Abstract
The intestinal microbiota is a dynamic community of bacteria, fungi, and viruses that mediates mucosal homeostasis and physiology. Imbalances in the microbiome and aberrant immune responses to gut bacteria can disrupt homeostasis and are associated with inflammatory bowel diseases (IBD) in humans and colitis in mice. We review genetic variants associated with IBD and their effects on the intestinal microbiome, the immune response, and disease pathogenesis. The intestinal microbiome, which includes microbial antigens, adjuvants, and metabolic products, affects the development and function of the intestinal mucosa and inflammatory responses in the gut. Strategies to manipulate the microbiome might therefore be used in treatment of IBD. We review microbe-based therapies for IBD and the potential to engineer patients’ intestinal microbiota. We discuss how studies of patients with IBD and mouse models have advanced our understanding of the interactions between genetic factors and the gut microbiome, and challenges to development of microbe-based therapies for IBD.
Inflammatory bowel diseases (IBD) include Crohn’s disease (CD) and ulcerative colitis (UC)— chronic diseases that develop via complex interactions among genetic, immune, environmental, and microbial factors.1–3 Dysregulation of any components of this network can result in intestinal inflammation and IBD. Genetic studies identified regulators of this network that are altered in patients with IBD—many of these control the immune response to microbes.4, 5 Variants associated with risk for IBD have been identified in NOD2, ATG16L1, CARD9, and CLEC7A.6 Variants in genes that control immune detection of and response to microbes can perturb intestinal homeostasis and promote intestinal inflammation. It is important to distinguish factors that mediate the immune response to pathogens from factors that control the overall microbial ecology, which can also be affected by environmental factors (diet, medications, geography).7 However, as we study the mechanisms by which genetic variants associated with IBD affect responses to microbes, we might learn more about environmental factors that also do so, and identify new targets for diagnosis and treatment of IBD.
Studies from model systems have indicated that the gut microbiome can be modified to increase or reduce the severity of intestinal inflammation. The gut microbiome can be altered by introduction of microbes or their effectors, such as lipids, small molecules, proteins, or sugars. Over the last decade, interest in microbe-based therapies has increased due to the number and perceived safety of these therapies, as well as the potential to correct one of the causes of a disease, rather than the symptoms. Increased interest in these therapies is partly due to insights from studies of antibiotics, probiotics, and more recently fecal microbial transplantation (FMT), for IBD and other disorders.8 Strategies to correct the microbiome or its functions in patients with UC or CD have produced inconsistent results, although antibiotics were found to be effective in patients with pouchitis, with an excellent safety profile.9–11 FMT was found to reduce symptoms in some patients with UC, although outcomes varied. Further studies are required to optimize selection of donors, determine the ability of the donated microbiota to engraft, and determine whether FMT might be better as an induction or maintenance therapy.
Studies are also needed to determine how variants in genes whose products function in microbe sensing pathways (such as NOD2) would affect microbial therapies. We review interactions between IBD-associated gene variants and the microbiome, and strategies to therapeutically target specific microbiome functions.9, 12 The growth of microbe-based therapies presents new challenges to drug development and regulatory approval.
Genes That Regulate the Microbiome
Genes encode many proteins that microbes are exposed to, as well as the availability of nutrients and the level of the immune response to microbes.13–16 Genome-wide association studies of patients with IBD have identified variants in genes that affect the intestinal response to microbes (Figure 1).6, 17–25
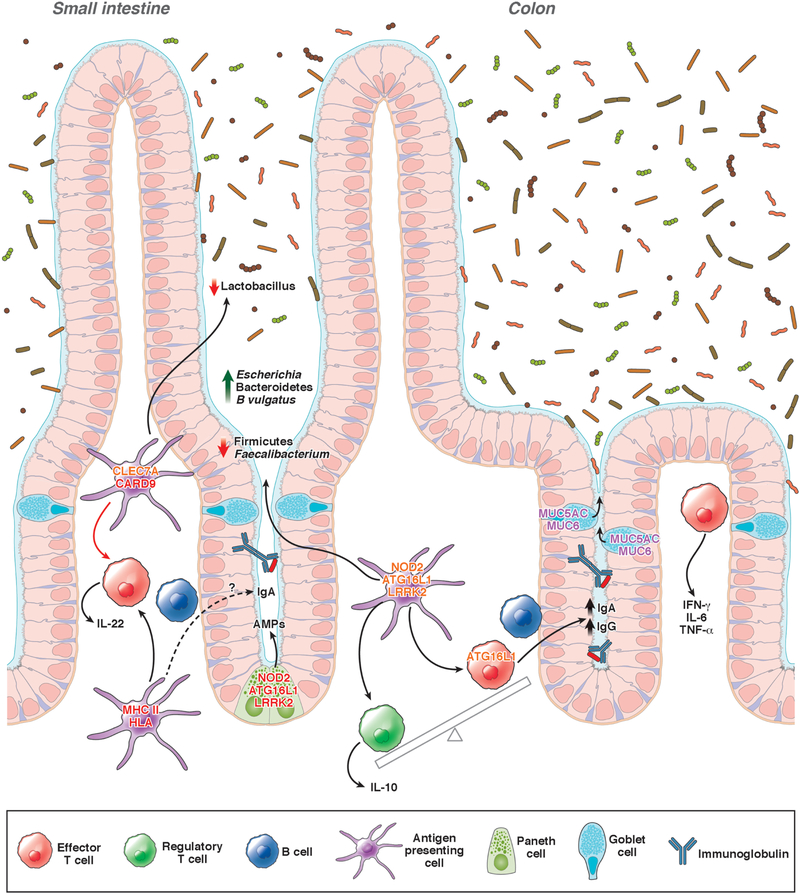
Figure 1. Genetic factors that affect the intestinal microbiome.
Variants genes that affect risk for IBD have been associated with alterations in the composition of the microbiome. Mutations in NOD2, ATG16L1, and LRRK2 reduce secretion of antimicrobial peptide (AMP) by Paneth cells. Variants in CLEC7A and CARD9 have been associated with decreased abundance of Lactobacillus, possibly due to altered activities of dendritic cells and macrophages. Variants in NOD2 are associated with increased abundance of Escherichia species and Bacteroides vulgatus and reductions in Faecalibacterium species. Impaired ATG16L1 signaling has been associated with increased production of IgG and IgA against commensal microbiota, resulting in a loss of tolerance to intestinal microbes. Polymorphisms in MHC class II or HLA genes affect production of IgA in response to microbes. Defects in mucus production alter the intestinal microbiome and increase susceptibility to colitis. Gene names in red have variants associated with CD and UC; gene names in orange have variants associated with only CD; and gene names in purple have variants associated with only UC.
Variants in NOD2
Variants in NOD2 were the first to be associated with risk of CD. NOD2 encodes an intracellular pattern recognition receptor, which interacts with peptidoglycan motifs of bacteria.26, 27,28 NOD2 helps control pathogenic bacteria through hematopoietic and non-hematopoietic cells. It was initially believed that individuals with some variants of NOD2 were unable to efficiently clear bacterial pathogens, leading to IBD pathology.29 This hypothesis is supported by the association in patient cohorts between gastrointestinal pathogens and IBD onset.30 However, it became apparent that NOD2 also mediates the immune response to non-pathogenic, commensal microbes. Patients with variants in NOD2 have microbiomes that are distinct from individuals without these variants, characterized by increased abundance of Escherichia species and reduced Faecalibacterium species, though this pattern can also be independent of NOD2 variants.15, 31–33 Nod2−/− mice have intestinal dysbiosis, which increase their susceptibility to colitis, compared with wild-type mice.32, 34–39 Researchers have identified commensal bacteria that are pathogenic in Nod2−/− mice (pathobionts), such as Bacteroides vulgatus. Mucosal barrier defects observed in Nod2−/− mice were linked to B vulgatus, including abnormalities in goblet cells, expression of inflammatory genes, and increased numbers of intraepithelial lymphocytes that express IFN gamma.37 Depletion of B vulgatus reversed the mucosal barrier defects in Nod2−/− mice, so targeted removal of organisms that exacerbate NOD2 signaling defects might restore intestinal barrier functions in patients with IBD.
Variants in ATG16L1 and autophagy
Several variants associated with risk of CD are in genes that regulate the autophagy pathway (such as ATG16L1, IGRM, and LRRK2). Autophagy has many functions, but one of its effects is to mediate lysosomal degradation and clearance of intracellular bacteria.18, 19, 40 Several studies have demonstrated that NOD2 interacts with ATG16L1 and that expression of CD-associated variants disrupts association between these proteins, impairing bacterial clearance and antigen presentation.41, 42 The variant encoding the T300A substitution in ATG16L1 increases susceptibility of the gene product to caspase-3 cleavage and reduces its function.43 Similar to Nod2−/− mice, mice hypomorphic for ATG16L1 have microbiota-dependent susceptibility to induction of colitis, as well as defects in toll-like receptor (TLR) signaling and production of antimicrobial peptides by Paneth cells. These abnormalities in TLR signaling and Paneth cell function have also been observed in patients with CD who are homozygous for the T300A substitution in ATG16L1.44, 45 In mice, disruption of the ATG16L1 gene affects CD4+ T cells, reducing numbers of intestinal Foxp3+ T-regulatory (Treg) cells and T-helper 2 (Th2) cell-mediated responses. These impaired T-cell functions contribute to disruption of the mucosal barrier, via loss of tolerance to intestinal antigens and increased production of IgG and IgA against commensal microbiota.46 Although many individuals carry IBD-associated variants in NOD2 and genes that regulate autophagy, only a small proportion develop IBD. Additional environmental factors and alterations to interactions between the intestinal epithelia and microbiota are therefore likely to be required for development of IBD.
Studies of mice and cells with deletion of ATG16L1 or NOD2 have found these proteins to mediate the effects of therapeutic microbes, by blocking immunomodulatory signals. For example, the common human commensal Bacteroides fragilis reduces colitis in mice by converting CD4+ T cells into Foxp3+ T-regulatory (Treg) cells that produce IL10.47, 48 This effect of B fragilis is lost when dendritic cells are defective in either NOD2 or ATG16L1 signaling.49 Human immune cells that express ATG16L1 T300A do not induce Treg cell development upon exposure to B fragilis.
Variants in CLEC7A and CARD9
Bacteria are the most well-defined microbes in the intestinal microbiota, but fungal communities are also altered in microbiomes of patients with IBD. This should not be surprising considering the associations between IBD and an aberrant immune response to fungal antigens, based on detection of antibodies to Saccharomyces cerevisiae.50 Similar to the intestinal bacterial communities in patients with IBD, the diversity of fungal microbiota is decreased and certain atypical phyla dominate, such Ascomycota and Basidiomycota.51 Fungal members of the gut microbiota interact with pattern recognition receptors such as CLEC7A (also called DECTIN1), a glycoprotein expressed by cells in the innate immune system that recognizes a beta-1,3-linked and beta-1,6-linked glucans from fungi. A single-nucleotide polymorphism in CLEC7A has been associated with IBD. Mice lacking DECTIN1 have increased susceptibility to colitis and an altered fungal community.52 Interestingly, some variants in CLEC7A have been associated with medically refractory ulcerative colitis; no other IBD-associated variants have been associated with response to therapy.52
DECTIN1 signals through the adaptor protein caspase recruitment domain containing protein 9 (CARD9).23 Card9−/− mice also have an altered fungal community structure with increased susceptibility to dextran sodium sulfate (DSS)-induced colitis.53 The fungal dysbiosis that results from loss of CARD9 in mice is associated with loss of Th17 cells, consistent with the importance of these cells to controlling fungal infections. Patients with a homozygous mutation in CARD9 (rs10781499) have severe mucocutaneous candiadiasis.53–55,56 DECTIN1 signaling via CARD9 might alter the immune response through changes in pathways regulated by NFκB, JNK, and MAP kinase.54
CARD9 also has a role in response to bacteria through its interaction with NOD2.57 A study of Card9−/− mice reported alterations in fungal and bacterial communities, but colitis susceptibility was dependent only on the bacterial community. Variations in phenotypes of knockout mice reveals the complexities of microbiome functions; many potential confounding variables affect host microbial interactions. In Card9−/− mice, bacterial tryptophan metabolites were found to account for some of variations in phenotypes.58 Bacterial tryptophan metabolites signal through human aryl hydrocarbon receptors (AHR), which are important for mucosal tolerance. Impaired microbial tryptophan metabolism in Card9−/− mice was associated with colitis susceptibility. Administration of Lactobacillus strains that metabolize tryptophan into AHR ligands was sufficient to reduce colitis in Card9−/− mice. Fecal samples from patients with CD or UC with IBD-associated polymorphisms in CARD9 lacked AHR ligands.
Human Leukocyte Antigens (HLAs)
HLA are encoded by genes in the major histocompatibility complex (MHC), among the most polymorphic in humans. Proteins encoded by the MHC locus mediate antigen presentation and coordination of the immune response. The diversity of HLAs allows the immune system to respond to a variety of pathogens, but certain polymorphisms increase risk for inappropriate responses to self-antigens.59 Variants in MHC class II genes have been associated with UC and CD and correlate with disease location.60 The heterozygosity of variants in class II genes is lower in patients with UC.61 These variants might reduce recognition of antigens on commensal microbes.61 Studies of mice and patients with other MHC class II-associated diseases, including rheumatoid arthritis, celiac disease, and ankylosing spondylitis,62–65,66–68 revealed a correlation between specific HLA alleles and distinct communities of microbes, which increase numbers of Th17 cells and intestinal permeability.69, 70 Studies of mice indicate that interactions between HLA polymorphisms and the intestinal microbiota are mediated by altered production of IgA and specific bacterial species, such as Bacteroides spp, which are sufficient to induce colitis.64, 71 Further research is needed into HLA variants and their associated microbial communities in patients with IBD.
Mucins
The intestinal mucus layer serves as another physical barrier that separates luminal microbes from the intestinal epithelium. The mucus layer comprises the glycoproteins MUC2, MUC5AC, MUC5B, and MUC6, which are secreted by goblet cells.72 Specific microbes degrade mucin glycoproteins, so microbial community structure corresponds with changes in mucin glycosylation. The diverse array of mucin glycans create a specific niche for specific intestinal bacteria that have evolved to bind these glycoproteins and use them as a carbon source. A disruption in the mucus barrier would therefore change bacterial ecology and deplete an important intestinal barrier function. In intestines of mice with colitis caused by administration of DSS or disruption of the Il10 gene, and in patients with UC, the mucus layer is thinner and highly penetrable to organisms that do not typically inhabit this niche.73, 74 Patients with UC have aberrant expression of MUC5AC, MUC6, and MUC2.75, 76,77, 78 Mice with deletion of MUC2 have increased susceptibility to colitis.79–81 Although variants in mucin genes have not been associated with IBD, their altered expression patterns in intestinal tissues from patients with IBD indicate that their activity is important for maintenance of the microbiome and prevention of inflammation.
Other genetic variants
Genome-wide association studies have identified over 200 loci with significant associations with IBD, but only a minority of these loci can be mapped to the 1 or 2 most likely alleles.82 These loci are generally enriched for protein-altering variants and proteins in cytokine pathways. For example, the IL23 signaling pathway includes IL23R, IL12B, TYK2, and JAK2. IL23 promotes development of Th17 cells, CD4+ T cells, Tc17 CD8+ T cells, and innate lymphoid cells, type 3. Agents that block IL12 and IL23 signaling have been approved by the Food and Drug Administration (FDA) for treatment of CD. However, cytokines produced by Th17 cells, including IL17 and IL22, protect against IBD.83, 84 In mice, IL17 prevents expansion of segmented filamentous bacteria (SFB), which promote colitis, via IL17 receptor (IL17R)-mediated epithelial cell signaling or by increasing neutrophil recruitment.85, 86 Some of the anti-inflammatory effects of IL22 are mediated by increased control of intestinal pathobionts and/or increased production of mucus.87, 88 Although many treatments for IBD aim to control cytokine production, dietary and bacterial metabolites can be ligands for G-protein coupled receptors (GPCR), which often activate anti-inflammatory signaling pathways.89, 90 Variants in GPCR genes have been associated with IBD. The GPCR GPR35 a receptor for the tryptophan intermediate kynurenic acid, which has been associated to IBD and primary sclerosing cholangitis.82, 89, 91
Interactions Between Genetic Variants and the Intestinal Microbiome
Altered interactions between the intestinal epithelium and the microbiota are an important step in IBD pathogenesis.92 These defective interactions might be corrected with microbes or microbial products. Many variants in genes associated with IBD affect responses of immune or intestinal cells to microbes, but do not affect the overall microbial ecology. However, we are currently able to assess only large changes in the overall microbiome, and we might miss changes in specific microbial populations or niches. We need to better understand interactions between genetic alterations and changes in specific populations of microbes. It is also important to conduct experiments with appropriate controls, because many factors can perturb the microbiome.93 The most powerful evidence for the mechanisms by which genetic variants alter the intestinal microbiome has come from studies of adoptive fecal transfer with littermate controls and careful analyses of knockout or knockdown mice, such as in the studies of NOD2.
Studies of mice have shown that alterations of the microbiota can promote colitis. Microbes interact with cell surface proteins, secreted metabolites, and other environmental substrates (Figure 2). It is not clear however, whether it is alterations in the microbes themselves or their effectors that promote development of IBD. Understanding how different intestinal microbes can cause different phenotypes of IBD could lead to development of microbe-based therapies.
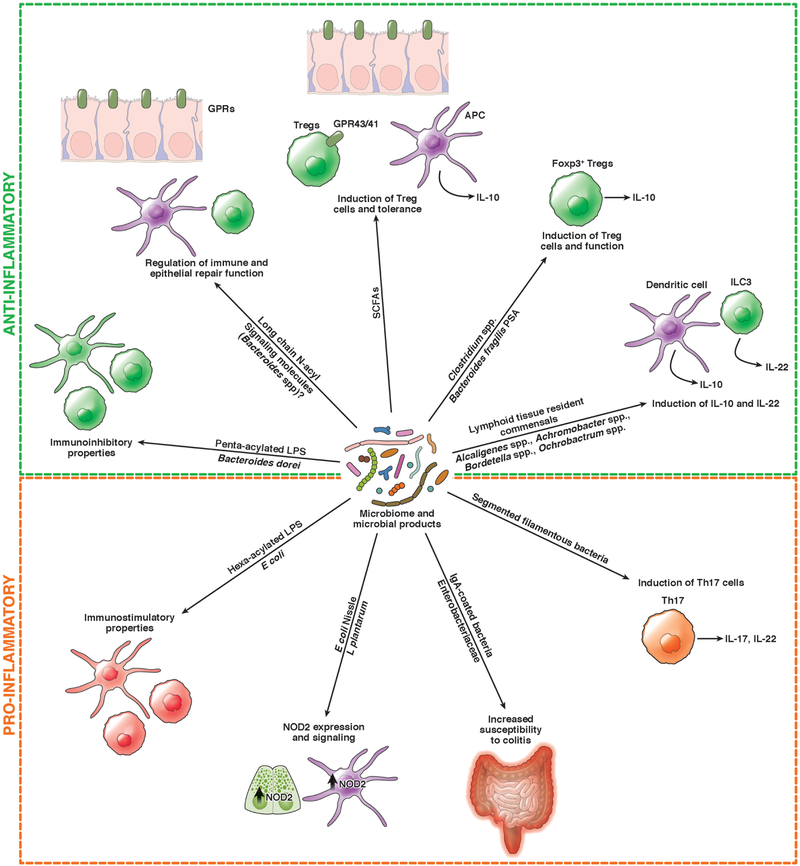
Figure 2. Effects of the Microbiome on Intestinal and Immune Cells.
The intestinal microbiome and its products modulate immune responses, via induction of dendritic cells (DCs) and lymphocytes (such as Th17 cells, Treg cells [Tregs in figure], and innate lymphoid cells (ILCs)), and cytokine production (IL10, IL22). Intestinal bacteria can also modulate immune signaling pathways, such as expression of NOD2, and epithelial repair. Specific microbes can increase susceptibility of mice to colitis.
Germ-free and gnotobiotic mice
Studies of germ-free and gnotobiotic mice have increased our understanding of interactions between intestinal cells, microbes, and development of the immune system. Mice raised under germ-free conditions have alterations in gut-associated lymphoid tissue, plasma cells, T cells, responses to microbial peptides, the crypt–villus architecture, and the mucus barrier.94, 95 Germ-free mice also have reduced expression of NOD2, indicating that its expression is regulated in response to microbes.34 Colonization of germ-free mice with a healthy microbiota restored intestinal homeostasis, although individual mucosal functions can be restored by organisms such as Lactobacillus plantarum or E coli Nissle 1917, which reactivates NOD2 signaling.34
Interestingly, infection with norovirus can restore most mucosal barrier abnormalities observed in germ-free mice.96 Experiments with germ-free and gnotobiotic mice established the role of the microbiota in development of colitis independent of genetic factors. In mice with T-cell transfer induced colitis, specific microbes promote development of colitis whereas others do not.97 Microbes can induce or reduce the severity of colitis in IL10-knockout mice or in mice given DSS.98–100 So, individual populations of microbes can either promote or prevent intestinal inflammation, depending on genotype; in IL10 knockout mice, most bacteria elicit colitis. Microbe-based therapies might therefore be selected based on a patient’s genotype, but not be effective in the entire population of patients with IBD. However, some microbiome therapies, such as those that increase mucosal barrier function, could have the widest applicability.
Interactions between microbes and intestinal cells
Intestinal microbes can alter the immune response. For example, commensal Clostridia strains promote accumulation of Foxp3+ Treg cells in the gut by inducing production of transforming growth factor beta and indoleamine 2,3-dioxygenase.101, 102 Treg cells downregulate inflammatory responses and mice colonized with specific species of Clostridia are resistant to induction of colitis.101 Although specific species of Clostridia can induce development of ROR0γt+ Treg cells, Clostridium ramosum also has this function.103, 104 Studies of gnotobiotic mice identified specific immune-modulatory effects of individual species of commensal bacteria and showed that specific types of immune cells, such as Treg cells can be induced by a wide range of bacteria whereas others appear to require specific microbes.105 Some microbes activate populations of immune cells that promote intestinal inflammation, such as Th17 cells.106 SFB activate Th17 cells in intestines of mice; adherent invasive E coli (AIEC) and Bifidobacteria adolescentis induce mucosal and systemic populations of Th17 cells in the gut.107–109 Reduction of Th17 cell-inducing bacteria can reduce the severity of colitis in mice.
An organism does not have to change its abundance in the population to have significant effects. SFB, which increases development of Th17 cells, also promotes T-cell dependent production of IgA.110 Coating of microbes by IgA has been proposed as a marker of immune activation by that microbe; IgA-coated bacteria, including certain Enterobacteriaceae, induce colitis in mice and have been associated with CD-associated spondyloarthritis.108, 110, 111 As IBD-promoting pathobionts may be specific to a gene or an individual, understanding IgA responses to microbes might help prevent the emergence of pathobionts or help us target pathobionts in specific individuals. The immune response to a microbe is likely specific to its niche. Alcaligenes, Achromobacter, Bordetella, and Ochrobactrum spp. specifically colonize lymphoid tissues, where they interact with innate lymphoid cells and dendritic cells to modulate IL10 production and intestinal repair mechanisms.112 The bioactivity of a microbe might require certain environmental signals, such as dietary metabolites, which can induce production or activity of bacterial effectors, or serve as metabolic substrates. Some Alisepes, Clostridium, and Bilophila spp can decrease production of tumor necrosis factor (TNF) by immune cells only in the presence of a certain diet.113,114
Specific species of Candida can cause colitis in mice; colonization can be inhibited by Bacteroides thetaiotamicron, which induce the production of antimicrobial peptide CRAMP.115 Interactions among organisms might affect the efficacy of microbiota-based therapies. Helminths are not considered commensals, but were prevalent during human history and are believed to have functions that affect microbiome development.116 Certain helminths induce responses of Th2 cells and increase IL10 secretion.117–119 Although helminths probably regulate these immune responses to promote their own infection of a patient, their functions might benefit patients with IBD, which has been demonstrated in phase 2 and 3 studies. The observation that the same organism can be beneficial or detrimental, depending on the patient or model, is not unique to helminths, but applies to many microbes. Viruses could have roles in IBD pathogenesis and norovirus is sufficient to restore mucosal barrier defects in germ-free mice, independent of microbiota.96, 120
Microbe effectors
Studies of model systems have identified organisms that induce specific cell responses, but these responses vary. Many complex factors mediate these interactions and we know little about the mechanisms by which bacteria alter the intestinal environment. Microbes interact with the intestinal epithelium or each other via secreted or cell-surface effectors. Identifying these effectors could help us learn more about the pathogenesis of IBD and lead to therapeutic strategies. Small molecules produced by microbes have tested for their therapeutic effects for decades.121 Studies are needed to identify the effectors produced by microbes that act on intestinal and immune cells.122
Short-chain fatty acids (SCFAs) reduce colitis, promote Treg-cell development, and downregulate of inflammatory signaling pathways.123, 124 Clostridia species produce SCFAs, which reach millimolar concentrations in the intestine and can activate GPCRs, inhibit histone deacetylases, and provide an energy source for colon epithelial cells.125–127 Polyamines such as putrescine or spermidine are virulence factors but also enhance intestinal barrier functions including mucus secretion, T-cell differentiation, and production of IgA.128–131 Bifidobacterium animalis increases polyamine levels, which correlates with decreased secretion of TNF and IL6 by myeloid cells.130 Interestingly, bacteria and human cells each produce polyamines, which might mediate some of their interactions.
Bacteria and human cells also each produce long-chain N-acyl signaling molecules that signal via specific GPCRs to regulate immunity, inflammation, and metabolism.90, 132 Structural similarity between human and bacterial signaling molecules is likely to be common, because bacteria are also able to synthesize the neurotransmitter GABA and certain Bacteroides metabolize tryptophan to tryptamine, a precursor to serotonin.133–135 Bacterial trypthophan metabolites are ligands for AHR, but the metabolism of tryptophan and other aromatic amino acids in Clostridia has been linked to intestinal barrier functions through the production of indoleproprionic acid.136 Bacterial metabolites of bile acids, such as the generation of taurine, might regulate inflammasome functions and increase microbial diversity.137
Bacteria interact through cell-surface effectors, including via secretion of outer membrane vesicles.138 Zwitterionic polysaccharide A (PSA), on the surface of B fragilis, regulates activity of Foxp3+ Treg cells in the gut.47 Administration of purified PSA, or B fragilis, is sufficient to activate intestinal Treg cells and reduce colitis in mice.48, 49, 139 Lipopolysaccharide (LPS), probably the most well-studied bacterial cell surface molecule, has countless variations in structure. Specific types of LPS, such as penta-acylated LPS produced by certain Bacteroides, can inhibit immune responses, in contrast to hexa-acylated LPS from E coli, which stimulates the immune response.140 Interestingly, in a mouse model of diabetes, this LPS from E coli reduced autoimmunity and development of diabetes. Therefore, bacteria can have different effects in different model systems, so it is important to understand all the effects of a microbe before it is included in a therapeutic strategy. Sphingolipids isolated from Bacteroides fragilis are similar to the human molecules and can regulate natural killer T cells.141
Challenges to Microbiome-based Therapeutics
Microbe-based therapeutic strategies can aim to alter the overall microbiome or its environment, introduce therapeutic microbes, or alter production of microbe effectors. Early studies focused on application of therapeutic microbes, despite the challenges of developing a drug that includes living organisms. Small-molecule development begins with basic research and discovery of bioactive molecules, followed by preclinical studies (formulation, toxicity, and pharmacokinetic analyses), followed by trials of safety and efficacy in patients. Development of microbe-based therapies has changed concepts of drug mechanisms, formulation, and monitoring, requiring new approaches for development and regulation (Figure 3). In 2012 the Center for Biologics Evaluation and Research issued guidelines to assist in therapeutic development of live organisms, which they classified as live biotherapeutic products (LBPs). LBPs are defined as biologic products that contain live organisms, such as bacteria, and that might be used in prevention, treatment, or cure of human diseases but are not vaccines. Development of prebiotics and bacterial effectors is likely to follow regular drug development pathways, but LBPs are the most pursued of the microbe-based therapies (Figure 3).
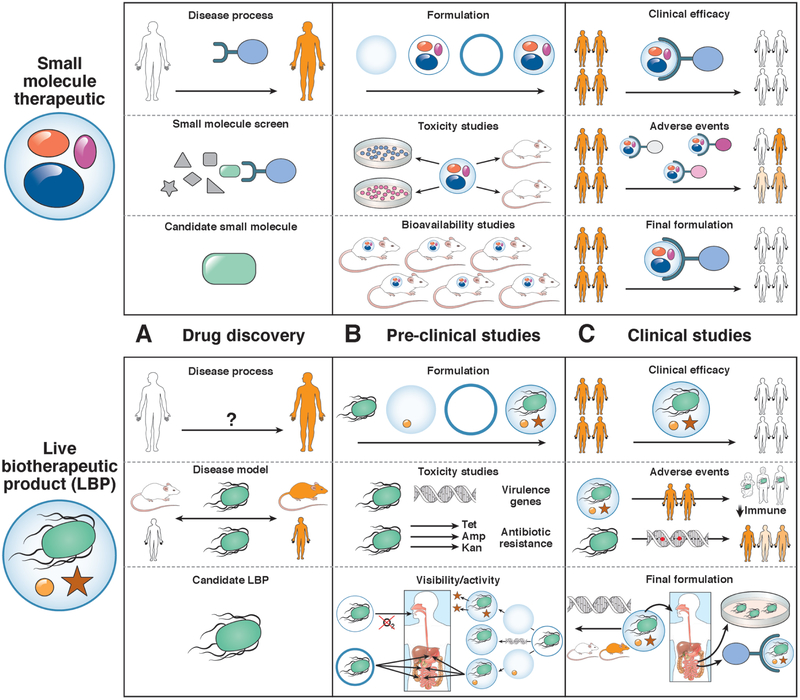
Figure 3. Challenges to Development of Therapeutic LBPs.
Compared with small molecules, LBPs have different challenges at each stage of drug development. Challenges are presented to drug discovery, preclinical studies, and clinical studies. A. In contrast to small molecules, which usually target a specific protein or class of feature of proteins, LBPs are identified based on their association with a disease phenotype in humans or mouse models. B. Small molecules require extensive toxicity studies, whereas LBPs are believed to be non-toxic but require assessments for virulence or antibiotic resistance. Preclinical studies of LBPs are not informative for bioavailability, but focus on viability or bioactivity, which can require specific encapsulation methods, adjuvants, or genetic modifications. C. Trials of LBPs require specific attention to adverse events related to transmission of the microbe, loss of its bioactivity, or off-target effects. Small molecules can also have off target effects, but these may be easier to predict, based on finding from preclinical studies. Early-phase studies of LPBs might be important for final formulation, because bioavailability and potential mechanisms can be assessed based on findings from small groups of patients.
FDA Regulation
In 2012 the FDA published regulations for development of LBPs, which were updated in 2016.142 It is important to distinguish LBPs from probiotics, which are organisms that have obtained the generally recognized as safe label and fall outside of this regulation. Trials of LBPs for treatment of diseases requires an investigational new drug (IND) application. An IND application can be waived for an LBP that is available in conventional foods or dietary supplements, in consultation with the FDA. FMTs were first performed without an IND because feces were considered to be widely available, but in 2012, regulation by the FDA changed— now an IND is required for studies of FMT in patients with IBD. However, use of FMT for recurrent C difficile infection (CDI) has discretionary regulation. The FDA does not require toxicity studies for trials of FMT, but it does require adequate characterization of microbe strains to be tested. Chemistry, manufacturing, and control data must include the historical context of the organism(s), the purity, and details about the presence of virulence factors, toxins or antibiotic resistance genes and the potential to spread these genes.
Whole-genome sequencing is performed for many LBPs to address these safety concerns. Genetically modified LBP require additional tests, to ensure the stability of genetic modifications. Antibiotic resistance genes may be present but require justification and are not acceptable for LBPs that could cause opportunistic infections. Phenotype-based antibiotic resistance testing for each LBP is required in addition to traditional toxicological profiling. Product release testing (the identity, viability, potency and purity of each LBP) is perhaps more important step, relative to small molecule therapies, because LBPs are a challenge to quantify and can change during production. Potency assays (such as colony-forming units) should be used to calculate the dose or release of a predefined product when mechanisms are well defined. Later stages of might require an assay that tests the agent’s mechanism, which might not be straightforward for a complex LBP. In addition, it is still not clear what level of evidence is needed to justify a specific selection of strains for an LBP. Regulations are likely to include standardization of these definitions among countries, because there is no equivalent definition of an LBP in the European Union, which has categorizations for therapeutic organisms not present in the United States.
Microbiota-based Therapeutics in Preclinical and Clinical Development
As the guiding framework for LBP has been clarified over last 6 years there has been an increase in companies developing discovery platforms and/or introducing candidate therapies into pre-clinical and clinical studies for the treatment of IBD (Table 1).
Table 1.
Microbiota-based Therapies in Development for IBD
| Therapy Type | Company | Location | Product* | Delivery* | Development Stage | Mechanism |
|---|---|---|---|---|---|---|
| Ecology modulator | Enterome | USA | EB-8018 small molecule | oral | phase 2 study of patients with CD | prevents pathobiont invasion by blocking FIMH |
| Immuron | Australia | N/A | oral | preclinical | antibodies and adjuvant to boost immune response to pathobionts | |
| BiomX | Israel | BX002 | N/A | preclinical | bacteriophage to deplete pathobionts | |
| Eligo | France | N/A | N/A | preclinical | Crispr-CAS to deplete pathobionts | |
| Intralytix | USA | EcoActive | oral | phase 1 and 2a study of patients with CD | bacteriophage to target AIEC | |
| Artizan Biosciences | USA | N/A | N/A | Preclinical | Subtractive therapy | |
| Live Biotherapeutic Product | Seres Therapeutics | USA | SER287 naturally derived community | oral | phase 2 study of patients with UC | not available |
| Seres Therapeutics Janssen (Vendanta License) | USA | SER301 defined community | oral | preclinical | not available | |
| VE202 defined community | oral | phase 1 study of patients with UC | Induce Treg cells | |||
| Rebiotix (acquired by Ferring) | USA | RBX2660 | enema | phase 1 in pediatric patients with UC | Restore microbiome composition | |
| 4D Pharma | UK | Thetanix B thetaiotamicron | oral | phase 1 study of patients with CD | not available | |
| 4D Pharma Osel | Rosburix R hominis | oral | preclinical | not available | ||
| USA | CBM588 C butyricum | oral | phase 1 | Increase SCFAs | ||
| ImmuneBiotech | Sweden | IB002 Lactobacilli | N/A | preclinical | not available | |
| Actobiotic | USA | AG-014 GMO | oral | phase 1 | heterologous expression of anti-TNF nanobody by Lactobacillus | |
| Rise Therapeutics | USA | R-3750 GMO | N/A | preclinical | Lactobacillus for heterologous expression | |
| Nordisk Rebalance | Denmark | Profermin L latarum oats phosphatidylcholine | oral | marketed as food for special medical purpose | phase 2 of patients with UC, a prebiotic that increases SCFAs | |
| Finch Therapeutics | USA | FIN524, defined community | oral | preclinical studies of colitis | not available | |
| Allergan | Ireland | ABI-M201,301 | oral | preclinical | licensed from Assembly Biosciences and uses Gemicel coating | |
| NextBiotix/Exeliom Biosciences | FRA | NBX-1650 | n/a | Preclinical | F. prausnitzii to treat inflammation | |
| ViThera | USA | VT301 | Oral | Preclinical | Modified Lactobacilli strains | |
| Chain Biotech | USA | CHN-1, CHN-2 | Oral | preclinical | anti-microbial peptide with Clostridium Assisted Drug Delivery | |
| PanTheryx | USA | PTX-400 | Oral | Preclinical | Medical food - prebiotic | |
| Microbial effectors | Host Therabiomics | UK/USA | L1173 | N/A | preclinical | platform to identify effectors |
| Second Genome | USA | SG-2–0776 protein | oral | preclinical | intestinal healing | |
| Symbiotix Biotherapies | USA | SYMB-104 polysaccharide A | N/A | preclinical | B fragilis-derived immune modulator | |
| Alma Bio Therapeutics | FRA | N/A Plasmid | Injection | Preclinical | Plasmids that produce Heat Shock Proteins | |
| Enterome | USA | EB110/EB220 | oral | preclinical | microbial metabolite associated with CD | |
| Formulation | Finch Therapeutics | USA | aquashell | oral | N/A | pH release polysaccharide |
| Intract Pharma | UK | phloral duocoat | oral | N/A | pH release | |
| Prodigest | Belgium | in vitro microbiome model | N/A | N/A | predict in vivo conditions | |
| Synlogic | USA | genetically modified organism | N/A | N/A | Not available | |
| Assembly | USA | gemicel capsule | oral | N/A | pH release | |
| Host Target | Second Genome | USA | SGM-1019 Small molecule | oral | phase 1 | modulates inflammasome |
N/A, not applicable because agent is in preclinical stage of development
Note: Companies were identified by the Janssen Human Microbiome Institute from public resources.
Modulators of the microbiome
The microbiome can be modified by improving mucosal barrier functions or depletion or enrichment of organisms linked with diseases, an increased or decreased immune response, or other outcomes. Zoenasa is a formulation of N-acetyl cysteine, phosphatidylcholine, and mesalamine that is believed to strengthen the mucosal barrier. Zoenasa has been formulated as a rectal gel or oral tablet and tested in a phase 2 study of patients with distal UC. Specific microbes or communities of microbes may be pathogens (pathobionts), and depletion of these microbes might be beneficial. A challenge to modulating the microbiome by depleting pathobionts is that these microbes may be specific to an individual based on their pathophysiology and/or genetics. Production of IgA in response to bacteria might be measured to identify specific pathobionts in individual patients.
Immuron is an oral immunotherapy (antigens, adjuvants, antibodies) designed to reduce or increase specific microbiota. IMM124E is an oral formulation of antibody against LPS and glycosphingolipid adjuvants that is preclinical studies but might be used to treat UC. BiomX directly targets pathobionts using bacteriophages and Eligo depletes bacteria by using CRISPR.143 The companies that are developing these agents have not revealed their specific target species, but Eligo has a platform to allow for an individualized assessment of potential pathobionts. Ecoactive has just entered phase 1 and 2 trials of patients with CD—it is an oral bacteriophage cocktail that depletes AIEC. AIEC is enriched by defects in NOD2 signaling in patients with CD.144 EB-8018 was designed to reduce AIEC by blocking fimH. EB-8018 is entering phase 2 studies of patients with CD and was found to be safe in a phase 1 study. A diagnostic assay (IBD-210) has been designed to measure fimH in fecal samples, to identify patients likely to respond to EB-8018.
LBPs
Studies of patients and animal models have led to the discovery of many LBPs. LBP are being developed using a variety of formulations, including naturally derived communities, defined communities, individual organisms, and genetically modified organisms. SER287 is a naturally derived community and SER301 a defined community based on human cohort studies whereas VE202 was developed as a synthetic community in mice, based on a targeted increases in Treg cells.101 In contrast to community LBP, Thetanix is a single strain of Bacteroides thetaiotamicron that is in a phase 1 trial of children with CD. B thetaiotamicron might have multiple mechanisms, including modulation of fungi, although there is also a negative association between B thetaiotamicron and infection with pathogenic strains of E coli.
The pleiotropic effects of LBP will warrant specific safety attention. A strain of Clostridium butyricum is in preclinical studies for IBD was found to be safe for treatment of CDI. C butyricum is believed to act specifically by increasing SCFAs, though other LBPs are also believed to increase SCFA.145 ImmuneBiotech has a narrow focus on a proprietary panel of lactobacilli, to which they assign immunomodulatory functions. Many lactobacilli carry generally recognized as safe designations, which will facilitate their approval process and are easy to manipulate in the laboratory for the development of genetically modified organisms. AG-014 is a lactobacillus engineered to produce a nanobody against TNF that is in phase 1 studies of patients with IBD.
Countries outside the United States, have an additional regulatory category, independent of dietary supplement and LBP, which is a food for special medical purposes. A food for special medical purposes is naturally found in the diet but can be marketed for the treatment of a disease. In Denmark, profermin is a food for special medical purposes—it is a combination of Lactobacillus plantarum, oats, and phosphatidylcholine. Each component of this pill has a separate effect as a prebiotic (oats), barrier modulator (phosphatidylcholine), and LBP (L plantarum) though it is unclear if there is interaction among components. Profermin is on the market and has been studied in small trials of patients with UC, in which it had moderate efficacy but without endoscopic endpoints.146, 147 Helminths, specifically Trichuris suis, showed efficacy in phase 2 studies of patients with CD but had limited efficacy in phase 3 studies.148–151
Microbe effectors
Microbe effectors have specific effects on cells and follow a traditional drug development strategy. Approval of microbial effectors by regulatory agencies might be straightforward, but there are few products in early stages of development, because we understand so little about them. EB110 is a microbe-derived metabolite identified in humans that has been associated with development of CD, via unknown mechanisms. SG-2–0776 is a microbe effector (protein) that promotes intestinal healing and is in preclinical studies for treatment of IBD. PSA has been one of the most extensively studied bacterial effectors and has a number of immune-regulatory properties.47, 152 However, IBD-associated variants in NOD2 and ATG16L1 could mitigate the effect of PSA and be used to identify patients not likely to respond.49 PSA is in preclinical development and it is unclear whether clinical trials will compare effects in patients with different genotypes.
Formulation strategies
Formulation is an important challenge for microbiome-based therapies. Formulation aims for reproducible effects among individuals and delivery of viable organisms to their niche. Many LBPs in early-stage trials are administered daily, because it is likely that LBPs do not incorporate into the microbiome and expand. However, they might be given in intermittent or even single doses, if our understanding of microbiome homeostasis improves, and we can more carefully select LBPs or use of adjuvants.
Companies have focused on formulations for better delivery an LBP to a niche. The Gemicel capsule was developed for colonic release of LBP, incorporating 2 separate pH dependent mechanisms. Aquashell is a pH-sensitive formulation for colonic delivery that incorporates a separate polysaccharide coating that is digested by colon microbes. Both formulation strategies have been used to encapsulate LBPs that are in trials of patients with IBD. Duocoat is optimized for duodenal release, using a pH-sensitive coat, and Phloral is optimized for colon release. Each of these encapsulation strategies necessitate specific attention to a strict anaerobic process that is unique for each LBP, because previous dietary probiotics (such as Lactobacillus or Bifidobacterium) were microaerophilic.153 Genetically modified organisms for treatment of IBD might be viewed as a type of formulation strategy to increase a therapeutic effect and mitigate variation. Genetic tools to manipulate human microbes are required. Synlogic programs internal circuits in E coli Nissle strains, to induce expression of effector genes in response to specific environmental signals. This technology is used to produce microbial effectors only in the correct environment. Synlogic has also developed chromosome markers for in vivo monitoring of LBPs, which will facilitate bioavailability studies.
Targeting interactions between intestinal cells and microbes
We have begun to identify cell signaling pathways that regulated by bacteria and might be therapeutically manipulated. SGM-1019 is a small molecule that affects the inflammasome, identified using a discovery platform. SGM-1019 has progressed through phase I studies and is being developed for treatment of IBD and non-alcoholic steatohepatitis, for which it is entering phase 2 studies.
Future Directions
Microbe-based therapies are becoming more diverse and effective as our understanding of the interactions between the microbiome and human cells increases. We have begun to better understand the effects of genetic and environmental factors on the microbiome and its products or effectors. Formulation strategies can be refined to address the primary challenge of diversity in the microbiomes among individuals and for treatment of specific diseases.
Additional challenges to development and use of microbe-based therapies involves issues regarding intellectual property. A full discussion of this topic is beyond the scope of this review, but one of the biggest problems is how to enforce patent laws for the composition of a natural product. LBPs that can be defined as natural products include isolated microbes or their effectors (such as metabolites or proteins). Natural products can be protected by method patents, which state their use for treatment of IBD, but these patents are not as easily enforced, which dissuades companies from developing these types of products. One strategy has been to genetically modify microbes or use them in combination with other microbes, as a genetically modified organism or a defined community that is not found in nature (not a natural product). Companies have obtained composition of matter patents for LBPs that are natural communities, so it will be important to see how these patents are enforced if these products come to market. Patent law protection is critical for development of microbe-based therapies; it is likely that the legal framework will change as it has in the past.
Funding Sources:
NIH K08 DK109287–01 (L.J.C.); NIH U01 DK62429, NIH U01 DK062422, NIH R01 DK092235, NIH R01 DK106593, Sanford J. Grossman Center for Integrative studies in Crohn’s disease (J.H.C.); NIH R00 DK110534 (H.C.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: D.G. is employed by Janssen which has invested or licensed products from Vedanta, BiomX and Enterome.
References
- 1.Abraham C, Medzhitov R. Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology 2011;140:1729–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell 2014;157:121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun M, He C, Cong Y, et al. Regulatory immune cells in regulation of intestinal inflammatory response to microbiota. Mucosal Immunol 2015;8:969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall AB, Tolonen AC, Xavier RJ. Human genetic variation and the gut microbiome in disease. Nat Rev Genet 2017;18:690–699. [DOI] [PubMed] [Google Scholar]
- 5.Luca F, Kupfer SS, Knights D, et al. Functional Genomics of Host-Microbiome Interactions in Humans. Trends Genet 2018;34:30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011;474:307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rothschild D, Weissbrod O, Barkan E, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018;555:210–215. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann DE, Palumbo FB, Ravel J, et al. A proposed definition of microbiota transplantation for regulatory purposes. Gut Microbes 2017;8:208–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez J, Grinspan A. Fecal Microbiota Transplantation for Inflammatory Bowel Disease. Gastroenterol Hepatol (N Y) 2016;12:374–9. [PMC free article] [PubMed] [Google Scholar]
- 10.Khan KJ, Ullman TA, Ford AC, et al. Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. Am J Gastroenterol 2011;106:661–73. [DOI] [PubMed] [Google Scholar]
- 11.Singh S, Stroud AM, Holubar SD, et al. Treatment and prevention of pouchitis after ileal pouch-anal anastomosis for chronic ulcerative colitis. Cochrane Database Syst Rev 2015:CD001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sartor RB, Wu GD. Roles for Intestinal Bacteria, Viruses, and Fungi in Pathogenesis of Inflammatory Bowel Diseases and Therapeutic Approaches. Gastroenterology 2017;152:327–339 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benson AK, Kelly SA, Legge R, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A 2010;107:18933–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodrich JK, Waters JL, Poole AC, et al. Human genetics shape the gut microbiome. Cell 2014;159:789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knights D, Silverberg MS, Weersma RK, et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med 2014;6:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blekhman R, Goodrich JK, Huang K, et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol 2015;16:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkes M, Barrett JC, Prescott NJ, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet 2007;39:830–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hampe J, Franke A, Rosenstiel P, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet 2007;39:207–11. [DOI] [PubMed] [Google Scholar]
- 19.Consortium WTCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet 2010;42:1118–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rivas MA, Beaudoin M, Gardet A, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nature genetics 2011;43:1066–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson CA, Boucher G, Lees CW, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet 2011;43:246–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu JZ, van Sommeren S, Huang H, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015;47:979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellinghaus D, Jostins L, Spain SL, et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet 2016;48:510–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 2001;411:599–603. [DOI] [PubMed] [Google Scholar]
- 27.Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 2001;411:603–6. [DOI] [PubMed] [Google Scholar]
- 28.Philpott DJ, Sorbara MT, Robertson SJ, et al. NOD proteins: regulators of inflammation in health and disease. Nat Rev Immunol 2014;14:9–23. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi KS, Chamaillard M, Ogura Y, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 2005;307:731–4. [DOI] [PubMed] [Google Scholar]
- 30.Molodecky NA, Kaplan GG. Environmental risk factors for inflammatory bowel disease. Gastroenterol Hepatol (N Y) 2010;6:339–46. [PMC free article] [PubMed] [Google Scholar]
- 31.Frank DN, Robertson CE, Hamm CM, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis 2011;17:179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rehman A, Sina C, Gavrilova O, et al. Nod2 is essential for temporal development of intestinal microbial communities. Gut 2011;60:1354–62. [DOI] [PubMed] [Google Scholar]
- 33.Imhann F, Vich Vila A, Bonder MJ, et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2018;67:108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petnicki-Ocwieja T, Hrncir T, Liu YJ, et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci U S A 2009;106:15813–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mondot S, Barreau F, Al Nabhani Z, et al. Altered gut microbiota composition in immune-impaired Nod2(−/−) mice. Gut 2012;61:634–5. [DOI] [PubMed] [Google Scholar]
- 36.Couturier-Maillard A, Secher T, Rehman A, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest 2013;123:700–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramanan D, Tang MS, Bowcutt R, et al. Bacterial sensor Nod2 prevents inflammation of the small intestine by restricting the expansion of the commensal Bacteroides vulgatus. Immunity 2014;41:311–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alnabhani Z, Hugot JP, Montcuquet N, et al. Respective Roles of Hematopoietic and Nonhematopoietic Nod2 on the Gut Microbiota and Mucosal Homeostasis. Inflamm Bowel Dis 2016;22:763–73. [DOI] [PubMed] [Google Scholar]
- 39.Al Nabhani Z, Lepage P, Mauny P, et al. Nod2 Deficiency Leads to a Specific and Transmissible Mucosa-associated Microbial Dysbiosis Which Is Independent of the Mucosal Barrier Defect. J Crohns [DOI] [PubMed] [Google Scholar]
- 40.Rioux JD, Xavier RJ, Taylor KD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet 2007;39:596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooney R, Baker J, Brain O, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med 2010;16:90–7. [DOI] [PubMed] [Google Scholar]
- 42.Travassos LH, Carneiro LA, Ramjeet M, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol 2010;11:55–62. [DOI] [PubMed] [Google Scholar]
- 43.Murthy A, Li Y, Peng I, et al. A Crohn’s disease variant in Atg16l1 enhances its degradation by caspase 3. Nature 2014;506:456–62. [DOI] [PubMed] [Google Scholar]
- 44.Saitoh T, Fujita N, Jang MH, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 2008;456:264–8. [DOI] [PubMed] [Google Scholar]
- 45.Cadwell K, Liu JY, Brown SL, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 2008;456:259–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kabat AM, Harrison OJ, Riffelmacher T, et al. The autophagy gene Atg16l1 differentially regulates Treg and TH2 cells to control intestinal inflammation. Elife 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008;453:620–5. [DOI] [PubMed] [Google Scholar]
- 48.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A 2010;107:12204–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chu H, Khosravi A, Kusumawardhani IP, et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 2016;352:1116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitsuyama K, Niwa M, Takedatsu H, et al. Antibody markers in the diagnosis of inflammatory bowel disease. World J Gastroenterol 2016;22:1304–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sokol H, Leducq V, Aschard H, et al. Fungal microbiota dysbiosis in IBD. Gut 2017;66:1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iliev ID, Funari VA, Taylor KD, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 2012;336:1314–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sokol H, Conway KL, Zhang M, et al. Card9 mediates intestinal epithelial cell restitution, T-helper 17 responses, and control of bacterial infection in mice. Gastroenterology 2013;145:591–601 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gross O, Gewies A, Finger K, et al. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature 2006;442:651–6. [DOI] [PubMed] [Google Scholar]
- 55.LeibundGut-Landmann S, Gross O, Robinson MJ, et al. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol 2007;8:630–8. [DOI] [PubMed] [Google Scholar]
- 56.Glocker EO, Hennigs A, Nabavi M, et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Engl J Med 2009;361:1727–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hsu YM, Zhang Y, You Y, et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat Immunol 2007;8:198–205. [DOI] [PubMed] [Google Scholar]
- 58.Lamas B, Richard ML, Leducq V, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Doherty PC, Zinkernagel RM. A biological role for the major histocompatibility antigens. Lancet 1975;1:1406–9. [DOI] [PubMed] [Google Scholar]
- 60.Cleynen I, Boucher G, Jostins L, et al. Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet 2016;387:156–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goyette P, Boucher G, Mallon D, et al. High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat Genet 2015;47:172–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolnick DI, Snowberg LK, Caporaso JG, et al. Major Histocompatibility Complex class IIb polymorphism influences gut microbiota composition and diversity. Mol Ecol 2014;23:4831–45. [DOI] [PubMed] [Google Scholar]
- 63.Lin P, Bach M, Asquith M, et al. HLA-B27 and human beta2-microglobulin affect the gut microbiota of transgenic rats. PLoS One 2014;9:e105684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kubinak JL, Stephens WZ, Soto R, et al. MHC variation sculpts individualized microbial communities that control susceptibility to enteric infection. Nat Commun 2015;6:8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Silverman M, Kua L, Tanca A, et al. Protective major histocompatibility complex allele prevents type 1 diabetes by shaping the intestinal microbiota early in ontogeny. Proc Natl Acad Sci U S A 2017;114:9671–9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.De Palma G, Capilla A, Nadal I, et al. Interplay between human leukocyte antigen genes and the microbial colonization process of the newborn intestine. Curr Issues Mol Biol 2010;12:1–10. [PubMed] [Google Scholar]
- 67.Rosenbaum JT, Davey MP. Time for a gut check: evidence for the hypothesis that HLA-B27 predisposes to ankylosing spondylitis by altering the microbiome. Arthritis Rheum 2011;63:3195–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Olivares M, Neef A, Castillejo G, et al. The HLA-DQ2 genotype selects for early intestinal microbiota composition in infants at high risk of developing coeliac disease. Gut 2015;64:406–17. [DOI] [PubMed] [Google Scholar]
- 69.Scher JU, Sczesnak A, Longman RS, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2013;2:e01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gomez A, Luckey D, Yeoman CJ, et al. Loss of sex and age driven differences in the gut microbiome characterize arthritis-susceptible 0401 mice but not arthritis-resistant 0402 mice. PLoS One 2012;7:e36095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rath HC, Herfarth HH, Ikeda JS, et al. Normal luminal bacteria, especially Bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human beta2 microglobulin transgenic rats. J Clin Invest 1996;98:945–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johansson ME, Hansson GC. Immunological aspects of intestinal mucus and mucins. Nat Rev Immunol 2016;16:639–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pullan RD, Thomas GA, Rhodes M, et al. Thickness of adherent mucus gel on colonic mucosa in humans and its relevance to colitis. Gut 1994;35:353–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Johansson ME, Gustafsson JK, Holmen-Larsson J, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut 2014;63:281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Van Klinken BJ, Van der Wal JW, Einerhand AW, et al. Sulphation and secretion of the predominant secretory human colonic mucin MUC2 in ulcerative colitis. Gut 1999;44:387–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Forgue-Lafitte ME, Fabiani B, Levy PP, et al. Abnormal expression of M1/MUC5AC mucin in distal colon of patients with diverticulitis, ulcerative colitis and cancer. Int J Cancer 2007;121:1543–9. [DOI] [PubMed] [Google Scholar]
- 77.Corfield AP, Myerscough N, Bradfield N, et al. Colonic mucins in ulcerative colitis: evidence for loss of sulfation. Glycoconj J 1996;13:809–22. [DOI] [PubMed] [Google Scholar]
- 78.Larsson JM, Karlsson H, Crespo JG, et al. Altered O-glycosylation profile of MUC2 mucin occurs in active ulcerative colitis and is associated with increased inflammation. Inflamm Bowel Dis 2011;17:2299–307. [DOI] [PubMed] [Google Scholar]
- 79.Van der Sluis M, De Koning BA, De Bruijn AC, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 2006;131:117–29. [DOI] [PubMed] [Google Scholar]
- 80.Johansson ME, Phillipson M, Petersson J, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 2008;105:15064–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wenzel UA, Magnusson MK, Rydstrom A, et al. Spontaneous colitis in Muc2-deficient mice reflects clinical and cellular features of active ulcerative colitis. PLoS One 2014;9:e100217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang H, Fang M, Jostins L, et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 2017;547:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O’Connor W Jr., Kamanaka M, Booth CJ, et al. A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat Immunol 2009;10:603–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zenewicz LA, Yancopoulos GD, Valenzuela DM, et al. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity 2008;29:947–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kumar P, Monin L, Castillo P, et al. Intestinal Interleukin-17 Receptor Signaling Mediates Reciprocal Control of the Gut Microbiota and Autoimmune Inflammation. Immunity 2016;44:659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Flannigan KL, Ngo VL, Geem D, et al. IL-17A-mediated neutrophil recruitment limits expansion of segmented filamentous bacteria. Mucosal Immunol 2017;10:673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hasegawa M, Yada S, Liu MZ, et al. Interleukin-22 regulates the complement system to promote resistance against pathobionts after pathogen-induced intestinal damage. Immunity 2014;41:620–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sugimoto K, Ogawa A, Mizoguchi E, et al. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest 2008;118:534–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thorburn AN, Macia L, Mackay CR. Diet, metabolites, and “western-lifestyle” inflammatory diseases. Immunity 2014;40:833–42. [DOI] [PubMed] [Google Scholar]
- 90.Cohen LJ, Esterhazy D, Kim SH, et al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature 2017;549:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ellinghaus D, Folseraas T, Holm K, et al. Genome-wide association analysis in primary sclerosing cholangitis and ulcerative colitis identifies risk loci at GPR35 and TCF4. Hepatology 2013;58:1074–83. [DOI] [PubMed] [Google Scholar]
- 92.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stappenbeck TS, Virgin HW. Accounting for reciprocal host-microbiome interactions in experimental science. Nature 2016;534:191–9. [DOI] [PubMed] [Google Scholar]
- 94.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol 2004;4:478–85. [DOI] [PubMed] [Google Scholar]
- 95.Maynard CL, Elson CO, Hatton RD, et al. Reciprocal interactions of the intestinal microbiota and immune system. Nature 2012;489:231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kernbauer E, Ding Y, Cadwell K. An enteric virus can replace the beneficial function of commensal bacteria. Nature 2014;516:94–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Garrett WS, Lord GM, Punit S, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 2007;131:33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kiesler P, Fuss IJ, Strober W. Experimental Models of Inflammatory Bowel Diseases. Cell Mol Gastroenterol Hepatol 2015;1:154–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cominelli F, Arseneau KO, Rodriguez-Palacios A, et al. Uncovering Pathogenic Mechanisms of Inflammatory Bowel Disease Using Mouse Models of Crohn’s Disease-Like Ileitis: What is the Right Model? Cell Mol Gastroenterol Hepatol 2017;4:19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Keubler LM, Buettner M, Hager C, et al. A Multihit Model: Colitis Lessons from the Interleukin-10-deficient Mouse. Inflamm Bowel Dis 2015;21:1967–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011;331:337–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013;500:232–6. [DOI] [PubMed] [Google Scholar]
- 103.Yissachar N, Zhou Y, Ung L, et al. An Intestinal Organ Culture System Uncovers a Role for the Nervous System in Microbe-Immune Crosstalk. Cell 2017;168:1135–1148 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sefik E, Geva-Zatorsky N, Oh S, et al. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science 2015;349:993–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Geva-Zatorsky N, Sefik E, Kua L, et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 2017;168:928–943 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Atarashi K, Tanoue T, Ando M, et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell 2015;163:367–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tan TG, Sefik E, Geva-Zatorsky N, et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci U S A 2016;113:E8141–E8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Viladomiu M, Kivolowitz C, Abdulhamid A, et al. IgA-coated E. coli enriched in Crohn’s disease spondyloarthritis promote TH17-dependent inflammation. Sci Transl Med 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009;139:485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bunker JJ, Flynn TM, Koval JC, et al. Innate and Adaptive Humoral Responses Coat Distinct Commensal Bacteria with Immunoglobulin A. Immunity 2015;43:541–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Palm NW, de Zoete MR, Cullen TW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014;158:1000–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fung TC, Bessman NJ, Hepworth MR, et al. Lymphoid-Tissue-Resident Commensal Bacteria Promote Members of the IL-10 Cytokine Family to Establish Mutualism. Immunity 2016;44:634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schirmer M, Smeekens SP, Vlamakis H, et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell 2016;167:1125–1136 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Devkota S, Wang Y, Musch MW, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 2012;487:104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fan D, Coughlin LA, Neubauer MM, et al. Activation of HIF-1alpha and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat Med 2015;21:808–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Helmby H Human helminth therapy to treat inflammatory disorders - where do we stand? BMC Immunol 2015;16:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schnoeller C, Rausch S, Pillai S, et al. A Helminth Immunomodulator Reduces Allergic and Inflammatory Responses by Induction of IL-10-Producing Macrophages. The Journal of Immunology 2008;180:4265–4272. [DOI] [PubMed] [Google Scholar]
- 118.Elliott DE, Summers RW, Weinstock JV. Helminths as governors of immune-mediated inflammation. Int J Parasitol 2007;37:457–64. [DOI] [PubMed] [Google Scholar]
- 119.Figueiredo CA, Barreto ML, Rodrigues LC, et al. Chronic intestinal helminth infections are associated with immune hyporesponsiveness and induction of a regulatory network. Infect Immun 2010;78:3160–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Norman JM, Handley SA, Baldridge MT, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015;160:447–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Eder J, Sedrani R, Wiesmann C. The discovery of first-in-class drugs: origins and evolution. Nat Rev Drug Discov 2014;13:577–87. [DOI] [PubMed] [Google Scholar]
- 122.Koppel N, Balskus EP. Exploring and Understanding the Biochemical Diversity of the Human Microbiota. Cell Chem Biol 2016;23:18–30. [DOI] [PubMed] [Google Scholar]
- 123.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 2016;16:341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013;341:569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shimazu T, Hirschey MD, Newman J, et al. Suppression of oxidative stress by beta-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science 2013;339:211–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Samuel BS, Shaito A, Motoike T, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proceedings of the National Academy of Sciences of the United States of America 2008;105:16767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kim MH, Kang SG, Park JH, et al. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 2013;145:396–406 e1–10. [DOI] [PubMed] [Google Scholar]
- 128.Chen J, Rao JN, Zou T, et al. Polyamines are required for expression of Toll-like receptor 2 modulating intestinal epithelial barrier integrity. Am J Physiol Gastrointest Liver Physiol 2007;293:G568–76. [DOI] [PubMed] [Google Scholar]
- 129.Dubin K, Callahan MK, Ren B, et al. Intestinal microbiome analyses identify melanoma patients at risk forcheckpoint-blockade-induced colitis. Nat Commun 2016;7:10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kibe R, Kurihara S, Sakai Y, et al. Upregulation of colonic luminal polyamines produced by intestinal microbiota delays senescence in mice. Sci Rep 2014;4:4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu L, Guo X, Rao JN, et al. Polyamines regulate E-cadherin transcription through c-Myc modulating intestinal epithelial barrier function. Am J Physiol Cell Physiol 2009;296:C801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cohen LJ, Kang HS, Chu J, et al. Functional metagenomic discovery of bacterial effectors in the human microbiome and isolation of commendamide, a GPCR G2A/132 agonist. Proc Natl Acad Sci U S A 2015;112:E4825–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pokusaeva K, Johnson C, Luk B, et al. GABA-producing Bifidobacterium dentium modulates visceral sensitivity in the intestine. Neurogastroenterol Motil 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Williams BB, Van Benschoten AH, Cimermancic P, et al. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe 2014;16:495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Strandwitz P, Kim KH, Terekhova D, et al. GABA-modulating bacteria of the human gut microbiota. Nat Microbiol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dodd D, Spitzer MH, Van Treuren W, et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 2017;551:648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Levy M, Thaiss CA, Zeevi D, et al. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell 2015;163:1428–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kaparakis-Liaskos M, Ferrero RL. Immune modulation by bacterial outer membrane vesicles. Nature Reviews Immunology 2015;15:375–387. [DOI] [PubMed] [Google Scholar]
- 139.Shen Y, Giardino Torchia ML, Lawson GW, et al. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 2012;12:509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Vatanen T, Kostic AD, d’Hennezel E, et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 2016;165:842–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Donia Mohamed S, Cimermancic P, Schulze Christopher J, et al. A Systematic Analysis of Biosynthetic Gene Clusters in the Human Microbiome Reveals a Common Family of Antibiotics. Cell 2014;158:1402–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dreher-Lesnick SM, Stibitz S, Carlson PE Jr. U.S. Regulatory Considerations for Development of Live Biotherapeutic Products as Drugs. Microbiol Spectr 2017;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bikard D, Euler CW, Jiang W, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol 2014;32:1146–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Paixao AC, Ferreira AC, Fontes M, et al. Detection of virulence-associated genes in pathogenic and commensal avian Escherichia coli isolates. Poult Sci 2016;95:1646–52. [DOI] [PubMed] [Google Scholar]
- 145.Yasueda A, Mizushima T, Nezu R, et al. The effect of Clostridium butyricum MIYAIRI on the prevention of pouchitis and alteration of the microbiota profile in patients with ulcerative colitis. Surg Today 2016;46:939–49. [DOI] [PubMed] [Google Scholar]
- 146.Krag A, Israelsen H, von Ryberg B, et al. Safety and efficacy of Profermin(R) to induce remission in ulcerative colitis. World J Gastroenterol 2012;18:1773–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Krag A, Munkholm P, Israelsen H, et al. Profermin is efficacious in patients with active ulcerative colitis--a randomized controlled trial. Inflamm Bowel Dis 2013;19:2584–92. [DOI] [PubMed] [Google Scholar]
- 148.Scholmerich J, Fellermann K, Seibold FW, et al. A Randomised, Double-blind, Placebo-controlled Trial of Trichuris suis ova in Active Crohn’s Disease. J Crohns Colitis 2017;11:390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Summers R Trichuris suis seems to be safe and possibly effective in the treatment of inflammatory bowel disease. Am J Gastroenterol 2003;98:2034–2041. [DOI] [PubMed] [Google Scholar]
- 150.Summers RW, Elliott DE, Urban JF, et al. Trichuris suis therapy for active ulcerative colitis: A randomized controlled trial. Gastroenterology 2005;128:825–832. [DOI] [PubMed] [Google Scholar]
- 151.Summers RW, Elliott DE, Urban JF Jr., et al. Trichuris suis therapy in Crohn’s disease. Gut 2005;54:87–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mazmanian SK, Liu CH, Tzianabos AO, et al. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005;122:107–18. [DOI] [PubMed] [Google Scholar]
- 153.Olle B Medicines from microbiota. Nat Biotechnol 2013;31:309–15. [DOI] [PubMed] [Google Scholar]