Abstract
BACKGROUND:
The defining feature of eukaryotic cells is the presence of membranebound organelles of diverse kinds, each with specialized functions. Most organelles have multiple copies in cells. In contrast, each cell contains only one endoplasmic reticulum (ER). However, the ER consists of an elaborated network of membrane cisternae and tubules that extends throughout the cell and occupies a large fraction of the cytoplasmic volume. While compartmentalization of biochemical reactions and processes in these organelles has obvious advantages, it also poses challenges for their coordinated activity, requiring mechanisms for regulated inter-organelle communication. However, these have remained elusive and the quintessential textbook cartoon still pictures organelles in isolation, floating in a cytoplasmic sea. The last decade radically changed this view and membrane contact sites (MCSs) between different organelles were brought to the center stage as prime, highly regulated routes for inter-organelle communication essential for cell homeostasis.
ADVANCES:
The presence of organelle contacts was recognized long ago. However, the significance of these structures remained unclear. Recent advances in the resolution of microscopy and the development of unique fluorophores have dramatically advanced our ability to study inter-organelle MCSs. The 3D structure of ER MCSs with other organelles and the plasma membrane can be visualized at nanometer resolution by electron microscopy (EM). Multi-spectral live-cell fluorescence microscopy displays the behavior of MCSs over time and in response to stimuli. Together these data have revealed the general features of MCSs. For example, EM has revealed that MCSs are closely opposed and tethered, but not fused membranes; MCSs are spaced at 10-30nm; and ribosomes are excluded from the ER surface at these sites. Fluorescence microscopy demonstrates that organelles can remain attached to ER tubules as they traffic along microtubules. The combinations of these tools with classical molecular biology and biochemical tools have identified molecules implicated in several MCSs and elucidated their functions, including lipid and ion transport between organelles and organelles positioning and division.
OUTLOOK:
MCSs are central to normal cell physiology. Moreover, several MCSs proteins are linked to various diseases: Seipin, Protrudin, and Spastin to hereditary spastic paraplegia; VAPA and VAPB to amyotrophic lateral sclerosis; Dnm2 and Mfn2 to charcot marie tooth; Stim1 and Orai1 to tubular aggregate myopathy; and ACBD5 to retinal dystrophy. Whether defects in MCSs functions cause these diseases directly or indirectly remain to be explored. Recent progress has begun to identify some of the molecular machineries that regulate MCSs formation. Dissecting roles of these factors will strengthen our understanding of the integrative nature of MCSs. The advancement of diverse microscopy techniques will allow us to track multiple factors at MCSs simultaneously in real time and in high resolution, and this may help us gain a more detailed view of MCSs biology and their related physiological processes.
Our textbook image of organelles has changed. Instead of isolated cellular compartments, the picture now emerging shows organelles as largely interdependent structures that can communicate through membrane contact sites (MCSs). MCSs are sites where opposing organelles are tethered but do not fuse. MCSs provide a hybrid location where the toolkits of two different organelles can work together to perform vital cellular functions, such as lipid and ion transfer, signaling, and organelle division. Here we concentrate on MCSs involving the endoplasmic reticulum (ER), an organelle forming an extensive network of cisternae and tubules. We will highlight how the dynamic ER network regulates a plethora of cellular processes through MCSs with various organelles and with the plasma membrane (PM).
Graphical Abstract
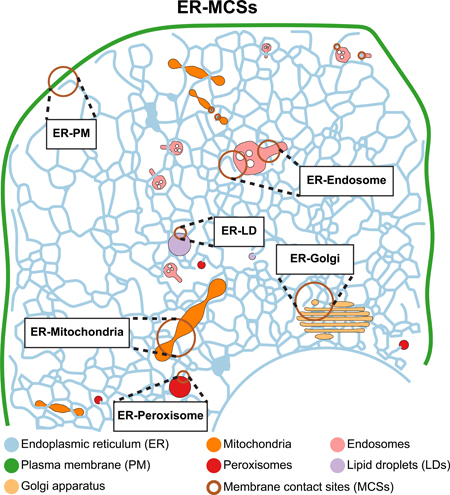
Fig. 0. Endoplasmic reticulum (ER) membrane contacts sites (MCSs) with other organelles and the plasma membrane (PM).
The ER forms MCSs with mitochondria, Golgi, endosomes, peroxisomes, lipid droplets and the PM. These MCSs are closely opposed but not fused membranes containing various molecular machineries. Factors localized to these MCSs mediate essential cellular processes including lipid and ion exchange, organelle positioning and biogenesis.
MCSs are conserved and remarkably extensive along the tubular ER membrane (Fig. 1A–B). Thus, it was postulated that MCSs must play key roles in cell physiology. The contacts between the membrane of the ER and other organelles were first appreciated decades ago, during the early days of electron microscopy (EM). But, what remained unclear was whether MCSs were short-lived interactions made for the quick transfer of cellular material, or stable sites of tethering. The advent of various technologies including the discovery of a vast spectrum of fluorescent proteins and the ability to image live cells at dramatically improved spatial and temporal resolution by microscopy have changed this view. Being able to image and track multiple dynamic organelles simultaneously over time has revealed the extent to which other organelles are tightly tethered to the ER (1) (Fig. 1C). Strikingly, MCSs with elastic ER tubules are maintained during trafficking, fusion and fission of the attached organelles. A major research focus in cell biology is to discover the factors establishing MCSs and how they regulate essential cellular processes from lipid and ion homeostasis to organelle division and distribution.
Fig. 1. Visualizing ER membrane contacts sites (MCSs) with other organelles.
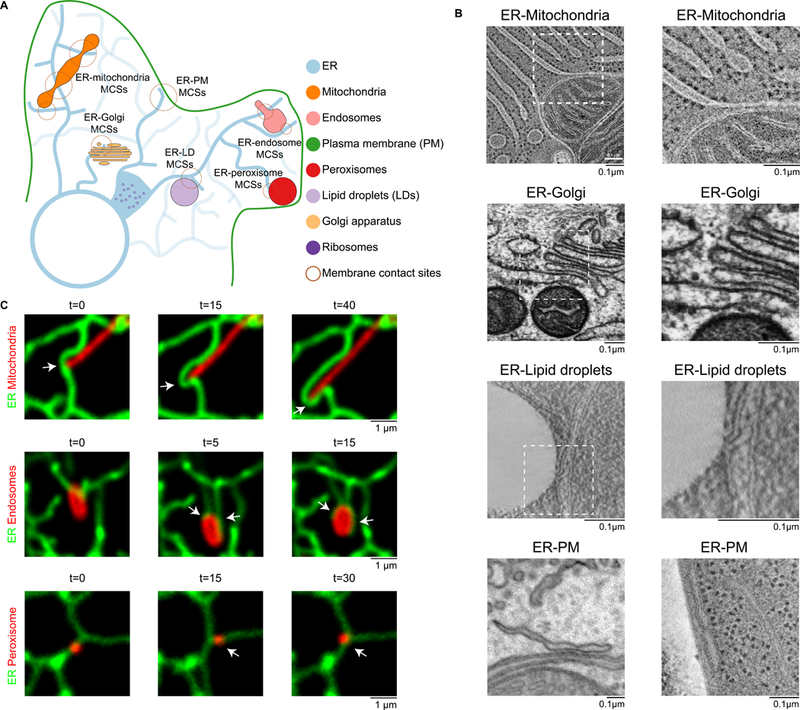
(A) Cartoon of the distribution and structure of the ER and the MCSs it forms with other organelles and with the plasma membrane (PM). (B) Electron micrographs of ER MCSs with mitochondria, Golgi, LDs, and the PM. Micrographs of ERmitochondria and ER-Golgi MCSs in rat epithelial cells were provided by M. Ladinsky. Micrographs of ER-LD MCSs in yeast cells were provided by M. Radulovic. ER-PM MCSs in a mouse neuron (left) were imaged by focused ion beam scanning electron microscopy, and the micrograph was provided by Y.Wu and P. De Camilli [adapted from (3)].The micrograph of ER-PM MCSs in yeast (right) was provided byM.West [reproduced from (83) (CCBY-NC-SA 3.0)]. Boxed areas correspond to images on the right. (C) Mitochondria, endosomes and peroxisomes remain tethered to the ER tubules as they traffic. Time-lapse fluorescent images of the ER (green) relative to mitochondria (red, top row), late endosomes (red, middle row), and peroxisomes (red, bottom row) in live COS-7 cells. Arrows denote MCSs. t, time in seconds.
Factors and Functions of ER-Mitochondria MCSs
From yeast to animal cells, live cell microscopy and EM tomography revealed that mitochondria are tightly associated with the tubular ER (Fig. 1C) (2). Together, these show that ER tubules wrap around the mitochondria to form MCSs that approach distances of 10 nm, are ribosome excluded (Fig. 1B), cover 2–5% of the mitochondrial surface area and radically influence mitochondrial dynamics (2, 3).
Early cell fractionation studies showed that certain ER components co-purified with mitochondria suggesting these MCSs could be biochemically isolated. These mitochondrial associated membranes (MAMs) were enriched in a subset of ER enzymes involved in lipid biosynthesis and Ca2+ signaling hinting at the functions of the ER MCSs with mitochondria (4). Indeed, ER-mitochondria MCSs can provide a conduit for transport of high concentrations of Ca2+ from the ER lumen to the mitochondrial matrix (5). Calcium is released from the ER through the tetrameric inositol 1,4,5-triphosphate receptor (IP3R) channel and funneled to the voltage-dependent anion-selective channel protein (VDAC) channel in the outer mitochondrial membrane (OMM) (6–10). Once Ca2+ traverses the OMM it can use the mitochondrial calcium uniporter to translocate across the IMM (inner mitochondrial membrane) into the matrix (11, 12). Grp75 is a cytosolic regulator of the IP3R-VDAC complex that promotes the interaction between the channels to increase the efficiency of mitochondrial Ca2+ uptake (8). The OMM protein mitofusin 2 (Mfn2) is one proposed tether for regulating this Ca2+ transport at ER-mitochondrial MCSs (13). Mfn2 is a dynamin-like protein that functions to tether mitochondria during homotypic membrane fusion, similar to its paralogue Mfn1 (14). Recently, an ER membrane protein PDZD8 was also implicated in ER-dependent mitochondria Ca2+ homeostasis (15). PDZD8 shares remarkable similarities to the yeast Mmm1, a subunit of the ERMES (ER-mitochondria encounter structure) complex (see below). PDZD8 concentrates at ER-mitochondria MCSs, although mechanistically it is still unclear how PDZD8 localization and its role in Ca2+ homeostasis are achieved.Both ER and mitochondria are required for lipid biosynthesis and various intermediate molecules must travel between these organelles, a shuttling likely to occur at MCSs. So far, the best candidate for this function is the ERMES complex, in yeast (16). ERMES has four subunits including Mmm1, Mdm10, Mdm12 and Mdm34. These subunits form a complex to bridge the ER and mitochondria. Three out of four ERMES subunits (Mmm1, Mdm12 and Mdm34) contain SMP (Synaptotagmin-like Mitochondrial lipid-binding Proteins) domains, which form a long hydrophobic cavity to transfer lipids between membranes (17). ERMES is proposed to transfer phosphatidylserine (PS) and phosphatidylcholine to the mitochondrial membrane (16, 18, 19). Although some crystal structures are available (19, 20), a structure of the whole ERMES complex is needed to mechanistically elucidate how exactly ERMES complex is involved in lipid transfer.
Genetic studies indicate that lipid transfer is quite pliable and may follow diverse routes. For example, only a marginal defect in cellular lipid composition is detected in ERMES mutants (16, 21). Under these conditions distinct MCSs between mitochondria and the vacuole, the vacuole and mitochondria patch (vCLAMP), become essential for lipid homeostasis. Highlighting the interdependence of these MCSs, ERMES becomes essential in cells lacking vCLAMP (22–24). vCLAMP MCSs involve the OMM Mcp1 and soluble proteins Vps13 and vacuolar Vps39, however, it is not clear how they cooperate in lipid transfer (22, 23, 25, 26).
Similarly, ERMES is also essential in cells lacking Lam6/Ltc1, a member of a recently identified family of Steroidogenic Acute Regulatory Transfer (StART)-like proteins (27). Lam6/Ltc1 is an ER membrane protein that localizes to ER-mitochondria MCSs through an interaction with OMM Tom70/71 (27, 28). Like other members of the lipid transfer proteins anchored at a membrane contact site (LAM) / lipid transfer at contact site (Ltc) family, Lam6/Ltc1 is capable of sterol transfer in vitro through the StART-like domain (27–29). However, further studies are required to test whether Lam6/Ltc1 lipid transfer activity is necessary at ER-mitochondria MCSs. So far there has not been any lipid trafficking machinery described in animal cells at ER- mitochondria MCSs.
Mitochondria are dynamic organelles that remodel their network to maintain the integrity of their genome and metabolic state through a balance of fission and fusion. Mitochondrial fission and fusion are elaborate processes tightly regulated by conserved molecular machineries. A surprising function for ER-mitochondria MCSs is that they define the sites of mitochondrial division, in yeast and animal cells (2) (Fig. 2A–B). This process has been described in detail and occurs sequentially (Fig. 2A). First, ER tubules wrap around mitochondria to form MCSs defining the fission position; subsequently, mitochondria constriction and division machineries are recruited to execute fission (2). Surprisingly, ER-associated IMM constriction occurs before OMM constriction (30). These data suggest that signals coming from the mitochondrial matrix initiate ER recruitment to the OMM in order to position the division machinery. That signal could be coming from the mtDNA, since actively replicating mtDNA nucleoids are present at sites of ER-associated mitochondrial constriction and division (Fig. 2A) (31, 32). In animal cells, mitochondria have wide diameters, and multiple machineries function in sequence to drive the process of ER-associated OMM constriction and division. First mitochondria are constricted by actin-myosin assemblies recruited to the MCSs by an ER-localized inverted formin (INF2) and a mitochondrial actin nucleator (Spire 1c) (33, 34). Next, the mitochondrial division dynamin Drp1 is recruited to ER- marked mitochondrial constrictions. Drp1 (Dnm1 in yeast) is a cytosolic GTPase and dynamin family member that oligomerizes around the OMM to drive mitochondrial constriction in a GTPase dependent manner (Fig. 2B) (35–37). Drp1 is recruited from the cytoplasm to the mitochondria at ER MCSs by adaptor proteins such as Mff, MiD49, and Mid51 in animal cells (38–42). Both Drp1 and the adaptors localize to ER-marked mitochondrial constrictions before division (2, 43). Drp1 can drive constriction of mitochondria down to <50nm at which point another dynamin family member, Dnm2, is recruited to complete fission at ER MCSs (Fig. 2A) (43).
Fig. 2. Organelle division by ER MCSs.
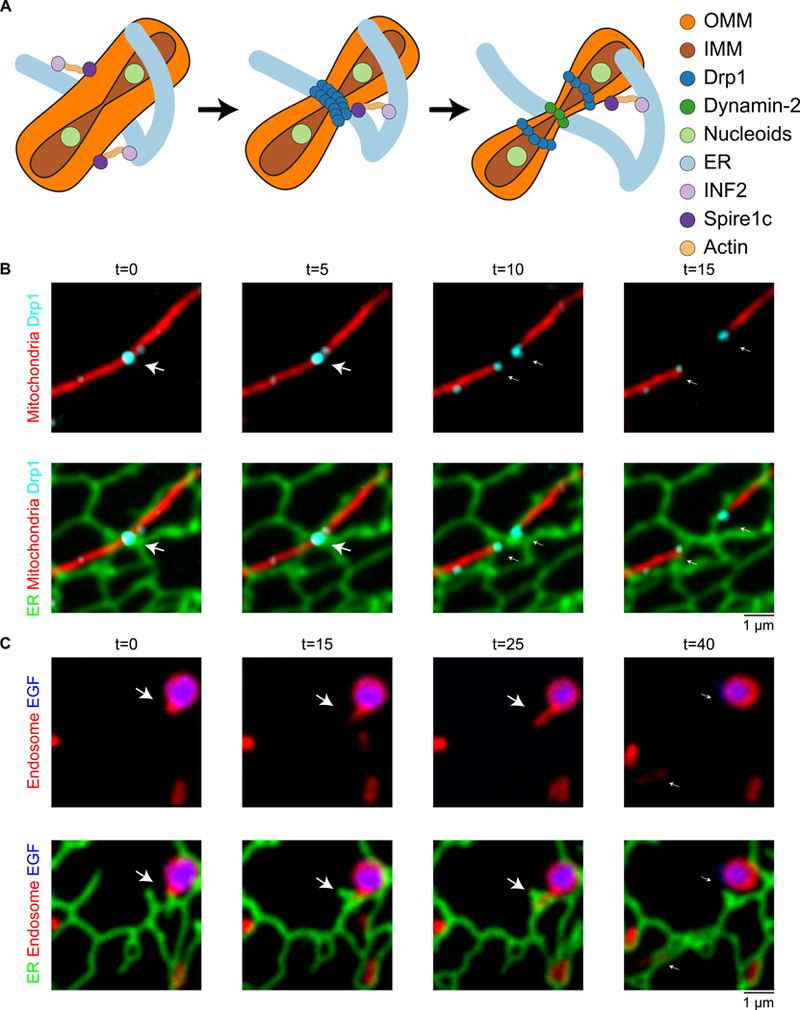
(A) Model of factors involved in ER-associated mitochondrial constriction and division in animal cells: ER MCSs and actively replicating nucleoids define where 1) inner mitochondrial membrane constriction followed by 2) outer mitochondrial membrane (OMM) constriction. OMM constriction requires the activities of INF2, Spirelc, and polymerized actin, followed by the sequential activity of Drp1 and Dnm2 to drive OMM constriction and division. (B) ER tubules define the position of mitochondrial constriction and division. Time-lapse live-cell imaging of the ER (green) and mitochondria (red) in a COS-7 cell shows the division machinery Drp1 (cyan) is localized to the position where an ER tubule crosses over a mitochondrial constriction (panel 1 and 2). As the mitochondria divides, the Drp1 punctum splits and the ER tubules bridge the gap to maintain contact with both Drp1-labeled ends on daughter mitochondria (arrow). Images provided by J. Lee. (C) Dynamic ER tubules are recruited and rearrange around endosome cargo sorting domains to promote endosome fission. Time-lapse live-cell imaging of the ER (green), late endosomes (red) and EGF cargo (blue) shows an endosome bud growing through an ER ring (arrow) and as the ring closes, the bud undergoes fission (compare 3rd and 4th time frame). Image reproduced from (61).
Factors and Functions of ER-Golgi MCSs
Between the ER and the Golgi, cargo traffics by both vesicular and non-vesicular routes. While proteins must be sorted into coated vesicles to traffic between these two organelles, lipids can take a more direct route. In fact, the general principle that lipids can be transferred at MCSs between the ER and other organelles was first demonstrated for the Golgi (44, 45). So far, VAPs (Vesicle-associated membrane protein-associated proteins) are the only ER proteins regulate non-vesicular lipid trafficking at ER-Golgi MCSs. VAPs are highly conserved integral ER membrane proteins (VAPA and VAPB in animals, Scs2 and Scs22 in yeast). VAPs localize throughout the ER, and they bridge contacts with various proteins to perform functions at multiple MCSs (Fig. 3). VAPs contain an MSP (Major sperm protein) domain that interacts with the FFAT (two phenylalanines (FF) in an Acidic Tract) motif of protein partners located on the opposing membrane (46). At the ER-trans Golgi network (TGN) MCSs, VAPs bridge contact with the FFAT motifs of three different lipid transfer proteins including: Nir2 (45), Ceramide transferase 1 (CERT) (44), and Oxysterol-binding protein (OSBP) (Fig. 3A–B) (47).
Fig. 3. The conserved ER proteins VAPA/B establish MCSs with multiple organelles.
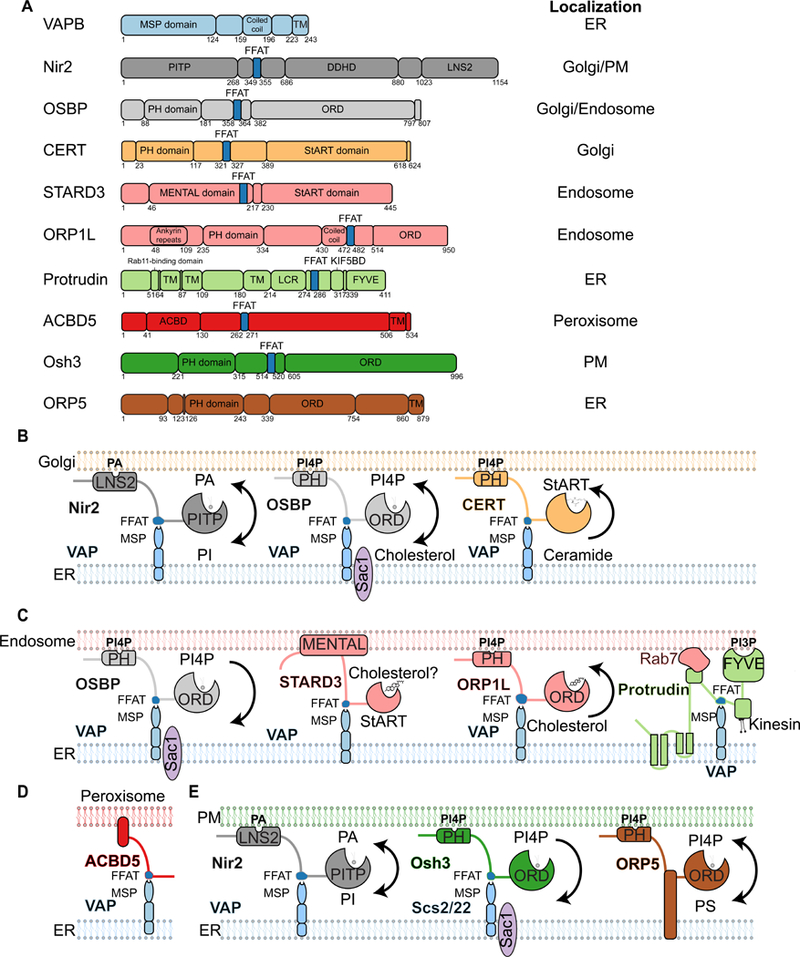
(A) Domain organization of ER-localized VAP and its binding partners. Some VAP binding partners have more than one location. VAPs are tail-anchored membrane proteins that contain an MSP domain, which interacts with the FFATmotif found on the other proteins listed. Orp5 is included as an exception because it is an ER-anchored lipid transfer protein that can regulate phospholipid trafficking at MCSs independently of VAPs. Other relevant domains are indicated, including PI transfer protein (PITP); DDHD, LNS2 (lipin/Nde1/Smp2), and PH domains; ORD; StART, MLN64 NH2-terminal (MENTAL),TM, and FYVE domains; the KIF5 binding domain (KIF5BD); the low- complexity region (LCR); and ACBD. Numbers indicate amino acid positions. (B) Diagram of VAP binding partners at ER-Golgi MCSs. Nir2 binds VAP, contains an LNS2 domain to bind to phosphatidic acid (PA) on the Golgi, and contains a PITP domain that may transfer PIs from the ER to the Golgi. OSBP binds VAP and contains a PH domain to bind PI4P at the Golgi and an ORD to transfer PI4P from the Golgi to the ER, where it can be processed by Sac1 in exchange for cholesterol. CERT binds VAP and contains a PH domain to bind PI4P at the Golgi and a StARTdomain to transfer ceramide from the ER to the Golgi. (C) Diagram of VAP binding partners at ER-endosome MCSs. OSBP binds VAP. OSBP contains a PH domain that binds PI4Pon endosomes, and its ORD transfers PI4P to the ER membrane to be processed by Sac1. STARD3 binds VAP and contains a StARTdomain to bind cholesterol.ORP1L binds VAP when endosomal cholesterol is low, and it contains a PH domain to bind PI4P on endosomes and an ORD to transfer cholesterol from the ER to endosomes. At high endosomal cholesterol levels, ORP1L dissociates from VAP and can be found instead in complex with dynein, resulting in retrograde trafficking of endosomes. Protrudin is an ER protein that binds VAP and Rab7 and contains a FYVE domain to bind to endosomal PI3P. The protrudin-VAP-Rab7-PI3P complex associates with kinesin and promotes anterograde trafficking of LEs on MTs. (D) Diagram of VAP binding partners at ER-peroxisome MCSs. ACBD5 is a tail-anchored peroxisomal protein that binds to VAP and may transfer lipids at these MCSs. (E) Diagram of VAP binding partners at ER-PM MCSs. Nir2 binds VAP and functions as depicted in (B) to transfer PI from the ER to the PM. Yeast Osh3 binds Scs2 and Scs22 (VAP orthologs) and contains a PH domain to bind PI4P on the PM and an ORD to transfer PI4P from the PM to the ER, where it can also be processed by Sac1. ORP5 is an example that does not bind VAP. It is a tail-anchored ER membrane protein that contains a PHdomain to bind PI4Pon thePM and anORD to exchange PI4Pon the PM with PS on the ER.
OSBP is the prototype member of the OSBP / OSBP-related proteins (ORPs) / oxysterol-binding homology (Osh) family. OSBP contains a pleckstrin homology (PH) domain that binds phosphatidylinositol 4-phosphate (PI4P) at the TGN and an FFAT motif to bind to ER-localized VAPs. This protein bridge stabilizes ER-TGN MCSs when TGN PI4P levels are high. Under these conditions, The OSBP-related domain (ORD) of OSBP facilitates the exchange of cholesterol with PI4P between the ER and the TGN. At ER-Golgi MCSs, PI4P levels are kept low by the ER phosphatase Sac1, which can convert PI4P into phosphatidylinositol (PI) (Fig. 3B)(47). The transfer of PI4P down its concentration gradient drives cholesterol transport against its concentration gradient (47, 48). This process may be facilitated by Nir2 which also binds VAPs through an MSP-FFAT interaction (49) and is proposed to supply PI from the ER to the Golgi at MCSs through its PI transfer domain (Fig. 3A–B) (45).
In animal cells, CERT transfers ceramide from the ER to the Golgi at ER-Golgi MCSs through its StART domain (44, 45). CERT also contains a PH domain to target it to the Golgi (44), and an FFAT motif to interact with ER-localized VAPs (Fig. 3A–B)(50). In yeast, the ER membrane protein Nvj2 promotes ceramide transport to the Golgi. Interestingly, Nvj2-dependent ceramide transport strongly increases during ER stress, preventing toxic ceramide accumulation in the ER (51). This conditional transport system involved Nvj2 relocalization and required its PH and SMP-like domains, implicated in lipid transfer at other MCSs. Curiously, Nvj2 mutants were partially suppressed by expression of its mammalian homologues suggesting that a similar CERT-independent ceramide transfer mechanism also operate in higher eukaryotes.
Factors and Functions of ER-endosome MCSs
Cargo from the PM is internalized into vesicles destined for the endocytic pathway. Endosomes sort cargo as they mature and traffic. Very early in their maturation, endosomes acquire ER MCSs so that most early and all late endosomes are bound to the tubular ER network (52, 53). Endosomes are so tightly tethered to the ER that they will pull ER tubules with them as they traffic with molecular motors along microtubules (MTs), as visualized by live cell imaging (Fig. 1C). Individual endosome can form several ER MCSs and cumulatively these contacts cover ~2–5% of their cytoplasmic surface (52, 54). The past decade has revealed several functions for ER MCSs with endosomes that include lipid trafficking, cargo sorting, endosome trafficking and fission (55).
VAPs regulate multiple functions at ER-endosome MCSs. In animal cells, VAPs interact with at least three endosome-localized FFAT-containing partners: OSBP, STARD3, and ORP1L (Fig. 3A and C). The VAP-ORP1L interaction is important to determine the direction of late endosomes (LEs) trafficking on MTs. ORP1L is an endosomal ORP, which is implicated in lipid homeostasis at ER MCSs (Fig. 3). Consistently, ORP1L contains a hydrophobic pocket that may be involved in cholesterol transport at ER-endosome MCSs (Fig. 3C) (56, 57). ORP1L is considered a sterol sensor because its binding to VAPs on the ER depends on endosomal cholesterol levels (56, 57). When high, ORP1L binds to cholesterol and the interaction with VAPs is precluded. Conversely, depletion of endosomal cholesterol frees ORP1L to interact with VAPs, which establishes a MCS between endosomes and the ER. Importantly, this MCS influences the direction of LEs trafficking on MTs. Rab7 GTPase also assembles into VAP-ORP1L complexes resulting in the displacement of the dynein from the LEs surface and halting their retrograde trafficking to the cell center (57). VAPs can also promote kinesin loading onto LEs when in complex with the ER membrane protein Protrudin. The VAP-Protrudin complex binds to Rab7 to recruit kinesin-1 to promote anterograde trafficking of LEs to the PM (Fig. 3A and C)(58). Perhaps Protrudin and VAPs work together with ORP1L and Rab7 to coordinate dynein dissociation and kinesin loading, resulting in anterograde trafficking of LEs in response to cholesterol levels.
VAPs also contribute to phospholipid homeostasis at ER-endosome MCSs. This is achieved through coordination with OSBP and the endosome-localized sorting nexin 2 (Snx2) to allow the ER-localized phosphatase Sac1 to process PI4P (Fig. 3C) (59). VAP depletion causes an increase in PI4P on early and late endosomal membranes and a ripple of downstream effects. The increase in endosomal PI4P boosts the levels of endosomal actin, which nucleates actin comets and alters endosomal motility and this also blocks cargo trafficking from the endosome to the TGN (59).
ER-endosome MCSs can regulate cargo sorting directly. At least one key growth factor receptor, epidermal growth factor receptor (EGFR), is dephosphorylated by an ER-localized phosphatase Ptp1B and this leads to its internalization and subsequent degradation. EGFR and Ptp1B can be co-localized by immuno-EM at MCSs and Ptp1b depletion by siRNA decreases the number of ER-endosome MCSs and reduces the number of intraluminal vesicles per multi-vesicular body (60). Finally, ER MCSs regulate cargo sorting by defining the position of endosome bud fission, in a manner similar to its role on mitochondria. Both early and late endosomes sort their cargo to be degraded away from material to be recycled into vacuolar versus budding domains, respectively. ER tubules are recruited to the saddle between these domains and fission follows MCSs formation (Fig. 2C) (61). It is not known what machinery tethers this interaction. However, an ER-localized isoform of the MT severing protein Spastin may drive the final step of bud fission at the MCSs. When Spastin is depleted, ER MCSs with endosome buds still form and accumulate but the efficiency of ER-associated bud fission is reduced (62).
Factors and functions of ER-peroxisome MCSs
The ER contacts most cellular organelles, and peroxisomes are no exception (Fig. 1C). These are ubiquitous organelles with essential roles in lipid synthesis, fatty acid turnover and detoxification of reactive oxygen species. Importantly, many of these metabolic functions of peroxisomes are carried out in partnership with the ER. For example, the synthesis of ether-linked phospholipids in mammalian cells is initiated in peroxisomes but completed in the ER (63). This process requires lipid intermediates to traffic between the two organelles. Similarly, the phospholipids essential for peroxisome growth and division originate in the ER. ER-peroxisome MCSs, likely facilitating this active trading of metabolites, have been described in a variety of cell types long ago. However, the identification of machineries involved has only recently begun.
Acyl-CoA Binding Domain protein 5 (ACBD5), a tail-anchored peroxisomal membrane protein, establishes ER-peroxisome MCSs by binding, through its FFAT motif, to VAPs in the ER (Fig. 3A and 3D) (64, 65). Supporting a function of this interaction in ER-peroxisomal tethering, overexpression of either ACBD5 or VAPA/B increases the number and surface of MCSs between the ER and peroxisomes, with maximal effect observed when both proteins were overexpressed (64, 65). Conversely, depletion of either ACBD5 or VAPA/B reduces ER-peroxisome MCSs. Depletion of ACBD5 and/or VAPA/B also prevent membrane expansion under conditions that favor peroxisome elongation and a reduction in total ether lipids and cholesterol (65, 66). These observations support the anticipated role of ER-peroxisome MCSs in lipid transfer, even if the direct transfer activity by VAP-ACBD5 has not yet been demonstrated.
Disruption of ER-peroxisome MCSs increases peroxisomal motility (64). While not much is known about how peroxisomal function depends on their spatial distribution, recent work uncovered a link between peroxisome positioning and cell fate decisions in skin epithelia (67). Thus, MCSs may also impact cellular homeostasis by controlling organelle positioning. The regulation of peroxisome positioning by MCSs has been investigated in greater detail in S. cerevisiae, where MCSs are essential for the proper partitioning of peroxisomes between mother and daughter cells in mitosis. In this case the MCS is composed of distinct Pex3 molecules, in ER and peroxisomal membranes, bridged by the soluble Inheritance of Peroxisomes (Inp1) protein (Fig. 4) (68). Besides VAP-ACBD5 and Pex3-Inp1, other proteins are likely to contribute to tether ER and peroxisomes in mammalian and yeast cells. Indeed, a peroxisomal ACBD4 isoform, which shares strong sequence similarity to ACBD5, interacts with VAPs and was suggested to also participate in ER-peroxisome MCSs (69).
Fig. 4. MCSs between the ER and ER-derived organelles.
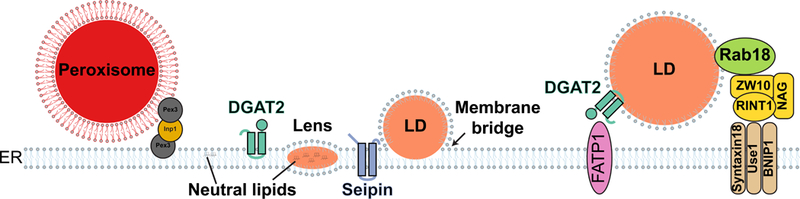
Peroxisomes form MCSs with the ER. VAP on the ER membrane interacts with ACBD5 on peroxisomes (see Fig. 3). Pex3 on both ER and peroxisomal membranes are bridged by Inp1. A lens of neutral lipids build up in the ER membrane to form a precursor of lipid droplets (LDs). Seipin and DGAT2 play important roles in LDs biogenesis. Membrane bridges exist between the continuous ER and LDs membranes. After budding off from the ER membrane, LDs and ER form canonical MCSs. FATP1 on the ER membrane binds DGAT2 on LDs. ER localized SNAREs (Syntaxin18, Use1 and BNIP1) form MCSs with Rab18 on LDs through NRZ complex (NAG, RINT1 and ZW10).
Factors and functions of ER-Lipid droplet MCSs
Lipid droplets (LDs) are storage organelles for neutral lipids such as triglycerides and steryl esters. LDs make frequent and conspicuous contacts with the ER (70). However, ER-LD contacts are one of a kind, often displaying membrane continuity between the two organelles (see Fig. 1B and Fig. 4), a feature that sets them apart from canonical MCSs. These unusual contacts, normally described as membrane bridges, are intimately linked to the unique structure and process of biogenesis of LDs (Fig. 4). Unlike the bilayer in most membrane-bound organelles, LDs contain a phospholipid monolayer surrounding a neutral lipid core. During LDs biogenesis, a lens of neutral lipids accumulates in between the leaflets of the ER bilayer. The expansion of the lens, due to neutral lipid synthesis, facilitates the emergence of LDs at the ER surface. Thus, the LD monolayer is literally derived and continuous with the cytoplasmic leaflet of ER membrane (Fig. 4). The ER-LD membrane bridge facilitates incorporation of neutral lipids into LDs as well as hairpin-containing membrane proteins (70). Many aspects of LD biogenesis remain enigmatic, but Seipin, an ER membrane protein sitting right at the ER-LD interface, clearly plays a central role in the process (71–73). LDs still form following Seipin depletion, however, ER-LD contacts are aberrant resulting in defective incorporation of both neutral lipids and proteins, and ultimately abnormal LD morphology (72–77). While diverse, non-mutually exclusive functions have been proposed for Seipin, the mechanisms by which it affects ER-LD contacts remains unresolved. In humans, Seipin is encoded by the Berardinelli-Seip congenital lipodystrophy 2 (BSCL2) gene, which is frequently mutated in patients with severe lipodystrophy. Lipodystrophy is a metabolic syndrome that is associated with a complete loss of adipose tissue. Whether and how the function of Seipin at ER-LD contacts leads to such a devastating pathology is a topic of very active research.
Membrane bridges, essential during biogenesis, often persist throughout the lifetime of LDs (78). However, in a population of LDs, the bridges dissolve and LDs completely detach from the ER. This appears to be a reversible process; as bridges may be re-established through a process involving components of the COPI coat, which is normally involved in trafficking between the Golgi apparatus and the ER (79).
Besides membrane bridges other ER-LDs tethers have been identified. These appear to be the canonical MCSs and act in parallel with membrane bridges to control different aspects of LD dynamics. Two protein complexes have been implicated in canonical MCSs between the ER and LDs. One of these consists of the ER localized acyl-CoA synthetase FATP1 and DGAT2, a diacylglycerol acyltransferase. While DGAT2 localizes both to the ER and LDs, the LD pool specifically interact with FATP1. Fatty acid activation by FATP1 coupled to DGAT2 acyltransferase activity results in local triglyceride synthesis and LD expansion (Fig. 4) (80).
The small GTPase Rab18 was identified as a key LDs regulator. In a GTP-dependent manner, Rab18 was recruited to the LDs surface and specifically to the ER-associated NAG-RINT1-ZW10 (NRZ) complex and SNARE proteins Syntaxin18, Use1 and BNIP1 (Fig. 4) (81). Consistent with the ER-LD linker activity, depletion of Rab18, NRZ complex components or associated SNAREs resulted in diminished LDs growth and triglyceride storage. Interestingly, the NRZ complex has long been studied for its tethering function of Golgi-derived COPI vesicles prior to their fusion with the ER (82). The connection between NRZ complex tethering to COPI vesicles and LDs is not yet clear. Similarly, SNARE proteins, normally involved in membrane fusion, may be acting differently in this ER-LD tethering complex. Rab18, NRZ and associated SNAREs functions as an ER-LD tether only in a subset of cell types. Curiously, the ability to assemble into a complex did not correlate with the expression of these proteins suggesting that additional posttranslational regulatory factors are at play (81). These observations also highlight that the regulation of MCSs is often dynamic and context-dependent.
Factors and Functions of ER-plasma membrane contact sites
In all eukaryotes, the ER forms extensive contacts with the PM. While the PM is not an organelle, ER-PM contacts share some functions (like lipid trafficking and homeostasis) and factors (like VAP and OSBP) with organellar MCSs. In yeast, ER-PM MCSs are substantial and cover up to 40% of the cytoplasmic surface of the PM (83, 84) (Fig. 1B). By comparison, ER-PM MCSs in animal cells only occupy ~2–5% of the cytoplasmic surface area (3, 85, 86) (Fig. 1B). Despite this difference, ER-PM contacts carry out conserved functions in lipid and Ca2+ homeostasis, which are discussed separately below.
Lipid trafficking at ER-PM MCSs
ER synthesized phospholipids, such as PI and PS, and sterols are transferred to the PM at ER-PM MCSs. ORP/Osh family members regulate phospholipid homeostasis at ER-PM MCSs. Like other family members, ORP5 and ORP8 contain both an ORD and a PH domain (Fig. 3A) (85). In vitro, the purified ORD of ORP8 can bind and transfer PS or PI4P between proteoliposomes, suggesting that it may also countertransport PS and PI4P at MCSs in cells (85). In animal cells, ORP5 and ORP8 are ER-localized lipid sensors that concentrate at ER-PM MCSs when PM PI4P levels are elevated (85). Because these ORPs are tail-anchored into the ER, they do not need to bind to VAP to regulate lipid trafficking (Fig. 3A and 3E). In yeast, Osh3 regulates PI4P metabolism at ER-PM MCSs. Osh3 similarly contains a PH domain that binds PI4P on the PM but requires an FFAT motif to bind to the ER-localized yeast VAP orthologues Scs2/Scs22 to bring it to the ER. The bridge between PI4P in the PM, Osh3, and Scs2/22 facilitates recruitment of the ER-localized phosphatase Sac1 to process PI4P at the MCSs (Fig. 3E) (87).
The extended synaptotagm in-like proteins (E-Syts) and their yeast homologues the tricalbins (Tcbs) are another conserved family of ER membrane proteins that regulate ER-PM MCSs formation and lipid transfer (Fig. 5) (86, 88, 89). There are three E-Syt paralogues in animal cells (E-Syt1–3) and three tricalbins in yeast (Tcb1–3) (89). Knockdown of E-Syts decreases the level of ER-PM MCSs in animal cells (86). The E-Syts/Tcbs are anchored to the ER membrane by an N-terminal reticulon-like hairpin transmembrane (TM) domain. Their cytoplasmic C-terminus contains an SMP domain and multiple (3–5) C2 domains (86, 90). The SMP domain can dimerize to form a long hydrophobic beta-barrel cavity, through which phospholipids may move between membranes (Fig. 5B) (17, 91). With a length of 10nm, this cavity is consistent with the minimum gaps measured between the ER and the PM (83, 91–93). Whether E-syts selectively transfer lipids in a certain direction remains unclear.
Fig. 5. Calcium-regulated ER-PM MCSs.
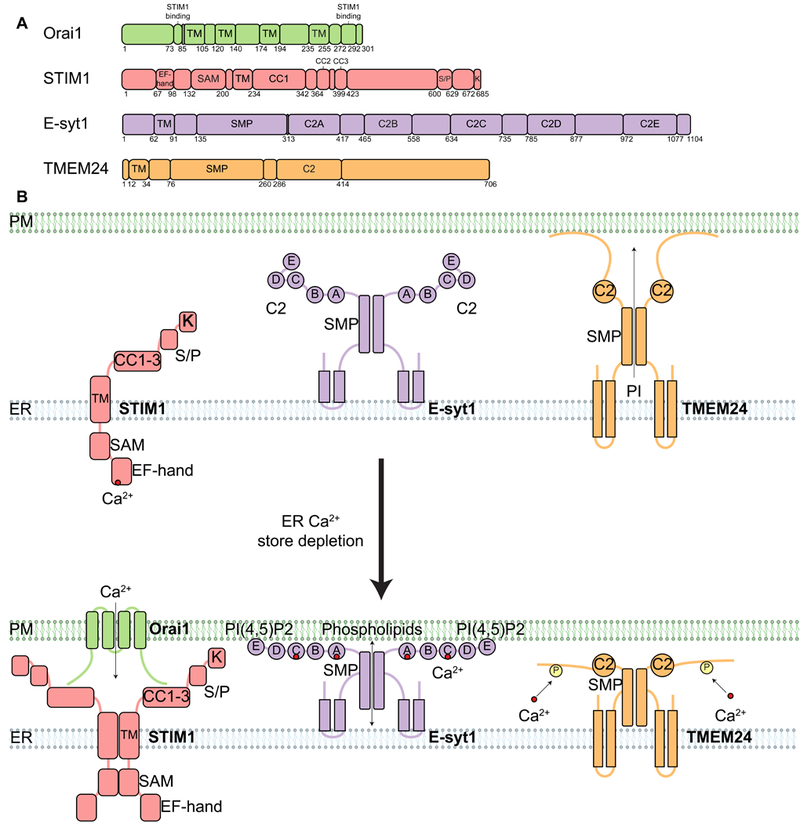
(A) Domain organization of proteins found at MCSs that are regulated by Ca2+. All examples are found at ER-PM MCSs. Relevant domains are indicated, including TM, EF-hand (binds Ca2+), C2 (binds Ca2+ and/or lipids), and SMP domains. SAM, sterile a motif; CC1 to CC3, coiled-coil domains 1 to 3; S/P, serine- and proline-enriched region; K, lysine-enriched region. (B) Diagram depicting the localization and oligomerization of ER MCS proteins that respond to calcium store depletion (compare top and bottom panels). The EF-hand domain of STIM1 is sensitive to calcium store depletion from the ER lumen, which leads to STIM1 oligomerization and translocation of STIM1 to ER-PM MCSs via its polybasic domain, allowing STIMI to bind and activate the Orail CRAC channel on the PM. Activation opens the Orail channel and funnels Ca2+ from the extracellular space back into the ER through the SERCA channel (not depicted). E-Syt1 is another ER membrane protein that translocates to ER-PM MCSs in response to ER calcium store depletion. E-Syt1 contains five C2 domains. ER calcium store depletion causes cytosolic Ca2+ to increase, the C2A and C2C domains bind this cytosolic Ca2+, and then C2E binds to PI(4,5)P2 at the PM, resulting in E-Syt1 translocation to ER-PM MCSs. E-Syt1 transfers phospholipids at ER-PM MCSs through the SMP domain. TMEM24 is an ER membrane protein with a C2 domain and an SMP domain. TMEM24 transfers PI from the ER to the PM through the SMP domain. However, ER calcium store depletion causes TMEM24 to bind Ca2+, become phosphorylated, and then dissociate from ER-PM MCSs.
Classically, C2 domains are capable of binding lipids and sensing Ca2+. E-Syt1 contains five C2 domains (Fig. 5A); two of which (C2A and C2C) can bind Ca2+ (90). E- Syt1 localization is sensitive to an increase in cytosolic Ca2+, which leads to its accumulation at ER-PM MCSs (86, 94). E-Syt2 and E-Syt3 contain three C2 domains of which C2A binds Ca2+ and C2C binds PI(4,5)P2 at the PM (86, 95). When PM PI(4,5)P2 is depleted, E-syt1 translocates to ER-PM MCSs. Interestingly, Nir2, and VAPs translocate to the same MCSs, presumably to replenish the PI (49, 96). Thus, Nir2 and VAPs appear to function similarly at both ER-PM and at ER-TGN MCSs to regulate PI trafficking (compare Fig. 3B and 3E). Depletion of cellular Nir2 decreases PI, PI(4)P, PI(4,5)P2 and PI(3,4,5)P3 levels at the PM (97, 98).
TMEM24 is an ER protein that regulates PI transfer at ER-PM MCSs in response to changes in cytosolic Ca2+ concentration. This protein’s mechanism of action reveals a link between lipid transfer and insulin secretion. TMEM24 contains an N-terminal TM domain, an SMP domain, a C2 domain, and a PM binding C-terminal domain (Fig. 5A) (99). Overexpression of TMEM24 promotes ER-PM MCSs formation, which is dependent on TMEM24’s ability to bind to the PM. TMEM24 preferentially transfers PI from the ER to the PM. TMEM24 knockout cells display decreased insulin secretion upon glucose stimulation. This decrease is directly related to the PI transfer ability of TMEM24 because TMEM24 lacking the SMP domain is unable to rescue insulin secretion deficiency (99). Recruitment of TMEM24 to ER-PM MCSs is regulated by both Ca2+ concentration and phosphorylation (Fig. 5B). Upon elevation of cytosolic Ca2+, TMEM24 is phosphorylated by PKC and it is depleted from the ER-PM MCSs. TMEM24 is recruited back to the PM upon its de-phosphorylation by protein serine/threonine phosphatase 2B (PP2B/calcineurin).
Lam1/Ysp1, Lam2/Ltc4/Ysp2, Lam3/Sip3 and Lam4/Ltc3, members of the Lam/Ltc protein family in yeast, were recently shown to localize to ER-PM MCSs (29). Among these, Lam2/Ltc4/Ysp2 and Lam4/Ltc3 directly transfer sterol between proteoliposomes through StART-like domains (29, 100, 101). Lam1/Ysp1, Lam2/Ltc4/Ysp2 and Lam3/Sip3 knockout cells are sensitive to PM sterol depletion, which indicates their involvement in sterol transfer (29). Indeed, mutations in Lam/Ltc family members such as Lam1/Ysp1, Lam2/Ltc4/Ysp2, and Lam3/Sip3 resulted in slowed sterol transfer from the PM to ER (29). How the directionality of sterol transfer is regulated is not known but Lam/Ltc proteins appear to play important roles at ER-PM MCSs. In mammalian cells, GRAMD1a and GRAMD2a, Lam/Ltc related proteins, also localize to ER-PM MCSs (102). However, co-localization experiments showed that GRAMD1a and GRAMD2a label distinct MCSs and are likely involved in different functions. Future studies will be required to test whether these proteins are also involved in sterol homeostasis.
Ca2+ regulation at ER-PM MCSs
In animal cells, the ER is a major storage site for Ca2+. When the ER is depleted of Ca2+, it relies on extracellular reservoirs to be replenished. The involvement of ER-PM MCSs in this process was proposed over 30 years ago (103). The best studied conduit for the influx of extracellular Ca2+ at ER-PM MCSs is the Orai1 channel (104–107). Orai1 is a hexameric Ca2+ release-activated Ca2+ (CRAC) channel on the PM that is required for store operated Ca2+ entry (108–110). Orai1 contains four TM domains with both its N- and C-terminus facing the cytosol. It contains an extracellular glutamate ring to select for Ca2+, and a basic region inside that regulates channel gating (Fig. 5A) (111).
STIM1 is an ER membrane protein, which following ER Ca2+ store depletion, oligomerizes and translocates to the ER-PM MCSs where it binds and activates Orai1 (Fig. 5) (112–114). This interaction guides Ca2+ into the ER lumen through the sarco/ER Ca2+-ATPase (SERCA) pump. STIM1 has a single TM domain, a cytosolic domain that includes a polybasic region responsible for PM lipid binding, three coiled-coil domains required for Orai1 activation and a luminal EF hand domain that senses ER Ca2+ concentration (107, 113, 115, 116). Upon Ca2+ store depletion from the ER lumen, the EF hand drives a conformational change to initiate STIM1 oligomerization (117).
The STIM1-Orai1 complex can be further regulated by the cytoplasmic, EF-Hand containing proteins CRACR2A and CRACR2B. Knockdown of CRACR2A decreases STIM1 recruitment and Orai1 clustering (118). Mutation of CRACR2A EF hand domain can lead to constitutive translocation of STIM1 to ER-PM MCSs. Several other regulators of STIM1-Orai1 function have been identified. Junctate is an ER membrane protein that interacts with STIM1 through its C-terminal luminal domain, which also contains a luminal EF-hand domain (119). Junctophilin-4 is a tail-anchored ER membrane protein which interacts with Junctacte and with the first two coiled-coil domains in the cytoplasmic region of STIM1. These interactions facilitate the translocation of STIM1 to the PM (120). The ER protein TMEM110/STIMATE also binds and promotes STIM1 translocation to ER-PM MCSs, and it is required for Orai1 channel activation (121, 122). There are also negative regulators of STIM1-Orai1 MCSs formation and function. SARAF is an ER membrane protein that translocates to STIM1- Orail and facilitates STIM1 from dissociating from ER-PM MCSs (123). These observations highlight the dynamic nature of MCSs and how they can be finely regulated in response to specific stimuli, such as Ca2+ concentration.
In skeletal muscle cells, ER-PM MCSs control Ca2+ flux to drive muscle contraction. Specialized membrane structures form in these cells to enhance Ca2+ flux: transverse tubules are invaginated structures of the PM that are physically opposed to terminal cisternae of the sarcoplasmic reticulum (SR, muscle cell ER). This MCS is composed of RyR1 (Ryanodine receptor) on the SR membrane and Cav1.1, a subunit of voltage-dependent calcium channel on the PM (124). Upon PM depolarization and action potential generation, Cav1.1 undergoes a conformational change, which allows Ca2+ release through RyR1 into the cytoplasm to trigger contraction (125). In cardiac muscle cells, a similar system including RyR2 and Cav1.2 establish ER-PM MCSs (126). In this case, however, action potential actually triggers Ca2+ influx into the cytosol from Cav1.2. This Ca2+ influx in turn triggers RyR2-mediated Ca2+ release from the SR. After muscle contractions are terminated, Ca2+ in the cytosol is recycled to the SR through the SERCA pump.
Conclusions
Some of the molecular machineries that regulate membrane tethering have now been identified and so MCSs can now be ascribed functions. Over the last years, the involvement of MCSs in lipid and ion transport was confirmed and some of the molecules and mechanisms involved in these processes could be pinpointed. Novel functions for MCSs were also identified, such as their crucial role in regulating organelle distribution and division. Moreover, it is becoming apparent that inter-organelle communication is highly integrated and subject of homeostatic regulation. For example, the establishment and regulation of MCSs between ER-mitochondria and mitochondria-vacuole is interdependent and appear to respond to nutritional cues (22, 25, 27, 29). Notably, MCSs are linked to human diseases. From the handful of proteins identified which specifically regulate MCSs functions, a high proportion is mutated in a variety of diseases (127–129).
ACKNOWLEDGMENTS
We thank M. Ladinsky, M. Radulovic, M. West, P. De Camilli, Y. Wu, P. Chitwood, and A. Rowland for images used here. We thank R. Salvador-Gallego for helpful comments on the manuscript.
Funding: P. Carvalho was supported by grants from the ERC (Starting grant DropFat; 309477) and The Wellcome Trust. H. Wu was supported by grants from the National Institutes of Health (T32 GM008759 and GM120998). G. Voeltz is an HHMI Faculty Scholar.
Footnotes
Competing interests: None declared.
REFERENCES
- 1.Valm AM et al. , Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature. 546, 162–167 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman JR et al. , ER Tubules Mark Sites of Mitochondrial Division. Science. 334, 358–362 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu Y et al. , Contacts between the endoplasmic reticulum and other membranes in neurons. Proc. Natl. Acad. Sci. U. S. A. 114, E4859–E4867 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vance JE, Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 265, 7248–56 (1990). [PubMed] [Google Scholar]
- 5.Cárdenas C et al. , Essential Regulation of Cell Bioenergetics by Constitutive InsP3 Receptor Ca2+ Transfer to Mitochondria. Cell. 142, 270–283 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rizzuto R et al. , Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 280, 1763–6 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Fan G et al. , Gating machinery of InsP3R channels revealed by electron cryomicroscopy. Nature. 527, 336–341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szabadkai G et al. , Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J. Cell Biol. 175, 901–11 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rapizzi E et al. , Recombinant expression of the voltage-dependent anion channel enhances the transfer of Ca2+ microdomains to mitochondria. J. Cell Biol. 159, 613–624 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shoshan-Barmatz V, Gincel D, The Voltage-Dependent Anion Channel: Characterization, Modulation, and Role in Mitochondrial Function in Cell Life and Death. Cell Biochem. Biophys. 39, 279–292 (2003). [DOI] [PubMed] [Google Scholar]
- 11.De Stefani D, Raffaello A, Teardo E, Szabò I, Rizzuto R, A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 476, 336–340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baughman JM et al. , Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 476, 341–345 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brito O. M.de, Scorrano L, Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 456, 605–610 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Chen H et al. , Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160, 189–200 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirabayashi Y et al. , ER-mitochondria tethering by PDZD8 regulates Ca2+ dynamics in mammalian neurons. Science. 358, 623–630 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kornmann B et al. , An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325, 477–481 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.A. P. AhYoung et al. , Conserved SMP domains of the ERMES complex bind phospholipids and mediate tether assembly. Proc. Natl. Acad. Sci. U. S. A. 112, E3179–88 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kojima R, Endo T, Tamura Y, A phospholipid transfer function of ER-mitochondria encounter structure revealed in vitro. Sci. Rep. 6, 30777 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeong H, Park J, Jun Y, Lee C, Crystal structures of Mmm1 and Mdm12- Mmm1 reveal mechanistic insight into phospholipid trafficking at ER-mitochondria contact sites. Proc. Natl. Acad. Sci. 114, E9502–E9511 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeong H, Park J, Lee C, Crystal structure of Mdm12 reveals the architecture and dynamic organization of the ERMES complex. EMBO Rep. 17, 1857–1871 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen TT et al. , Gem1 and ERMES Do Not Directly Affect Phosphatidylserine Transport from ER to Mitochondria or Mitochondrial Inheritance. Traffic. 13, 880–890 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hönscher C et al. , Cellular Metabolism Regulates Contact Sites between Vacuoles and Mitochondria. Dev. Cell. 30, 86–94 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Lang AB, John Peter AT, Walter P, Kornmann B, ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J. Cell Biol. 210, 883–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan T, Ozbalci C, Brügger B, Rapaport D, Dimmer KS, Mcp1 and Mcp2, two novel proteins involved in mitochondrial lipid homeostasis. J. Cell Sci. 126, 3563–74 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Elbaz-Alon Y et al. , A Dynamic Interface between Vacuoles and Mitochondria in Yeast. Dev. Cell. 30, 95–102 (2014). [DOI] [PubMed] [Google Scholar]
- 26.John Peter AT et al. , Vps13-Mcp1 interact at vacuole-mitochondria interfaces and bypass ER-mitochondria contact sites. J. Cell Biol. 216, 3219–3229 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murley A et al. , Ltc1 is an ER-localized sterol transporter and a component of ER-mitochondria and ER-vacuole contacts. J. Cell Biol. 209, 539–48 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elbaz-Alon Y et al. , Lam6 Regulates the Extent of Contacts between Organelles. Cell Rep. 12, 7–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gatta AT et al. , A new family of StART domain proteins at membrane contact sites has a role in ER-PM sterol transport. Elife. 4, e07253 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho B et al. , Constriction of the mitochondrial inner compartment is a priming event for mitochondrial division. Nat. Commun. 8, 15754 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis SC, Uchiyama LF, Nunnari J, ER-mitochondria contacts couple mtDNA synthesis with Mitochondrial division in human cells. Science. 353, aaf5549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murley A et al. , ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. Elife. 2, e00422 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Korobova F, Ramabhadran V, Higgs HN, An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 339, 464–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manor U et al. , A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. Elife. 4, e08828 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bleazard W et al. , The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1, 298–304 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM, A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 143, 351–8 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM, elegans C dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell. 4, 815–26 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Gandre-Babbe S, van der Bliek AM, The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell. 19, 2402–12 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otera H et al. , Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 191, 1141–58 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J et al. , Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 30, 2762–2778 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmer CS et al. , MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 12, 565–573 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osellame LD et al. , Cooperative and independent roles of the Drp1 adaptors Mff, MiD49 and MiD51 in mitochondrial fission. J. Cell Sci. 129, 2170–2181 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JE, Westrate LM, Wu H, Page C, Voeltz GK, Multiple dynamin family members collaborate to drive mitochondrial division. Nature. 540, 139–143 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanada K et al. , Molecular machinery for non-vesicular trafficking of ceramide. Nature. 426, 803–809 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Peretti D, Dahan N, Shimoni E, Hirschberg K, Lev S, Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol. Biol. Cell. 19, 3871–84 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loewen CJR, Roy A, Levine TP, A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 22, 2025–35 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mesmin B et al. , A Four-Step Cycle Driven by PI(4)P Hydrolysis Directs Sterol/PI(4)P Exchange by the ER-Golgi Tether OSBP. Cell. 155, 830–843 (2013). [DOI] [PubMed] [Google Scholar]
- 48.de Saint-Jean M et al. , Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J. Cell Biol. 195, 965–978 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amarilio R, Ramachandran S, Sabanay H, Lev S, Differential Regulation of Endoplasmic Reticulum Structure through VAP-Nir Protein Interaction. J. Biol. Chem. 280, 5934–5944 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Kawano M, Kumagai K, Nishijima M, Hanada K, Efficient trafficking of ceramide from the endoplasmic reticulum to the Golgi apparatus requires a VAMP-associated protein-interacting FFAT motif of CERT. J. Biol. Chem. 281, 30279–88 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Liu L-K, Choudhary V, Toulmay A, Prinz WA, An inducible ER-Golgi tether facilitates ceramide transport to alleviate lipotoxicity. J. Cell Biol. 216, 131–147 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Friedman JR, Dibenedetto JR, West M, Rowland AA, Voeltz GK, Endoplasmic reticulum-endosome contact increases as endosomes traffic and mature. Mol. Biol. Cell. 24, 1030–40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zajac AL, Goldman YE, Holzbaur ELF, Ostap EM, Local cytoskeletal and organelle interactions impact molecular-motor- driven early endosomal trafficking. Curr. Biol. 23, 1173–80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alpy F et al. , STARD3 or STARD3NL and VAP form a novel molecular tether between late endosomes and the ER. J. Cell Sci. 126, 5500–5512 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Phillips MJ, Voeltz GK, Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol. 17, 69–82 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Kant R, Zondervan I, Janssen L, Neefjes J, Cholesterol-binding molecules MLN64 and ORP1L mark distinct late endosomes with transporters ABCA3 and NPC1. J. Lipid Res. 54, 2153–2165 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rocha N et al. , Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150Glued and late endosome positioning. J. Cell Biol. 185, 1209–1225 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raiborg C et al. , Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature. 520, 234–238 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Dong R et al. , Endosome-ER Contacts Control Actin Nucleation and Retromer Function through VAP-Dependent Regulation of PI4P. Cell. 166, 408–423 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eden ER, White IJ, Tsapara A, Futter CE, Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nat. Cell Biol. 12, 267–72 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Rowland AA, Chitwood PJ, Phillips MJ, Voeltz GK, ER contact sites define the position and timing of endosome fission. Cell. 159, 1027–1041 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Allison R et al. , Defects in ER-endosome contacts impact lysosome function in hereditary spastic paraplegia. J. Cell Biol. 216, 1337–1355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lodhi IJ, Semenkovich CF, Peroxisomes: A Nexus for Lipid Metabolism and Cellular Signaling. Cell Metab. 19, 380–392 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Costello JL et al. , ACBD5 and VAPB mediate membrane associations between peroxisomes and the ER. J. Cell Biol. 216, 331–342 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hua R et al. , VAPs and ACBD5 tether peroxisomes to the ER for peroxisome maintenance and lipid homeostasis. J. Cell Biol. 216, 367–377 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Herzog K et al. , Functional characterisation of peroxisomal β-oxidation disorders in fibroblasts using lipidomics. J. Inherit. Metab. Dis. 41, 479–487 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Asare A, Levorse J, Fuchs E, Coupling organelle inheritance with mitosis to balance growth and differentiation. Science. 355, eaah4701 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Knoblach B et al. , An ER-peroxisome tether exerts peroxisome population control in yeast. EMBO J. 32, 2439–53 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Costello JL, Castro IG, Schrader TA, Islinger M, Schrader M, Peroxisomal ACBD4 interacts with VAPB and promotes ER-peroxisome associations. Cell Cycle. 16, 1039–1045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walther TC, Chung J, Farese RV, Lipid Droplet Biogenesis. Annu. Rev. Cell Dev. Biol. 33, 491–510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Magré J et al. , Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat. Genet. 28, 365–370 (2001). [DOI] [PubMed] [Google Scholar]
- 72.Szymanski KM et al. , The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. U. S. A. 104, 20890–5 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fei W et al. , Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180, 473–82 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang C-W, Miao Y-H, Chang Y-S, Control of lipid droplet size in budding yeast requires the collaboration between Fld1 and Ldb16. J. Cell Sci. 127, 1214–1228 (2014). [DOI] [PubMed] [Google Scholar]
- 75.Grippa A et al. , The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J. Cell Biol. 211, 829–44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang H et al. , Seipin is required for converting nascent to mature lipid droplets. Elife. 5, e16582 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Salo VT et al. , Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J. 35, 2699–2716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jacquier N et al. , Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Sci. 124, 2424–37 (2011). [DOI] [PubMed] [Google Scholar]
- 79.Wilfling F et al. , Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. Elife. 3, e01607 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu N et al. , The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J. Cell Biol. 198, 895–911 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu D et al. , Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions. J. Cell Biol. 217, 975–995 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hirose H et al. , Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J. 23, 1267–1278 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.West M, Zurek N, Hoenger A, Voeltz GK, A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J. Cell Biol. 193, 333–346 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schuck S, Prinz WA, Thorn KS, Voss C, Walter P, Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J. Cell Biol. 187, 525–36 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chung J et al. , PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science. 349, 428–432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Giordano F et al. , PI(4,5)P2-Dependent and Ca2+-Regulated ER-PM Interactions Mediated by the Extended Synaptotagmins. Cell. 153, 1494–1509 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stefan CJ et al. , Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 144, 389–401 (2011). [DOI] [PubMed] [Google Scholar]
- 88.Manford AGG, Stefan CJJ, Yuan HLL, Macgurn JAA, Emr SDD, ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell. 23, 1129–40 (2012). [DOI] [PubMed] [Google Scholar]
- 89.Toulmay A, Prinz WA, A conserved membrane-binding domain targets proteins to organelle contact sites. J. Cell Sci. 125, 49–58 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Min S-W, Chang W-P, Sudhof TC, E-Syts, a family of membranous Ca2+-sensor proteins with multiple C2 domains. Proc. Natl. Acad. Sci. 104, 3823–3828 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schauder CM et al. , Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature. 510, 552–555 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Orci L et al. , STIM1-induced precortical and cortical subdomains of the endoplasmic reticulum. Proc. Natl. Acad. Sci. 106, 19358–19362 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fernandez-Busnadiego R, Saheki Y, De Camilli P, Three-dimensional architecture of extended synaptotagmin-mediated endoplasmic reticulum-plasma membrane contact sites. Proc. Natl. Acad. Sci. U. S. A. 112, E2004–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Idevall-Hagren O, Lu A, Xie B, De Camilli P, Triggered Ca2+ influx is required for extended synaptotagmin 1-induced ER-plasma membrane tethering. EMBO J. 34, 2291–2305 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu J et al. , Structure and Ca2+-Binding Properties of the Tandem C2 Domains of E-Syt2. Structure. 22, 269–280 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Saheki Y et al. , Control of plasma membrane lipid homeostasis by the extended synaptotagmins. Nat. Cell Biol. 18, 504–515 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim YJ, Guzman-Hernandez M-L, Wisniewski E, Balla T, Phosphatidylinositol-Phosphatidic Acid Exchange by Nir2 at ER-PM Contact Sites Maintains Phosphoinositide Signaling Competence. Dev. Cell. 33, 549–561 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim S et al. , The phosphatidylinositol-transfer protein Nir2 binds phosphatidic acid and positively regulates phosphoinositide signalling. EMBO Rep. 14, 891–899 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lees JA et al. , Lipid transport by TMEM24 at ER-plasma membrane contacts regulates pulsatile insulin secretion. Science. 355, eaah6171 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Horenkamp FA, Valverde DP, Nunnari J, Reinisch KM, Molecular basis for sterol transport by StART-like lipid transfer domains. EMBO J. 37, e98002 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tong J, Manik MK, Im YJ, Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites. Proc. Natl. Acad. Sci. U. S. A. 115, E856–E865 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Besprozvannaya M et al. , GRAM domain proteins specialize functionally distinct ER-PM contact sites in human cells. Elife. 7, e31019 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Putney JW, A model for receptor-regulated calcium entry. Cell Calcium. 7, 1–12 (1986). [DOI] [PubMed] [Google Scholar]
- 104.Prakriya M et al. , Orai1 is an essential pore subunit of the CRAC channel. Nature. 443, 230–233 (2006). [DOI] [PubMed] [Google Scholar]
- 105.Vig M et al. , CRACM1 multimers form the ion-selective pore of the CRAC channel. Curr. Biol. 16, 2073–9 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yeromin AV et al. , Molecular identification of the CRAC channel by altered ion selectivity in a mutant of Orai. Nature. 443, 226–229 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Park CY et al. , STIM1 Clusters and Activates CRAC Channels via Direct Binding of a Cytosolic Domain to Orai1. Cell. 136, 876–890 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Feske S et al. , A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 441, 179–185 (2006). [DOI] [PubMed] [Google Scholar]
- 109.Vig M et al. , CRACM1 Is a Plasma Membrane Protein Essential for Store¬Operated Ca2+ Entry. Science. 312, 1220–1223 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang SL et al. , Genome-wide RNAi screen of Ca2+ influx identifies genes that regulate Ca2+ release-activated Ca2+ channel activity. Proc. Natl. Acad. Sci. U. S. A. 103, 9357–62 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hou X, Pedi L, Diver MM, Long SB, Crystal Structure of the Calcium Release-Activated Calcium Channel Orai. Science. 338, 1308–1313 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Roos J et al. , STIM1, an essential and conserved component of store-operated Ca 2+ channel function. J. Cell Biol. 169, 435–445 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liou J et al. , STIM Is a Ca2+ Sensor Essential for Ca2+-Store-Depletion-Triggered Ca2+ Influx. Curr. Biol. 15, 1235–1241 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang X, Jin H, Cai X, Li S, Shen Y, Structural and mechanistic insights into the activation of Stromal interaction molecule 1 (STIM1). Proc. Natl. Acad. Sci. U. S. A. 109, 5657–62 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou Y et al. , Initial activation of STIM1, the regulator of store-operated calcium entry. Nat. Struct. Mol. Biol. 20, 973–981 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yuan JP et al. , SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat. Cell Biol. 11, 337–343 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stathopulos PB, Zheng L, Li G-Y, Plevin MJ, Ikura M, Structural and Mechanistic Insights into STIM1-Mediated Initiation of Store-Operated Calcium Entry. Cell. 135, 110–122 (2008). [DOI] [PubMed] [Google Scholar]
- 118.Srikanth S et al. , A novel EF-hand protein, CRACR2A, is a cytosolic Ca2+ sensor that stabilizes CRAC channels in T cells. Nat. Cell Biol. 12, 436–446 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Srikanth S et al. , Junctate is a Ca2+-sensing structural component of Orai1 and stromal interaction molecule 1 (STIM1). Proc. Natl. Acad. Sci. 109, 8682–8687 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Woo JS et al. , Junctophilin-4, a component of the endoplasmic reticulum-plasma membrane junctions, regulates Ca2+ dynamics in T cells. Proc. Natl. Acad. Sci. 113, 2762–2767 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Quintana A et al. , TMEM110 regulates the maintenance and remodeling of mammalian ER-plasma membrane junctions competent for STIM-ORAI signaling. Proc. Natl. Acad. Sci. 112, 201521924 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jing J et al. , Proteomic mapping of ER-PM junctions identifies STIMATE as a regulator of Ca2+ influx. Nat. Cell Biol. 17, 1339–1347 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Palty R, Raveh A, Kaminsky I, Meller R, Reuveny E, SARAF Inactivates the Store Operated Calcium Entry Machinery to Prevent Excess Calcium Refilling. Cell. 149, 425–438 (2012). [DOI] [PubMed] [Google Scholar]
- 124.Grabner M, Dirksen RT, Suda N, Beam KG, The II-III loop of the skeletal muscle dihydropyridine receptor is responsible for the Bi-directional coupling with the ryanodine receptor. J. Biol. Chem. 274, 21913–9 (1999). [DOI] [PubMed] [Google Scholar]
- 125.Hernandez-Ochoa EO, Pratt SJP, Lovering RM, Schneider MF, Critical Role of Intracellular RyR1 Calcium Release Channels in Skeletal Muscle Function and Disease. Front. Physiol. 6, 420 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Scriven DRL, Asghari P, Moore EDW, Microarchitecture of the dyad. Cardiovasc. Res. 98, 169–176 (2013). [DOI] [PubMed] [Google Scholar]
- 127.Nunnari J, Suomalainen A, Mitochondria: in sickness and in health. Cell. 148, 1145–59 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mannan AU et al. , ZFYVE27 (SPG33), a novel spastin-binding protein, is mutated in hereditary spastic paraplegia. Am. J. Hum. Genet. 79, 351–7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Blackstone C, Hereditary spastic paraplegia. Handb. Clin. Neurol. 148, 633–652 (2018). [DOI] [PubMed] [Google Scholar]


