Abstract
Objective
To assess the effects of dystrophia myotonica protein kinase (DMPK) DNA methylation (DNAme) epivariation on muscular and respiratory profiles in patients with myotonic dystrophy type 1 (DM1).
Methods
Phenotypes were assessed with standardized measures. Pyrosequencing of bisulfite-treated DNA was used to quantify DNAme levels in blood from 90 patients with DM1 (adult form). Modal CTG repeat length was assessed using small-pool PCR. The presence of Acil-sensitive variant repeats was also tested.
Results
DNAme levels upstream of the CTG expansion (exon and intron 11) were correlated with modal CTG repeat length (rs = −0.224, p = 0.040; rs = −0.317, p = 0.003; and rs = −0.241, p = 0.027), whereas correlations were observed with epivariations downstream of the CTG repeats (rs = 0.227; p = 0.037). The presence of a variant repeat was associated with higher DNAme levels at multiple CpG sites (up to 10% higher; p = 0.001). Stepwise multiple linear regression modeling showed that DNAme contributed significantly and independently to explain phenotypic variability in ankle dorsiflexor (3 CpGs: p = 0.001, 0.013, and 0.001), grip (p = 0.089), and pinch (p = 0.028) strengths and in forced vital capacity (2 CpGs: p = 0.002 and 0.021) and maximal inspiratory pressure (p = 0.012).
Conclusions
In addition to the CTG repeat length, DMPK epivariations independently explain phenotypic variability in DM1 and could thus improve prognostic accuracy for patients.
Myotonic dystrophy type 1 (DM1) is an autosomal dominant multisystemic1 disorder caused by a CTG repeat expansion in the 3'untranslated region of the dystrophia myotonica protein kinase (DMPK) gene.2–4 DM1 affects all systems including muscular, cardiac, respiratory, endocrine, and CNS. The hallmark feature of DM1 is the involvement of the muscular system including presence of myotonia, progressive skeletal muscle impairment with a distal to proximal pattern, and respiratory muscle. The phenotypic variability observed in terms of age at onset, clinical presentation, and severity has led to a clinical classification that has evolved over time going from 35 to 5 phenotypes recognized today.6 Although patients born with a larger expansion usually present a more severe phenotype than those with a smaller one, the CTG repeat length only partially explains this large phenotypic variability.
DNA methylation (DNAme) regulates gene expression7 and alternative splicing8 without modifying the nucleic acid sequence of the genes. DNAme occurs mainly on cytosines upstream of a guanine (CpG dinucleotides). The DMPK CTG expansion locus harbors a long CpG island, a genomic structure enriched in CpGs prone to DNAme. This led to hypothesize that DNAme contributes to disease development and progression and explains part of the phenotypic variability reported in DM1.9–16
Only a few studies have been conducted, and all have considerable limitations such as small sample size and inclusion of multiple DM1 phenotypes, limiting comparison between studies and the conclusions, which can be drawn. None tested associations with DM1 clinical phenotypes, except for the Muscular Impairment Rating Scale (MIRS) in 1 study.12 Therefore, we have assessed DNAme levels at the DMPK gene locus and tested for association with muscular and respiratory impairments in the largest number of patients with DM1 with the adult phenotype.
Methods
Standard protocol approvals, registrations, and patient consents
The study was approved by the Ethics Review Board of the Centre de santé et de services sociaux de Chicoutimi, Canada, and all participants provided written consent.
Participants
In 2002, 200 of the 416 patients with DM1 followed at the Neuromuscular Clinic of the Centre de santé et des services sociaux de Jonquière (Québec, Canada) were randomly selected and recruited in a longitudinal study aimed at identifying the determinants of social participation. Selection criteria were (1) age ≥18 years, (2) molecular confirmation of diagnosis (late-onset vs adult phenotypes), and (3) capacity to provide informed consent. The adult phenotype was assigned to participants who had at least 2 of the following characteristics: (1) number of CTG repeats >200 in the standard diagnostic test, (2) Muscular Impairment Rating Score (MIRS) ≥3, and (3) age at symptom onset <40 years.17 Age at onset was defined as the age at which the participant presented his first symptom. To reduce phenotypic variability that is present between patients with DM1 presenting different phenotypes of the disease, only 90 participants presenting the adult form of the disease were kept for the analysis. Patients with other forms of muscular dystrophies including congenital and childhood DM1 were thus excluded.
Muscle strength measurements
An isometric make test using a MicroFET2 dynamometer (Hoggan Health Industries, West Jordan, UT) was performed twice by a trained physiotherapist to measure the strength of the shoulder abductors, knee extensors, and ankle dorsiflexors. The participant's position was standardized for each muscle group. If there was more than 10% discrepancy between 2 assessments for a muscle group, a third measure was performed, and the mean of the 2 closest values was kept as the final result. The maximal isometric torque was computed by multiplying strength units (N) by length of the corresponding lever arm (m).18 A Jamar dynamometer (Asimow Engineering Co, Sequim, WA) was used to measure grip strength.19 Three measures were performed with a standardized protocol, and the mean was used as the final result. A B & L Pinch Gauge (Fabrication Enterprises Inc, New York, NY) was used to measure lateral pinch strength. Again, 3 measures were performed with a standardized protocol, and the mean was used as the final result.19
Respiratory profile assessment
Forced vital capacity, peak expiratory flow, and maximal inspiratory and expiratory pressure (cm H2O) were measured by a respiratory therapist at the Jonquiere Hospital following a standardized protocol. Percentage of predicted values for forced vital capacity and peak expiratory flow (% predicted) were calculated using the equation as in reference 20. Smoking status was determined using a life habit questionnaire.
DNA extraction and methylation analyses
Blood was collected in EDTA tubes during the same visit muscular and respiratory profiles were assessed. Buffy coats were kept at −80°C until DNA extraction using the Gentra Puregene Blood Kit (Qiagen).
DNA methylation analysis takes advantage of the chemical propriety of unmethylated cytosines, which are sensitive to sodium bisulfite treatment (NaBis; EpiTect Bisulfite Kit, Qiagen). Treatment of unmethylated cytosines thus generates a C→T (U) transition, whereas methylated cytosines are protected from deamination to uracils in the presence of NaBis, thus remaining cytosines. Combined with NaBis treatment, pyrosequencing is a quantitative real-time sequencing technology that measures the site-specific DNA methylation percentage for each CpG of a given sequence. PCR and sequencing primers were designed using Pyromark Assay Design 2.0 (Qiagen). Primer sequences are provided in table e-1 (links.lww.com/NXG/A157).
The PyroMark PCR Kit (Qiagen) was used for amplification of bisulfite-treated DNA. A Pyromark model q24 (Qiagen) was used to quantify DNA methylation levels.
CTG repeat length analysis and variant repeat analysis
Small-pool PCR and Southern blot hybridization were used to measure CTG repeat length. PCR amplification of the DMPK CTG repeat was performed using the flanking primers DM-C and DM-DR, as previously described in reference 21. The PCR buffer was Custom PCR Master Mix-No Taq (Thermo Scientific #SM 0005) supplemented with 69 mM 2-mercaptoethanol. Taq polymerase (Sigma-Aldrich, United Kingdom) was used at 1 unit per 10 μL. Where required, reactions were supplemented with 10% dimethyl sulfoxide (Sigma-Aldrich), and the annealing temperature reduced from 68°C to 63.5°C. PCR products were digested with Acil (New England Biolabs UK Ltd) in accordance with the manufacturer's instructions. PCR-amplified DM1 alleles were run on 1% agarose gels in 0.5X tris-borate-EDTA, blotted, and hybridized as described in reference 21. α-32P-dCTP (Perkin Elmer UK) and the Invitrogen Random Primers DNA Labeling System (Thermo Fisher Scientific UK) were used to label the molecular weight marker (Invitrogen 1 kb+ ladder, Thermo Fisher Scientific UK) and DNA probes. Flanking primers DM-C and DM-DR and a patient with DM1 with 56 CTG repeats were used to amplify the CTG probe. After autoradiography, blot images were scanned, and then, estimated progenitor allele length and baseline modal allele lengths were estimated from the lower boundary21 and the densest part of the expanded alleles,22 respectively. Allele length was estimated by comparison to the 1 kb+ ladder, using CLIQS 1D gel analysis software (TotalLabs UK). Somatic instability was computed as modal allele length minus progenitor allele length.
Statistical analyses
Normality of distribution was tested using the Kolmogorov-Smirnov test and then visually confirmed on histograms. The following variables were rank transformed to respect the normality condition of the regression analysis: age, modal and progenitor CTG repeat length, DNA methylation levels, smoking, weight, shoulder abductors, knee extensors, ankle dorsiflexors, grip and pinch strengths, percentage of predicted forced vital capacity, percentage of predicted peak expiratory flow, maximal expiratory and inspiratory pressure, age at symptom onset, and instability. Spearman correlation coefficients were used to assess the association between DNAme and CTG repeat length. Mann-Whitney U tests were then applied on residual scores to assess the differences in DNAme between patients with and without repeat variants. For Spearman and Mann-Whitney U analysis, linear regression and resultant unstandardized residuals were used to adjust DNAme levels for sex and age of the participants. Stepwise multiple linear regression analyses were applied to identify variables independently contributing to explain muscular- and respiratory-related phenotypes. Confounding factors were added to each model based on their association with the corresponding phenotype. The 4 CpGs correlated with CTG repeat length without correction for age and sex were selected to be included into the stepwise regression model. We used relaxed 0.1 and 0.15 cutoff p values, respectively, for entry and removal of variables within the regression. Significance levels were set to p < 0.05. Bonferroni correction was applied to adjust for multiple testing. Statistical analyses were performed on IBM SPSS statistics version 24 (IBM) software.
Data availability
The data sets generated and/or analyzed during the current study are not publicly available for ethical reasons, but are available from the corresponding author on reasonable request.
Results
Description of study participants and overall DNAme profile within the DMPK locus
The participants are, on average, middle aged and normal weight, with women being slightly overrepresented. Participants were aged 41.0 years (±7.7) and 603.4 repeats (±267.9) on average (table 1). Eight of the 90 participants were carriers of a CTG repeat variant (8.9%).
Table 1.
Characteristics of the sample
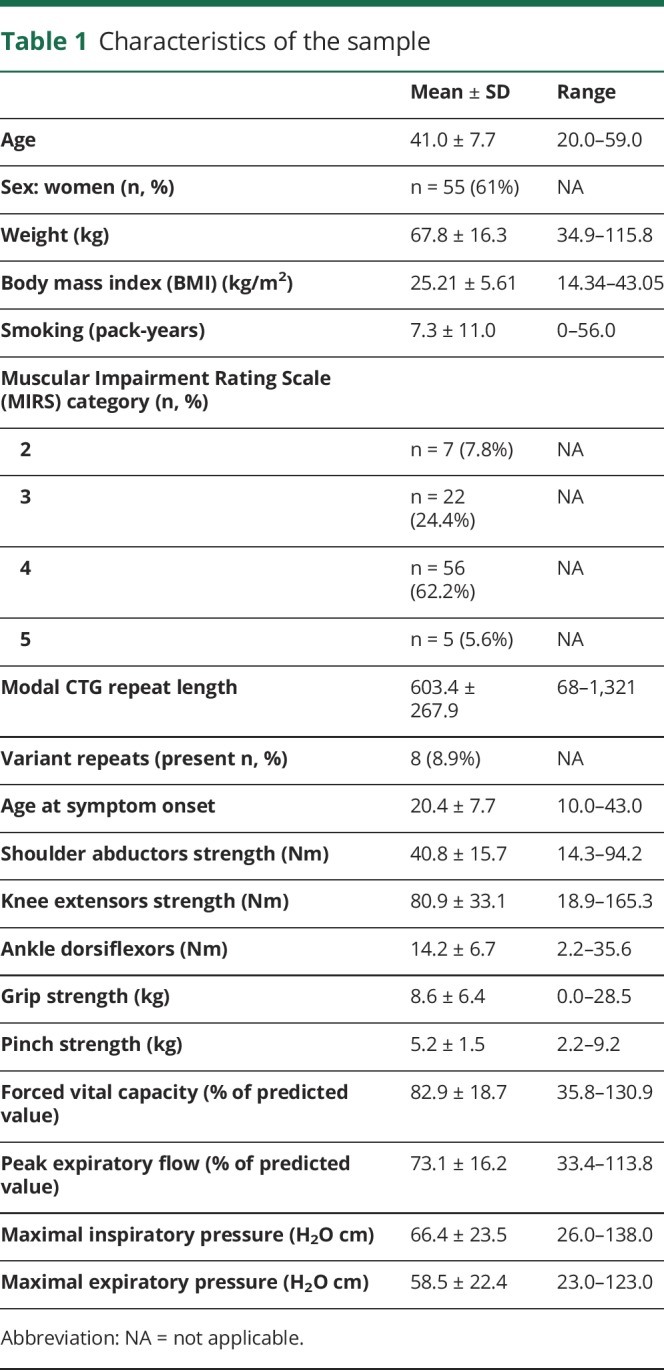
DNAme levels were quantified at 8 different loci (L1 to L8) within the CpG island covering 35 and 17 CpG sites upstream and downstream of the CTG expansion, respectively (figure e-1, links.lww.com/NXG/A154). Some of these CpGs/loci have previously been studied in patients with DM1 or in mouse models.9–16 Upstream of the CTG expansion, DNAme profiles are multidimensional with those proximal to the CTG expansion (L3, 4, and 5 in intron 14 and exon 15) being unmethylated (<5%) and those more distally located showing increasing levels of DNAme from ∼50% methylation, at L2 (intron 13) to close to fully methylated CpGs at site L1 (exon 11). Downstream of the CTG expansion, the CpG sites tested showed low (L6; <10%) to very low (L7 and L8; unmethylated to <5%) levels of DNAme.
Associations between DNAme and CTG repeats
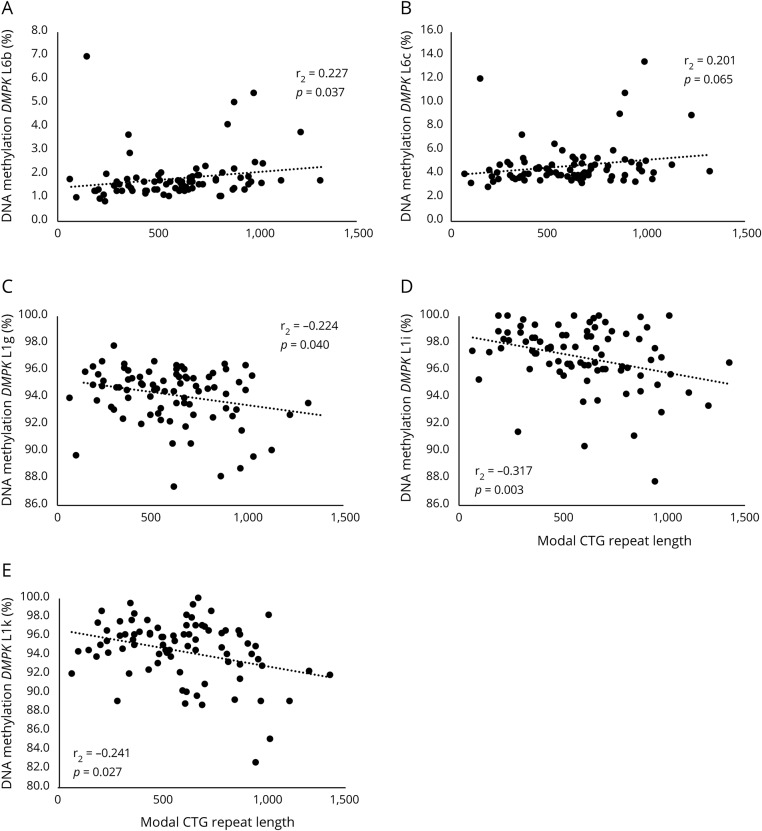
Although the coefficients are similar, we observed that DNAme levels were correlated with the CTG repeat length (figure 1) in opposite directions depending on whether they were upstream or downstream of the CTG expansion. Briefly, DNAme levels in exon and intron 11 were correlated with modal CTG repeat length after correction for age and sex (L1g [rs = −0.224; p = 0.040], L1i [rs = −0.317; p = 0.003] and L1k [rs = −0.241; p = 0.027]), whereas those downstream showed a correlation (L6b [rs = 0.227; p = 0.037] and L6c [rs = 0.201; p = 0.065]). The results remain significant with slightly stronger correlation coefficients after excluding patients with variant repeat.
Figure 1. Spearman correlation between DMPK DNA methylation and modal CTG repeat length in patients with myotonic dystrophy type 1 (DM1).
Each panel (A to E) represents a different CpG. Results corrected for age and sex.
We then assessed the links between DNAme and presence or absence of a variant repeat within the CTG expansion. We found that the presence of a variant repeat was associated with higher DNAme levels only with CpGs just downstream of the repeat. On average, DNAme levels at any of the 5 CpGs tested were 3–4 times higher (L6; figure e-2, links.lww.com/NXG/A155). For example, we observed that the mean DNAme level at L6c for patients with a variant repeat was 15.5%, whereas it was 4.7% for patients without Acil-sensitive sequence (p < 0.001), even after correction for modal CTG and progenitor allele length. Although a nonparametric statistical test was applied, these associations seem primarily driven by 3 participants. We have carefully reviewed their familial, metabolic, and genetic characteristics, and the 2 patients with the highest levels of DNAme are siblings. No associations were found between the presence of a variant repeat and DNAme levels upstream of the CTG expansion.
Stepwise multiple linear regression analyses
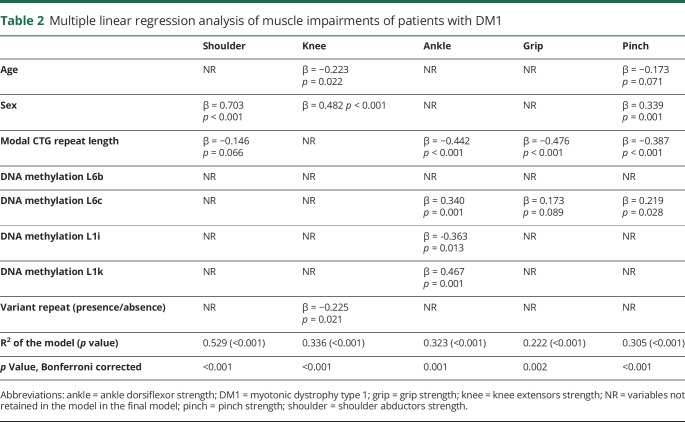
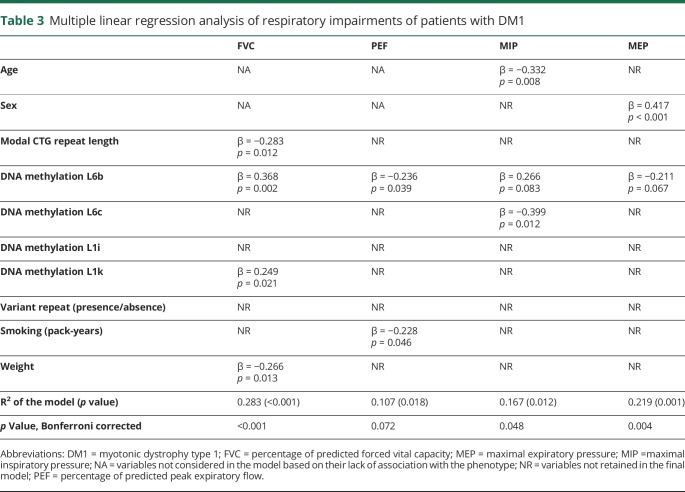
We then assessed whether DNAme levels measured at the DMPK gene locus were associated with muscular strength and respiratory profile. Using multiple linear regression models and stepwise procedures, we found that DNAme contributed to explain variability in muscular and respiratory profiles independently of the CTG repeat length. These results are shown in detail in table 2 for tested muscular strength phenotypes and table 3 for respiratory measures. Very briefly, DNAme levels at one or more loci contributed significantly to every DM1-phenotype tested, except for knee and shoulder strengths and peak expiratory flow. Of interest, robust and persistent associations were observed between DNAme levels at L6c and all 3 muscular-related phenotypes tested, whereas our results suggest a similar pattern of associations between DNAme levels measured at L6b and all respiratory-related phenotypes.
Table 2.
Multiple linear regression analysis of muscle impairments of patients with DM1
Table 3.
Multiple linear regression analysis of respiratory impairments of patients with DM1
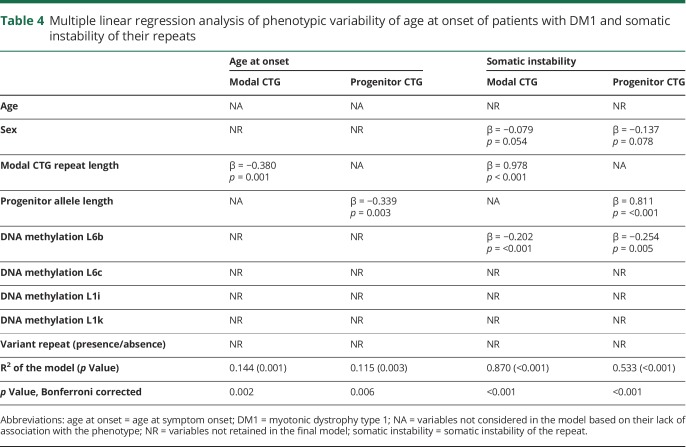
DNAme levels were also tested for associations with somatic instability of the CTG repeat and age at symptom onset. We found that DNAme levels were associated with variability in somatic instability (differences between progenitor and average repeat length) of the CTG repeat, but not with age at symptom onset (table 4). All models except that for peak expiratory flow remain significant after Bonferroni correction. In addition to DNAme levels at other loci, age, sex, and smoking status contributed to explain one or more of the phenotypes tested.
Table 4.
Multiple linear regression analysis of phenotypic variability of age at onset of patients with DM1 and somatic instability of their repeats
Discussion
Although epigenetics (DNAme) has been a mechanism of interest in DM1 since 1993, only 7 studies have been published so far. Of them, 5 focused on the DM1 congenital phenotype.9,11,13–15 Two other studies reported DNAme analyses related to DMPK and SIX5 gene expression,10,16 and only one tested association between a clinical phenotype, namely the MIRS.12 We are the first to study and demonstrate that DNAme at the DMPK gene locus contributes to variability of both muscular strengths and respiratory profiles. Our results also support that DNAme contributes to DM1 phenotypic variability independently of the CTG repeat length. Our study has been conducted exclusively in 90 patients with the adult form of the disease, which we believe reduces the confounding effects of extreme phenotypic and age at sampling variability that characterizes broader DM1 cohorts and allows us to reveal new insights into the role of DNAme in DM1.
Figure e-3 (links.lww.com/NXG/A156) summarizes the results published so far. Our results for sites L1 and L2 are in line with some studies15,16 showing constitutive DNAme in intron 11, but not with 1 study,13 although we report differences in DNAme levels between individuals at these CpG sites. Our results are also in agreement with those previously published13,15,16 showing that the L3 region is unmethylated, although they found some DNAme when patients with DM1 had more than 300 CTG repeats.13,15,16 For the L4 and L5 regions, hypomethylation of the region has been observed in other studies,9,12–15 although 1 study12 found DNAme at various degrees in patients with pure CTG repeats, as did another.11 For L6, the region has been found to present some variability in our study and in mouse,10,11 in patients with variant repeats,12 and patients with DM1.9 This region has been associated with hypomethylation in patients with DM111,13,15 and patients with DM1 with a pure CTG tract.12 The L7 and L8 sites have been found to be hypomethylated both by our group and in a previous study.13 The studies published so far are of small to very small sample size. Also, inconsistencies between the studies might be explained by the different DNAme quantification methods, which are in some cases far from ideal. For example, methylation-sensitive restriction enzymes have been routinely used, but do not allow detection of small changes in DNAme unlike pyrosequencing. These studies have also analyzed a broader range of DM1 phenotypes than in our study. The other DM1 phenotypes should thus be also investigated.
Of interest, we also report that the CTG repeat length was correlated with DNAme, but in opposite directions depending on which side of the CTG expansion DNAme levels were assessed. We report a correlation with DNAme measured in intron 11. Some groups reported similar associations for DNAme measured in closer vicinity of the CTG expansion,9,10,12,15,16 although discrepancies exist.11,14 Overall, ours and previous results support the hypothesis that DNAme and modal CTG repeat length are correlated with each other. However, we cannot yet conclude which of the 2 influences the other.
Of interest, we also found that presence of a variant repeat within the CTG expansion was associated with higher DNAme levels downstream of the CTG repeat (L6). Similar results have been reported showing an association driven by participants who had more “CG” sequences in their repeat.12 The “CG” content has not been assessed in our study, precluding us from drawing a clear conclusion. However, we found that 2 of the 3 patients with higher DNAme levels were siblings. Variant repeats were first observed in a small number of families,24,25 suggesting that they might be inherited. As such, the pattern of each variant repeat is also probably inherited and similar within families.
Next, we tested whether DNAme was also associated with key impairments in DM1. DNAme at 2 different CpG sites, both within the same L6 region downstream of the CTG expansion, contributed independently of the CTG repeat length to variability of muscular- (L6c; ankle dorsiflexors, handgrip, and pinch strength) and respiratory-related phenotypes (L6b; forced vital capacity, maximal inspiratory pressure, and maximal expiratory pressure). This suggests that DNAme at those CpG sites might affect distinctive muscle groups by changing affinity for different transcription factors. However, no specific transcription factor consensus sequences covering each of the CpG sites were found in the ENCODE database. Also, we did not find that DNAme contributes to strength variability of proximal muscle groups (shoulder abductors and knee extensors), which could be explained by several reasons including measurement errors, muscle impairment level, and other factors.
At least 3 hypotheses could explain how DNAme might drive clinical variability in DM1. First, DNAme might block CTCFs binding to one or both of the CTCF consensus sequences located on each side of the CTG expansion, leading to CTCF insulator disruption ability between DMPK and SIX5.26 Second, DNAme might reduce the mRNA levels of DMPK and SIX5 and thus reducing the number of DMPK and SIX5 protein.10,16 Third, DNAme might alter CTG repeat instability 27,28 through disruption of CTCF binding,28 although another study reported that it was DNA demethylation that was promoting repeat instability27 with another one showing no link between DNAme and CTG repeat instability.14 Of interest, we found that increased DNAme was associated with lower somatic instability. Over time, this might be beneficial for the patients, as longer repeats usually means a worse prognosis.
Strengths and limitations
Our study is the largest in terms of number of patients with DM1 with the same phenotype (adult form) and the second largest overall. Our participants were extensively phenotyped and genotyped, including measures of CTG repeat length and occurrence of CTG variant repeats. DNAme was measured in blood only (this could also be seen as a limitation) and using pyrosequencing, the gold standard method to assess CpG-specific DNAme levels, thus reducing potential technical variability. One limitation is that we only covered a small part of the DM1 locus (<3%), leaving many other CpG sites to be tested. Another limitation is that we did not have access to non-DM1 participants, and thus, comparisons with what could be considered a normal DNAme profile were not possible. However, the DMPK region has previously been found to be hypomethylated near the CTG repeat and hypermethylated in the region of exons 11 and 12 in controls and the general population in various studies.9,11,12,29 No new data on the methylation status of the DMPK region are available on the epigenome roadmap than the data already published.29
DNAme at the DMPK gene locus contributes to specific clinical phenotypes (muscular strengths and respiratory profiles) in DM1. These associations are independent of the CTG repeat length. Our results thus provide evidence that measuring DNAme might help to predict progression of the disease and to establish a more reliable prognosis for those patients. We nevertheless agree that further studies, including longitudinal studies and in other DM1 forms, are needed before strong conclusions can be drawn and clinically applied.
Acknowledgment
Pascal-Denys Grenier performed the quantitative muscle testing during a single visit. Céline Bélanger revised the language of the manuscript.
Glossary
- DM1
dystrophy type 1
- DMPK
dystrophia myotonica protein kinase
- MIRS
Muscular Impairment Rating Scale
Appendix. Authors
Study funding
This project has been supported by the Marigold Foundation, the Canadian Institutes of Health Research (CIHR) (#JNM-108412), the Fondation du grand défi Pierre Lavoie, and Muscular Dystrophy UK.
Disclosure
Disclosures available: Neurology.org/NG.
References
- 1.Thornton CA. Myotonic dystrophy. Neurol Clin 2014;32:705–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brook JD, McCurrach ME, Harley HG, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3' end of a transcript encoding a protein kinase family member. Cell 1992;68:799–808. [DOI] [PubMed] [Google Scholar]
- 3.Fu YH, Pizzuti A, Fenwick RG Jr, et al. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science 1992;255:1256–1258. [DOI] [PubMed] [Google Scholar]
- 4.Mahadevan M, Tsilfidis C, Sabourin L, et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3' untranslated region of the gene. Science 1992;255:1253–1255. [DOI] [PubMed] [Google Scholar]
- 5.New nomenclature and DNA testing guidelines for myotonic dystrophy type 1 (DM1). The International Myotonic Dystrophy Consortium (IDMC). Neurology 2000;54:1218–1221. [DOI] [PubMed] [Google Scholar]
- 6.De Antonio M, Dogan C, Hamroun D, et al. Unravelling the myotonic dystrophy type 1 clinical spectrum: a systematic registry-based study with implications for disease classification. Revue neurologique 2016;172:572–580. [DOI] [PubMed] [Google Scholar]
- 7.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 2002;16:6–21. [DOI] [PubMed] [Google Scholar]
- 8.Shukla S, Kavak E, Gregory M, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 2011;479:74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbe L, Lanni S, Lopez-Castel A, et al. CpG methylation, a parent-of-origin effect for maternal-biased transmission of congenital myotonic dystrophy. Am J Hum Genet 2017;100:488–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brouwer JR, Huguet A, Nicole A, Munnich A, Gourdon G. Transcriptionally repressive chromatin remodelling and CpG methylation in the presence of expanded CTG-repeats at the DM1 locus. J Nucleic Acids 2013;2013:567435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez Castel A, Nakamori M, Tome S, et al. Expanded CTG repeat demarcates a boundary for abnormal CpG methylation in myotonic dystrophy patient tissues. Hum Mol Genet 2011;20:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santoro M, Fontana L, Masciullo M, et al. Expansion size and presence of CCG/CTC/CGG sequence interruptions in the expanded CTG array are independently associated to hypermethylation at the DMPK locus in myotonic dystrophy type 1 (DM1). Biochim Biophys Acta 2015;1852:2645–2652. [DOI] [PubMed] [Google Scholar]
- 13.Shaw DJ, Chaudhary S, Rundle SA, et al. A study of DNA methylation in myotonic dystrophy. J Med Genet 1993;30:189–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spits C, Seneca S, Hilven P, Liebaers I, Sermon K. Methylation of the CpG sites in the myotonic dystrophy locus does not correlate with CTG expansion size or with the congenital form of the disease. J Med Genet 2010;47:700–703. [DOI] [PubMed] [Google Scholar]
- 15.Steinbach P, Glaser D, Vogel W, Wolf M, Schwemmle S. The DMPK gene of severely affected myotonic dystrophy patients is hypermethylated proximal to the largely expanded CTG repeat. Am J Hum Genet 1998;62:278–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yanovsky-Dagan S, Avitzour M, Altarescu G, et al. Uncovering the role of hypermethylation by CTG expansion in myotonic dystrophy type 1 using mutant human embryonic stem cells. Stem Cel Rep 2015;5:221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gagnon C, Mathieu J, Noreau L. Life habits in myotonic dystrophy type 1. J Rehabil Med 2007;39:560–566. [DOI] [PubMed] [Google Scholar]
- 18.Petitclerc E, Hebert LJ, Mathieu J, Desrosiers J, Gagnon C. Lower limb muscle strength impairment in late-onset and adult myotonic dystrophy type 1 phenotypes. Muscle Nerve 2017;56:57–63. [DOI] [PubMed] [Google Scholar]
- 19.American Society of Hand T. Clinical Assessment Recommendations. Chicago: (401 N. Michigan Ave, Chicago 60611-4267): The Society, 1992. [Google Scholar]
- 20.Quanjer PH, Tammeling GJ, Cotes JE, Pedersen OF, Peslin R, Yernault JC. Lung volumes and forced ventilatory flows. Eur Respir J 1993;6(suppl 16):5–40. [DOI] [PubMed] [Google Scholar]
- 21.Monckton DG, Wong LJ, Ashizawa T, Caskey CT. Somatic mosaicism, germline expansions, germline reversions and intergenerational reductions in myotonic dystrophy males: small pool PCR analyses. Hum Mol Genet 1995;4:1–8. [DOI] [PubMed] [Google Scholar]
- 22.Morales F, Couto JM, Higham CF, et al. Somatic instability of the expanded CTG triplet repeat in myotonic dystrophy type 1 is a heritable quantitative trait and modifier of disease severity. Hum Mol Genet 2012;21:3558–3567. [DOI] [PubMed] [Google Scholar]
- 23.Raymond K, Levasseur M, Mathieu J, Desrosiers J, Gagnon C. A 9-year follow-up study of the natural progression of upper limb performance in myotonic dystrophy type 1: a similar decline for phenotypes but not for gender. Neuromuscul Disord 2017;27:673–682. [DOI] [PubMed] [Google Scholar]
- 24.Braida C, Stefanatos RK, Adam B, et al. Variant CCG and GGC repeats within the CTG expansion dramatically modify mutational dynamics and likely contribute toward unusual symptoms in some myotonic dystrophy type 1 patients. Hum Mol Genet 2010;19:1399–1412. [DOI] [PubMed] [Google Scholar]
- 25.Musova Z, Mazanec R, Krepelova A, et al. Highly unstable sequence interruptions of the CTG repeat in the myotonic dystrophy gene. Am J Med Genet A 2009;149A:1365–1374. [DOI] [PubMed] [Google Scholar]
- 26.Filippova GN, Thienes CP, Penn BH, et al. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat Genet 2001;28:335–343. [DOI] [PubMed] [Google Scholar]
- 27.Gorbunova V, Seluanov A, Mittelman D, Wilson JH. Genome-wide demethylation destabilizes CTG.CAG trinucleotide repeats in mammalian cells. Hum Mol Genet 2004;13:29792989. [DOI] [PubMed] [Google Scholar]
- 28.Libby RT, Hagerman KA, Pineda VV, et al. CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination. Plos Genet 2008;4:e1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buckley L, Lacey M, Ehrlich M. Epigenetics of the myotonic dystrophy-associated DMPK gene neighborhood. Epigenomics 2016;8:13–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets generated and/or analyzed during the current study are not publicly available for ethical reasons, but are available from the corresponding author on reasonable request.