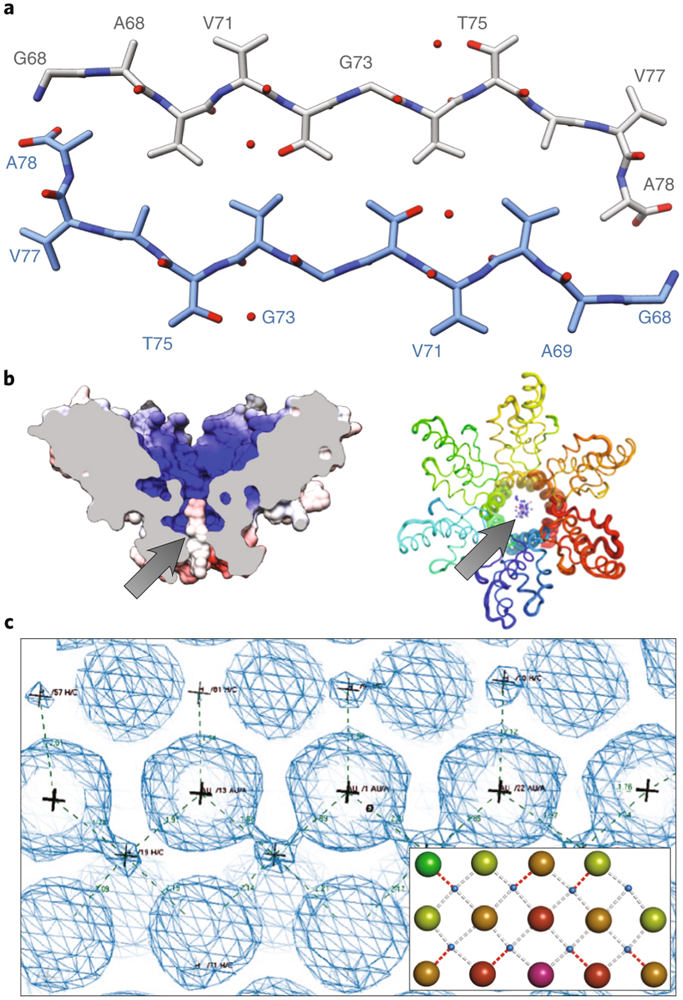
(A) The NACore fragment from α-synuclein was determined to 1.4 Å resolution. (B) Gag-bevirimat MicroED structures (side and top views shown with a surface and ribbon representation, respectively), where the location of the bevirimat (arrows) and its interactions with the Gag complex could be determined. (C) 2Fo-Fc density map (contoured at 2.0σ) from MicroED data of Au146(p-MBA)57 where rows of ligand stabilized gold atoms can be clearly seen. The modeled atoms are shown in the inset.
Table 1.
Examples of MicroED structures
| Sample | Year | Resolution | Ref. | Commentary | |
|---|---|---|---|---|---|
| Lysozyme (14.4 kDa) |
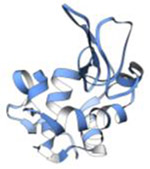 |
2013 | 2.9 Å | 7 | First structure determined by electron diffraction of 3D crystals. PDB ID: 3J4G |
| Lysozyme (14.4 kDa) |
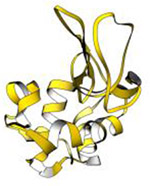 |
2014 | 2.5 Å | 8 | First structure determined by continuous rotation data collection, which currently is the standard MicroED method of data collection. PDB ID: 3J6K |
| Catalase (245 kDa) |
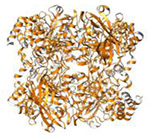 |
2014 | 3.2Å | 33 | These microcrystals resisted structure determination for several decades. PDB ID: 3J7B |
| Sup35 prion protein core (905 Da) |
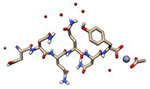 |
2016 | 1.0 Å | 34 | One of the first structures with phases determined by direct methods. PDB ID: 5K2E |
| Trypsin (23.4 kDa) |
 |
2017 | 1.7 Å | 24 | Microcrystals were generated by fragmenting larger crystals. PDB ID: 5K7R |
| Xylanase (21.1 kDa) |
 |
2017 | 2.3 Å | 24 | Microcrystals were generated by fragmenting larger crystals. PDB ID: 5K7P |
| Thaumatin (22.2 kDa) |
 |
2017 | 2.5 Å | 24 | Microcrystals were generated by fragmenting larger crystals. PDB ID: 5K7Q |
| TGF-b/TbRII complex (22.9 kDa) |
 |
2017 | 2.9 Å | 24 | First structure of a protein complex determined by MicroED. PDB ID: 5TY4 |
| Au146(p-MBA)57 nanoparticle (37.5 kDa) |
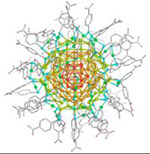 |
2017 | 0.85 Å | 36 | Structure of a gold nanoparticle determined by MicroED |
| Proteinase K (28.9 kDa) |
 |
2018 | 1.7 Å | 83 | Ultra-low dose MicroED structure. PDB ID: 6CL7 |
| Bank vole prion protein segment (1.1 kDa) |
 |
2018 | 0.75 Å | 39 | One of the highest resolution MicroED structures to date. PDB ID: 6AXZ |
| Carbamazepine (236 Da) |
 |
2018 | 0.85 Å | 39, 67, 91 | Structure of the drug, carbamazepine determined by MicroED |
MicroED is different than other electron diffraction techniques that have been reported over the years such as automated diffraction tomography (ADT)27 and rotation electron diffraction (RED)28. ADT uses discrete tilting of the goniometer together with beam precession27, while RED uses discrete goniometer tilts combined beam tilting to sample reciprocal space, and both techniques require specialized software to control data collection and process the data that are collected. These techniques were developed and used on a variety of inorganic and organic samples from materials science29-36, which can generally tolerate a higher electron dose than biological materials. In contrast, MicroED was developed for structure determination of proteins and radiation sensitive biological material. MicroED uses very low dose rates (~0.01 e-/Å2/s) and relies only on stage rotation to collect data in a manner analogous to rotation method in X-ray crystallography37, which facilitates data processing by standard crystallographic programs38. Although MicroED was initially developed for biological material it has now also been used successfully on small molecules39, 40 and inorganic material41.
X-ray crystallography methods and MicroED.
The growth of large and well-ordered crystals represents the largest bottleneck for structure determination by crystallographic approaches, especially for difficult samples such as membrane proteins and protein complexes42. Therefore, new methods that are capable of producing diffraction data from smaller and smaller crystals are of great interest to the structural biology community. Additionally, it has been shown that for some systems small crystals are better ordered and produced higher quality diffraction data than larger crystals. One approach relies on the more intense and smaller microfocus beamlines at synchrotron facilities, which allows the use of samples smaller than tens of microns43; however, the high-flux of the microfocus beam can cause issues with radiation damage from these small crystals44. X-ray Free Electron Lasers (XFEL) use a much brighter beam than traditional synchrotrons and as a result meaningful data can be obtained from crystals down to ~1 μm, after which the small crystals are usually destroyed following only a single exposure45. However, because the pulse is so short, in the femtosecond range, the diffraction data is collected before the results of damage are seen. In this method, each crystal only produces one image; therefore, XFEL data usually consists of partially recorded reflections and data from thousands of crystals must be collected and merged to yield a complete data set. While XFELs are producing exciting and impactful results46, the hardware required is extremely expensive to build, maintain, and use, limiting the accessibility of this technique.
Relative to X-rays, electrons deposit significantly less damaging energy into a crystal per useful elastic scattering event47. For this reason, electron diffraction data can be collected from extremely small micro and nanocrystals at an ultra-low dose that allows many exposures from a single crystal. MicroED data collection, described in more detail below, typically allows for the collection of up to a 140° wedge of data from a single crystal, and the data recorded are of mostly fully measured intensities. The measurement of full reflections leads to improved data processing and high-quality final data relative to the collection of only partial reflections8. If the crystal orientation and symmetry are favorable, data collection from a single crystal can be enough to determine a high-quality structure8, 48. The ability to determine structures from a single crystal, or from merging a small number of crystals greatly enhances the throughput of MicroED relative to other multi-crystal X-ray techniques. In cases where crystals diffract well in the cryo-EM, the entire process of data collection and structure solution can be performed in under an hour. Moreover, the hardware required for a MicroED experiment is orders of magnitude cheaper than for XFELs, the cost per hour of operation and the amount of sample needed are likewise orders of magnitude lower. The resolutions obtained from several samples39, 41, 49-52 have been higher than with XFELs, showing that MicroED is a competitive method in microcrystallography.
MicroED data collection and analysis
The workflow for MicroED has elements similar to both cryo-EM and X-ray crystallography. The electron microscopist will be familiar with the grid preparation and operation of the cryo-EM, while X-ray crystallographers will be accustomed to the procedures of the crystal growth, data processing and refinement steps of MicroED. Data collection methods are also similar to the rotation method used in X-ray crystallography. In depth protocols on MicroED data collection and processing have been published previously37, 38, and readers are encouraged to also consult these for guidance on using MicroED.
As with any crystallographic technique, the initial step of MicroED is the identification of diffraction quality crystals and sample preparation for irradiation. Light microscopes can be used to identify conditions that produce nanocrystals but with difficulty. This is because in certain cases the nanocrystals used for MicroED may be ‘invisible’ when viewed by a conventional light microscope. Improved methods for crystal identification involve the use of UV fluorescence (Fig. 5) or in some cases Second-Order Nonlinear Optical Imaging of Chiral Crystals (SONICC)53, which are each capable of identifying potential crystals in the nanometer scale. If conditions are identified with potential microcrystals, negative stain electron microscopy can be used to confirm the presence and quality of the microcrystals present in the drop54-56. However, it is important to note that the negative stain may be incompatible with certain crystallization buffers or may deteriorate the quality of the crystals. Therefore, visualizing high-quality microcrystals by negative stain indicates that MicroED studies are likely to be successful, but the absence of well-ordered crystals does not necessarily indicate the quality of the crystals is poor. A useful alternative is to freeze a crystal slurry on a sample loop and utilize powder-type X-ray diffraction at a synchrotron. No coherent lattice will be observed but if diffraction rings are seen, the presence of protein nanocrystals in the solution as well as the potential resolution can be confirmed. Additionally, it has been shown that larger, imperfect crystals can be broken down by a variety of methods (e.g vortexing, sonication or vigorous pipetting) to produce crystal fragments of suitable size for MicroED24.
Figure 5: Crystal identification and sample preparation for MicroED.
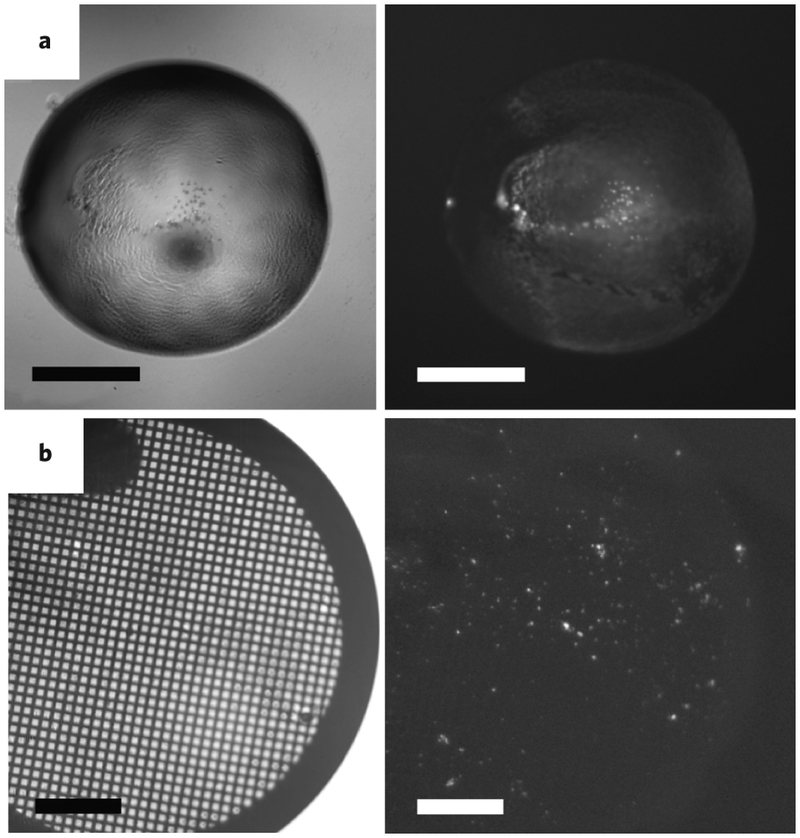
(A) Frequently, identification of microcrystals in drops that appear to have cloudy precipitates is difficult by visible light (left panel); however, when the drops are imaged using UV fluorescence, the presence of microcrystals is clearly seen by sharp glowing spots (right panel). (B) UV fluorescence can also be used to visualize the presence of microcrystals deposited on the EM grid during sample preparation for EM. Scale bars represent 500 μm.
Once nanocrystals suitable for MicroED are identified, samples are prepared by vitrification methods analogous to those used for other cryo-EM techniques. As with all cryo-EM modalities, grid preparation for imaging is difficult and empirical. Grid preparation and optimization can often be the most delicate and time-consuming step of a cryo-EM experiment, especially for sensitive samples, and generalized procedures for difficult targets (e.g. membrane proteins) are still an area of development. Samples must be blotted until they are thin enough for the electron beam to penetrate, yet the sample must also not be too disturbed during the process. Blotting times and often the use of low viscosity cryo-protectants (e.g. 2-Methyl-2,4-pentanediol) must be optimized for MicroED experiments. Generally, samples are applied to a holey carbon grid that has been glow-discharged on both sides. The grids are then blotted and vitrified in liquid ethane using either a manual or automated plunge-freezing device. Even though holey carbon grids are used, unlike for single particle cryo-EM, it is not necessary that the crystals be in the holes. Indeed, most MicroED data are collected from crystals on the carbon surface rather than in the holes. For MicroED, these carbon grids are used to help with efficient blotting rather than imaging in the diffraction experiments. Following vitrification, cryo-EM grids may be stored indefinitely under liquid nitrogen until the user is ready to perform MicroED.
An alternative promising method for preparing appropriate crystalline specimens for MicroED is to use a focused ion beam under cryogenic conditions (cryo-FIB) to mill thicker specimens down to the hundreds of nanometer thicknesses for diffraction data collection. For this approach, nanocrystals are placed directly on the EM grid without blotting, the grids plunged into ethane and crystals milled using the FIB to the desired thickness (Fig. 6). After milling, the grids are transferred into a TEM for MicroED data collection. Because cryo-FIB milling would help alleviate the challenges associated with the use of viscous samples and with sample damage during blotting, once the availability of cryo-FIB systems become more widespread, we predict that this will be a standard method by which samples are prepared for MicroED in the future57.
Figure 6. Cryo-FIB milling of a thick crystal.
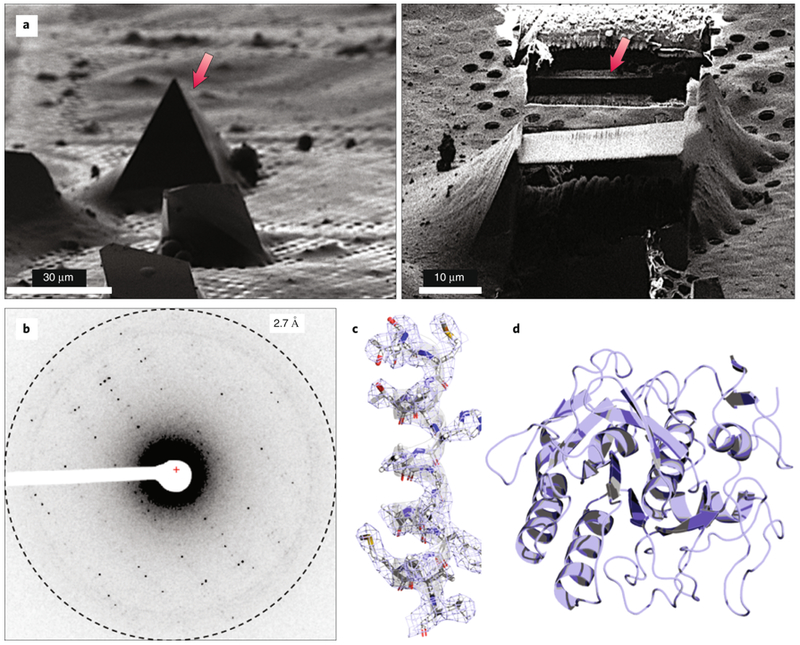
(A) Sample preparation for MicroED using a cryo-FIB to mill down thick crystals to a few hundred nanometers (left and right). (B) Following cryo-FIB milling, the grid would be loaded into the TEM and diffraction data would be collected from the thin lamella. (C-D) Final structure of proteinase K.
The cryo-TEM must be set up appropriately for MicroED data collection. Most modern cryo-TEMs equipped with quality detectors for diffraction (described more below) are capable of collecting MicroED data, and as with all other cryo-EM techniques, the cryo-TEM must be well-aligned and calibrated. A Field Emission Gun (FEG) as an electron source is highly desirable for increased beam coherence, together with a well calibrated sample stage. An energy filter helps by cutting out the inelastic scattering, which effectively reduces noise and helps identify faint reflections from weakly diffracting crystals. As such, the energy filter is important not only for data collection but also for screening (Fig. 7).
Figure 7: Comparison of Proteinase K data collected with and without an energy filter.
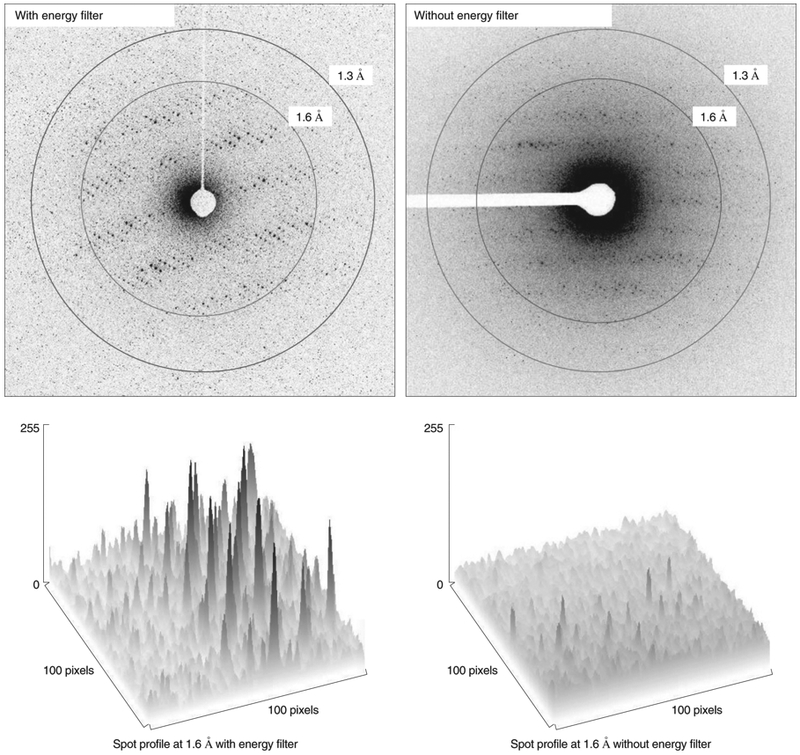
The zero-loss data collected on a cryo-TEM equipped with an in-column energy filter shows much less diffuse scattering at lower resolutions.
Setting up the microscope for MicroED data collection has been described previously37.Following this, the next step is to search the grid at low magnification to identify the location of suitable crystals on the grid. High-quality crystals show clear defined edges and are well-separated from other crystals on the grid. If a promising crystal is identified, a test diffraction pattern should be collected to determine the diffraction quality of that crystal: high-quality crystals will diffract to high-resolution and show well-separated reflections. Identifying well diffracting crystals and trouble-shooting has been described before in detail58. We recommend that new MicroED users become familiar with the method and what is considered high-quality diffraction data by testing standard samples. Common protein samples that have used to benchmark MicroED instruments and protocols are lysozyme, catalase, and proteinase K 8, 23, 48. Of these samples, we prefer to use proteinase K as a higher number of crystals on the grid show high-quality diffraction.
A diffraction data set is next collected from crystals that were deemed high quality. The first step is to set the eucentric height accurately to ensure that the crystal remains in the beam as the sample is tilted throughout the rotation range of the stage. For a well aligned sample stage the crystal can remain in the beam over an entire 140° rotation (+/− 70°). To collect continuous rotation data, the stage is smoothly and uni-directionally rotated at a constant rate while the detector constantly collects the diffraction data as a movie. The detector readout system must be fast enough so that the delay between writing out the frames of the movie is negligible. The camera used for most MicroED structures is the CMOS based TVIPS F416 while other CMOS detectors with fast readouts such as the Thermo Fisher CetaD, Gatan OneView, or TVIPS XF416 are also suitable and have been used. These cameras are widely available and are much less expensive than the direct electron detectors used for imaging.
After MicroED data are collected, the data are integrated, scaled and merged using software developed for X-ray crystallography, which make it relatively straightforward for users with this experience. We recommend that users perform MicroED data integration with whatever program they feel most comfortable. The following suites have been tested and used: MOSFLM59, 60, XDS61, HKL200062 and DIALS63, 64. It should be noted that the initial indexing of the crystal is different in MicroED than in X-ray crystallography. Because the de Broglie wavelength of the electrons is significantly shorter than the wavelength for X-rays (e.g. 0.025 Å for electrons in a 200kV TEM vs. 1.54 Å for X-rays from a copper source), the Ewald sphere is essentially flat at the resolutions typical for protein crystallography. This produces diffraction patterns that sample planes of reciprocal space, with very little information about the 3rd dimension in a single image. Therefore, MicroED requires multiple diffraction patterns that span a tilt range of at least 20° to obtain the 3D information necessary to index and determine the crystal orientation successfully without any prior knowledge. Following data integration, merging, and scaling, standard crystallographic software suites such as CCP465 and PHENIX66 are used to phase the data and refine the model. Aside from high-resolution peptides, small molecules and metals, which can be phased by direct methods, all MicroED structures have been phased thus far by molecular replacement. Refinement, typically performed with REFMAC67 or phenix.refine66, is done with the same programs and procedures used for X-ray crystallographic data, with the exception of the use of electron scattering factors rather than X-ray scattering factors. The ability to use electron scattering factors within REFMAC and phenix.refine is already a feature that has been added by the program developers.
MicroED structure determination highlights.
Since its inception, MicroED has been used to determine several structures that were previously unsolved despite major effort, for example, the structures of fragments derived from amyloidogenic proteins6. The hallmark of amyloid diseases is the formation of amyloid fibrils, which are composed of proteins (e.g. Aβ in Alzheimer’s and α-synuclein in Parkinson’s) that have adopted an aggregation prone β-sheet morphology. Determining the structure of these fibrils and their oligomeric precursors is key to the understanding of the molecular mechanisms of amyloid disease progression and the design of novel therapeutics. Peptide fragments from amyloidogenic proteins composed of 4 to 7 residues could produce large crystals suitable for analysis by X-ray crystallography68-70. However, as the number of residues increases the size of the crystals is reduced and limited presumably due to the increased strain associated with the twisting of the β-sheets. The 11-residue fragment from α-synuclein, which is called the NACore, yielded crystals which were only a few nanometers in size and had resisted structure determination for over a decade. Multiple researchers over the years were unsuccessful at growing these nanocrystals to a size that would allow X-ray diffraction. Milliliters of these nanocrystals were sent for analysis at XFELs, but even XFELs failed to deliver a structure. These α-synuclein nanocrystals were the first novel samples investigated by MicroED, yielding a 1.4 Å data set and a fully refined structure in just a few days71 (Fig. 4A). Since this first structure of the NACcore was published, MicroED has been used to solve a number of other amyloidogenic structures using crystals too small for other techniques, including those from IAPP, Aβ, Tau, and TDP-43 and the low complexity motif in FUS24, 25, 39, 49-52, 72, 73.
More recently MicroED was used to shed light on HIV maturation, which involves the cleavage of the Gag capsid protein by protease74. As gag is cleaved, the capsid undergoes a conformational change from a sphere to a bullet shape, a hallmark indicator of a fully infectious HIV particle. Bevirimat is a known maturation inhibitor that binds to HIV gag and prevents the cleavage by protease. However, the effectiveness of bevirimat is low and efforts at further optimizing the drug binding to HIV Gag were hindered by a lack of understanding of how bevirimat binds Gag and how it prevents protease cleavage. The structure of HIV-1 Gag was determined by X-ray crystallography producing a 3.2 Å structure75; however, co-crystallization with bevirimat failed to produce crystals and incubation of pre-formed crystals with the drug led to a severe deterioration of the crystals preventing structure determination by X-ray crystallography. In contrast, nanocrystals of Gag diffracted in MicroED to 2.8 Å and addition of bevirimat to these pre-formed nanocrystals likewise yielded a 2.8 Å resolution data set. The MicroED structure of Gag with and without bevirimat indicate that a single bevirimat molecule bound at the 6-fold axis of Gag acts as an allosteric inhibitor preventing cleavage of Gag by protease (Fig. 4B). Drug optimization can now begin to make bevirimat a better binder in the 6-fold symmetric binding pocket in order to increase the effectiveness of this inhibitor.
MicroED has also been applied to the study of inorganic nanomaterials to determine the structure of a ligand capped gold nanoparticle41. Such gold clusters have important applications in nanotechnology, cancer therapy and targeted drug delivery. The structure of the gold cluster Au146(p-MBA)57 was determined by MicroED to 0.85 Å resolution and the structure confirmed by X-ray analysis, although at a lower resolution of 1.2 Å (XFELs delivered 1.5 Å only). The structure showed that the gold nanoparticle is organized as a twinned FCC cluster and revealed how the ligands interact with and stabilize the surface. Ordered rows of gold-hydrogen bonds were observed shedding light on the mechanism by which this nanocage is stabilized (Fig. 4C). This work also demonstrated the importance of collecting samples under cryogenic conditions in a frozen-hydrated state when hydration is important, even for materials science samples. When the nanoparticle crystals were dehydrated prior to data collection, it was not possible to collect diffraction data from them. However, when the samples were prepared in the frozen-hydrated state by standard MicroED sample preparation techniques, the crystals diffracted to very high-resolution and the structure could be determined.
Opportunities for future development
Time resolved structural studies:
A significant advantage to using nanocrystals in crystallography is that time-resolved studies that involve the diffusion of molecules into the crystal is feasible and limited only by diffusion rates76. While diffusion of small molecules into large crystals is inefficient (on the time scale of seconds when crystals have dimensions in the hundreds of μm) resulting in low occupancy and lack of detail in density maps, diffusion into nanocrystals is extremely efficient (microsecond time scale) resulting in high occupancy and density maps with excellent details. Protein dynamics in electron crystallography have been performed for decades using 2D crystals14, 77-79 and similar protocols can be used for MicroED. The time-resolution for these studies is limited to the microsecond time scales by the rate of vitrification. While XFELs can look at reactions with femtosecond time resolution80, 81, many biological processes are much slower, in the microsecond to second time scales (e.g. ligand binding and enzymatic reactions) providing an immense opportunity for time resolved studies by MicroED. Synchrotrons can also be used to look at dynamics as this time scale as well, however diffusion of ligand into large crystals is inefficient and slow so using extremely small crystals and MicroED is a more attractive alternative. New sample preparation tools and methods, which allow fine control over the timing of sample mixing, application, and blotting will greatly facilitate time-resolved MicroED studies, and these are ongoing areas of methods development. The ability of MicroED to determine structures from a small number of crystals would make these kinds of time-resolved studies much more efficient and feasible for systems where the quantity of available sample is limiting. Additionally, because electrons are sensitive to chemical bonding82 MicroED could one day be used to directly visualize the bonding interactions in a time-dependent experiment.
Conformational changes have been observed in MicroED experiments in at least two examples. For Proteinase K, dynamics were observed in response to increasing exposure to the electron beam83. At the start of the experiment intact disulfide bridges were observed, but as the exposures increased these bonds broke apart, accompanied with a movement of the cysteine side chains away from one another (Fig. 8), which mirrors the observations seen in X-ray crystallography84. In another example, also described above, the HIV gag protein structure was determined with and without the maturation inhibitor bevirimat74. This small molecule was added immediately prior to vitrification and could diffuse efficiently into preformed Gag nanocrystals before damaging the lattice and allowing the structure of both gag alone as well as gag-bevirimat to be determined to help with future drug optimization efforts.
Figure 8: Dynamics probed in response to radiation damage.
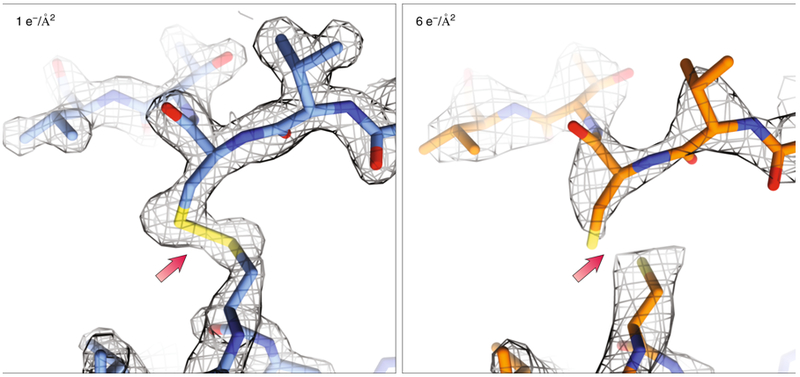
When less than 1 e−/Å2 (left) was used for structure determination of Proteinase K (1.7Å, PDB ID: 6CL7), local radiation damage was minimal. When higher doses (right) were used (2.8Å, PDB ID: 6CLA) the damaging effects of the beam can already be seen with the breakage of disulfide bonds and the lower attainable resolution. Density maps (2Fo-Fc) in gray are contoured at 1.5σ.
Realizing the damaging effects of the electron beam:
Recently a thorough analysis of radiation damage in cryo-EM was performed using MicroED83 (Fig. 8). Because sufficient data can be collected using exposures to ultralow electron doses, multiple complete high-resolution data sets can be collected from single crystals. Using very well diffracting crystals of Proteinase K, it was shown that specific radiation damage was already observed with extremely low total exposures of ~1 e-/Å2. A challenge to the future will be the creation of even more sensitive detectors and new electron sources to collect data faster such that the additive effects of radiation damage become less significant, while at the same time providing the necessary high-resolution data for required for both cryo-EM imaging and diffraction techniques.
Phasing methods in MicroED:
Most MicroED structures have been determined using molecular replacement with homologous proteins as search models8, 23-25, 48, 71, 83, 85. Direct methods were also successful in phasing MicroED data when the obtained resolution nears 1 Å39, 49, 72 using SHELX86; however, it is only applicable in the cases where the samples are exceptionally well-diffracting. Therefore, the development of experimental phasing methods that can be used to determine novel structures where molecular replacement is not possible, but where the obtainable resolution is limited to ~2–3 Å, will be highly welcomed. Imaging of the crystals or tomographic reconstructions are obvious steps forward, taking advantage of the unique properties of the TEM. By using images, a moderate resolution molecular envelop of the sample within the crystal could be determined, which then could be used to phase the MicroED data. These efforts are currently underway in several laboratories87, 88. Heavy metal phasing is another avenue of active research. While this approach has been extremely successful in X-ray crystallography, no anomalous signal nor absorption edge exists for compounds at the wavelengths of the electron microscope; therefore, only isomorphous replacement can be used for MicroED.
High-throughput small molecule structure determination:
As with biological molecules, electron diffraction can be extended to the study of organic molecules that form very small microcrystals40, 82, 89-91. Recently, a study was presented which used MicroED to determine the atomic resolution structures of 11 small molecules40. These structures were determined directly from powders with no additional crystallization optimization, and the process from sample preparation to structure solution was very quick. Following the deposition of the samples on to the grid, data collection and processing was done in the same manner as described for biological crystals. The extension of MicroED to organic molecules offers a new analytical tool for studying the synthesis of new molecules. Future work that focuses on integrating the MicroED and organic syntheses pipelines promises to make this a routine method in chemistry labs.
Facilities, cameras and automation:
As the use of cryo-EM methods such as MicroED become increasingly widespread, the need for additional centralized centers and facilities will increase. The equipment used for MicroED is essentially the same as for other cryo-EM modalities. For protein work it is important to have an FEG installed, a sensitive and fast camera, and if possible, an energy filter. Camera technology is quickly progressing and the use of direct electron detectors for MicroED is already starting. Cameras such as the Falcon3 should provide ultimate flexibility since this camera could be used for all modalities in cryo-EM including MicroED, while the other cameras appropriate for diffraction such as the hybrid pixel detectors (e.g. Timepix and EIGER90-92) in their current configuration are not well suited for imaging and single particle EM.
Automation is incredibly important and will be vital for MicroED as it has greatly improved productivity for single particle EM. Protocols and software have been already developed for MicroED automation, including an adaptation to SerialEM93, but a complete suite with a friendly GUI is still lacking and is currently being developed by several laboratories. In the near future, users could ship samples to the user facilities, and following sample loading, could screen and collect MicroED data remotely from their home institutions similar to the way most X-ray synchrotron facilities operate. With the current implementation of SerialEM more than 300 data sets can be collected overnight automatically93. Because of the synergy among all modalities in cryo-EM user facilities, we expect that MicroED will have a broad impact on structural biology in the coming decades.
Acknowledgements
We would like to thank all of our collaborators and trainees who have contributed either directly or indirectly to the development of MicroED. We thank Guillermo Calero (U. Pittsburgh) for providing figures 4c and 5b. The Gonen laboratory is supported by funding from the Howard Hughes Medical Institute. The Nannenga laboratory is supported by the National Institutes of Health (R01GM124152). MicroED was developed at the Janelia Research Campus of HHMI using HHMI funds and Janelia Visitor Program funds.
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Cheng Y, Grigorieff N, Penczek PA & Walz T A primer to single-particle cryo-electron microscopy. Cell 161, 438–449 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck M & Baumeister W Cryo-Electron Tomography: Can it Reveal the Molecular Sociology of Cells in Atomic Detail? Trends Cell Biol 26, 825–837 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Wisedchaisri G, Reichow SL & Gonen T Advances in structural and functional analysis of membrane proteins by electron crystallography. Structure 19, 1381–1393 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glaeser RM Review: electron crystallography: present excitement, a nod to the past, anticipating the future. J Struct Biol 128, 3–14 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Nannenga BL & Gonen T MicroED opens a new era for biological structure determination. Curr Opin Struct Biol 40, 128–135 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez JA, Eisenberg DS & Gonen T Taking the measure of MicroED. Curr Opin Struct Biol 46, 79–86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi D, Nannenga BL, Iadanza MG & Gonen T Three-dimensional electron crystallography of protein microcrystals. Elife 2, e01345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- *8.Nannenga BL, Shi D, Leslie AG & Gonen T High-resolution structure determination by continuous-rotation data collection in MicroED. Nat Methods 11, 927–930 (2014).This work introduced the “continuous rotation” method of MicroED data collection, which is currently the standard procedure for data collection.
- 9.Gonen T, Sliz P, Kistler J, Cheng Y & Walz T Aquaporin-0 membrane junctions reveal the structure of a closed water pore. Nature 429, 193–197 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Gonen T et al. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature 438, 633–638 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henderson R & Unwin PN Three-dimensional model of purple membrane obtained by electron microscopy. Nature 257, 28–32 (1975). [DOI] [PubMed] [Google Scholar]
- 12.Henderson R et al. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol 213, 899–929 (1990). [DOI] [PubMed] [Google Scholar]
- 13.Grigorieff N, Ceska TA, Downing KH, Baldwin JM & Henderson R Electron-crystallographic refinement of the structure of bacteriorhodopsin. J Mol Biol 259, 393–421 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Subramaniam S & Henderson R Crystallographic analysis of protein conformational changes in the bacteriorhodopsin photocycle. Biochim Biophys Acta 1460, 157–165 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Kuhlbrandt W, Wang DN & Fujiyoshi Y Atomic model of plant light-harvesting complex by electron crystallography. Nature 367, 614–621 (1994). [DOI] [PubMed] [Google Scholar]
- 16.Unwin PN & Henderson R Molecular structure determination by electron microscopy of unstained crystalline specimens. J Mol Biol 94, 425–440 (1975). [DOI] [PubMed] [Google Scholar]
- 17.Dorset DL & Parsons DF Electron-Diffraction from Single, Fully-Hydrated, Ox Liver Catalase Microcrystals. Acta Crystallographica Section A A 31, 210–215 (1975). [Google Scholar]
- 18.Dorset DL & Parsons DF Thickness Measurements of Wet Protein Crystals in Electron-Microscope. J Appl Crystallogr 8, 12–14 (1975). [Google Scholar]
- 19.Taylor KA & Glaeser RM Electron microscopy of frozen hydrated biological specimens. J Ultrastruct Res 55, 448–456 (1976). [DOI] [PubMed] [Google Scholar]
- 20.Jiang LH, Georgieva D, Zandbergen HW & Abrahams JP Unit-cell determination from randomly oriented electron-diffraction patterns. Acta Crystallogr D 65, 625–632 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nederlof I, van Genderen E, Li YW & Abrahams JP A Medipix quantum area detector allows rotation electron diffraction data collection from submicrometre three-dimensional protein crystals. Acta Crystallogr D Biol Crystallogr 69, 1223–1230 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iadanza MG & Gonen T A suite of software for processing MicroED data of extremely small protein crystals. J Appl Crystallogr 47, 1140–1145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hattne J, Shi D, de la Cruz MJ, Reyes FE & Gonen T Modeling truncated pixel values of faint reflections in MicroED images. J Appl Crystallogr 49, 1029–1034 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- *24.de la Cruz MJ et al. Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED. Nat Methods 14, 399–402 (2017).Here several methods for the fragmentation of larger, and in some cases poorly ordered, crystals into microcrystals suitable for MicroED are described.
- 25.Luo F et al. Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation. Nat Struct Mol Biol 25, 341–346 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Duyvesteyn HME et al. Machining protein microcrystals for structure determination by electron diffraction. Proc Natl Acad Sci U S A 115, 9569–9573 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolb U, Mugnaioli E & Gorelik TE Automated electron diffraction tomography – a new tool for nano crystal structure analysis. Crystal Research and Technology 46, 542–554 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Wan W, Sun JL, Su J, Hovmoller S & Zou XD Three-dimensional rotation electron diffraction: software RED for automated data collection and data processing. J Appl Crystallogr 46, 1863–1873 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mugnaioli E et al. Ab Initio Structure Determination of Vaterite by Automated Electron Diffraction. Angew Chem Int Edit 51, 7041–7045 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Feyand M et al. Automated diffraction tomography for the structure elucidation of twinned, sub-micrometer crystals of a highly porous, catalytically active bismuth metal-organic framework. Angew Chem Int Ed Engl 51, 10373–10376 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Jiang JX et al. Synthesis and Structure Determination of the Hierarchical Meso-Microporous Zeolite ITQ-43. Science 333, 1131–1134 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Gorelik TE, Stewart AA & Kolb U Structure solution with automated electron diffraction tomography data: different instrumental approaches. J Microsc 244, 325–331 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Mugnaioli E, Gorelik T & Kolb U “Ab initio” structure solution from electron diffraction data obtained by a combination of automated diffraction tomography and precession technique. Ultramicroscopy 109, 758–765 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Simancas J et al. Ultrafast Electron Diffraction Tomography for Structure Determination of the New Zeolite ITQ-58. Journal of the American Chemical Society 138, 10116–10119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C et al. An Extra-Large-Pore Zeolite with 24×8×8-Ring Channels Using a Structure-Directing Agent Derived from Traditional Chinese Medicine. Angewandte Chemie International Edition 57, 6486–6490 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Zhang YB et al. Single-Crystal Structure of a Covalent Organic Framework. Journal of the American Chemical Society 135, 16336–16339 (2013). [DOI] [PubMed] [Google Scholar]
- *37.Shi D et al. The collection of MicroED data for macromolecular crystallography. Nat Protoc 11, 895–904 (2016).Detailed protocols for the collection of MicroED data. New users are encouraged to read these protocols prior to using MicroED.
- 38.Hattne J et al. MicroED data collection and processing. Acta Crystallogr A Found Adv 71, 353–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallagher-Jones M et al. Sub-angstrom cryo-EM structure of a prion protofibril reveals a polar clasp. Nat Struct Mol Biol 25, 131–134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones CG et al. The CryoEM Method MicroED as a Powerful Tool for Small Molecule Structure Determination. ACS Central Science (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vergara S et al. MicroED Structure of Au146(p-MBA)57 at Subatomic Resolution Reveals a Twinned FCC Cluster. J Phys Chem Lett (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fromme P & Spence JC Femtosecond nanocrystallography using X-ray lasers for membrane protein structure determination. Curr Opin Struct Biol 21, 509–516 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith JL, Fischetti RF & Yamamoto M Micro-crystallography comes of age. Curr Opin Struct Biol 22, 602–612 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto M et al. Protein microcrystallography using synchrotron radiation. Iucrj 4, 529–539 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin-Garcia JM, Conrad CE, Coe J, Roy-Chowdhury S & Fromme P Serial femtosecond crystallography: A revolution in structural biology. Arch Biochem Biophys 602, 32–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spence JCH X-ray lasers for structure and dynamics in biology. IUCrJ 5, 236–237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henderson R The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q Rev Biophys 28, 171–193 (1995). [DOI] [PubMed] [Google Scholar]
- 48.Nannenga BL, Shi D, Hattne J, Reyes FE & Gonen T Structure of catalase determined by MicroED. Elife 3, e03600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sawaya MR et al. Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED. Proc Natl Acad Sci U S A (2016).The first structure determined by MicroED that was phased experimentally. In this study, the high-resolution and MicroED data quality allowed structure solution by direct methods
- 50.Krotee P et al. Atomic structures of fibrillar segments of hIAPP suggest tightly mated beta-sheets are important for cytotoxicity. Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seidler PM et al. Structure-based inhibitors of tau aggregation. Nat Chem 10, 170–176 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hughes MP et al. Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks. Science 359, 698–701 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kissick DJ, Wanapun D & Simpson GJ Second-order nonlinear optical imaging of chiral crystals. Annu Rev Anal Chem (Palo Alto Calif) 4, 419–437 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stevenson HP et al. Use of transmission electron microscopy to identify nanocrystals of challenging protein targets. Proc Natl Acad Sci U S A 111, 8470–8475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barnes CO et al. Assessment of microcrystal quality by transmission electron microscopy for efficient serial femtosecond crystallography. Arch Biochem Biophys 602, 61–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stevenson HP et al. Transmission electron microscopy for the evaluation and optimization of crystal growth. Acta Crystallogr D Struct Biol 72, 603–615 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martynowycz MW, Zhao W, Hattne J, Jensen GJ & Gonen T Collection of Continuous Rotation MicroED Data from Ion Beam-Milled Crystals of Any Size. Structure 27, 545–548.e542 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gonen T The collection of high-resolution electron diffraction data. Methods Mol Biol 955, 153–169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leslie AGW & Powell HR Processing diffraction data with mosflm. 245, 41–51 (2007). [Google Scholar]
- 60.Battye TG, Kontogiannis L, Johnson O, Powell HR & Leslie AG iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67, 271–281 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kabsch W Xds. Acta Crystallogr D Biol Crystallogr 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Otwinowski Z & Minor W [20] Processing of X-ray diffraction data collected in oscillation mode. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 63.Waterman DG et al. The DIALS framework for integration software. CCP4 Newsl, 16–19 (2013). [Google Scholar]
- 64.Clabbers MTB, Gruene T, Parkhurst JM, Abrahams JP & Waterman DG Electron diffraction data processing with DIALS. Acta Crystallogr D Struct Biol 74, 506–518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winn MD et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Murshudov GN, Vagin AA & Dodson EJ Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 68.Balbirnie M, Grothe R & Eisenberg DS An amyloid-forming peptide from the yeast prion Sup35 reveals a dehydrated beta-sheet structure for amyloid. Proc Natl Acad Sci U S A 98, 2375–2380 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nelson R et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature 435, 773–778 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sawaya MR et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 447, 453–457 (2007). [DOI] [PubMed] [Google Scholar]
- *71.Rodriguez JA et al. Structure of the toxic core of alpha-synuclein from invisible crystals. Nature (2015).First novel structure determined by MicroED. This work paved the way for the many new peptide structures from amyloidogenic proteins.
- 72.Guenther EL et al. Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation. Nat Struct Mol Biol 25, 463–471 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krotee P et al. Common fibrillar spines of amyloid-beta and human islet amyloid polypeptide revealed by microelectron diffraction and structure-based inhibitors. J Biol Chem 293, 2888–2902 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Purdy MD et al. MicroED structures of HIV-1 Gag CTD-SP1 reveal binding interactions with the maturation inhibitor bevirimat. Proceedings of the National Academy of Sciences, 201806806 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wagner JM et al. Crystal structure of an HIV assembly and maturation switch. Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schmidt M Mix and Inject: Reaction Initiation by Diffusion for Time-Resolved Macromolecular Crystallography. Adv Cond Matter Phys (2013). [Google Scholar]
- 77.Unwin N & Fujiyoshi Y Gating movement of acetylcholine receptor caught by plunge-freezing. J Mol Biol 422, 617–634 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Berriman J & Unwin N Analysis of transient structures by cryo-microscopy combined with rapid mixing of spray droplets. Ultramicroscopy 56, 241–252 (1994). [DOI] [PubMed] [Google Scholar]
- 79.Unwin N Acetylcholine receptor channel imaged in the open state. Nature 373, 37–43 (1995). [DOI] [PubMed] [Google Scholar]
- 80.Nogly P et al. Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser. Science 361, eaat0094 (2018). [DOI] [PubMed] [Google Scholar]
- 81.Pande K et al. Femtosecond structural dynamics drives the trans/cis isomerization in photoactive yellow protein. Science 352, 725–729 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu JS & Spence JC Structure and bonding in alpha-copper phthalocyanine by electron diffraction. Acta Crystallogr A 59, 495–505 (2003). [DOI] [PubMed] [Google Scholar]
- 83.Hattne J et al. Analysis of Global and Site-Specific Radiation Damage in Cryo-EM. Structure 26, 759–766 e754 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Garman EF Radiation damage in macromolecular crystallography: what is it and why should we care? Acta Crystallogr D Biol Crystallogr 66, 339–351 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nannenga BL, Iadanza MG, Vollmar BS & Gonen T Overview of electron crystallography of membrane proteins: crystallization and screening strategies using negative stain electron microscopy. Curr Protoc Protein Sci Chapter 17, Unit17 15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sheldrick G A short history of SHELX. Acta Crystallographica Section A 64, 112–122 (2008). [DOI] [PubMed] [Google Scholar]
- 87.Nederlof I, Li YW, van Heel M & Abrahams JP Imaging protein three-dimensional nanocrystals with cryo-EM. Acta Crystallogr D Biol Crystallogr 69, 852–859 (2013). [DOI] [PubMed] [Google Scholar]
- 88.van Genderen E, Li YW, Nederlof I & Abrahams JP Lattice filter for processing image data of three-dimensional protein nanocrystals. Acta Crystallographica Section D-Structural Biology 72, 34–39 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gorelik TE, van de Streek J, Kilbinger AF, Brunklaus G & Kolb U Ab-initio crystal structure analysis and refinement approaches of oligo p-benzamides based on electron diffraction data. Acta Crystallogr B 68, 171–181 (2012). [DOI] [PubMed] [Google Scholar]
- 90.Gruene T et al. Rapid structure determination of microcrystalline molecular compounds using electron diffraction. Angewandte Chemie 0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.van Genderen E et al. Ab initio structure determination of nanocrystals of organic pharmaceutical compounds by electron diffraction at room temperature using a Timepix quantum area direct electron detector. Acta Crystallogr A Found Adv 72, 236–242 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Clabbers MTB et al. Protein structure determination by electron diffraction using a single three-dimensional nanocrystal. Acta Crystallogr D Struct Biol 73, 738–748 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jason de la Cruz M, Martynowycz MW, Hattne J & Gonen T MicroED data collection with SerialEM. Ultramicroscopy (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Subramanian G, Basu S, Liu H, Zuo JM & Spence JC Solving protein nanocrystals by cryo-EM diffraction: multiple scattering artifacts. Ultramicroscopy 148, 87–93 (2015). [DOI] [PubMed] [Google Scholar]
- 95.Vincent R & Midgley PA Double conical beam-rocking system for measurement of integrated electron diffraction intensities. Ultramicroscopy 53, 271–282 (1994). [Google Scholar]
- 96.Midgley PA & Eggeman AS Precession electron diffraction - a topical review. IUCrJ 2, 126–136 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gjonnes J et al. Structure Model for the Phase AlmFe Derived from Three-Dimensional Electron Diffraction Intensity Data Collected by a Precession Technique. Comparison with Convergent-Beam Diffraction. Acta Crystallographica Section A 54, 306–319 (1998). [Google Scholar]
- 98.Fernández-Busnadiego R et al. Insights into the molecular organization of the neuron by cryo-electron tomography. Journal of Electron Microscopy 60, S137–S148 (2011). [DOI] [PubMed] [Google Scholar]
- 99.Bartesaghi A et al. 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor. Science 348, 1147–1151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- *100.Liu S & Gonen T MicroED structure of the NaK ion channel reveals a Na+ partition process into the selectivity filter. Communications Biology 1, 38 (2018).The first membrane protein determined by continuous rotation MicroED.


