Abstract
All proteins end with a carboxyl terminus that has unique biophysical properties and is often disordered. Although there are examples of important C-termini functions, a more global role for the C-terminus is not yet established. In this review, we summarize research on C-termini, a unique region in proteins that cells exploit. Alternative splicing and proteolysis increase the diversity of proteins and peptides in cells with unique C-termini. The C-termini of proteins contain minimotifs, short peptides with an encoded function generally characterized as binding, posttranslational modifications, and trafficking. Many of these activities are specific to minimotifs on the C-terminus. Approximately 13% of C-termini in the human proteome have a known minimotif, and the majority, if not all of the remaining termini have conserved motifs inferring a function that remains to be discovered. C-termini, their predictions, and their functions are collated in the C-terminome, Proteus, and Terminus Oriented Protein Function INferred Database (TopFIND) database/web systems. Many C-termini are well conserved, and some have a known role in health and disease. We envision that this summary of C-termini will guide future investigation of their biochemical and physiological significance.
Keywords: C-terminus, minimotifs, C-terminome, short linear motifs, posttranslational modification, trafficking, C-terminal minimotifs
C-termini background
The carboxyl (C-) termini of proteins are often disordered and solvent exposed (Lobanov et al. 2010). This conformational flexibility of the C-terminus enables induced fit interactions through one or more C-terminal minimotifs. Unlike longer and more complex protein domains with stable 3D folds, shorter minimotifs can have secondary structures that drive weak, transient, and dynamic protein interactions (Neduva and Russell 2005; Sargeant et al. 2012). Minimotifs (also known as short linear motifs [SLiMs]) are much shorter (2–15 residues) functional components of proteins. Fundamentally, minimotifs are the targets of protein domains for protein–protein interactions, are modified by co- and post-translational modifications (PTMs), and are necessary and sufficient for trafficking of proteins to specific sub-cellular localizations (Balla et al. 2006; Rajasekaran et al. 2009; Vyas et al. 2009; Fortelny et al. 2015). The C-terminal minimotifs take advantage of the additional specificity of molecular recognition through the charged and polar backbone carboxyl moiety.
We have previously cataloged 3593 minimotifs in the human proteome that are anchored at the C-terminus and amalgamated in our C-terminome database/web-application (Sharma et al. 2016). For these experimentally verified C-terminal minimotifs, 82% are PTMs, 18% bind protein domains, and less than 1% are for protein trafficking (Sharma et al. 2016). For instance, the C-terminus of soluble proteins retained in the endoplasmic reticulum contains a KDEL > minimotif (letters are for the single letter amino acid code; “>” indicates the C-terminus of the proteins) (Cabrera et al. 2003; Sharma et al. 2016). Type II transmembrane proteins targeted to the plasma membrane often contains a CaaX > minimotif that enzymes recognize and prenylate the Cys thiol in the minimotif, “a” is an aliphatic amino acid, and “x” represents redundancy for any of the 20 amino acids (Sharma et al. 2016; Wang and Casey 2016). Lipidation modifications, like prenylation anchor proteins to membranes by insertion of the hydrophobic fatty acid chains into the lipid bilayer. Similarly, PDZ domains, bind to other proteins containing [DE]x[LV]>, x[ST[x[LV]>, and xϕxϕ> minimotifs at their C-terminus, where “ɸ” represents hydrophobic residues and residues within brackets indicate positional degeneracy (Beuming et al. 2005; Sharma et al. 2016). There are approximately 500 proteins with at least one PDZ domain and approximately 3,000 instances of C-terminal PDZ minimotifs in the human proteome (Letunic and Bork 2018). Such a large number of C-terminal minimotif instances for one type of minimotif: protein domain interaction supports a generalizable use of C-terminal minimotifs for regulating cellular processes.
Early studies on the C-termini of proteins examined their role in protein translation termination, folding, and enzymatic activities. Several reviews have focused on these aspects of protein C-termini. Marino et al. and Tanco et al. reviewed the contemporary proteomics techniques to study C-terminal modifications (Marino et al. 2015; Tanco et al. 2015), and Carugo covered the biophysical properties of both the N- and the C-protein termini (Carugo 2017). However, while Chung et al. summarized the molecular functions of protein C-termini (Chung et al. 2002), there is no recent comprehensive review of the C-terminal region of proteins and C-terminal minimotifs in the last 15 years.
Herein, we cover the biophysical properties of the C-terminus; different cellular mechanisms that produce a staggering number of C-termini, the C-terminal sequence patterns, their conservation, and evolution. We next, introduce the concept of minimotifs and subsequently, detail the molecular function of the C-terminal minimotifs. We also address the biochemical and bioinformatics approaches that have identified C-terminal sequences, sequence patterns, and functions. We also cover the databases dedicated to C-termini and their role in disease.
Biophysical properties of the carboxyl terminus
Because C-termini are polar, like charged amino acids, they are generally solvent exposed and available for binding to and modification by enzymes. To assess if C-terminal residues are solvent-exposed, Jacob and Unger calculated the distance of the terminal residues from the center of mass for 425 small monomeric proteins. The C-terminal residues were more distant from the COM than other residues including the charged residues (Jacob and Unger 2007). On average, ~87% of the C-terminal residues were solvent exposed, consistent with most solved proteins structures in the PDB. Given the polarity of the C-terminus, its solvent exposure is generally consistent with thermodynamics constraints (Jacob and Unger 2007).
While C-termini are on the surface, the last 5–10 amino acids lack significant electron density in structures determined by X-ray diffraction or inter-residue NOEs in structures determined by NMR. These disordered regions, generally are flexible with no stable structure, have a high net charge, low sequence complexity, and contain fewer aromatic residues (Lobanov et al. 2010). Several lines of evidence support disordered carboxyl termini. First, the C-terminal amino acid has fewer constraints, thus making it more flexible than other residues (Lobanov et al. 2010). Second, 30% of the disordered residues in proteins with solved structures are in the last 10 amino acids of the C-terminus (Lobanov et al. 2010). We and others have previously presented an in-depth discussion of the heterotypic nature and classification of disorder, an important consideration for C-terminal motifs (Sargeant et al. 2012; van der Lee et al. 2014). Because it is disordered, the C-terminus can assume different conformations a concept referred to as molecular recognition features or elements (MoRFs or MoREs) in intrinsically disordered proteins (Mohan et al. 2006; Hsu et al. 2013).
The disordered C-terminus binds to other proteins with an induced fit. Therefore, the MoRFs minimotifs with several different structures in the same protein can engage binding distinct partners with several different structural modes of interaction. Several disordered minimotifs assume α-helices or β-strands upon binding a target, whereas other disordered regions assume irregular secondary structures (Lobanov et al. 2010). Although disordered minimotifs can be present anywhere in a protein, they are particularly prevalent at the C-terminus. Notably, crystallizing a protein with its full C-terminus is difficult unless it is bound to its target (Laio and Micheletti 2006). Therefore, crystallographers often truncate the proteins at the C-terminus to reduce disorder, rendering the protein more prone to crystallization (Carugo 2017).
Despite the disorderedness, the C-terminus has local contacts and more often adopts an α-helix rather than a b-turn or coiled-coiled secondary structure (Laio and Micheletti 2006). Laio and Micheletti evaluated 373 monomeric proteins and 85 multidomain proteins from Protein Data Bank. By calculating the average gyration radius of the last 15 amino acids from the C-terminus, and the average number of contacts that the residues in any protein sequence make to the terminal residues, the authors found the C-terminal residues to be more locally organized and compact when compared to the N-terminus. The motif at the C-terminus is similar to an α-helix pattern (Laio and Micheletti 2006). An additional study identified a 1.3-fold preference for α-helices in the C-terminus over the N-terminus (Krishna and Englander 2005).
Although not relevant to minimotifs, there are examples where C-termini are also important for protein folding and are good epitopes for raising peptide-based antibodies (Asami-Odaka et al. 2005; Krishna and Englander 2005).
Cellular processes increasing the number of carboxyl termini
Approximately, 22,000 protein-encoding human genes are transcribed into mRNAs are subsequently translated into proteins with synthesis progressing from the amino to carboxyl terminus (Figures 1 and 2). There are several cellular processes that can produce many proteins with alternative C-termini from the same gene: splicing of pre-mRNA, translation termination by suppressor tRNAs, proteolytic processing of immature and mature proteins, degradation of C-termini by exonucleases, and PTMs. These processes create multiple C-terminal minimotifs in proteins produced from the same gene.
Figure 1.
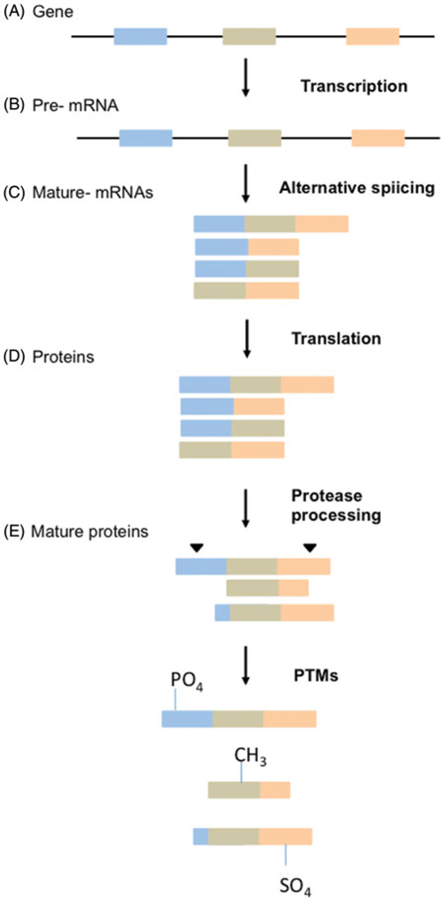
Cellular processes creating a diverse C-terminal end. A protein-coding gene with three exons is transcribed into a pre-mRNA, alternative spliced, and each transcript is translated into a unique protein. Proteases cleave each protein and PTMs are attached creating new protein species.
Figure 2.
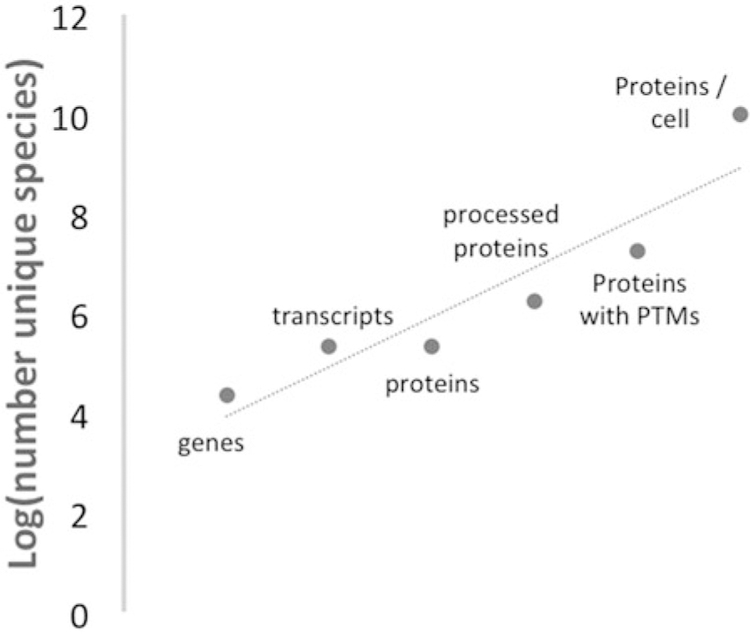
Estimate of factors contributing to numbers of C-terminal ends in the human proteome. The human genome has approximately 22,000 protein-coding genes. Each gene produces five alternatively spliced transcripts, with approximately 110,000 transcripts encoding the same number of proteins. Assuming each protein is proteolyzed during its life producing eight proteins then there are 1.8 M proteins with c-termini. With approximately 10 PTMs per protein, there are 17.6 M unique possible proteins present in a cell assuming one modification per protein (Jensen 2004; de Klerk and ‘t Hoen 2015; Tress et al. 2017). If we consider all PTM permutations, there could be nearly a quadrillion unique protein molecules, far exceeding the estimated 10 billion proteins per cell.
Alternative splicing of a pre-mRNA at differing intron-exon junctions produces multiple spliced mature mRNA molecules from the same gene. Approximately, 95% of the human genes are alternative spliced (Stamm et al. 2005; Wang et al. 2008). RNA-Seq analysis suggests that most genes produce, on average >10 unique transcripts (Tress et al. 2017). Approximately, 5% of the protein-coding human genes are more extensively spliced, expressing about 100 putative transcripts. An analysis of 1421 splice variants from 404 genes with minimotifs showed that minimotifs are enriched in splice variants (Weatheritt et al. 2012). Both PTM and binding minimotifs were identified in these splice variants supporting a well-defined role for molecular functions in C-termini. Based on these observations, we estimate about 220,000 unique C-termini from alternative splicing.
Another process increasing the number and diversity of the C-termini is alternative termination of protein translation by suppressor tRNAs. Suppressor tRNAs are tRNAs having mutations in anticodons. These mutations allow suppressor tRNAs to bind stop codons, add an amino acid and continue to elongate polypeptides in lieu of termination. Thus, this process extends the C-terminus of proteins. Additionally, deficiency of eukaryotic release factors (eRF1 and eRF3) generates alternative C-termini from premature translational termination through peptidyl-tRNA hydrolysis. It is clear that in some human cells premature translational termination and alternative termination of protein translation produces new C-termini (Janzen and Geballe 2004). Stop codon tRNAs can encode other amino acids express proteins with a different C-termini (Lee et al. 1990; Swart et al. 2016). The extent of this phenomenon at the human proteome level remains to be determined.
Precursor proteins are frequently proteolyzed creating new C-termini in mature proteins. This is prevalent in peptide hormones, precursor proteins, and zymogens. Exopeptidases and endopeptidases can produce different C-termini from the same protein substrate. Where exopeptidases trim proteins, one amino acid at a time from either protein termini, endopeptidases cleave proteins somewhere in the middle of their sequence. Both proteases produce new C-termini with minimotifs that change a protein’s function, stability, or location. It is challenging to biochemically identify the C-terminus of proteins biochemically as the C-terminal is relatively less reactive than the amino terminus (Marino et al. 2015). Nevertheless, the TopFIND group improvised MS techniques, such as C-terminal amine-based isotope labeling of substrates (C-TAILS) and combined fractional diagonal chromatography to identify 130,182 neo C-termini (Bonetto et al. 1997; Sechi and Chait 2000; Colaert et al. 2013; Nika et al. 2013; Fortelny et al. 2015; Zhang et al. 2018). Proteomic analysis of Escherichia coli, Mus musculus, and Homo sapiens identified that >10% of C-termini are derived from proteolysis, thus we estimate that endoproteolysis produces at least 2000 new human C-termini (Lange and Overall 2013). Although the large-scale impact of the exoprotease-generated C-terminal ends is not known, we extrapolate from a small set of proteins. Exoproteolysis produces an average of eight new C-terminal ends per protein for Fibrinogen-a C3f, C4a, and IYIH4 (Villanueva et al. 2006). Therefore, as many as 176,000 C-terminal ends may be present and available for C-terminal minimotif-driven interactions.
Nearly, all proteins have multiple PTMs. C-termini with PTMs can be considered chemically distinct entities, even though the sequence does not change. PTMs are added to growing polypeptides during the protein translation. An example is that Ser/Thr glycosylation is critical for protein folding quality control in the endoplasmic reticulum. PTMs on C-termini regulate the activity and localize proteins in a signal-dependent spatiotemporal manner. Although there are approximately 470 types of PTMs, only 1% of PTMs are thought to occur in vivo (Lobanov et al. 2010). Thus, PTM validation needs to be rigorous. The C-terminome database has approximately 3000 instances of PTMs at carboxyl termini.
Clearly, many types of transcriptional, translational, and post-translational events increase the number and diversity of C-termini and cells may have a total of 100,000–500,000 unique C-termini at a unique time. Many of these ends are likely to have functions.
Sequence conservation and composition of carboxyl-termini
Analysis of 1000 genomes project data revealed that 83% of the known C-terminal minimotifs are fixed in the human population with much of the variation in the PTM C-terminal minimotifs. Moreover, a global analysis done on the human proteome indicated that the C-terminal minimotifs are more constrained than the entire genome and protein-coding regions, indicating that the C-terminal minimotifs are likely to have important functional significance (Sharma et al. 2016).
The determinants of protein interactions and PTMs can be represented as sequence consensus motifs with a combination of fixed and degenerate positions. To predict and evaluate new motifs it is important to know whether the carboxyl termini in proteomes have altered amino acid composition or positional biases. Berezovsky et al. analyzed the last 10 amino acids of 1000 s of proteins: 1918 Escherichia coli, 3303 Saccharomyces cerevisiae, and 3243 Homo sapiens sequences (Berezovsky et al. 1997). In all three species, the positively charged K and R amino acids were favored in the two positions juxtaposed to the terminus. G residues were least common at the C-terminus (Berezovsky et al. 1997). However, a more recent, proteome-wide analysis indicated that the overall amino acid composition in the C-terminus (last 10 amino acids) is similar to that of the human proteome (Sharma et al. 2016). Certain residues, such as R, C, Q, H, K, P, and S, notably charged and polar amino acids, were slightly over-represented in the C-terminus (Sharma et al. 2016).
There are several biological processes that can contribute to bias in the amino acid composition of C-termini. Since many globular proteins begin to fold co-translationally, the C-terminus may contribute to folding and structural stability. In Escherichia coli, the last and the penultimate residue from the C-terminus of a growing polypeptide cooperatively enhance translation termination (Björnsson et al. 1996). The ordered last residues (an α-helix or β-strand) and K, R, and Q residues at the penultimate position correlated with an efficient translation termination for UGA stop codon. The last residue at the C-terminus may be important for the efficiency of translation termination and thus gene expression. In Escherichia coli, efficiently terminated genes have a K, whereas inefficiently terminated proteins have Thr or Pro as the last residue (Björnsson 1996).
Mutation of stop codons or a frameshift mutation that causes read-through of a stop codon introduces new C-termini. This is pervasive, as identified in human, mouse, rat, yeast, and Drosophila strains (Lobanov et al. 2010). In yeast new C-termini, are not yet fixed in the population indicating that they are likely still under selection (Giacomelli et al. 2007). Whether or not the alternative termini identified in these studies are of functional significance, will require further investigation.
Introduction to minimotifs
The term “motif” first described restriction enzyme recognition sites in nucleic acids in the literature (Smith and Wilcox 1970). One of the first functional protein motifs and the corresponding concept of a consensus sequence were identified for spore endoproteolysis of a protease recognition site in 1980s (Setlow et al. 1980). In 1987, the first trafficking motif, KDEL > was discovered and later identified as a receptor binding motif (Munro and Pelham 1987; Vitale and Denecke 1999). However, for many years, varied nomenclature of minimotifs existed in the literature. The conceptual term loosely included both structural and sequence motifs (Hodgman 1989; Thornton and Gardner 1989; Seto et al. 1990; Sheridan and Venkataraghavan 1992; Bork and Koonin 1996). In 1990, Tim Hunt unified the minimotif concept, calling these functional elements “short linear motifs” (Dice 1990). Eventually, it was the minimotif miner (MnM) and eukaryotic linear motif (ELM) resource groups that formalized a minimotif definition and model (Balla et al. 2006; Rajasekaran et al. 2009; Vyas et al. 2009; Dinkel et al. 2012; Mi et al. 2012; Lyon et al. 2018).
A minimotif has four attributes: a source protein, a target, an activity, and a secondary structure of the minimotif sequence (Vyas et al. 2009; Sargeant et al. 2012). A source is a protein that contains a minimotif sequence, having one or more known activities (binding, modification, and trafficking) and requires another molecule, a target, for its activity. Although structure is apparent in minimotif interaction with its targets, it is more complex often driven through induced-fit disorder to secondary structure transitions. Minimotifs are present in both disordered regions of proteins and also can occupy any of the 32 protein secondary structures (Vyas et al. 2009; Sargeant et al. 2012). An analysis of a nonrandom sample of 241,990 minimotifs in the MnM3 database quantified a distribution of 28% of the minimotifs in the intrinsically disordered regions, 27% in the folded regions, and 45% in the hybrid regions of proteins (Sargeant et al. 2012).
Minimotifs are modeled as consensus sequences, position specific-scoring matrices (PSSMs), and instances. Instances are contiguous amino acid sequences in a peptide or protein with that have an established molecular function. Sets of minimotif instances in several proteins can be aligned to obtain consensus sequences or PSSMs where degenerate substitutions at certain positions are modeled, generally by frequency. Consensus sequences and PSSMs infer new minimotifs instances that can be tested for prediction accuracy.
The majority of the known molecular functions and sequence specificity of minimotifs comes from the low-throughput experimentation examining a small number of instances. SPOT arrays, phage display, and affinity MS/MS, because of their throughput, are tools to more broadly assess minimotif specificity (Sundell and Ivarsson 2014; Liu 2017). Large numbers of PTM minimotifs are also discovered through affinity mass-spectrometry studies. Despite these advancements, determinants of minimotif specificity are incomplete. One likely explanation is that secondary structure is an essential component of specificity. However, because minimotif interactions are generally transient, reversible and have weak affinities (Kd = 1–10 μM), it may be difficult to resolve the complete specificity of minimotifs through high-throughput approaches that do not assess the relative strengths of the interactions (Neduva and Russell 2005).
C-terminal minimotifs
Minimotifs can be located at the C- or N-terminus, or at any position throughout a protein. The termini have unique chemistries and some minimotifs are specific to the termini. More than 3500 C-terminal minimotifs were identified from 73,532 human minimotifs from the MnM3 database (Sharma et al. 2016).
C-terminal minimotifs can have sequences with a consensus residue exactly on the C-terminus (anchored) or a short distance away from the last residue of the C-terminus (non-anchored). There are several examples of non-anchored C-terminal minimotifs. The FCYENEV minimotif in the Kir2.1 channel is both necessary and sufficient to export of Kir2.1 from the ER to Golgi complex (Ma et al. 2001; Stockklausner et al. 2001). Many G-protein-coupled receptors (GPCRs) contain a conserved hydrophobic minimotif, FxxxFxxxF, near its C-terminus (Bermak et al. 2001). DRiP78, an ER resident protein, binds to this minimotif and is essential for trafficking D1 GPCRs. The VxxSL motif in Kv1 channels is required for its trafficking to the cell surface (Levitan and Takimoto 2000).
Types of C-terminal minimotifs
Binding C-terminal minimotifs
At least 650 C-terminal minimotifs have a binding activity for protein interactions often driving large macromolecular complexes and cell signaling. Minimotif-domain interactions complement other modes of protein interactions, such as domain-domain interactions. At least 63 unique protein domains bind to specific minimotifs and several key examples are shown in Table 1 (Stein et al. 2011; Dinkel et al. 2012). Given a large number of minimotifs and their generally low complexity when compared to protein domains, many minimotif interactions are promiscuous engaging multiple target domains. However, the physical separation of proteins in different sub-cellular compartments, different phases of the cell cycle, and differential tissue-specific expression impart some specificity to minimotif interactions.
Table 1.
Examples of the C-terminal minimotifs and protein domain binding.
| Domains | # H. sapiensa | # All species | Minimotif sequence | #Minimotifs | Reference |
|---|---|---|---|---|---|
| PDZ | 666 | 333,040 | Class I: x[S/T[x[L/V]>; Class II: xfs and protein domain | 2849 | Hung and Sheng (2002) |
| TPR | 330 | 1,309,452 | EEVD> | 22 | Brinker et al. (2002) |
| ZNF_UBD | 11 | – | GG> | – | Reyes-Turcu et al. (2006) |
| 14_3_3 | 22 | 6,285 | YDI>; YTV> | 4 | Fuglsang et al. (2003) |
Data from SMART databases.
14–3-3 proteins binding minimotifs.
The 14–3-3 family of proteins interact with phospho-proteins where the Ser and Thr residues within the minimotif sequence are phosphorylated (Coblitz et al. 2006). 14–3-3 domains can interact with both C-terminal and internal consensus sequences. In plants, 14–3-3 domains bind the YTV > motif in the Hþ-ATPase, present in the plasma membrane activating an auto-inhibited ATPase (Fuglsang et al. 2003). This instance of a 14–3-3 minimotif does not require phosphorylation to drive the interaction, although Ser/Thr phosphorylation is required for most 14–3-3 domains (Fuglsang et al. 2003; Madeira et al. 2015).
PDZ domain-binding minimotifs.
Proteins with PDZ domains function in signal transduction, trafficking, and are scaffolds for large molecule complexes (Kim and Sheng 2004; Lee and Zheng 2010). PDZ domains (80–100 residues) have only one binding site that binds to proteins with a C-terminal minimotif; however, there are exceptions for binding non-C-terminal motifs (Wong et al. 2003; Zhang et al. 2009). PDZ minimotifs are classified based on the consensus recognition patterns: class I: x[S/T[x[L/V]>, class II: xϕxϕ>, and class III: xDxV> (Hung and Sheng 2002). 591 C-terminal human minimotifs interact with PDZ domains (Sharma et al. 2016). In Homo sapiens, 546 proteins in the SMART database contain PDZ domains (Letunic and Bork 2018). Upon binding to the PDZ domain, the C-terminal minimotif assumes a b-strand conformation and extends a b-sheet in the PDZ domain, an example of a secondary structure determinant (Laskowski et al. 2005; Lee and Zheng 2010). Minimotifs for phosphorylation of Ser, Thr, and Tyr residues in, or juxtaposed to a PDZ binding motif, regulate binding of C-terminal minimotifs to PDZ domains. However, there is no consensus as to whether phosphorylation inhibits or enhances binding (Lee and Zheng 2010). PDZ-binding minimotifs in the C-terminus are well-conserved among vertebrates and invertebrates (Chimura et al. 2011).
Transducin-like enhancer (TLE) binding minimotifs.
There are 57 TLE of split domains in co-repressors bind to and repressor proteins that inactivate promoters (Fisher et al. 1996; Aronson et al. 1997). There are 11 human proteins that have the WRP[YW] TLE consensus binding motif. The core binding factor acute myeloid leukemia (AML1/CBF) a-subunit contains a C-terminal WRPY > minimotif that binds to transcriptional corepressors with TLE domains. Together, TLE domain proteins and their binding partners are required for development. TLE1 inhibits the trans-activation of T cell receptor enhancers (Levanon et al. 1998). Other C-terminal sequences in the AML1 are also required for these interactions (Fisher et al. 1996; Aronson et al. 1997). Hairy family factors contain a WRPW > minimotif, which is both necessary and sufficient for the binding of Groucho to the Hairy-related transcription factors, which contains a TLE domain (Chen and Courey 2000).
Tandem tetratricopeptide repeat (TPR) binding minimotifs.
TPRs are domains that interact with minimotifs in multiprotein complexes (Zeytuni and Zarivach 2012). TPRs are 34 amino acid helix-turn-helix structural motif with 1–16 repeat in proteins (Cortajarena and Regan 2006). Although there is no known general consensus minimotif sequence for TPRs, the TPRs in Hop proteins bind to the EEVD > minimotifs in the Hsp70 and Hsp90 chaperones. These minimotifs bind in an extended coil conformation (Brinker et al. 2002; Zeytuni and Zarivach 2012). There are 273 TPR domain-containing proteins and 22 EEVD > minimotifs in the human proteome (Sharma et al. 2016).
Ubiquitin-binding (UBP) minimotifs.
Isopeptidase T contains a zinc-finger ubiquitin-binding domain (ZnF UBP) that binds the GG > minimotif of ubiquitin (Reyes-Turcu et al. 2006). Although many UBP domains exist, the general specificity for GG > remains to be determined, as this minimotif may bind exclusively to ZnF UBP (Hurley et al. 2006).
Trafficking C-terminal minimotifs
Proteins trafficking to specific cellular compartments are important for correct function. Several trafficking signals are located at the C-terminus of proteins and masking of such signals by fusing an epitope tag often leads to their mislocalization. There are 44 trafficking C-terminal minimotif consensi and instances that are most often necessary and sufficient for localization (Sharma et al. 2016). Many proteins destined to be in the secretory pathway and endomembrane system contains minimotif signals at their C-termini (Yofe et al. 2016). Aberrant mislocalization to the wrong subcellular compartment can lead to disease. Standard techniques to determine protein localization, and thereby the trafficking determinants include immunostaining, confocal microscopy, Fösters resonance energy transfer (FRET) microscopy, fluorescence life-time imaging microscopy, and various ultracentrifugation fractionation approaches (Teesalu et al. 2009). Several examples of trafficking C-terminal minimotifs are summarized below:
Endoplasmic reticulum (ER) retention and retrieval minimotifs.
Protein sorting between the ER and Golgi-apparatus is mostly dependent upon C-terminal minimotifs (Teasdale and Jackson 1996). Soluble proteins synthesized in the ER lumen contain an ER retention minimotif, KDEL> (Gomez et al. 2000). Type I transmembrane proteins contain dilysine retrieval minimotifs, KKx(1,3), KxKx>, and KxKxx> (Jackson et al. 1990; Gomez et al. 2000). These Lys residues are critical for the functioning and cannot be replaced with other charged residues such as Arg and His (Jackson et al. 1990).
Internalization minimotifs.
The C-end rule (also known as C-endR) peptides have sequence (R/K)xx(R/K)> (Teesalu et al. 2009). These peptides are tissue penetrating internalization peptides that bind to Neuropilin-1 (NRP-1), a cell surface receptor that is essential for angiogenesis and nervous system development (Teesalu et al. 2009; Zanuy et al. 2013). A screen for proteins that bind C-endR peptide minimotifs identified NRP-1 (Teesalu et al. 2009).
Peroxisome targeting minimotif.
Peroxisomes are small organelles that are present in all eukaryotic cells and contain hydrolytic enzymes. Peroxisomes contain Pex5, a soluble import receptor expressed on peroxisomal membranes. Pex5 binds to proteins with the SKL > minimotif on the proteins that are imported into the peroxisome lumen. The SKL > minimotif is in 34 proteins and is called the PTS1 signal. Another peroxisome import signal, PTS2 has a minimotif on the N-terminus of imported proteins (Smith and Aitchison 2013).
Post-translation modification of C-terminal minimotifs
Translation is only the start of protein a proteins life. Co- and PTMs are the 100 s of covalent attachments of molecules to polypeptides changing their physiochemical properties. Broadly, there are two main types of PTMs: 1. The exoproteolysis of polypeptides removes small stretches of residues from either termini. Endoproteolysis of a polypeptide hydrolyzes an internal peptide bond producing two smaller polypeptide chains. 2. The reversible or the irreversible addition or removal of small chemical moieties, isomerizations, or covalent attachment of other proteins. PTMs can occur at multiple sites in a protein and/or multiple PTMs can occur sequentially in a protein regulating functional outputs. There are approximately 469 different types of modifications in eukaryotes (Khoury et al. 2011; UniProt Consortium 2015). The majority of known C-terminal minimotifs are PTM minimotifs. The major experimentally verified C-terminal modifications include phosphorylation, acetylation, ubiquitylation, proteolysis, amidation, and lipidation with another 18 PTMs occurring less frequently in a limited set of C-terminal minimotifs (Sharma et al. 2016).
Amidation.
The C-terminal a-amidation of bioactive neuropeptides and hormones catalyzed by peptidylglycine a-amidating mono-oxygenase (PAM) is one of the most common PTMs and is the 4th most common in Swiss-Prot (Marino et al. 2015). Amidated peptides are produced by sequential degradation of a C-terminal Gly residue. Peptidylglycine a-hydroxylating monooxygenase (PHM) and peptidyl-a-hydroxyglycine a-amidating lyase (PAL), the two catalytic domains of PAM amidates peptides. The amide group neutralizes the negative charge of the carboxyl end, thus changing its chemical properties (Marino et al. 2015). In addition to many bioactive peptides, the C-terminus of the heavy chain of many monoclonal antibody-based drugs is amidated, although its significance is not yet known (Tsubaki et al. 2013). Since cells contain exopeptidases, the C-terminus of the cellpenetrating peptides is often amidated, which increases their half-life (Soleymani-Goloujeh et al. 2017).
Crotonylation.
Crotonylation is the addition of crotonyl group to Lys side chains, regulating gene expression by modifying histones in promoters or enhancer nucleosomes (Tan et al. 2011; Ju and He 2017; Wei et al. 2017). Although not exclusive to the C-terminus, KxxK, and PxK minimotifs have a higher propensity for crotonylation (Wei et al. 2017). Histone proteins (H2A1H, H2A1B, H2A1, H2A1D, H2A2A, H2A3, and H2A1C) with conserved C-termini are crotonylated (Tan et al. 2011; Sharma et al. 2016).
Glycosylation and glycation.
Glycosylation is an enzyme-catalyzed covalent addition of sugars, both co- and post-translationally (Spiro 2002). Glycation, on the other hand, is the non-enzymatic reaction of adding sugars. Sugars are added primarily on the newly synthesized unfolded polyproteins resulting in the proper folding of proteins. Glycosylation usually occurs on the membrane-bound and soluble proteins of the secretory pathway. Most often, glycosylation occurs in the ER proteins, but several examples of proteins synthesized in the cytosol also exist. Several types of glycosylation exist based on the position of linkage, with N-linked and O-linked glycosylation being the most common. N-glycosylation forms a covalent amide link-age to the side chain of Asn in Nx[ST] minimotifs and O-glycosylation occurs on Ser and Thr residue with no clear specificity. While most glycosylation occurs throughout proteins, there are at least ten N-glycosylation, three O-glycosylation, and two glycation minimo-tif instances in the C-terminus (Sharma et al. 2016).
Lipidation.
Lipidation is the addition of different types of fatty acyl or polyisoprenyl groups moieties to either termini. Lipids are attached to the side chains of Cys, Ser, and Lys and target proteins to a lipid bilayer. Cholesterol esterification and glycosylphosphatidylinositol anchorage are two other types of C-terminal lipid modifications on extracellular membrane-bound proteins (Jiang et al. 2018). The two sub-types of prenylation modification are farnesylation (three isoprene groups) and geranylgeranylation (four isoprene groups) on the Cys residues near the C-termini. A total of 45 prenylation instances (six farnesylation and two geranylgeranylations) are noted at the C-termini (Jiang et al. 2018).
The substrates of farnesyltransferase and geranylgeranyl transferase contain Caax > minimotif. In Ras proteins and lamin B, after prenylation of C in Caax>, the aax > sequence gets removed and another enzyme carboxymethylates the prenylated Cys C-terminus. Rab geranylgeranyl transferase adds two geranylgeranyl groups at the CC > or CxC > minimotifs of Rab proteins (Sharma et al. 2016). N-terminal lipidation modification is also known to occur at Gly in the consensus sequence < MGxxx[S/T] (Jiang et al. 2018).
Methylation and demethylation.
There are two types of biological protein methylation: 1. methylation of the C-terminal protein carboxyl group; and 2. that on side chains of internal residues such as Arg or Lys. Carboxyl methylation neutralizes the charge of the C-terminus (Kawata et al. 1990; Favre et al. 1994; Ghomashchi et al. 1995; Choy et al. 1999; Brahms et al. 2000; Kowluru 2000; Ahmad et al. 2002; Winter-Vann et al. 2003; Ong et al. 2004; Zhang et al. 2008; Ren et al. 2010). Carboxymethylation is most often attached to the terminal Cys carboxylate during C-terminal protein lipidation. One of the classical examples is the methylation of phosphoprotein phosphatase 2 A which along with protein phosphatase 1 (PP1) accounts for more than 90% of the phosphatase activity in mammalian cells (Tolstykh et al. 2000). The TPDYFL > minimotif is methylated at the Leu carboxyl. While side chain methylation can occur throughout a protein, there are 43 methylation instances (mono-and di-) of methylation of these amino acids in the C-terminus. Methylated Arg or Lys residues drive protein-protein interactions.
Phosphorylation and dephosphorylation.
Phosphorylation, the most predominant form of PTM, is the reversible covalent attachment of phosphate groups, on Ser, Thr, and Tyr residues, and in the case of prokaryotes on His and Asp. The interplay of kinases phosphorylating proteins and phosphatases removing the phosphate moieties act as an “on” and “off” switch in signaling pathways and almost all cellular processes. Of 2096 C-terminal minimotifs are phosphorylated with ~73% modifications at Ser and the rest on Thr and Tyr (Karve and Cheema 2011). Given that Ser is the most common residue in the proteome, it is likely that the phosphoproteome studies are biased in predicting Ser to be more commonly phosphorylated residue when compared to Thr and Tyr (Sharma et al. 2016).
Proteolysis.
Exo- and endo-peptidases catalyze an irreversible hydrolysis of peptide bonds. There are ~130,000 C-terminal instances of proteins reported due to limited proteolysis (Fortelny et al. 2015, p. 0; Sharma et al. 2016). The solvent-exposed C-terminus is often prone to proteolytic cleavage. This may be because the C-terminus often does not assume a stable structure. The C-terminus is often removed from proteins to enhance their crystallization and also often does not produce a signal in electron density maps for protein structure determination by X-ray crystallography nor NOEs in structures determined by nuclear magnetic resonance spectrometry (Laio and Micheletti 2006). Dedicated sub-fields of proteomics termed as degradomics or terminomics have emerged to study proteolysis. The current methods for identifying peptides produced by proteolysis were recently reviewed (Rogers and Overall 2013).
Prohormones are expressed in an inactive form. Likewise, many enzymes, including proteases are expressed as inactive zymogens. Both are activated upon endoproteolytic processing, releasing the bioactive molecule and producing new C-termini. Carboxypeptidases hydrolyze a peptide bond at the C-terminus of proteins, and have a role in blood clotting, wound healing, and many other cellular processes (Wallace et al. 1982; Lyons and Fricker 2010; Tanco et al. 2015).
In addition to prohormones, the ubiquitin/SUMO/NEDD family members are expressed in an inactive form having an extended C-terminal tail. Many proteases cleave ubiquitin unmasking a GG motif, then recognized for attachment to Lys residues (Grou et al. 2015). The SENP (sentrin/SUMO-specific protease) family of proteases cleave the inactive form of small ubiquitinlike modifier (SUMO) exposing a di-glycine motif recognized by SUMO-E1 ligases (Reyes-Turcu et al. 2006). Similarly, NEDP1 or UCHL3 proteases cleave the C-terminus of Nedd8, another ubiquitin-like protein, activation it for recognition by Nedd8 E1 ligases (Zheng and Shabek 2017). The ligases bind the functionally active forms of ubiquitin and SUMO catalyzing formation of an isopeptide bond with the Lys of their target substrates. Some additional examples are listed in Table 2.
Table 2.
Examples proteins activated upon proteolysis.
| Precursor protein | Mature protein | Protease | Function | PubMed |
|---|---|---|---|---|
| Precursor-small ubiquitin like modifier (SUMO) | SUMO-1, SUMO-2, and SUMO-3 | SENP (sentrin/SUMO-specific protease) family of proteins | Mark for degradation, localization, etc. | van der Veen and Ploegh (2012) |
| Precursor polyubiquitin protein | Ubiquitin | Many proteases including UCHL3, USP9X, USP7, and USP5 | Mark for degradation, localization, etc. | Kimura and Tanaka (2010); Grou et al. (2015) |
| Neural precursor cell expressed developmentally down-regulated protein 8 (precursor-Nedd8) | Nedd8 | NEDP1 (DEN, SEN8); UCHL3 | Cell cycle progression and survival | Wada et al. (1998); van der Veen and Ploegh (2012) |
| Fau with ribosomal S30 | Ubiquitin-like protein FUBI | – | ERK-MAPK pathway and LPS/interferon-induced apoptosis | Watanabe et al. (2013) |
| Ubiquitin-fold modifier (UFM-1) precursor | Ufm1 | UfSP1 and UfSP2 | Nuclear receptors mediated transcription and cellular response to ER stress | van der Veen and Ploegh (2012); Yoo et al. (2014) |
| Interferon-stimulated protein 15 precursor | ISG15 | UBP43 | Immunity | Jeon et al. (2010); Rahnefeld et al. (2014) |
| Proinsulin | Insulin | PC1 and PC2 | Metabolism | Orci et al. (1986) |
| Angiotensinogen | Angiotensin I and Angiotensin II | Renin and Angiotensin-converting enzyme | Increase vasopressin production in CNS, Adhesion and aggregation of platelets, etc. | Velez et al. (2012) |
| Pro-caspase 8a | Caspase p30 | Not known yet | Sensitization of cells toward apoptosis | Hoffmann et al. (2009) |
Sulfation.
Only a few proteins in the secretory pathway are sulfated on tyrosine as recently reviewed (Kehoe and Bertozzi 2000; Stone et al. 2009; Yang et al. 2015). Tyrosine sulfation drives protein–protein interactions, primarily of several extracellular and secreted proteins (Kehoe and Bertozzi 2000; Yang et al. 2015). For example, cholecystokinin (CCKN) contains two C-terminal sulfated tyrosines (Vishnuvardhan and Beinfeld 2000). The inhibition of sulfation in precursor CCKN had no effect on the processing of precursor CCKN but increased the amount of CCKN secretion in endocrine cells. Therefore, it seems that tyrosine sulfation renders proteins more stable and soluble. However, the implications of such mutations in vivo still need to be established. The C-terminal sulfated tyrosines are also present in the fibrin binding domain of fibronectin (Liu and Suiko 1987; Meh et al. 2001).
Ubiquitylation and sumoylation.
Ubiquitylation is a reversible modification that targets proteins for degradation in different proteolytic structures, such as proteasome, vacuoles, and lysosomes; it is also initiates cell signaling (Komander and Rape 2012; Herhaus and Dikic 2015). In addition, new functions of ubiquitin addition in transcription, translation, and DNA repair are reported recently (Weissman 2001). Ubiquitin is an 8.5 kDa protein that is conjugated to Lys on other proteins. Polyubiquitin chains are built by sequential ubiquitylation of several Lys residues in ubiquitin. Subtle differences in the Lys residue positions that are ubiquitylated, types of ubiquitin chains (mono-versus multi-) and intra-cellular location of ubiquitylated proteins determine the fate of a protein. Like ubiquitin modification, sumoylation also regulates proteins in different cellular processes. SUMO proteins conjugate to the C-terminal Lys residues. Both ubiquitylation and sumoylation minimotifs are located throughout proteins with several C-terminal minimotifs including sumoylated Lys residues in Axin1 and p53 and an additional 215 instances of ubiquitylated C-terminal minimotifs (Gostissa et al. 1999; Muller et al. 2000; Rui et al. 2002; Li et al. 2006). Although the frequency of Lys at the C-terminus is higher than in the overall proteome (Sharma et al. 2016), based on current studies it is unlikely that all C-terminal Lys residues are ubiquitylated and sumoylated and that those modifications target proteins for degradation.
Identifying new carboxyl terminal minimotif activities
Despite significant bioinformatics advancements, computational methods to predict functional C-terminal sequence patterns with high sensitivity and specificity are rather limited. Therefore, the identification of C-terminal minimotifs thus far largely relies on low-throughput experiments to discover new or validate predicted C-terminal functions. PTM minimotifs are identified primarily by mass-spectrometry. Binding minimotifs for specific protein domains are identified through spot peptide array, phage display libraries, and other low-throughput assays. Proteome-wide studies to identify trafficking minimotifs do not yet exist.
Analysis of the C-termini of proteins has identified many over-represented C-terminal sequences and patterns in proteomes from a variety of organisms (Attwood et al. 1997; Gostissa et al. 1999; Gatto and Berg 2003; Prakash et al. 2004; Bahir and Linial 2005, 2006; Austin et al. 2007; Lam et al. 2010; Fortelny et al. 2015; Sharma et al. 2016). In 2003, analysis of six diverse proteomes identified numerous overrepresented C-terminal tripeptide pattern sequences (Gatto and Berg 2003). In 2007, sequences for seven eukaryotic proteomes were analyzed and identified several known true positives and several new anchored C-terminal tripeptide minimotif that were statistically enriched (Austin et al. 2007). In ProTeUS, a set of the over-represented unanchored C-terminal patterns was identified (Bahir and Linial 2005). Although these studies give insight into the potential for modular functions for C-termini, they examined a small fraction of human proteome. In 2015, a comprehensive analysis of the human proteome identified overrepresented sequence patterns up to 10 amino acids long with up to five degenerate positions (Sharma et al. 2016). Published molecular functions for 13% of human C-termini are known and additional functional inferences were based on rodent proteome minimotifs and matches to C-terminal consensus sequences (Sharma et al. 2016). The C-terminome datbase of known C-terminal minimotfs can serve as a benchmark and new sequence patterns are a foundation to explore and discover functions and targets for new C-terminal minimotifs.
C-termini databases
Known C-terminal minimotifs can be specifically identified from larger minimotif databases, such as in MnM and ELMs (Lyon et al. 2015; Gouw et al. 2018). Likewise, potential C-terminal minimotifs can be predicted from comprehensive peptide signatures (CoPS), and MOTIPS, utilities built to identify the signature sequences throughout a given protein sequence (Prakash et al. 2004; Lam et al. 2010). More recently, several databases have emerged that are dedicated to the C-terminus such as the C-terminome, TopFIND, and ProTEUS (Table 3) (Sharma et al. 2016). Although not specifically designed to the C-terminus, many specialized databases focus on specific molecular functions, such as O-Glycbase, PHOSIDA, Phosho3D, PhosphoSite-Plus, dbPTM, Human Histone modification databases, MEROPS, CutDB, and TOPPR for proteolytic sites; PDZBase and Binding DB for binding minimotifs (Gupta et al. 1999; Beuming et al. 2005; Igarashi et al. 2007; Liu et al. 2007; Zanzoni et al. 2007; Gnad et al. 2011; Colaert et al. 2013; Lu et al. 2013; Rawlings et al. 2014; Hornbeck et al. 2015; El Kennani et al. 2017).
Table 3.
List of minimotifs based databases.
| Web-applications | C-term only | Species | Type of dataset | # of functions | Reference |
|---|---|---|---|---|---|
| C-terminome | Yes | Homo sapiens | C-terminal minimotifs and Proteins | 3546 verified and ⁓9 million predicted | Sharma et al. (2016) |
| TopFIND3.0 | No | 5 species | Neo C-termini as from mass-spectrometry | 128 modifications, 130,182 proteolytic ends | Fortelny et al. (2015) |
| ProTeus | Yes | None specified | Signature groups in Swiss-Prot | Unknown (all predicted) | Gnad et al. (2012) |
| ELM | No | All | Short linear motifs anywhere in proteins | >275 motif classes and >300 motif instances | Dinkel et al. (2012) |
| MnM | No | All | Minimotifs anywhere in proteins | 1,060,436 instances | Lyon et al. (2018) |
| DILIMOT | No | None specified | Discovery-based tool to identify motifs in a protein sequence based on homology | None specified | Neduva and Russell (2006) |
| Proteolysis | |||||
| MEROPS | No | Many | Peptidase, peptidase substrates | 92,216 cleaved products; 908,326 peptidases | Rawlings et al. (2014) |
| CutDB | No | All | Proteolytic events, both actual and predicted | 3070 proteolytic events; 470 proteases | Igarashi et al. (2007) |
| TOPPR | No | human and mouse | Published proteolytically processed sites in human and mouse proteins based on MS/MS | Proteolysis sites | Colaert et al. (2013) |
| Binding | |||||
| PDZBase | Yes | None specified | PDZ-domain interaction with protein s | 300 interactions | Beuming et al. (2005) |
| Binding DB | No | None specified | Protein: drug molecules interactions | 1,391,403 binding data, for 7225 protein targets and 621,060 small molecules | Liu et al. (2007) |
| Post-translational modification0073 | |||||
| SwissProt | No | All | PTM, protein sequences | 556,196 entries on protein sequences | Boeckmann et al. (2003) |
| O-Glycbase | No | All | O-linked glycosylation sites | 242 glycoprotein entries | Gupta et al. (1999) |
| PHOSIDA | No | 9 species | Phosphorylation, acetylation and N-glycosylation site database | more than 60,000 sites | Gnad et al. (2011) |
| Phospho3D | No | All | 3D structures for phosphorylation site database | 5314 phosphorylation sites in 1805 sequences | Zanzoni et al. (2007) |
| PhosphoSite-Plus | No | Mammal | PTMs | 330,000 non-redundant | Hornbeck et al. (2015) |
| dbPTM | No | None specified | Integrated resource from 14 public databases on PTMs | 610,037 experimental and 546,911 putative PTM sites | Lu et al. (2013) |
| Human Histone Modification Database | No | Human | Histone modification database. | 43 modification types | Zhang et al. (2010) |
The C-terminome database contains the known experimentally verified C-terminal minimotifs, inferences based on the rodent C-terminal minimotifs, predictions of molecular functions for C-terminal sequences based on consensus sequences, and bioinformatic predictions of C-terminal sequences and patterns that are overrepresented in the human proteome when compared to that of random proteomes (Sharma et al. 2016). C-terminome can be searched or browsed for any human protein or C-terminal minimotifs.
The Terminus Oriented Protein Function Inferred Database (TopFIND) includes some published PTMs literature and 130,182 proteolytically derived neo-C-terminal ends (Fortelny et al. 2015, p. 0). The neo-C-terminal ends arise by in vivo proteolytic processing are identified through a mass-spectrometry-based approach, C-TAILS. TopFIND version 3.0 includes the data on five diverse species. The ProTEUS database has both predicted anchored and non-anchored C-terminal sequences allowing one position of degeneracy in the last 10 amino acids of proteins (Bahir and Linial 2005; Gnad et al. 2012). ProTEUS recognizes the terminal minimotifs based on a small set of approximately 100 known functional C-terminal minimotifs.
Carboxyl termini in diseases and medicine
The carboxyl end of proteins and C-terminal minimotifs are involved in several cellular processes including translation termination, protein folding, protein turnover, protein trafficking, and cellular signaling (Birrane et al. 2003; Chong et al. 2005; Millhouse and Manley 2005; Coblitz et al. 2006; Chinnadurai 2007; Wang and Malbon 2012; Lübkemeier et al. 2013; Sucic et al. 2013; Krishnan et al. 2014; Sun et al. 2014; Liu et al. 2015; Patil et al. 2015; Ryan et al. 2016; Wu et al. 2016). Consequently, the deletion of the C-terminus could compromise any of these processes by losing a C-terminal motif or creating a new one (Table 4). For instance, the 104-amino acid C-terminal truncation in the human ether-a-go-go-related gene (HERG) channels causes long QT syndrome type 2 (LQT2), a cardiac disorder (Kupershmidt et al. 2002). Because of this truncation, a gain-of-function endoplasmic reticulum retention signal RGR > is exposed reducing the trafficking of HERG to the cell surface where it functions. Several other examples are provided.
Table 4.
C-terminal minimotifs in human disease.
| Protein | Minimotif | General function | Disease | Reference |
|---|---|---|---|---|
| HERG channelsa | RGR> | Traffic | Long QT syndrome type 2 | Kupershmidt et al. (2002) |
| Rhodopsin | VAPA> | Traffic | Vision loss | Deretic et al. (2005) |
| MNK | LL> | Traffic | Menkes disease | Petris et al. (1998) |
| NKCC2 | LL> | Traffic | Bartter syndrome | Zaarour et al. (2012) |
| Bile salt export pump | YYKLV> | Traffic | Primary familial intrahepatic cholestasis type 2 | Lam et al. (2012) |
| SANS gene | TEL> | Bind | Usher syndrome type I | Reiners et al. (2006) |
| Claudin-16 | TRV> | Bind | Nephrocalcinosis | Müller et al. (2003) |
| NaV1.5 channel | SIV> | Bind | Cardiac disease | Shy et al. (2013) |
Gain of function.
Apolipoprotein E (Apo E) binds amyloid b peptides of plaques and forms neurofibrillary tangles with hyperphosphorylated tau. Amyloid plaques and neurofibrillary tangles are the hallmarks of Alzheimer’s disease (AD), a progressive neurodegenerative disorder and ApoE are a risk factor for late-onset disease. Chymotrypsin-like serine proteases preferentially cleave the E4 haplotype of ApoE over the E3 haplotype. The resulting ApoE4 fragment (d272–299) is neurotoxic in the transgenic mice leading to neuronal pathology in AD (Harris et al. 2003). The deletion of the C-terminus of ApoE4 unmasks a lipid-binding function in ApoE (244–272) and also interacts with amyloid beta peptides.
The binding of calmodulin to the C-terminus of aquaporin-O (residues 225–263 of a 263-residue protein) inhibits water permeability leading to the progression of cataract and vision impairment (Schey et al. 1999; Gold et al. 2012). Protein kinase A phosphorylates Ser-235 of aquaporin-O preventing its interaction with CaM. Thus, a mutation that eliminates Ser-235 can be detrimental to the vision (Lin et al. 2007).
Missense mutations can disrupt a C-terminal minimotif critical for protein function leading to diseases (Table 4). For instance, mutations in the VAPA > trafficking minimotif in Rhodopsin cause autosomal dominant retinitis pigmentosa (Deretic et al. 1998, 2005). Rhodopsin interacts with ADP-ribosylation factor 4 (ARF-4), a rhodopsin transport carrier, through its VAPA > minimotif. VAPA > is required for Rhodopsin trafficking to the photoreceptor rod outer segments. Some other examples are provided.
Several examples of mutations in a PDZ domainbinding minimotif in diseases exist. For instance, frameshift mutations in the SANS gene cause Usher syndrome type I, an autosomal recessive genetic neurodegenerative condition with hearing and vision loss (Verpy et al. 2000; Weil et al. 2003). SANS protein contains a PDZ domain-binding motif that interacts with the PDZ domain of the harmonin and scaffold protein (Reiners et al. 2006). Compromise of similar protein networks based on PDZ domain binding in photoreceptor cells may also lead to Usher syndrome. Similarly, NaV1.5 channel proteins contain an SxV > PDZ binding minimotif, that binds the PDZ domains of syntrophins and SAP97 in cardiomyocytes (Shy et al. 2014). The mutation of this motif mislocalizes the NaV1.5 channel and causes cardiac disease. A mutation in the PDZ binding motif of Claudin-16 abrogates its interaction with the PDZ domain of ZO-1, a scaffolding protein. This mutation mislocalizes Claudin-16 to lysosomes instead of tight junctions in kidney epithelial cells causes familial hypomagnesemia, hypercalciuria, and nephrocalcinosis (Müller et al. 2003) (Table 3).
Mutations in some C-terminal di-leucine trafficking minimotifs cause Bartter’s syndrome (Zaarour et al. 2012). Di-leucine minimotifs are evolutionarily conserved in solute carrier family 12 (SLC12A) members. Electroneutral cation-chloride co-transporter family members are therapeutic drug targets. One example of this family of proteins is Na-K-2Cl co-transporter (NKCC2) proteins. NKCC2 proteins can cause type I Bartter’s syndrome, a life-threatening disease caused by the abnormal electrolyte absorption. The conserved C-terminal di-leucine motifs in NKCC2 gets glycosylated, aid in protein maturation, and act as ER export signals for proteins. However, the LL mutants of NKCC2 get trapped in ER (Zaarour et al. 2012). An additional 1048LI1049 motif was also identified in NKCC2 that disrupts ER exit. Similarly, a defect in dileucine minimotif of MNK can cause Menkes disease, an X-linked recessive disorder, and disrupting copper homeostasis. A mutation in the 1487LL1488 minimotif in the transmembrane copper-transporting P-type ATPase (MNK) protein blocks trafficking of MNK from the plasma membrane to the trans-Golgi network (Petris et al. 1998). MNK is a Cuþ2 importer in the secretory pathway.
Several instances of C-terminal peptides (some modified) are at the stage of preclinical testing for new therapeutics (Schiöth et al. 2006; Tsubaki et al. 2013). For example, although the molecular mechanism of activation is not clear, it is known that the KPV > peptide of α-melanocyte stimulating hormone (α-MSH) contains both strong inflammatory properties and have antipyretic effects (Richards and Lipton 1984; Schiöth et al. 2006; Luger and Brzoska 2007).
Conclusions
Cellular processes, such as alternative splicing and proteolysis increase the number of C-terminal ends in the proteome. The negatively charged, often disordered and flexible C-termini have C-terminal minimotifs with a bind, traffic, or PTM function (Sharma et al. 2016). Given that 13% of the human proteome has known C-terminal motifs, the C-termini may have a generalizable function in all proteins. The further study of C-terminal minimotifs will most certainly enhance our understanding of hereditary disease etiology. Mutations in C-terminal minimotifs causes vision loss, Menkes disease, Usher syndrome, and other diseases (Petris et al. 1998; Verpy et al. 2000; Deretic et al. 2005; Puckerin et al. 2016). There are now several web systems for C-termini, the most comprehensive for function being the C-terminome (Sharma et al. 2016).
Acknowledgments
Funding
The National Institutes of Health under grants R15-GM107983-01 and P20-GM121325-01A1; and the Nevada Governor’s Office of Economic Development supported this research.
Footnotes
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Ahmad M, Zhang Y, Zhang Y, Papharalambus C, Alexander RW. 2002. Role of isoprenylcysteine carboxyl methyltransferase in tumor necrosis factor-alpha stimulation of expression of vascular cell adhesion molecule-1 in endothelial cells. Arter Thromb Vasc Biol 22:759–764. [DOI] [PubMed] [Google Scholar]
- Aronson BD, Fisher AL, Blechman K, Caudy M, Gergen JP. 1997. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol Cell Biol 17: 5581–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami-Odaka A, Obayashi-Adachi Y, Matsumoto Y, Takahashi H, Fukumoto H, Horiguchi T, Suzuki N, Shoji M. 2005. Passive immunization of the Abeta42(43) C-terminalspecific antibody BC05 in a mouse model of Alzheimer’s disease. Neurodegener Dis 2:36–43. [DOI] [PubMed] [Google Scholar]
- Attwood TK, Avison H, Beck ME, Bewley M, Bleasby AJ, Brewster F, Cooper P, Degtyarenko K, Geddes AJ, Flower DR, et al. 1997. The PRINTS database of protein fingerprints: a novel information resource for computational molecular biology. J Chem Inf Comput Sci 37:417–424. [DOI] [PubMed] [Google Scholar]
- Austin RS, Provart NJ, Cutler SR. 2007. C-terminal motif prediction in eukaryotic proteomes using comparative genomics and statistical over-representation across protein families. BMC Genomics 8:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahir I, Linial M. 2006. Functional grouping based on signatures in protein termini. Proteins 63:996–1004. [DOI] [PubMed] [Google Scholar]
- Bahir I, Linial M. 2005. ProTeus: identifying signatures in protein termini. Nucleic Acids Res 33:W277–W280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla S, Thapar V, Verma S, Luong T, Faghri T, Huang CH, Rajasekaran S, del Campo JJ, Shinn JH, Mohler WA, et al. 2006. Minimotif Miner: a tool for investigating protein function. Nat Meth 3:175–177. [DOI] [PubMed] [Google Scholar]
- Berezovsky IN, Kilosanidze GT, Tumanyan VG, Kisselev L. 1997. COOH-terminal decamers in proteins are nonrandom. FEBS Lett 404:140–142. [DOI] [PubMed] [Google Scholar]
- Bermak JC, Li M, Bullock C, Zhou QY. 2001. Regulation of transport of the dopamine D1 receptor by a new membrane-associated ER protein. Nat Cell Biol 3:492–498. [DOI] [PubMed] [Google Scholar]
- Beuming T, Skrabanek L, Niv MY, Mukherjee P, Weinstein H. 2005. PDZBase: a protein-protein interaction database for PDZ-domains. Bioinformatics 21:827–828. [DOI] [PubMed] [Google Scholar]
- Birrane G, Chung J, Ladias JAA. 2003. Novel mode of ligand recognition by the Erbin PDZ domain. J Biol Chem 278: 1399–1402. [DOI] [PubMed] [Google Scholar]
- Björnsson A, Mottagui-Tabar S, Isaksson LA. 1996. Structure of the C-terminal end of the nascent peptide influences translation termination. EMBO J 15:1696–1704. [PMC free article] [PubMed] [Google Scholar]
- Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O’Donovan C, Phan I, et al. 2003. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 31:365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetto V, Bergman A-C, Jörnvall H, Sillard R. 1997. C-terminal sequence analysis of peptides and proteins using carboxypeptidases and mass spectrometry after derivatization of Lys and Cys residues. Anal Chem 69:1315–1319. [DOI] [PubMed] [Google Scholar]
- Bork P, Koonin EV. 1996. Protein sequence motifs. Curr Opin Struct Biol 6:366–376. [DOI] [PubMed] [Google Scholar]
- Brahms H, Raymackers J, Union A, de Keyser F, Meheus L, Lührmann R. 2000. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J Biol Chem 275:17122–17129. [DOI] [PubMed] [Google Scholar]
- Brinker A, Scheufler C, Von Der Mulbe F, Fleckenstein B, Herrmann C, Jung G, Moarefi I, Hartl FU. 2002. Ligand discrimination by TPR domains. Relevance and selectivity of EEVD-recognition in Hsp70 x Hop x Hsp90 complexes. J Biol Chem 277:19265–19275. [DOI] [PubMed] [Google Scholar]
- Cabrera M, Muñiz M, Hidalgo J, Vega L, Martín ME, Velasco A. 2003. The retrieval function of the KDEL receptor requires PKA phosphorylation of its C-terminus. Mol Biol Cell 14:4114–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carugo O 2017. Protein termini. Curr Protein Pept Sci 18:211–216. [DOI] [PubMed] [Google Scholar]
- Chen G, Courey AJ. 2000. Groucho/TLE family proteins and transcriptional repression. Gene 249:1–16. [DOI] [PubMed] [Google Scholar]
- Chimura T, Launey T, Ito M. 2011. Evolutionarily conserved bias of amino-acid usage refines the definition of PDZ-binding motif. BMC Genomics 12:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnadurai G 2007. Transcriptional regulation by C-terminal binding proteins. Int J Biochem Cell Biol 39:1593–1607. [DOI] [PubMed] [Google Scholar]
- Chong YP, Mulhern TD, Cheng HC. 2005. C-terminal Src kinase (CSK) and CSK-homologous kinase (CHK)–endogenous negative regulators of Src-family protein kinases. Growth Factors 23:233–244. [DOI] [PubMed] [Google Scholar]
- Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, Ivanov IE, Philips MR. 1999. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell 98:69–80. [DOI] [PubMed] [Google Scholar]
- Chung JJ, Shikano S, Hanyu Y, Li M. 2002. Functional diversity of protein C-termini: more than zipcoding? Trends Cell Biol 12:146–150. [DOI] [PubMed] [Google Scholar]
- Coblitz B, Wu M, Shikano S, Li M. 2006. C-terminal binding: an expanded repertoire and function of 14–3-3 proteins. FEBS Lett 580:1531–1535. [DOI] [PubMed] [Google Scholar]
- Colaert N, Maddelein D, Impens F, Van Damme P, Plasman K, Helsens K, Hulstaert N, Vandekerckhove J, Gevaert K, Martens L. 2013. The online protein processing resource (TOPPR): a database and analysis platform for protein processing events. Nucleic Acids Res 41:D333–D337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortajarena AL, Regan L. 2006. Ligand binding by TPR domains. Protein Sci 15:1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Klerk E, ‘t Hoen PAC. 2015. Alternative mRNA transcription, processing, and translation: insights from RNA sequencing. Trends Genet 31:128–139. [DOI] [PubMed] [Google Scholar]
- Deretic D, Schmerl S, Hargrave PA, Arendt A, McDowell JH. 1998. Regulation of sorting and post-Golgi trafficking of rhodopsin by its C-terminal sequence QVS(A)PA. Proc Natl Acad Sci USA 95:10620–10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic D, Williams AH, Ransom N, Morel V, Hargrave PA, Arendt A. 2005. Rhodopsin C terminus, the site of mutations causing retinal disease, regulates trafficking by binding to ADP-ribosylation factor 4 (ARF4). Proc Natl Acad Sci USA 102:3301–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice JF. 1990. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci 15: 305–309. [DOI] [PubMed] [Google Scholar]
- Dinkel H, Michael S, Weatheritt RJ, Davey NE, Van Roey K, Altenberg B, Toedt G, Uyar B, Seiler M, Budd A, et al. 2012. ELM–the database of eukaryotic linear motifs. Nucleic Acids Res 40:D242–D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kennani S, Adrait A, Shaytan AK, Khochbin S, Bruley C, Panchenko AR, Landsman D, Pflieger D, Govin J. 2017. MS_HistoneDB, a manually curated resource for proteomic analysis of human and mouse histones. Epigenetics Chromatin 10:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre B, Zolnierowicz S, Turowski P, Hemmings BA. 1994. The catalytic subunit of protein phosphatase 2A is carboxylmethylated in vivo. J Biol Chem 269:16311–16317. [PubMed] [Google Scholar]
- Fisher AL, Ohsako S, Caudy M. 1996. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol Cell Biol 16:2670–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortelny N, Yang S, Pavlidis P, Lange PF, Overall CM. 2015. Proteome TopFIND 3.0 with TopFINDer and PathFINDer: database and analysis tools for the association of protein termini to pre- and post-translational events. Nucleic Acids Res 43:D290–D297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang AT, Borch J, Bych K, Jahn TP, Roepstorff P, Palmgren MG. 2003. The binding site for regulatory 14–3-3 protein in plant plasma membrane H-ATPase: involvement of a region promoting phosphorylation-independent interaction in addition to the phosphorylation-dependent C-terminal end. J Biol Chem 278:42266–42272. [DOI] [PubMed] [Google Scholar]
- Gatto GJ, Berg JM. 2003. Nonrandom tripeptide sequence distributions at protein carboxyl termini. Genome Res 13:617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghomashchi F, Zhang X, Liu L, Gelb MH. 1995. Binding of prenylated and polybasic peptides to membranes: affinities and intervesicle exchange. Biochemistry 34:11910–11918. [DOI] [PubMed] [Google Scholar]
- Giacomelli MG, Hancock AS, Masel J. 2007. The conversion of 3’ UTRs into coding regions. Mol Biol Evol 24:457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, Estrada J, Gunawardena J. 2012. Proteus: a web-based, context-specific modelling tool for molecular networks. Bioinformatics 28:1284–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, Gunawardena J, Mann M. 2011. PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res 39:D253–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold MG, Reichow SL, O’Neill SE, Weisbrod CR, Langeberg LK, Bruce JE, Gonen T, Scott JD. 2012. AKAP2 anchors PKA with aquaporin-0 to support ocular lens transparency. EMBO Mol Med 4:15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez M, Scales SJ, Kreis TE, Perez F. 2000. Membrane recruitment of coatomer and binding to dilysine signals are separate events. J Biol Chem 275:29162–29169. [DOI] [PubMed] [Google Scholar]
- Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwarz SE, Scheffner M, Del Sal G. 1999. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J 18: 6462–6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouw M, Michael S, Sámano-Sánchez H, Kumar M, Zeke A, Lang B, Bely B, Chemes LB, Davey NE, Deng Z, et al. 2018. The eukaryotic linear motif resource – 2018 update. Nucleic Acids Res 46:D428–D434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grou CP, Pinto MP, Mendes AV, Domingues P, Azevedo JE. 2015. The de novo synthesis of ubiquitin: identification of deubiquitinases acting on ubiquitin precursors. Sci Rep 5: 12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Birch H, Rapacki K, Brunak S, Hansen JE. 1999. O-GLYCBASE version 4.0: a revised database of O-glycosylated proteins. Nucleic Acids Res 27:370–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris FM, Brecht WJ, Xu Q, Tesseur I, Kekonius L, Wyss-Coray T, Fish JD, Masliah E, Hopkins PC, Scearce-Levie K, et al. 2003. Carboxyl-terminal-truncated apolipoprotein E4 causes Alzheimer’s disease-like neurodegeneration and behavioral deficits in transgenic mice. Proc Natl Acad Sci USA 100:10966–10971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herhaus L, Dikic I. 2015. Expanding the ubiquitin code through post-translational modification. EMBO Rep 16:1071–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgman TC. 1989. The elucidation of protein function by sequence motif analysis. Comput Appl Biosci 5:1–13. [DOI] [PubMed] [Google Scholar]
- Hoffmann JC, Pappa A, Krammer PH, Lavrik IN. 2009. A new C-terminal cleavage product of procaspase-8, p30, defines an alternative pathway of procaspase-8 activation. Mol Cell Biol 29:4431–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. 2015. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res 43:D512–D520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu WL, Oldfield CJ, Xue B, Meng J, Huang F, Romero P, Uversky VN, Dunker AK. 2013. Exploring the binding diversity of intrinsically disordered proteins involved in one-to-many binding. Protein Sci 22:258–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung AY, Sheng M. 2002. PDZ domains: structural modules for protein complex assembly. J Biol Chem 277: 5699–5702. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G. 2006. Ubiquitin-binding domains. Biochem J 399:361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi Y, Eroshkin A, Gramatikova S, Gramatikoff K, Zhang Y, Smith JW, Osterman AL, Godzik A. 2007. CutDB: a proteolytic event database. Nucleic Acids Res 35:D546–D549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MR, Nilsson T, Peterson PA. 1990. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J 9:3153–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob E, Unger R. 2007. A tale of two tails: why are terminal residues of proteins exposed? Bioinformatics 23: e225–e230. [DOI] [PubMed] [Google Scholar]
- Janzen DM, Geballe AP. 2004. The effect of eukaryotic release factor depletion on translation termination in human cell lines. Nucleic Acids Res 32:4491–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen ON. 2004. Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr Opin Chem Biol 8:33–41. [DOI] [PubMed] [Google Scholar]
- Jeon YJ, Yoo HM, Chung CH. 2010. ISG15 and immune diseases. Biochim Biophys Acta 1802:485–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Zhang X, Chen X, Aramsangtienchai P, Tong Z, Lin H. 2018. Protein lipidation: occurrence, mechanisms, biological functions, and enabling technologies. Chem Rev 118:919–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju Z, He JJ. 2017. Prediction of lysine crotonylation sites by incorporating the composition of k-spaced amino acid pairs into Chou’s general PseAAC. J Mol Graph Model 77: 200–204. [DOI] [PubMed] [Google Scholar]
- Karve TM, Cheema AK. 2011. Small changes huge impact: the role of protein posttranslational modifications in cellular homeostasis and disease. J Amino Acids 2011:207691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata M, Farnsworth CC, Yoshida Y, Gelb MH, Glomset JA, Takai Y. 1990. Posttranslationally processed structure of the human platelet protein SMG p21B: evidence for geranylgeranylation and carboxyl methylation of the C-terminal cysteine. Proc Natl Acad Sci USA 87:8960–8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe JW, Bertozzi CR. 2000. Tyrosine sulfation: a modulator of extracellular protein-protein interactions. Chem Biol 7:R57–R61. [DOI] [PubMed] [Google Scholar]
- Khoury GA, Baliban RC, Floudas CA. 2011. Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci Rep 1:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Sheng M. 2004. PDZ domain proteins of synapses. Nat Rev Neurosci 5:771–781. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Tanaka K. 2010. Regulatory mechanisms involved in the control of ubiquitin homeostasis. J Biochem 147: 793–798. [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M. 2012. The ubiquitin code. Annu Rev Biochem 81:203–229. [DOI] [PubMed] [Google Scholar]
- Kowluru A 2000. Evidence for the carboxyl methylation of nuclear lamin-B in the pancreatic beta cell. Biochem Biophys Res Commun 268:249–254. [DOI] [PubMed] [Google Scholar]
- Krishna MMG, Englander SW. 2005. The N-terminal to C-terminal motif in protein folding and function. Proc Natl Acad Sci USA 102:1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N, Koveal D, Miller DH, Xue B, Akshinthala SD, Kragelj J, Jensen MR, Gauss CM, Page R, Blackledge M, et al. 2014. Targeting the disordered C-terminus of PTP1B with an allosteric inhibitor. Nat Chem Biol 10:558–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupershmidt S, Yang T, Chanthaphaychith S, Wang Z, Towbin JA, Roden DM. 2002. Defective human Ether-à-go-go-related gene trafficking linked to an endoplasmic reticulum retention signal in the C terminus. J Biol Chem 277:27442–27448. [DOI] [PubMed] [Google Scholar]
- Laio A, Micheletti C. 2006. Are structural biases at protein termini a signature of vectorial folding? Proteins 62:17–23. [DOI] [PubMed] [Google Scholar]
- Lam HYK, Kim PM, Mok J, Tonikian R, Sidhu SS, Turk BE, Snyder M, Gerstein MB. 2010. MOTIPS: automated motif analysis for predicting targets of modular protein domains. BMC Bioinformatics 11:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P, Xu S, Soroka CJ, Boyer JL. 2012. A C-terminal tyrosine-based motif in the bile salt export pump directs clathrin-dependent endocytosis. Hepatology 55:1901–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange PF, Overall CM. 2013. Protein TAILS: when termini tell tales of proteolysis and function. Curr Opin Chem Biol 17:73–82. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Chistyakov VV, Thornton JM. 2005. PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res 33:D266–D268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BJ, Rajagopalan M, Kim YS, You KH, Jacobson KB, Hatfield D. 1990. Selenocysteine tRNA[Ser]Sec gene is ubiquitous within the animal kingdom. Mol Cell Biol 10: 1940–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Zheng JJ. 2010. PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun Signal 8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon D, Goldstein RE, Bernstein Y, Tang H, Goldenberg D, Stifani S, Paroush Z, Groner Y. 1998. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc Natl Acad Sci USA 95:11590–11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitan ES, Takimoto K. 2000. Surface expression of Kv1 voltage-gated K channels is governed by a C-terminal motif. Trends Cardiovasc Med 10:317–320. [DOI] [PubMed] [Google Scholar]
- Li T, Santockyte R, Shen RF, Tekle E, Wang G, Yang DCH, Chock PB. 2006. Expression of SUMO-2/3 induced senescence through p53- and pRB-mediated pathways. J Biol Chem 281:36221–36227. [DOI] [PubMed] [Google Scholar]
- Lin H, Hejtmancik JF, Qi Y. 2007. A substitution of arginine to lysine at the COOH-terminus of MIP caused a different binocular phenotype in a congenital cataract family. Mol Vis 13:1822–1827. [PubMed] [Google Scholar]
- Liu BA. 2017. Characterizing SH2 domain specificity and network interactions using SPOT peptide arrays. Methods Mol Biol 1555:357–373. [DOI] [PubMed] [Google Scholar]
- Liu MC, Suiko M. 1987. Tyrosine sulfation site is located in the C-terminal fibrin-binding domain in secreted fibronectin from rat embryo fibroblasts, line 3Y1. Arch Biochem Biophys 255:162–167. [DOI] [PubMed] [Google Scholar]
- Liu R, van Veldhoven JPD, IJzerman AP. 2015. The role of the C-terminus of the human hydroxycarboxylic acid receptors 2 and 3 in G protein activation using Ga-engineered yeast cells. Eur J Pharmacol 770:70–77. [DOI] [PubMed] [Google Scholar]
- Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. 2007. BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res 35:D198–D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanov MY, Furletova EI, Bogatyreva NS, Roytberg MA, Galzitskaya OV. 2010. Library of disordered patterns in 3D protein structures. PLoS Comput Biol 6:e1000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CT, Huang KY, Su MG, Lee TY, Bretan~a NA, Chang WC, Chen YJ, Chen YJ, Huang HD. 2013. DbPTM 3.0: an informative resource for investigating substrate site specificity and functional association of protein post-translational modifications. Nucleic Acids Res 41:D295–D305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lübkemeier I, Requardt RP, Lin X, Sasse P, Andrié R, Schrickel JW, Chkourko H, Bukauskas FF, Kim JS, Frank M, et al. 2013. Deletion of the last five C-terminal amino acid residues of connexin43 leads to lethal ventricular arrhythmias in mice without affecting coupling via gap junction channels. Basic Res Cardiol 108:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger TA, Brzoska T. 2007. alpha-MSH related peptides: a new class of anti-inflammatory and immunomodulating drugs. Ann Rheum Dis 66: iii52–iii55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon KF, Cai X, Young RJ, Mamun AA, Rajasekaran S, Schiller MR. 2018. Minimotif Miner 4: a million peptide minimotifs and counting. Nucleic Acids Res 46:D465–D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon KF, Strong CL, Schooler SG, Young RJ, Roy N, Ozar B, Bachmeier M, Rajasekaran S, Schiller MR. 2015. Natural variability of minimotifs in 1092 people indicates that minimotifs are targets of evolution. Nucleic Acids Res 43: 6399–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons PJ, Fricker LD. 2010. Substrate specificity of human carboxypeptidase A6. J Biol Chem 285:38234–38242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Zerangue N, Lin YF, Collins A, Yu M, Jan YN, Jan LY. 2001. Role of ER export signals in controlling surface potassium channel numbers. Science 291:316–319. [DOI] [PubMed] [Google Scholar]
- Madeira F, Tinti M, Murugesan G, Berrett E, Stafford M, Toth R, Cole C, MacKintosh C, Barton GJ. 2015. 14–3-3-Pred: improved methods to predict 14–3-3-binding phosphopeptides. Bioinformatics 31:2276–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Eckhard U, Overall CM. 2015. Protein termini and their modifications revealed by positional proteomics. ACS Chem Biol 10:1754–1764. [DOI] [PubMed] [Google Scholar]
- Meh DA, Siebenlist KR, Brennan SO, Holyst T, Mosesson MW. 2001. The amino acid sequence in fibrin responsible for high affinity thrombin binding. Thromb Haemost 85: 470–474. [PubMed] [Google Scholar]
- Mi T, Merlin JC, Deverasetty S, Gryk MR, Bill TJ, Brooks AW, Lee LY, Rathnayake V, Ross CA, Sargeant DP, et al. 2012. Minimotif Miner 3.0: database expansion and significantly improved reduction of false-positive predictions from consensus sequences. Nucleic Acids Res 40:D252–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millhouse S, Manley JL. 2005. The C-terminal domain of RNA polymerase II functions as a phosphorylation-dependent splicing activator in a heterologous protein. Mol Cell Biol 25:533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. 2006. Analysis of molecular recognition features (MoRFs). J Mol Biol 362:1043–1059. [DOI] [PubMed] [Google Scholar]
- Müller D, Kausalya PJ, Claverie-Martin F, Meij IC, Eggert P, Garcia-Nieto V, Hunziker W. 2003. A novel claudin 16 mutation associated with childhood hypercalciuria abolishes binding to ZO-1 and results in lysosomal mistargeting. Am J Hum Genet 73:1293–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Berger M, Lehembre F, Seeler JS, Haupt Y, Dejean A. 2000. c-Jun and p53 activity is modulated by SUMO-1 modification. J Biol Chem 275:13321–13329. [DOI] [PubMed] [Google Scholar]
- Munro S, Pelham HR. 1987. A C-terminal signal prevents secretion of luminal ER proteins. Cell 48:899–907. [DOI] [PubMed] [Google Scholar]
- Neduva V, Russell RB. 2006. DILIMOT: discovery of linear motifs in proteins. Nucleic Acids Res 34:W350–W355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neduva V, Russell RB. 2005. Linear motifs: evolutionary interaction switches. FEBS Lett 579:3342–3345. [DOI] [PubMed] [Google Scholar]
- Nika H, Nieves E, Hawke DH, Angeletti RH. 2013. C-terminal protein characterization by mass spectrometry using combined micro scale liquid and solid-phase derivatization. J Biomol Tech 24:17–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Mittler G, Mann M. 2004. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat Meth 1:119–126. [DOI] [PubMed] [Google Scholar]
- Orci L, Ravazzola M, Amherdt M, Madsen O, Perrelet A, Vassalli JD, Anderson RG. 1986. Conversion of proinsulin to insulin occurs coordinately with acidification of maturing secretory vesicles. J Cell Biol 103:2273–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil NA, Bathgate RAD, Kocan M, Ang SY, Tailhades J, Separovic F, Summers R, Grosse J, Hughes RA, Wade JD, et al. 2015. The C-terminus of the B-chain of human insulin-like peptide 5 is critical for cognate RXFP4 receptor activity. Amino Acids 48:987–992. [DOI] [PubMed] [Google Scholar]
- Petris MJ, Camakaris J, Greenough M, LaFontaine S, Mercer JF. 1998. A C-terminal di-leucine is required for localization of the Menkes protein in the trans-Golgi network. Hum Mol Genet 7:2063–2071. [DOI] [PubMed] [Google Scholar]
- Prakash T, Khandelwal M, Dasgupta D, Dash D, Brahmachari SK. 2004. CoPS: comprehensive peptide signature database. Bioinformatics 20:2886–2888. [DOI] [PubMed] [Google Scholar]
- Puckerin A, Aromolaran KA, Chang DD, Zukin RS, Colecraft HM, Boutjdir M, Aromolaran AS. 2016. hERG 1a LQT2 C-terminus truncation mutants display hERG 1b-dependent dominant negative mechanisms. Heart Rhythm 13:1121–1130. [DOI] [PubMed] [Google Scholar]
- Rahnefeld A, Klingel K, Schuermann A, Diny NL, Althof N, Lindner A, Bleienheuft P, Savvatis K, Respondek D, Opitz E, et al. 2014. Ubiquitin-like protein ISG15 (interferon-stimulated gene of 15 kDa) in host defense against heart failure in a mouse model of virus-induced cardiomyopathy. Circulation 130:1589–1600. [DOI] [PubMed] [Google Scholar]
- Rajasekaran S, Balla S, Gradie P, Gryk MR, Kadaveru K, Kundeti V, Maciejewski MW, Mi T, Rubino N, Vyas J, et al. 2009. Minimotif miner 2nd release: a database and web system for motif search. Nucleic Acids Res 37:D185–D190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Waller M, Barrett AJ, Bateman A. 2014. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucl Acids Res 42:D503–D509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiners J, Nagel-Wolfrum K, Jürgens K, Märker T, Wolfrum U. 2006. Molecular basis of human Usher syndrome: deciphering the meshes of the Usher protein network provides insights into the pathomechanisms of the Usher disease. Exp Eye Res 83:97–119. [DOI] [PubMed] [Google Scholar]
- Ren J, Wang Y, Liang Y, Zhang Y, Bao S, Xu Z. 2010. Methylation of ribosomal protein S10 by protein-arginine methyltransferase 5 regulates ribosome biogenesis. J Biol Chem 285:12695–12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. 2006. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell 124:1197–1208. [DOI] [PubMed] [Google Scholar]
- Richards DB, Lipton JM. 1984. Effect of alpha-MSH 11–13 (lysine-proline-valine) on fever in the rabbit. Peptides 5:815–817. [DOI] [PubMed] [Google Scholar]
- Rogers LD, Overall CM. 2013. Proteolytic post-translational modification of proteins: proteomic tools and methodology. Mol Cell Proteomics 12:3532–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui HL, Fan E, Zhou HM, Xu Z, Zhang Y, Lin SC. 2002. SUMO-1 modification of the C-terminal KVEKVD of Axin is required for JNK activation but has no effect on Wnt signaling. J Biol Chem 277:42981–42986. [DOI] [PubMed] [Google Scholar]
- Ryan E, Shen D, Wang X. 2016. Structural studies reveal an important role for the pleiotrophin C terminus in mediating interactions with chondroitin sulfate. FEBS J 283:1488–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargeant DP, Gryk MR, Maciejewski MW, Thapar V, Kundeti V, Rajasekaran S, Romero P, Dunker K, Li SC, Kaneko T, et al. 2012. Secondary structure, a missing component of sequence-based minimotif definitions. PLoS One 7:e49957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schey KL, Fowler JG, Shearer TR, David L. 1999. Modifications to rat lens major intrinsic protein in selenite-induced cataract. Invest Ophthalmol Vis Sci 40:657–667. [PubMed] [Google Scholar]
- Schiöth HB, Muceniece R, Mutule I, Wikberg JES. 2006. New melanocortin 1 receptor binding motif based on the C-terminal sequence of alpha-melanocyte-stimulating hormone. Basic Clin Pharmacol Toxicol 99:287–293. [DOI] [PubMed] [Google Scholar]
- Sechi S, Chait BT. 2000. A method to define the carboxyl terminal of proteins. Anal Chem 72:3374–3378. [DOI] [PubMed] [Google Scholar]
- Setlow P, Gerard C, Ozols J. 1980. The amino acid sequence specificity of a protease from spores of Bacillus megaterium. J Biol Chem 255:3624–3628. [PubMed] [Google Scholar]
- Seto Y, Ikeuchi Y, Kanehisa M. 1990. Fragment peptide library for classification and functional prediction of proteins. Proteins 8:341–351. [DOI] [PubMed] [Google Scholar]
- Sharma S, Toledo O, Hedden M, Lyon KF, Brooks SB, David RP, Limtong J, Newsome JM, Novakovic N, Rajasekaran S, et al. 2016. The functional human C-terminome. PLoS One 11:e0152731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan RP, Venkataraghavan R. 1992. A systematic search for protein signature sequences. Proteins 14:16–28. [DOI] [PubMed] [Google Scholar]
- Shy D, Gillet L, Abriel H. 2013. Cardiac sodium channel NaV1.5 distribution in myocytes via interacting proteins: the multiple pool model. Biochim Biophys Acta 1833: 886–894. [DOI] [PubMed] [Google Scholar]
- Shy D, Gillet L, Ogrodnik J, Albesa M, Verkerk AO, Wolswinkel R, Rougier JS, Barc J, Essers MC, Syam N, et al. 2014. PDZ domain-binding motif regulates cardiomyocyte compartment-specific NaV1.5 channel expression and function. Circulation 130:147–160. [DOI] [PubMed] [Google Scholar]
- Smith HO, Welcox KW. 1970. A restriction enzyme from Hemophilus influenzae. I. Purification and general properties. J Mol Biol 51:379–391. [DOI] [PubMed] [Google Scholar]
- Smith JJ, Aitchison JD. 2013. Peroxisomes take shape. Nat Rev Mol Cell Biol 14:803–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleymani-Goloujeh M, Nokhodchi A, Niazi M, Najafi-Hajivar S, Shahbazi-Mojarrad J, Zarghami N, Zakeri-Milani P, Mohammadi A, Karimi M, Valizadeh H. 2017. Effects of N-terminal and C-terminal modification on cytotoxicity and cellular uptake of amphiphilic cell penetrating peptides. Artif Cells Nanomed Biotechnol 46: 91–103. [DOI] [PubMed] [Google Scholar]
- Spiro RG. 2002. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12:43R–56R. [DOI] [PubMed] [Google Scholar]
- Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. 2005. Function of alternative splicing. Gene 344:1–20. [DOI] [PubMed] [Google Scholar]
- Stein A, Céol A, Aloy P. 2011. 3did: identification and classification of domain-based interactions of known three-dimensional structure. Nucleic Acids Res 39:D718–D723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockklausner C, Ludwig J, Ruppersberg JP, Klöcker N. 2001. A sequence motif responsible for ER export and surface expression of Kir2.0 inward rectifier K( ) channels. FEBS Lett 493:129–133. [DOI] [PubMed] [Google Scholar]
- Stone MJ, Chuang S, Hou X, Shoham M, Zhu JZ. 2009. Tyrosine sulfation: an increasingly recognised post-translational modification of secreted proteins. New Biotechnol 25:299–317. [DOI] [PubMed] [Google Scholar]
- Sucic S, Koban F, El-Kasaby A, Kudlacek O, Stockner T, Sitte HH, Freissmuth M. 2013. Switching the clientele: a lysine residing in the C terminus of the serotonin transporter specifies its preference for the coat complex II component SEC24C. J Biol Chem 288:5330–5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Huang C, He J, Lamb KL, Kang X, Gu T, Shen WH, Yin Y. 2014. PTEN C-terminal deletion causes genomic instability and tumor development. Cell Rep 6:844–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundell GN, Ivarsson Y. 2014. Interaction analysis through proteomic phage display. BioMed Res. Int 2014:176172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart EC, Serra V, Petroni G, Nowacki M. 2016. Genetic codes with no dedicated stop codon: context-dependent translation termination. Cell 166:691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. 2011. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146:1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanco S, Gevaert K, Van Damme P. 2015. C-terminomics: targeted analysis of natural and posttranslationally modified protein and peptide C-termini. Proteomics 15:903–914. [DOI] [PubMed] [Google Scholar]
- Teasdale RD, Jackson MR. 1996. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the Golgi apparatus. Annu Rev Cell Dev Biol 12: 27–54. [DOI] [PubMed] [Google Scholar]
- Teesalu T, Sugahara KN, Kotamraju VR, Ruoslahti E. 2009. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc Natl Acad Sci USA 106:16157–16162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JM, Gardner SP. 1989. Protein motifs and data-base searching. Trends Biochem Sci 14:300–304. [DOI] [PubMed] [Google Scholar]
- Tolstykh T, Lee J, Vafai S, Stock JB. 2000. Carboxyl methylation regulates phosphoprotein phosphatase 2A by controlling the association of regulatory B subunits. EMBO J 19:5682–5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tress ML, Abascal F, Valencia A. 2017. Alternative splicing may not be the key to proteome complexity. Trends Biochem Sci 42:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubaki M, Terashima I, Kamata K, Koga A. 2013. C-terminal modification of monoclonal antibody drugs: amidated species as a general product-related substance. Int J Biol Macromol 52:139–147. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium. 2015. UniProt: a hub for protein information. Nucleic Acids Res 43:D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT, et al. 2014. Classification of intrinsically disordered regions and proteins. Chem Rev 114:6589–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen AG, Ploegh HL. 2012. Ubiquitin-like proteins. Annu Rev Biochem 81:323–357. [DOI] [PubMed] [Google Scholar]
- Velez JCQ, Ierardi JL, Bland AM, Morinelli TA, Arthur JM, Raymond JR, Janech MG. 2012. Enzymatic processing of angiotensin peptides by human glomerular endothelial cells. Am J Physiol Renal Physiol 302:F1583–F1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verpy E, Leibovici M, Zwaenepoel I, Liu XZ, Gal A, Salem N, Mansour A, Blanchard S, Kobayashi I, Keats BJ, et al. 2000. A defect in harmonin, a PDZ domain-containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C. Nat Genet 26:51–55. [DOI] [PubMed] [Google Scholar]
- Villanueva J, Shaffer DR, Philip J, Chaparro CA, Erdjument-Bromage H, Olshen AB, Fleisher M, Lilja H, Brogi E, Boyd J, et al. 2006. Differential exoprotease activities confer tumor-specific serum peptidome patterns. J Clin Invest 116:271–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnuvardhan D, Beinfeld MC. 2000. Role of tyrosine sulfation and serine phosphorylation in the processing of procholecystokinin to amidated cholecystokinin and its secretion in transfected AtT-20 cells. Biochemistry 39: 13825–13830. [DOI] [PubMed] [Google Scholar]
- Vitale A, Denecke J. 1999. The endoplasmic reticulum— gateway of the secretory pathway. Plant Cell 11:615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas J, Nowling RJ, Maciejewski MW, Rajasekaran S, Gryk MR, Schiller MR. 2009. A proposed syntax for Minimotif Semantics, version 1. BMC Genomics 10:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Kito K, Caskey LS, Yeh ET, Kamitani T. 1998. Cleavage of the C-terminus of NEDD8 by UCH-L3. Biochem Biophys Res Commun 251:688–692. [DOI] [PubMed] [Google Scholar]
- Wallace EF, Evans CJ, Jurik SM, Mefford IN, Barchas JD. 1982. Carboxypeptidase B activity from adrenal medulla-is it involved in the processing of proenkephalin? Life Sci 31:1793–1796. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456:470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Malbon CC. 2012. Dishevelled C-terminus: prolyl and histidinyl motifs. Acta Physiol (Oxf) 204:65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Casey PJ. 2016. Protein prenylation: unique fats make their mark on biology. Nat Rev Mol Cell Biol 17:110–122. [DOI] [PubMed] [Google Scholar]
- Watanabe J, Nakagawa M, Watanabe N, Nakamura M. 2013. Ubiquitin-like protein MNSFb covalently binds to Bcl-G and enhances lipopolysaccharide/interferon c-induced apoptosis in macrophages. Febs J 280:1281–1293. [DOI] [PubMed] [Google Scholar]
- Weatheritt RJ, Davey NE, Gibson TJ. 2012. Linear motifs confer functional diversity onto splice variants. Nucleic Acids Res 40:7123–7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Mao A, Tang B, Zeng Q, Gao S, Liu X, Lu L, Li W, Du JX, Li J, et al. 2017. Large-scale identification of protein crotonylation reveals its role in multiple cellular functions. J Proteome Res 16:1743–1752. [DOI] [PubMed] [Google Scholar]
- Weil D, El-Amraoui A, Masmoudi S, Mustapha M, Kikkawa Y, Lainé S, Delmaghani S, Adato A, Nadifi S, Zina ZB, et al. 2003. Usher syndrome type I G (USH1G) is caused by mutations in the gene encoding SANS, a protein that associates with the USH1C protein, harmonin. Hum Mol Genet 12:463–471. [DOI] [PubMed] [Google Scholar]
- Weissman AM. 2001. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol 2:169–178. [DOI] [PubMed] [Google Scholar]
- Winter-Vann AM, Kamen BA, Bergo MO, Young SG, Melnyk S, James SJ, Casey PJ. 2003. Targeting Ras signaling through inhibition of carboxyl methylation: an unexpected property of methotrexate. Proc Natl Acad Sci USA 100: 6529–6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HC, Bourdelas A, Krauss A, Lee HJ, Shao Y, Wu D, Mlodzik M, Shi DL, Zheng J. 2003. Direct binding of the PDZ domain of Dishevelled to a conserved internal sequence in the C-terminal region of Frizzled. Mol Cell 12: 1251–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Leng T, Jing L, Jiang N, Chen D, Hu Y, Xiong ZG, Zha XM. 2016. Two di-leucine motifs regulate trafficking and function of mouse ASIC2a. Mol Brain 9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YS, Wang CC, Chen BH, Hou YH, Hung KS, Mao YC. 2015. Tyrosine sulfation as a protein post-translational modification. Molecules 20:2138–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yofe I, Weill U, Meurer M, Chuartzman S, Zalckvar E, Goldman O, Ben-Dor S, Schütze C, Wiedemann N, Knop M, et al. 2016. One library to make them all: streamlining the creation of yeast libraries via a SWAp-Tag strategy. Nat Methods 13:371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HM, Kang SH, Kim JY, Lee JE, Seong MW, Lee SW, Ka SH, Sou YS, Komatsu M, Tanaka K, et al. 2014. Modification of ASC1 by UFM1 is crucial for ERa transactivation and breast cancer development. Mol Cell 56:261–274. [DOI] [PubMed] [Google Scholar]
- Zaarour N, Demaretz S, Defontaine N, Zhu Y, Laghmani K. 2012. Multiple evolutionarily conserved Di-leucine like motifs in the carboxyl terminus control the anterograde trafficking of NKCC2. J Biol Chem 287:42642–42653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanuy D, Kotla R, Nussinov R, Teesalu T, Sugahara KN, Alemán C, Haspel N. 2013. Sequence dependence of C- end rule peptides in binding and activation of neuropilin-1 receptor. J Struct Biol 182:78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanzoni A, Ausiello G, Via A, Gherardini PF, Helmer-Citterich M. 2007. Phospho3D: a database of three-dimensional structures of protein phosphorylation sites. Nucleic Acids Res 35:D229–D231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeytuni N, Zarivach R. 2012. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure 20:397–405. [DOI] [PubMed] [Google Scholar]
- Zhang CE, Tian Q, Wei W, Peng JH, Liu GP, Zhou XW, Wang Q, Wang DW, Wang JZ. 2008. Homocysteine induces tau phosphorylation by inactivating protein phosphatase 2A in rat hippocampus. Neurobiol Aging 29:1654–1665. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Appleton BA, Wiesmann C, Lau T, Costa M, Hannoush RN, Sidhu SS. 2009. Inhibition of Wnt signaling by Dishevelled PDZ peptides. Nat Chem Biol 5:217–219. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li Q, Huang J, Wu Z, Huang J, Huang L, Li Y, Ye J, Zhang X. 2018. An approach to incorporate multi-enzyme digestion into C-TAILS for C-terminomics studies. Proteomics 18:1700034. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lv J, Liu H, Zhu J, Su J, Wu Q, Qi Y, Wang F, Li X. 2010. HHMD: the human histone modification database. Nucleic Acids Res 38:D149–D154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Shabek N. 2017. Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem 86:129–157. [DOI] [PubMed] [Google Scholar]