Abstract
Calcium ions are the principal second messenger in the control of gene expression by electrical activation of neurons. However, the full complexity of calcium-signaling pathways leading to transcriptional activation and the cellular machinery involved are not known. Using the c-fos gene as a model system, we show here that the activity of its complex promoter is controlled by three independently operating signaling mechanisms and that their functional significance is cell type-dependent. The serum response element (SRE), which is composed of a ternary complex factor (TCF) and a serum response factor (SRF) binding site, integrates two calcium-signaling pathways. In PC12 cells, calcium-regulated transcription mediated by the SRE requires the TCF site and is not inhibited by expression of the dominant-negative Ras mutant, RasN17, nor by the MAP kinase kinase 1 inhibitor PD 98059. In contrast, TCF-dependent transcriptional regulation by nerve growth factor or epidermal growth factor is mediated by a Ras/MAP kinases (ERKs) pathway targeting the TCF Elk-1. In AtT20 cells and hippocampal neurons, calcium signals can stimulate transcription via a TCF-independent mechanism that requires the SRF binding site. The cyclic AMP response element (CRE), which cooperates with the TCF site in growth factor-regulated transcription, is a target of a third calcium-regulated pathway that is little affected by the expression of RasN17 or by PD 98059. Thus, calcium can stimulate gene expression via a TCF-, SRF-, and CRE-linked pathway that can operate independently of the Ras/MAP kinases (ERKs) signaling cascade in a cell type-dependent manner.
Keywords: gene regulation, c-fos, calcium signaling, cyclic AMP response element, serum response element, ternary complex factor
Changes in gene expression are important mechanisms by which neurons transform short-lasting electrical events into long-lasting functional and morphological alterations that may be responsible for plasticity-related events such as memory formation (for review, see Sheng and Greenberg, 1990; Morgan and Curran, 1991). Calcium entry into neurons through voltage or ligand-gated ion channels serves as the trigger for electrical activity-dependent transcriptional responses (for review, see Ghosh and Greenberg, 1995). Signal transduction cascades initiated by increases in intracellular calcium concentrations control the transcription rate of inducible genes by modifying transcription factors that interact with specific cis-acting DNA regulatory elements in the promoter of the gene. However, the exact signal transduction mechanisms involved in calcium-regulated transcription are unknown. To analyze this, we have used the c-fos gene, a strongly calcium-inducible gene, as a model system. Previous work has identified two control regions in the c-fos promoter, the cyclic AMP response element (CRE) and the serum response element (SRE), as calcium-responsive promoter/enhancer elements (Sheng et al., 1988, 1990; Bading et al., 1993; Misra et al., 1994; Miranti et al., 1995; Robertson et al., 1995). Our recent study demonstrates that CRE-dependent transcriptional activation is controlled by increases in nuclear calcium concentrations (Hardingham et al., 1997), suggesting a model in which a nuclear calcium-responsive enzyme, possibly calcium/calmodulin (CaM)-dependent protein kinase IV (Jensen et al., 1991), mediates this response. Consistent with this hypothesis is the finding that CaM kinases can phosphorylate the CRE binding protein (CREB), which can mediate calcium-activated transcription of c-fos (Sheng et al., 1990, 1991). This phosphorylation occurs on residue serine 133 that is critical for CREB to function as a transcriptional activator (Gonzales and Montminy, 1989; Sheng et al., 1991; Matthews et al., 1994; Sun et al., 1994).
Calcium signaling at the c-fos SRE is still an enigma. Mechanisms of calcium-activated transcription by the SRE can function independently of increases in nuclear calcium concentrations (Hardingham et al., 1997), suggesting that a signal transduction machinery localized to the cytoplasm can control this response. A candidate signaling cascade is the Ras/MAP kinases (ERKs) pathway, which is activated in response to calcium signals (Bading and Greenberg, 1991; Rosen et al., 1994; Rusanescu et al., 1995). Indeed, a recent study suggested that, in rat cortical neurons, transcriptional induction of a c-fos-based model reporter gene that follows calcium entry through the NMDA receptor occurs via a MAP kinase-dependent pathway targeting the SRE-interacting protein ternary complex factor (TCF) Elk-1 (Xia et al., 1996). However, analysis of similar model reporter genes in the rat pheochromocytoma cell line, PC12, indicates that calcium activates transcription by a TCF-independent pathway involving the principal SRE binding protein, the serum response factor (SRF), and CaM kinases (Misra et al., 1994;Miranti et al., 1995).
In this study we have focused on the analysis of the role of the Ras/MAP kinases (ERKs) signaling cascade in calcium-activated transcription via the SRE and the CRE. Because SRE and CRE function may depend on their context within the c-fos promoter, we constructed mutants of the human c-fos gene by altering these regulatory sites without disturbing surrounding sequences and relative spacing. These in-context mutations allow us, first, to determine the contribution of an individual element to the overall transcriptional response elicited by the intact c-fospromoter and, second, to dissect the multitude of signaling pathways targeting the c-fos promoter and to investigate their nature. We used DNA transfection and microinjection techniques to introduce these plasmids into cells, analyzed them for transcriptional inducibility by calcium signals, and compared them with inducibility by nerve growth factor (NGF) and epidermal growth factor (EGF), two classical activators of the Ras/MAP kinases (ERKs) pathway (Thomas et al., 1992; Wood et al., 1992) (for review, see Marshall, 1994, 1995). Because the functional significance of signal transduction pathways targeting the CRE or the SRE may be cell type-dependent, we have studied calcium-activated transcription in PC12 cells, in the mouse pituitary cell line AtT20, and in primary rat hippocampal neurons.
MATERIALS AND METHODS
Cell culture and transfection. PC12 cells were grown on collagen-coated dishes in DMEM containing 10% heat-inactivated horse serum, 5% fetal calf serum (FCS), 100 U/ml penicillin G, and 100 μg/ml streptomycin (PC12 media) at 37°C in an atmosphere consisting of 10% CO2/90% air. AtT20 cells were grown in DMEM containing 10% FCS, 100 U/ml penicillin G, and 100 μg/ml streptomycin at 37°C in 5% CO2/95% air. For DNA transfection, cells were transferred to poly-d-lysine-coated dishes 1–2 d before transfection. Transfection of PC12 cells [20–30 μg of human c-fosplasmid and 1.5 μg of α-globin plasmid (pSVα1) (Sheng et al., 1988) per 100 mm dish] was performed by the calcium phosphate method (Sheng et al., 1988) with the following modifications: cells were kept in DMEM containing 10% FCS at 37°C in 5% CO2/95% air for 1 hr before transfection. After the addition of calcium phosphate precipitates, cells were incubated in DMEM containing 10% FCS at 37°C in 5% CO2/95% air for 5–6 hr. Cells were shocked with 25% (v/v) glycerol in HEPES-buffered saline, pH 7.05, for 2 min and washed three times with PBS, followed by incubation in PC12 media at 37°C in 10% CO2/90% air until stimulation. AtT20 cells and, in some experiments, PC12 cells were transfected with Lipofectamine (Life Technologies, Gaithersburg, MD).
Analysis of gene expression. At 40–48 hr after transfection, cells were stimulated with NGF (100 ng/ml NGF-7S or 50 ng/ml NGF-2.5S; Sigma, Poole, UK; no difference was observed between the two NGF preparations) for 30–40 min or with 3 ng/ml EGF (Sigma) for 30–40 min or 10 μm forskolin (Calbiochem, Lucerne, Switzerland) and 0.5 mm IBMX (3-isobutyl-1-methylxanthine; Sigma) for 50 min or by adding to the medium 0.41 volume of KCl depolarization solution [containing (in mm) 10 HEPES, pH 7.2, 170 KCl, 1 MgCl2, and 2 CaCl2] for 50 min. Calcium influx into AtT20 cells was induced by adding to the medium 0.41 volume of KCl depolarization solution containing a 5 μm concentration of the L-type calcium channel agonist FPL 64176 (Zheng et al., 1991). Extraction of total RNA and RNase protection assay were as described (Treisman, 1985; Sheng et al., 1988;Bading et al., 1993). In each reaction 10–20 μg of total RNA was hybridized to the c-fos and α-globin antisense probe. Data were quantitated by PhosphorImager (Molecular Dynamics, Sunnyvale, CA) or densitometer analysis and normalized for transfection efficiency by reference to expression of the α-globin gene. CAT and β-galactosidase assays were performed as described (Ausubel et al., 1987); pCH110 containing the β-galactosidase gene under the control of an SV40 enhancer was used as a reference plasmid.
Western blot analysis. Lysis of cells, separation of proteins by SDS-PAGE (10% polyacrylamide), and blotting onto polyvinylidene fluoride membranes was done as described (Bading and Greenberg, 1991). Immunoblot analysis with phospho-MAP kinase/ERK or MAP kinase/ERK-specific antibodies (New England Biolabs, Beverly, MA) and signal detection by chemiluminescence were performed according to the manufacturer’s instruction (New England Biolabs).
Plasmids. The wild-type c-fos plasmid, pF711, contains 711 base pairs (bp) of 5′ flanking sequences; pF222 is identical to pF711 except that it contains only 222 bp of 5′ flanking sequences (Treisman, 1985). All c-fos mutants are derivatives of pF711 and were named according to the particular regulatory element mutated. The following mutants have been described previously: pFosΔTCF (Hill et al., 1994), pFosΔSRF (Hill and Treisman, 1995), and pFosΔSIF (Hill and Treisman, 1995). Plasmids MLV128β, NL.Elk, NL.Elk307Δ, NL.Elk383/389, RSVβ128, RSVrasN17, (Gal)2-TATA-CAT, Gal-ElkC, and Gal-ElkC383/389 also have been described previously (Hill et al., 1993,1995; Marais et al., 1993). Plasmid pFosΔCRE was made by cloning theNotI fragment of pF711 containing the CRE sequence into pGEM13Zf(+) (Promega, Madison, WI). TheNarI/BssH2 fragment containing the CRE was replaced with annealed oligonucleotides 1 and 2 (see below) in which the CRE sequence was replaced with a Gal4 binding site. The resulting plasmid contained a point mutation that was repaired by PCR mutagenesis. Then the NotI fragment containing the Gal4 binding site was cloned into pF711 to create pFosΔCRE. By swapping the BssH2/NcoI fragment of pFosΔCRE into plasmids containing mutations of the SRF binding site, TCF binding site, or SIE, we generated plasmids pFosΔCREΔSRF, pFosΔCREΔTCF, pFosΔCREΔSIF, pFosΔCREΔSRFΔSIF, and pFosΔCREΔTCFΔSIF. The c-fos genomic constructs were tagged with the 9E10 myc epitope (EQKLISEEDL, single letter code) by inserting annealed oligonucleotides 3 and 4 (see below) into the NcoI site located in the fourth exon of the c-fos gene. All DNA manipulations were performed by standard techniques, and plasmids were verified by sequencing.
Oligonucleotides. Oligonucleotides used included the following: (1) CG-CGCCACCCCTCTGGCGCCACCGTGGTTGACGGAGTACTGTCCT-CCGTCATTCATAAAACGCTTGTTATAAAAGCAGTGGCTGCGG (top strand), (2) CGCCGCAGCCACTGCTTTTATAACAAGCGTTTT-ATGAATGACGGAGGACAGTACTCCGTCAACCACGGTGGCGCC-AGAGGGGTGG (bottom strand), (3), CATGAAGCTTGAGCAGAAG-CTGATCAGCGAGGAAGATCTGGC (top strand), and (4) CATGGC-CAGATCTTCCTCGCTGATCAGCTTCTGCTCAAGCTT (bottom strand).
Microinjection of primary hippocampal neurons and immunofluorescence. Hippocampal neurons were isolated and cultured as described (Bading and Greenberg, 1991). At 9 d after plating the growth medium was replaced by transfection medium (Bading et al., 1993). Neurons were injected 11–12 d after plating, using a Zeiss Microinjection Workstation (Oberkochen, Germany). Plasmids were injected into the nucleus at 100 μg/ml in half-strength PBS containing 0.9% (w/v) Texas Red-conjugated 70 kDa dextran (Molecular Probes, Eugene, OR) as an injection marker. At 24 hr after microinjection, neurons were stimulated with 20 μmglutamate. Glutamate stimulation was terminated after 10 min by adding 1 mm sodium kynurenate and 11 mmMgCl2 to the medium, as described (Bading et al., 1993). Cells were fixed 2 hr after stimulation in 3% paraformaldehyde in PBS containing 4% sucrose for 20 min, washed twice with 10 mmglycine in PBS for 10 min, permeabilized in 0.5% NP-40 in PBS for 5 min, and then incubated in blocking solution (1.5% normal goat serum in PBS) for 20 min. All steps were at room temperature. Incubation with the anti-myc monoclonal antibody 9E10 (Santa Cruz Biotechnology, Tebu, France) (1:200 dilution in PBS) was overnight at 4°C. Incubations with biotinylated anti-mouse antibody (Jackson ImmunoResearch, West Grove, PA) and with fluorescein–avidin (Vector Laboratories, Burlingame, CA), both diluted 1:200 in PBS, were for 1 hr each at room temperature. Cells were washed with PBS and mounted in Vectashield (Vector). Immunofluorescence was quantitated by an MRC 600 confocal laser scanning microscope.
RESULTS
c-fos promoter in-context mutations
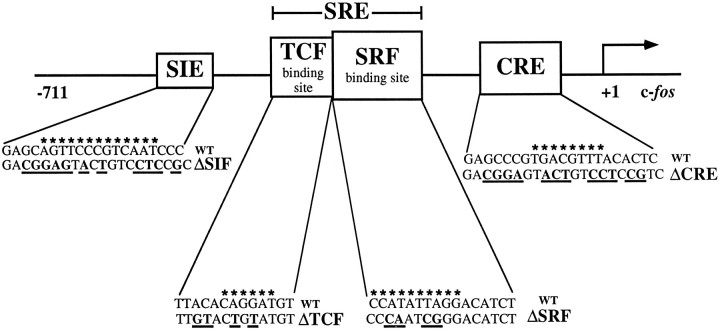
To study the function of the SRE and the CRE in the context of the intact c-fos gene, we introduced specific mutations into the promoter of the human c-fos gene by changing the key native regulatory sequences to heterologous DNA binding sites for the yeast proteins Gal4 or MCM1 or to a half-site for the bacterial LexA protein (see Fig. 1 for schematic representation). The parental plasmid pF711 contains the entire transcribed region of the c-fos gene and 711 bp of upstream regulatory sequence (Treisman, 1985). The SRE, centered on nucleotide −310 relative to the transcription start site, was changed to an MCM1 site that abrogates binding of SRF (plasmid pFosΔSRF; Hill and Treisman, 1995). SRF is the principal transcription factor that interacts with the SRE (Prywes and Roeder, 1987; Schröter et al., 1987; Treisman, 1987; Norman et al., 1988). At the c-fos SRE, SRF can form a ternary complex with Ets domain proteins such as Elk-1 or SAP-1 [also termed ternary complex factors (TCFs)] that are transcription factor targets of the Ras/MAP kinases (ERKs) pathway and contain growth factor-regulated transcriptional activation domains (Shaw et al., 1989;Hipskind et al., 1991; Dalton and Treisman, 1992; Gille et al., 1992;Hill et al., 1993; Janknecht et al., 1993; Marais et al., 1993) (for review, see Treisman, 1994). Recruitment of Elk-1 or SAP-1 to the SRE/SRF complex requires the TCF site (Dalton and Treisman, 1992;Treisman et al., 1992), which is an Ets motif located 5′ to the SRF binding site and critical for c-fos activation by certain stimuli (Shaw et al., 1989; Graham and Gilman, 1991; Hill and Treisman, 1995). To disrupt ternary complex formation at the c-fosSRE, we replaced the TCF site by a LexA half-site (plasmid pFosΔTCF;Hill et al., 1994). The CRE, located at nucleotide −60 relative to the transcription start site, was changed to a Gal4 binding site (plasmid pFosΔCRE; see Materials and Methods). The sis-inducible element (SIE), which is located at nucleotide −345 and plays a role in growth factor and cytokine-activated gene expression (Hayes et al., 1987;Wagner et al., 1990; Fu and Zhang, 1993; Sadowski et al., 1993; Zhong et al., 1994) (for review, see Darnell et al., 1994; Ihle et al., 1994), also was changed to a Gal4 binding site (plasmid pFosΔSIF;Hill and Treisman, 1995). Additional constructs contain combinations of these mutations.
Fig. 1.
Schematic representation of the human c-fos promoter illustrating the in-context mutations. All c-fos gene constructs contain 711 bp of c-fos upstream regulatory sequence and the entire transcribed region. The start of transcription is indicated by anarrow. The base changes used to create the promoter mutants are shown in bold below the wild-type sequence. The ΔCRE and ΔSIF mutations both generate Gal4 binding sites and abrogate the binding of CREB/ATF and sis-inducible factor (SIF)/STAT, respectively. TheΔTCF mutation generates a LexA half-site and blocks ternary complex formation; the ΔSRF mutation creates an MCM1 site that cannot bind SRF. The consensus binding sites for STAT factors, TCFs, SRF, and CREB/ATF are indicated byasterisks.
c-fos induction by NGF and EGF is mediated by a Ras/MAP kinases (ERKs) signaling pathway targeting the TCF Elk-1
To establish our experimental system, we first tested in DNA transfection experiments the wild-type and mutant c-fos gene constructs in PC12 cells for inducibility by NGF and EGF treatment, which is known to signal to the nucleus via the Ras/MAP kinases (ERKs) signaling pathway (Thomas et al., 1992; Wood et al., 1992). Expression of the endogenous rat c-fos (c-fosR) and the transfected human c-fos (c-fosH) was measured by RNase protection assay. A plasmid containing the human α-globin gene under the control of the SV40 enhancer was cotransfected to normalize for transfection efficiency. The result of a representative experiment is shown in Figure 2. The basal level of expression of the endogenous rat c-fos gene and the transfected wild-type and mutant human c-fos gene constructs was low (Fig. 2A, lanes 1, 4, 7, 10, 13,and 16). Stimulation of PC12 cells with NGF and EGF resulted in robust transcriptional induction of the endogenous and transfected c-fos genes (Fig. 2A, lanes 2and 3). Mutation of the SRF binding site (plasmid pFosΔSRF) reduced inducibility by NGF and EGF treatment to 23 ± 1% (n = 3) and 18 ± 1% (n = 3) (Fig. 2A, lanes 5 and 6), respectively, demonstrating that the SRF binding site is critical for transcriptional regulation by NGF and EGF. We next investigated whether the SRF binding site or the adjacent TCF binding site (which is nonfunctional in the absence of SRF binding) is the site of regulation by NGF and EGF signaling pathways. We tested the c-fos gene construct pFosΔTCF that contains an intact SRF binding site but no longer allows ternary complex formation (Hill et al., 1993, 1994). Mutation of the TCF site reduced the growth factor-induced transcriptional responses to 27 ± 6% (NGF; n = 5) and 31 ± 4% (EGF; n = 5) (Fig.2A, lanes 14 and 15). This demonstrates that, in the context of the intact c-fos gene, the SRF binding site is necessary, but not sufficient, for transcriptional induction by NGF and EGF and that ternary complex formation is critical for transcriptional regulation by NGF and EGF signaling pathways. Mutation of the SIE (plasmid pFosΔSIF) did not reduce c-fos inducibility after growth factor treatment (NGF: 91 ± 19%, n = 3; EGF: 86 ± 14%,n = 3) (Fig. 2A, lanes 8 and9). Double mutations of the SRF binding site and the SIE (plasmid pFosΔSRFΔSIF; Fig. 2A, lanes 11 and12) or of the TCF site and the SIE (plasmid pFosΔTCFΔSIF; Fig. 2A, lanes 17 and18) resulted in levels of transcriptional induction similar to those seen with the TCF site or SRE mutations alone. Therefore, the SIE appears to play only a minor role in transcriptional induction by NGF and EGF. Mutation of the CRE reduced the growth factor-stimulated transcriptional responses to 45 ± 5% (NGF; n = 3) and 46 ± 8% (EGF; n = 5) of the induction observed with the c-fos wild-type gene (Fig.2B, lanes 5 and 6). Double mutation of the CRE and either the SRF binding site or the TCF site resulted in a virtually complete loss of inducibility (pFosΔCREΔSRF: 6 ± 1%, n = 2, NGF; 5 ± 1%, n = 2, EGF; pFosΔCREΔTCF: 8 ± 1%, n = 2, NGF; 12 ± 2%, n = 3, EGF) (Fig. 2B, lanes 8 and 9, 11 and 12). These results indicate that the CRE is required for a maximal growth factor response and suggest a mechanism by which TCFs cooperate with CRE-interacting proteins in NGF and EGF-regulated c-fostranscription.
Fig. 2.
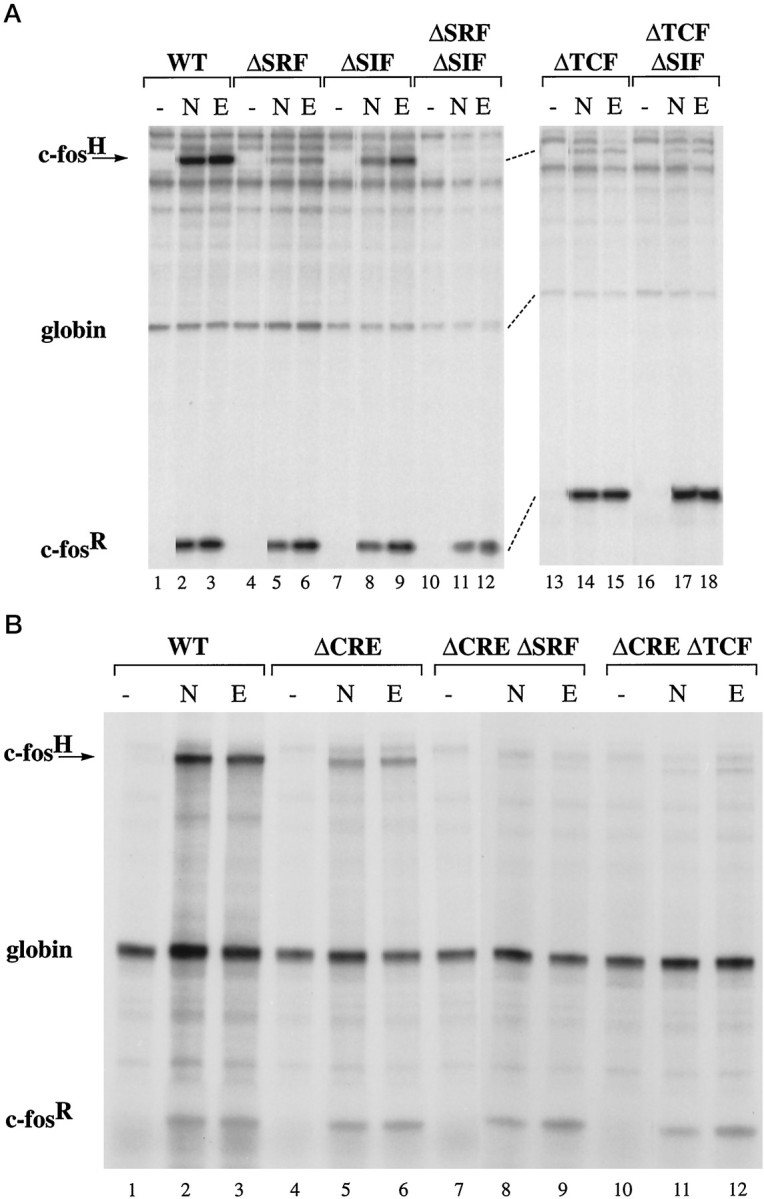
The TCF site and the CRE control c-fos transcriptional induction in PC12 cells after NGF and EGF treatment. RNase protection analysis was used to measure expression of the endogenous rat c-fos gene (c-fosR), the transfected human α-globin gene (globin; to indicate transfection efficiency), and human c-fos gene (c-fosH) after transfection of plasmid pF711 (wild-type c-fos gene;A, lanes 1–3; B, lanes 1–3) or one of the following plasmids containing in-context mutations: pFosΔSRF (A, lanes 4–6), pFosΔSIF (A, lanes 7–9), pFosΔSRFΔSIF (A, lanes 10–12), pFosΔTCF (A, lanes 13–15), pFosΔTCFΔSIF (A, lanes 16–18), pFosΔCRE (B, lanes 4–6), pFosΔCREΔ-SRF (B, lanes 7–9), or pFosΔCREΔTCF (B, lanes 10–12). RNA was isolated from unstimulated cells (lanes marked −) or cells stimulated with NGF (N) or EGF (E).
Having established that the TCF site is a critical target for NGF and EGF signaling pathways, we next tested the function of TCF Elk-1 as an NGF- and EGF-responsive transcription factor. Mutation of the TCF site introduces a half-site for the bacterial LexA protein into the c-fos promoter, which allowed us to determine whether Elk-1 can restore NGF and EGF inducibility by expressing a LexA-Elk-1 fusion protein (NL.Elk) in PC12 cells. LexA-Elk-1 binds to the ΔTCF mutation in a SRF-dependent manner (Hill et al., 1993). As shown in Figure3A, expression of NL.Elk restored inducibility of pFosΔTCF to 81 ± 14% (NGF; n = 3) and 60 ± 14% (EGF; n = 4), respectively, of the induction observed with the c-fos wild-type pF711 (Fig.3A, compare lanes 2 and 3 withlanes 8 and 9; 3B). Expression of a truncated form of Elk-1 that lacks C-terminal sequences (NL.Elk307Δ), including phosphorylation sites or the mutation of two MAP kinase/ERK phosphorylation sites on Elk-1 (serine 383 and serine 389; NL.Elk383/389) previously shown to be critical for Elk-1 activity (Hill et al., 1993; Janknecht et al., 1993; Marais et al., 1993), renders Elk-1 functionally inactive (Fig. 3A, lanes 11 and12, 14 and 15; 3B). To investigate whether Elk-1 is sufficient to mediate this transcriptional response, we examined the ability of a Gal4-Elk-1 fusion protein (Gal-ElkC) to transactivate a Gal4-CAT reporter construct in NGF-treated PC12 cells. The basal level of expression of the Gal4-CAT reporter gene in unstimulated cells was low (Fig. 3C, lanes 1, 3, and 5). NGF stimulation of PC12 cells cotransfected with the Gal-ElkC expression plasmid, but not with the vector control, resulted in a robust transactivation of the Gal4-CAT reporter gene (Fig. 3C, lanes 2 and 4). Mutations of serine 383 and serine 389 on Elk-1 (Gal-ElkC383/389) abolished its ability to transactivate the reporter gene (Fig. 3C, lane 6). Similar results were obtained with other reporter gene constructs, using EGF as the stimulus (data not shown). These results demonstrate that Elk-1 is sufficient for mediating NGF- and EGF-induced transcriptional responses in PC12 cells.
Fig. 3.
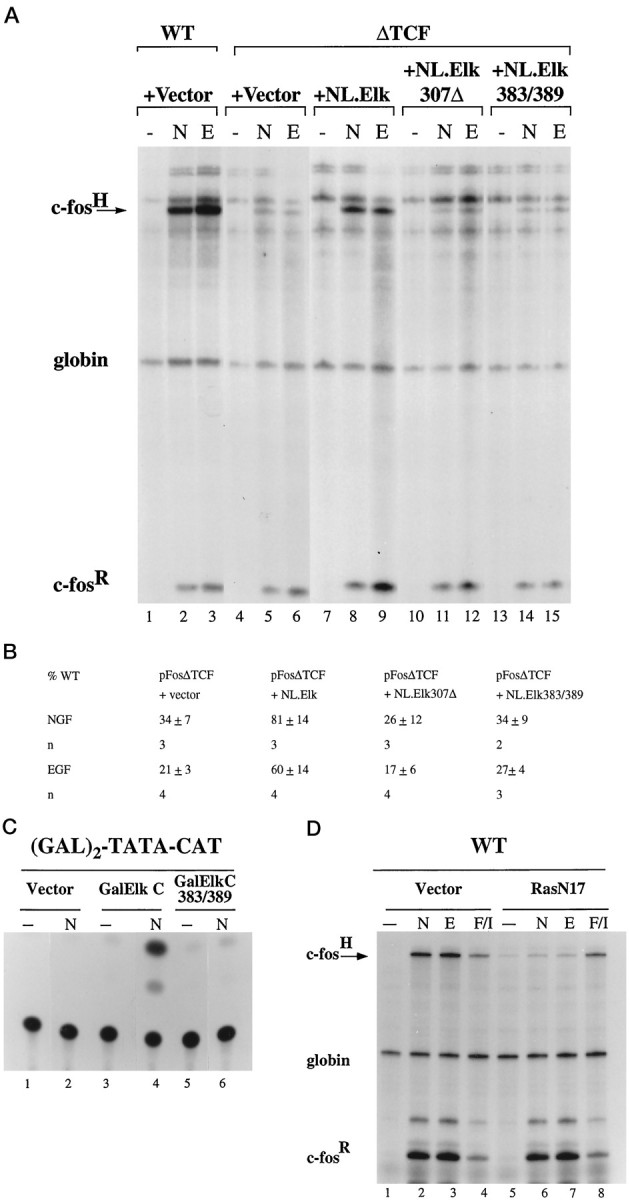
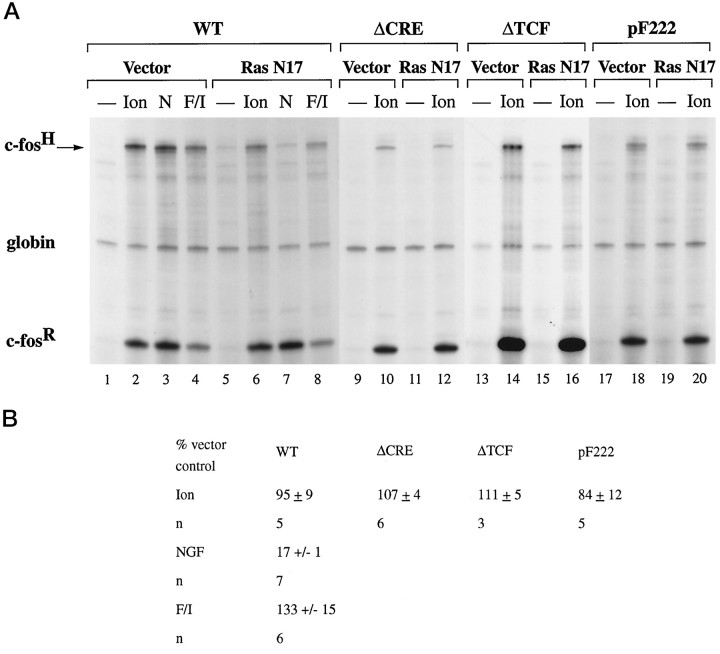
NGF and EGF treatment of PC12 cells activates c-fos expression by a Ras/MAP kinases (ERKs) signaling pathway targeting TCF Elk-1. A, RNase protection analysis was performed as described in Figure 2. PC12 cells were transfected either with 30 μg of plasmid pF711 (wild-type c-fos gene, lanes 1–3) or with 30 μg of pFosΔTCF (lanes 4–15) plus 1 μg of vector plasmid (MLV128β, lanes 4–6), plasmid NL.Elk, which expresses an altered binding specificity Elk-1 protein that can bind the ΔTCF mutation (lanes 7–9; Hill et al., 1994), plasmid NL.Elk307Δ (lanes 10–12), or NL.Elk383/389 (lanes 13–15). Cells were stimulated with NGF (N) or EGF (E).B, Quantitation of RNase protection experiments by PhosphorImager and densitometer analysis. The levels of c-fos mRNA transcribed from plasmidpFosΔTCF, normalized for transfection efficiency to that of the α-globin gene, are expressed as a percentage of the level of mRNA produced by the transfected wild-type c-fos gene construct pF711, normalized to α-globin expression, in response to the same stimulus. The mean ± SEM of three independent experiments is shown. C, NGF-regulated transcriptional activation by Gal4-Elk-1 fusion proteins measured with a (Gal)2-TATA-CAT reporter gene construct. PC12 cells were transfected with 2 μg of plasmid (Gal)2-TATA-CAT and 1 μg of vector (pUC19; lanes 1 and 2), Gal-ElkC expression plasmid (lanes 3 and4), or Gal-ElkC383/389 expression plasmid (lanes 5 and 6). Transfections also included 0.5 μg of a reference plasmid, pCH110, containing the β-galactosidase gene under the control of the SV40 enhancer. Extracts were prepared from unstimulated cells (lanes marked −), and cells were stimulated with 100 ng/ml NGF-7S (N) for 8 hr and analyzed for CAT activity. Data were quantified by the PhosphorImager and normalized for transfection efficiency to expression of the β-galactosidase reference gene. NGF-induced levels of CAT activities, relative to that obtained with PC12 cells expressing GalElk C, which was taken as 100%, were 6% (Vector) and 5% (Gal-ElkC383/389). Similar results were obtained in other experiments. D, RNase protection analysis was performed as described in Figure 2. PC12 cells (60 mm dish) were transfected by using Lipofectamine with 4 μg of plasmid pF711 (wild-type c-fos gene) plus 1 μg of either vector plasmid (RSVβ128, lanes 1–4) or plasmid RSVrasN17 (lanes 5–8; Hill et al., 1995). Transfections also included 0.75 μg of a reference plasmid pSVα1. Cells were stimulated with NGF (N), EGF (E), or forskolin/IBMX (F/I) or were left unstimulated (lanes marked −). NGF-induced levels of c-fos mRNA transcribed from pF711, normalized for transfection efficiency to the levels of α-globin mRNA, in cells expressing RasN17 are 17 + 1% (n = 7) of that obtained in cells transfected with the vector control. Similar results were obtained in EGF-treated cells. cAMP-induced expression of the pF711, normalized to the levels of α-globin mRNA, in cells expressing RasN17 is 133 ± 15% (n = 6) of that in cells transfected with the vector control.
We next directly tested the role of the Ras/MAP kinases (ERKs) signaling pathway in NGF- and EGF-activated transcription. Expression of a dominant interfering Ras mutant, RasN17 (Feig and Cooper, 1988), blocked NGF- and EGF-induced expression of the wild-type c-fos gene construct pF711 (Fig. 3D, comparelanes 2 and 3 with lanes 6 and7). In contrast, expression of RasN17 did not reduce c-fos induction in response to increased intracellular levels of cAMP after treatment of the cells with a combination of forskolin (stimulator of adenylate cyclase) and IBMX (inhibitor of cAMP phosphodiesterase) (Fig. 3D, compare lanes 4 and8).
This analysis of NGF- and EGF-induced gene expression, which established a direct link between a growth factor receptor signal, a particular signal transduction cascade, and a transcription factor target, illustrates that the c-fos gene constructs when used in conjunction with other genetic or pharmacological means are powerful tools to investigate the mechanisms of signal-regulated transcription. We next applied this experimental approach to the analysis of gene expression controlled by calcium signals.
In PC12 cells, calcium-signaling pathways control transcription via CRE- and TCF-dependent mechanisms
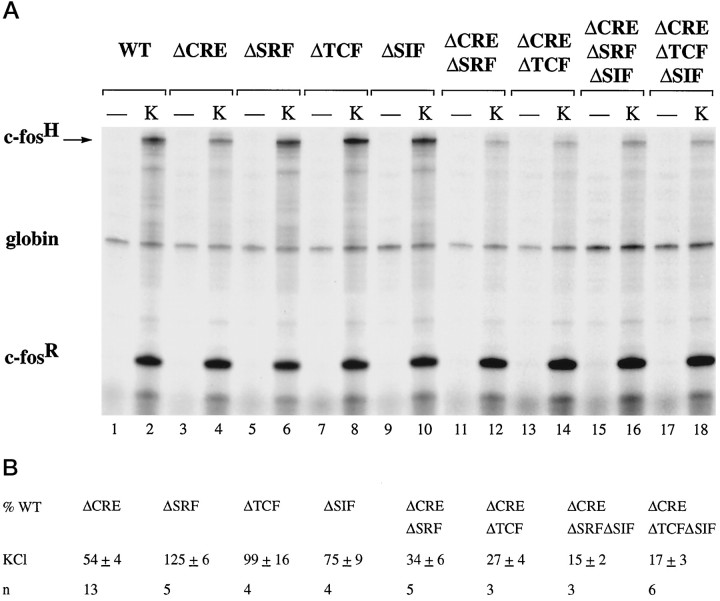
To activate calcium-signaling pathways in PC12 cells, we exposed cells to elevated extracellular KCl, which causes membrane depolarization and the opening of L-type voltage-gated calcium channels. In contrast to NGF and EGF stimulation, induction of c-fos transcription by KCl treatment was not reduced by mutations of either the TCF site or the SRF binding site (Fig.4A, compare lane 2 withlanes 6 and 8; 4B). Because this is most likely caused by the presence of the CRE (at position −60 relative to the start site of transcription), which previously has been shown to function as a calcium-responsive element (Sheng et al., 1988,1990; Bading et al., 1993), we tested c-fos gene constructs that lack this CRE. In-context mutation of the CRE reduced c-fos induction after KCl treatment to 54 ± 3% (n = 13) (Fig. 4A, lane 4; 4B), demonstrating the importance of the CRE in calcium-activated transcription. However, significant induction still occurred in the absence of the CRE, indicating the existence of at least one other calcium-responsive element in the c-fospromoter. To determine whether CRE-independent activation of the c-fos promoter by calcium signals is mediated by the SRE, we transfected the c-fos gene construct pFosΔCREΔSRF in which both the CRE and the SRF binding site had been mutated. Compared with construct pFosΔCRE, transcriptional induction of pFosΔCREΔSRF is reduced further (Fig. 4A, comparelanes 4 and 12; 4B), indicating that binding of SRF is required for CRE-independent calcium-activated transcription. We next investigated whether this response is TCF-dependent and tested the c-fos gene construct pFosΔCREΔTCF that contains an intact SRF binding site but does not allow for ternary complex formation. Mutation of the TCF site resulted in a reduction of the CRE-independent calcium response that was similar to that seen with the mutation of the SRF binding site (Fig.4A, compare lanes 12 and 14; 4B). This indicates that, in the absence of the CRE, calcium-signaling pathways can stimulate transcription via a TCF-dependent mechanism. To identify transcription factors involved in this response, we used the same strategy that was used to investigate NGF- and EGF-regulated transcription factors. Similar to the experiments described in Figure 3A, we used a LexA-Elk-1 fusion protein to test whether Elk-1 can mediate the TCF-dependent calcium response. Expression of NL.Elk resulted in only a very small increase in calcium-dependent inducibility of the c-fos gene constructs pFosΔCREΔTCF and pFosΔCREΔ-TCFΔSIF (data not shown). Although this may indicate that TCFs other than Elk-1 function as a calcium-activated transcription factor, it is also possible that, in the absence of the CRE, TCF-LexA fusion proteins are unable to reconstitute the complex required to activate transcription in response to calcium-signaling pathways.
Fig. 4.
The TCF site and the CRE are targets of calcium-signaling pathways in PC12 cells. A, RNase protection analysis was performed as described in Figure 2. PC12 cells were transfected with plasmid pF711 (wild-type c-fosgene, lanes 1 and 2) or one of the following plasmids containing in-context mutations: pFosΔCRE (lanes 3 and 4), pFosΔSRF (lanes 5 and 6), pFosΔTCF (lanes 7 and 8), pFosΔSIF (lanes 9 and 10), pFosΔCREΔSRF (lanes 11 and 12), pFosΔCREΔTCF (lanes 13 and 14), pFosΔCREΔSRFΔSIF (lanes 15 and 16), or pFosΔCREΔTCFΔSIF (lanes 17 and 18). RNA was isolated from unstimulated cells (lanes marked −) or cells stimulated with KCl (K).B, Quantitation of RNase protection experiments by PhosphorImager analysis. The levels of c-fos mRNA transcribed from the indicated plasmids and normalized for transfection efficiency to the level of α-globin mRNA are expressed as a percentage of the amount of mRNA produced by the transfected wild-type c-fos gene construct pF711, normalized to α-globin expression, in response to the same stimulus. The mean ± SEM is shown. Compared with the wild-type construct pF711, mutation of the SRF binding site consistently caused a small but significant (p < 0.001; paired t test) enhancement of the KCl-induced transcriptional response.
Double mutations of either the CRE and TCF site or the CRE and SRE abolish most of the calcium-activated transcriptional response. However, a small portion of the induction, ∼25–35%, still remains. Introduction of an SIE mutation further reduced this remaining response to 15 ± 2% (pFosΔCREΔSRFΔSIF, n = 3) and to 17 ± 3% (pFosΔCREΔ-TCFΔSIF, n = 6) (Fig. 4A, lanes 16 and 18; 4B), This indicates that the SIE can, in the absence of the CRE and SRE, make a small contribution to transcriptional activation by calcium signal.
In AtT20 cells, calcium-signaling pathways control transcription via CRE and TCF-independent SRF-linked mechanisms
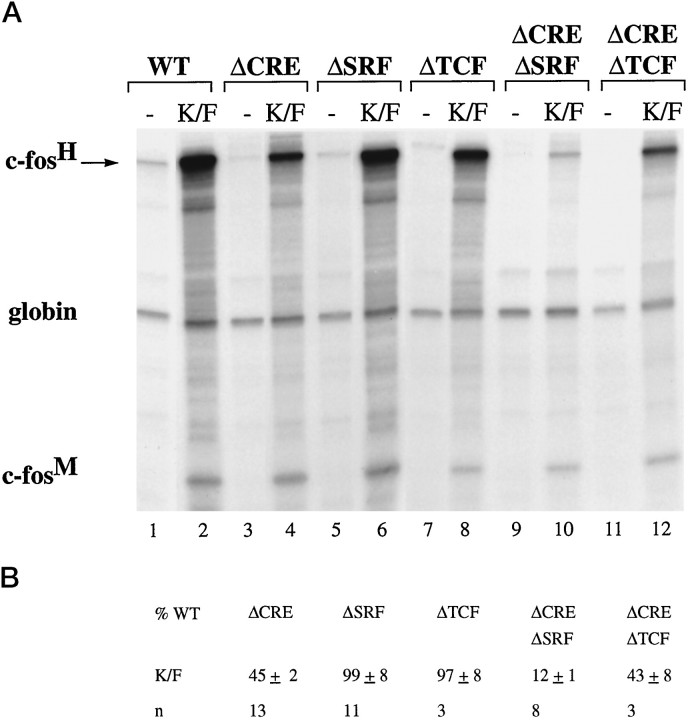
AtT20 cells were stimulated by exposing them to elevated extracellular KCl in the presence of a 5 μm concentration of the L-type calcium channel agonist FPL 64176 (Zheng et al., 1991) (KCl/FPL stimulation). KCl treatment alone gives rise to only small increases in intracellular calcium concentrations, insufficient to activate gene transcription (Hardingham et al., 1997). Similar to the results obtained with PC12 cells, mutation of either the SRF binding site or the TCF site did not affect KCl/FPL-induced c-fosexpression (Fig. 5A, compare lane 2 with lanes 6 and 8; 5B), and mutation of the CRE reduced the response to 45 ± 2% (n = 13) (Fig. 5A, lane 4; 5B) of that obtained with pF711. However, in contrast to PC12 cells, calcium-activated transcription via the SRE (plasmid pFosΔCRE: 45 ± 2%, n = 13) was reduced by a mutation of the SRF binding site (plasmid pFosΔCREΔSRF: 12 ± 1%, n = 8), but not by a mutation of the TCF site (plasmid pFosΔCREΔTCF: 43 ± 8%, n = 3) (Fig.5A, compare lane 4 with lanes 10 and12; 5B). This indicates that, in AtT20 cells, SRE-mediated calcium-activated transcription is controlled by a TCF-independent SRF-linked pathway.
Fig. 5.
The SRF-binding site and the CRE are targets of calcium-signaling pathways in AtT20 cells. A, RNase protection analysis was performed as described in Figure 2. AtT20 cells were transfected with plasmid pF711 (wild-type c-fosgene, lanes 1 and 2) or one of the following plasmids containing in-context mutations: pFosΔCRE (lanes 3 and 4), pFosΔSRF (lanes 5 and 6), pFosΔTCF (lanes 7 and 8), pFosΔCREΔSRF (lanes 9 and 10), or pFosΔCREΔTCF (lanes 11 and 12). RNA was isolated from unstimulated cells (lanes marked −) or cells stimulated with KCl/FPL 64176 (K/F). B, Quantitation of RNase protection experiments by the PhosphorImager. Analysis was done as described in Figure 4B.
NMDA receptor activation can mediate transcriptional induction by a TCF-independent mechanism in hippocampal neurons
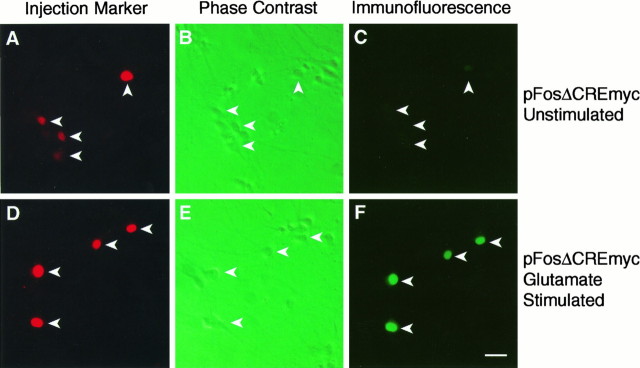
The NMDA receptor is a major site for calcium entry into hippocampal neurons and is the principal ion channel responsible for transcriptional regulation by the excitatory neurotransmitter glutamate (Cole et al., 1989; Wisden et al., 1990; Lerea et al., 1992; Bading et al., 1993, 1995; Lerea and McNamara, 1993). A previous study has shown that the c-fos SRE can function as an NMDA receptor-responsive element (Bading et al., 1993). However, it is unknown whether NMDA receptor-induced transcription is mediated by the SRF binding site or requires, in addition, ternary complex formation (as is the case for calcium, NGF, and EGF induction of c-fosexpression in PC12 cells; see Figs. 2, 4). We initially addressed this issue in DNA transfection and RNase protection experiments, similar to those described above that used PC12 cells. However, the basal level of expression of the transfected c-fos gene constructs pF711 and pFosΔCRE in unstimulated hippocampal neurons was very high and only moderately induced on NMDA receptor activation (data not shown), which made the interpretation of the data very difficult. In the same experiments, the endogenous c-fos gene exhibited low basal expression levels and was robustly induced after NMDA receptor activation, suggesting that deregulation of the transfected c-fos constructs may be a result of the transfection procedure. We, therefore, established a microinjection technique as an alternative means of introducing DNA into hippocampal neurons. To allow transcriptional induction of the injected c-fos gene constructs to be measured at the single cell level, we inserted an oligonucleotide encoding the 9E10 myc epitope in frame into the c-fos coding region. Expression of the myc-tagged c-fos protein was detected by immunocytochemical methods, using the 9E10 antibody, and quantified by the confocal laser scanning microscope. Similar to the DNA transfection experiments, microinjection into primary hippocampal neurons of the myc-tagged wild-type c-fos gene, pF711myc, gave rise to high basal levels of expression in many unstimulated cells (data not shown). However, in-context mutation of the CRE reduced this basal level of expression, providing an assay for analyzing mechanisms of NMDA receptor-regulated gene expression at the single-cell level. To determine whether ternary complex formation is critical for transcriptional regulation by the NMDA receptor pathway, we tested the myc-tagged constructs pFosΔCREmyc, pFosΔCREΔ-SRFmyc, and pFosΔCREΔTCFmyc (Fig.6, Table 1). Expression of pFosΔCREmyc was very low in the majority (91%) of unstimulated cells and in most cells induced to moderate or high levels in response to glutamate treatment, which was used to activate NMDA receptors. We observed, however, a small number of unstimulated neurons that express pFosΔCREmyc at moderate and even high levels. Although this may be attributable to spontaneous electrical activation of the neurons, as described previously for the endogenous c-fos gene (Bading et al., 1995), it is also possible that the stress associated with microinjection of DNA constructs triggers a transcriptional response in some hippocampal neurons, giving rise to false-positive cells. In a fraction of the neurons (28%) expression of pFosΔCREmyc was not induced after glutamate stimulation, which resembles results obtained previously with the endogenous c-fos gene (Bading et al., 1995) and may reflect a lack of functional NMDA receptors. Glutamate regulation of pFosΔCREΔTCFmyc was virtually identical to that of pFosΔCREmyc, with most cells showing moderate or strong levels of induction and a fraction of neurons (27%) with no detectable transcriptional response. In contrast, mutation of the SRE (pFosΔCREΔSRFmyc) resulted in a loss of inducibility in most neurons (74%). These results indicate that the NMDA receptor signaling pathway can activate transcription by a TCF-independent mechanism that requires the SRF binding site. Table 1 summarizes our microinjection experiments; Figure 6 shows representative examples.
Fig. 6.
Analysis of NMDA receptor regulation of c-fos expression in primary hippocampal neurons by microinjection technique. The example shown illustrates neurons that were injected with plasmid pFosΔCREmyc. Similar experiments were performed with other c-fos gene constructs, the results of which are summarized in Table 1. Injected neurons, left unstimulated (A–C) or stimulated with 20 μmglutamate (D–F), are identified by the fluorescence of the Texas Red-conjugated dextran (A andD) present in the injection solution and are indicated with arrows in the phase-contrast photographs (B and E). Expression of myc-tagged c-fos protein in unstimulated neurons (C; positions of injected cells are marked by arrows) and glutamate-stimulated neurons (F) was detected by immunofluorescence technique with the 9E10 monoclonal antibody. Scale bar, 50 μm.
Table 1.
Expression of different c-fos gene constructs injected into primary hippocampal neurons
| c-fos gene construct | Stimulation | No expression (%) | Moderate expression (%) | Strong expression (%) |
|---|---|---|---|---|
| pFosΔCREmyc | — | 91 | 3 | 6 |
| pFosΔCREmyc | Glutamate | 28 | 38 | 34 |
| pFosΔCREΔTCFmyc | Glutamate | 27 | 35 | 27 |
| pFosΔCREΔSRFmyc | Glutamate | 74 | 16 | 10 |
Hippocampal neurons (30–50) were injected for each construct in four independent experiments. Cells were classified as neurons on the basis of morphology and expression of neuron-specific markers. Immunofluorescence was quantified on a confocal laser scanning microscope. Cells with an immunofluorescence signal of <10%, between 10 and 40%, and >40% relative to the brightest cell in an experiment were categorized as showing no, moderate, and strong expression, respectively. Numbers indicate the percentage of the total number of neurons injected.
Role of Ras/MAP kinases (ERKs) pathway in calcium-activated transcription
Expression of a dominant-negative form of Ras, RasN17, previously has been shown to block calcium activation of MAP kinase kinase (MEK) and MAP kinases/ERK (Rosen et al., 1994; Rusanescu et al., 1995). To investigate the role of MAP kinases/ERK in the regulation of gene expression by calcium signals, we tested the effect of expression of RasN17 on CRE and TCF-mediated calcium-activated transcription in PC12 cells. Because expression of RasN17 may affect the function of L-type voltage-gated calcium channels (Rosen et al., 1994), we activated calcium-signaling pathways in PC12 cells by adding 10 μmionomycin to the medium, which, similar to KCl treatment, activated c-fos expression via a CRE-dependent and TCF-dependent mechanism (data not shown). Expression of RasN17 had very little effect on calcium-induced transcription from the transfected wild-type gene construct pF711, the CRE-dependent gene construct pFosΔSRF, and the TCF-dependent gene construct pFosΔCRE (Fig.7A, compare lanes 2 and6, 10 and 12, 14 and16; 7B). Calcium-activated transcription of another CRE-dependent c-fos gene construct, pF222, which contains a deletion of upstream regulatory sequence 5′ to nucleotide −222 and therefore lacks the SRE, also was affected only very little by expression of RasN17 (Fig. 7A, compare lanes 18 and 20; 7B). cAMP-induced transcription from pF711 after treatment of the cells with forskolin and IBMX also was not inhibited by the expression of RasN17 (Fig. 7A, compare lanes 4 and 8; 7B), whereas NGF-induced transcription from pF711 was blocked by RasN17 (Fig.7A, compare lanes 3 and 7; 7B). These results indicate that calcium can induce CRE-mediated and TCF-mediated transcription independently of Ras activation. Because Ras activation is critical for the stimulation of the MAP kinases (ERKs) signaling cascade, our results suggest that calcium-activated transcription operates independently of the Ras/MAP kinases (ERKs) pathway. To test this hypothesis directly, we treated PC12 cells with the MAP kinase kinase (MEK) 1 inhibitor, PD 98059. This treatment blocked KCl-induced and ionomycin-induced MAP kinases/ERKs activation as assessed by Western blot analysis, using antibodies specific for the phosphorylation sites on MAP kinases/ERKs that are indicative of stimulation of their enzymatic activity (Fig.8A, compare lanes 2 and3 with 5 and 6). In contrast, similar to the results obtained in the RasN17 expression experiments, PD 98059 treatment had little effect on calcium-induced transcriptional activation of the transfected wild-type gene construct pF711, the TCF-dependent construct pFosΔCRE, and the CRE-dependent constructs pFosΔTCF and pF222 (Fig. 8B, compare lanes 2 and 3, 7 and 8, 10and 11, 13 and 14; 8C). In contrast, NGF-induced transcription from pF711 was inhibited by PD 98059 (Fig. 8B, lanes 4, 5; 8C). This result provides further evidence against a critical role of the Ras/MAP kinases (ERKs) signaling cascade in calcium-regulated transcription.
Fig. 7.
Effect of expression of RasN17 on CRE-dependent and TCF-dependent calcium-activated transcription in PC12 cells.A, RNase protection analysis was performed as described in Figure 2. PC12 cells were transfected with plasmid pF711 (wild-type c-fos gene, lanes 1–8), pFosΔ-CRE (lanes 9–12), pFosΔTCF (lanes 13–16), or pF222 (lanes 17–20) plus either vector plasmid (RSVβ128, lanes 1–4, 9 and10, 13 and 14,17 and 18) or plasmid RSVrasN17 (lanes 5–8, 11 and 12, 15and 16, 19 and 20). Cells were stimulated with ionomycin (Ion), NGF (N), and forskolin/IBMX (F/I) or were left unstimulated (lanes marked −). B, Quantitation of RNase protection experiments by the PhosphorImager. The levels of c-fos mRNA transcribed from the indicated plasmids, normalized for transfection efficiency to the level of α-globin mRNA, in cells expressing RasN17 are shown as a percentage of the amount of c-fos mRNA, normalized to α-globin expression, produced by cells transfected with the vector control in response to the same stimulus. The mean ± SEM is shown.
Fig. 8.
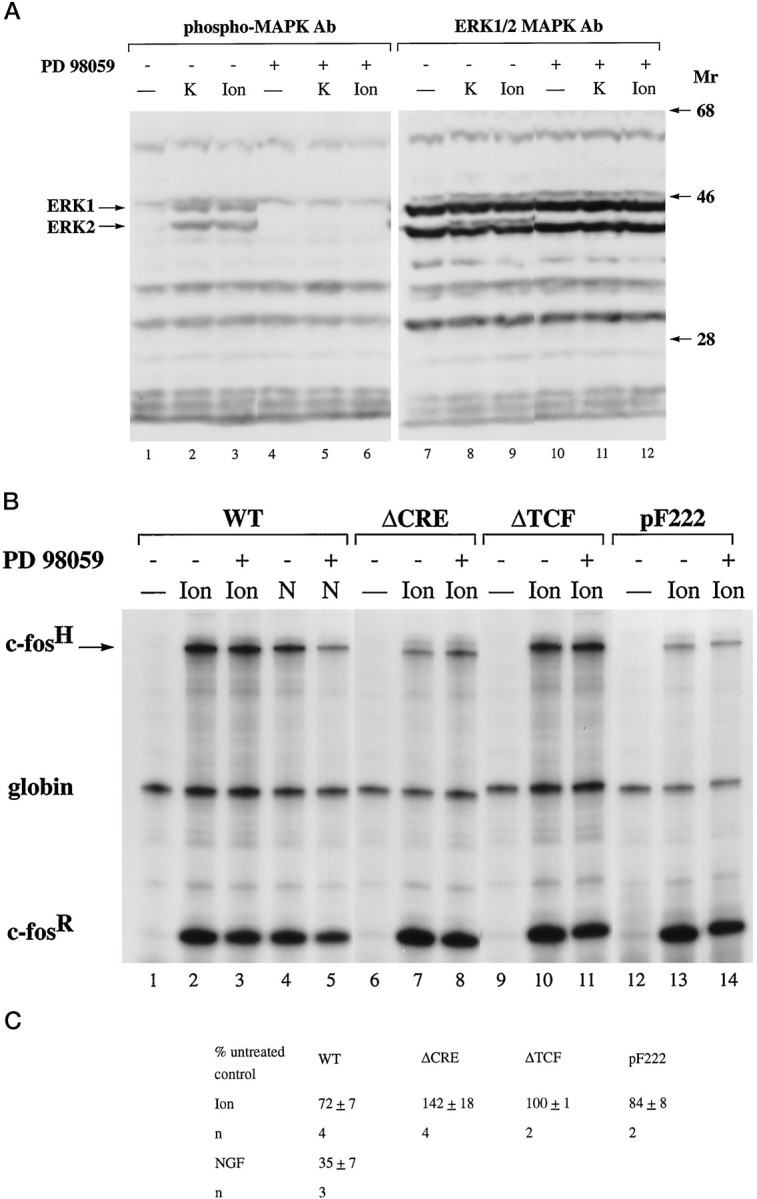
Effect of the MAP kinase kinase 1 inhibitor PD 98059 on MAP kinases/ERKs activation (A) and CRE-dependent and TCF-dependent calcium-activated transcription in PC12 cells (B, C). A, Immunoblot analysis of lysates from PC12 cells before (lanes 1, 4, 7, 10) and 5 min after either KCl stimulation (K; lanes 2, 5, 8, 11) or ionomycin treatment (Ion; lanes 3, 6, 9, 12), using antibodies specific for either phospho-MAP kinases/ERKs (lanes 1–6) or for MAP kinases/ERKs (lanes 7–12) to control for even protein loading. Treatment of the cells with 50 μm PD 98059 (lanes 4–6 and10–12) was done for 60 min before stimulation.Mr, Protein molecular weight standards (× 10−3). B, RNase protection analysis was performed as described in Figure 2. PC12 cells were transfected with plasmid pF711 (wild-type c-fosgene, lanes 1–5), pFosΔCRE (lanes 6–8), pFosΔTCF (lanes 9–11), and pF222 (lanes 12–14). RNA was isolated from unstimulated cells (lanes marked −) or cells stimulated with ionomycin (Ion) or NGF (N). Treatment of the cells with 50 μm PD 98059 (lanes 3, 5, 8, 11, 14) was done for 60 min before stimulation. Similar results were obtained with PD 98059 on KCl-induced TCF-dependent and CRE-dependent transcriptional activation (data not shown).C, Quantitation of RNase protection experiments by the PhosphorImager. The levels of c-fos mRNA transcribed from the indicated plasmids and normalized for transfection efficiency to the level of α-globin mRNA in cells treated with PD 98059 are expressed as a percentage of the amount of c-fos mRNA, normalized to α-globin expression, produced by untreated cells in response to the same stimulus. The mean ± SEM is shown.
We next investigated, using AtT20 cells, the involvement of MAP kinases/ERKs in TCF-independent SRF-linked calcium-activated transcription. We were unable to perform experiments with the RasN17 expression vector because nontoxic concentrations of ionomycin, which may be necessary to activate calcium entry into RasN17-expressing cells (see above), only poorly induces c-fos expression (our unpublished observation). However, we performed experiments with the MEK 1 inhibitor PD 98059. Treatment of AtT20 cells with PD 98059 blocked KCl/FPL-induced activation of MAP kinases (ERKs) (Fig.9A, compare lanes 2 and4) but decreased inducibility of the transfected constructs pF711, pFosΔCRE, and pFosΔSRF to only ∼65–75% (Fig.9B, compare lanes 2 and 3,5 and 6, 8 and 9; 9C). A slightly larger effect of PD 98059 was seen on KCl/FPL inducibility of transcription from pF222, which when compared with cells not treated with PD 98059 was reduced to 44 ± 1% (n = 3) (Fig. 9B, compare lanes 11 and 12; Fig. 9C).
Fig. 9.
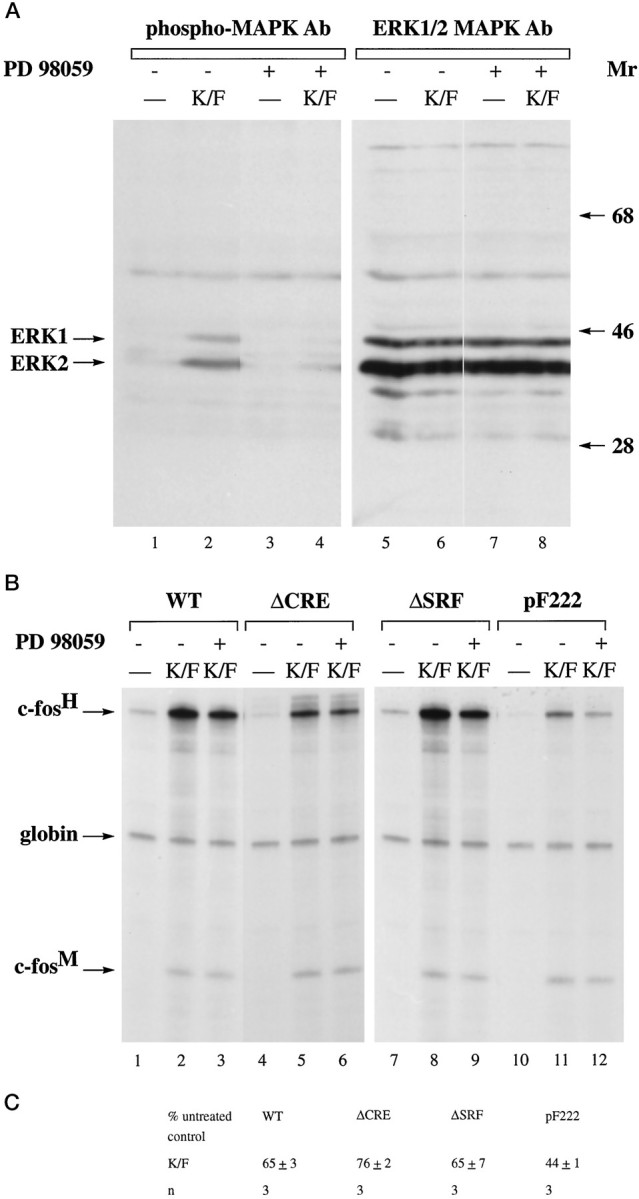
Effect of the MAP kinase kinase 1 inhibitor PD 98059 on MAP kinases/ERKs activation (A) and on CRE-dependent and SRF-linked calcium-activated transcription in AtT20 cells (B, C). A, Immunoblot analysis of lysates from AtT20 cells before (lanes 1, 3, 5, 7) and 5 min after KCl/FPL stimulation (K/F; lanes 2, 4, 6, 8), using antibodies specific either for phospho-MAP kinases/ERKs (lanes 1–4) or for MAP kinases/ERKs (lanes 5–8) to control for even protein loading. Treatment of the cells with 50 μm PD 98059 (lanes 3, 4, 7, 8) was done for 60 min before stimulation.Mr, Protein molecular weight standards (× 10−3). B, RNase protection analysis was performed as described in Figure 5. AtT20 cells were transfected with plasmid pF711 (wild-type c-fosgene, lanes 1–3), pFosΔCRE (lanes 4–6), pFosΔSRF (lanes 7–9), and pF222 (lanes 10–12). RNA was isolated from unstimulated cells (lanes marked −) or cells stimulated with KCl/FPL (K/F). Treatment of the cells with 50 μm PD 98059 (lanes 3, 6, 9, 12) was done for 60 min before stimulation. C, Quantitation of RNase protection experiments by the PhosphorImager. Analysis was done as described in Figure 8C.
These results indicate that the propagation of calcium signals to the nucleus and calcium-dependent transcriptional activation can function independently of the Ras/MAP kinases (ERKs) signaling cascade.
DISCUSSION
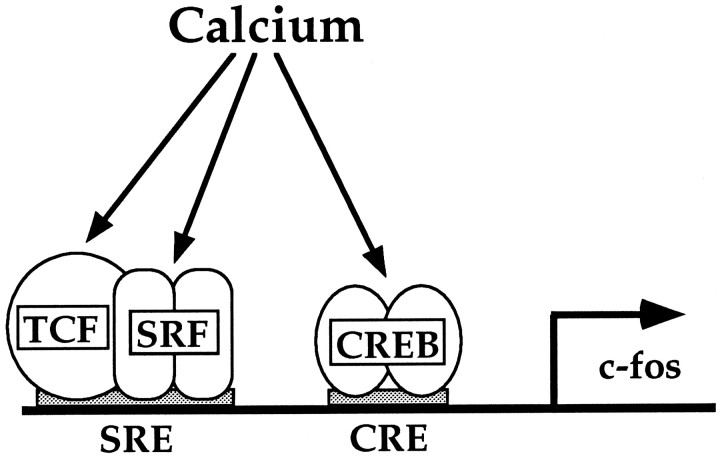
In this study we have investigated the complexity of calcium-signaling mechanisms to the nucleus, using the c-fospromoter as a model system. We demonstrate that calcium controls gene expression by multiple mechanistically distinct pathways that operate in a cell type-dependent manner. Calcium activates transcription via the CRE, and, at the SRE, via a TCF-dependent and a TCF-independent SRF-linked mechanism. A model illustrating calcium signaling to the c-fos promoter is shown in Figure 10.
Fig. 10.
Schematic model of calcium-signaling pathways to the c-fos promoter.
CRE-mediated calcium-activated transcription
The presence of an intact CRE in the c-fos promoter can, in the absence of other regulatory sequences such as the SRE, confer a calcium-induced transcriptional response that is similar in magnitude to that obtained with the wild-type c-fos gene. This confirms previous observations that the CRE is a potent mediator of transcriptional activation by calcium-signaling pathways (Sheng et al., 1988, 1990; Bading et al., 1993). However, we recently demonstrated that increases in cytoplasmic calcium concentrations are not sufficient to activate a CRE and that increased nuclear calcium concentrations are required (Hardingham et al., 1997). This finding, together with our observation that CRE-mediated transcription is largely independent of the Ras/MAP kinases (ERKs) cascade, suggests that a nuclear localized mechanism directly activated by calcium controls gene expression via the CRE. This has important implications for gene expression by electrical activation of neurons. Calcium signals, associated with electrical activity, per se will not necessarily activate CRE-dependent gene expression, although they may activate cytoplasmic signaling pathways such as the Ras/MAP kinases (ERKs) cascade. Only stimuli that give rise to increases in nuclear calcium will cause CRE-mediated transcriptional responses. This provides a possible explanation for the finding that CRE-mediated gene expression can be induced by multiple trains of high-frequency electrical stimulation of neurons, but not by a single high-frequency stimulus (Impey et al., 1996). The former, but not the latter, stimulus may be associated with a sustained calcium influx into the cell, giving rise to larger increases in nuclear calcium. In contrast, SRE-dependent transcription (discussed below) can be stimulated independently of increases in nuclear calcium by a rise in cytoplasmic calcium (Hardingham et al., 1997). This may explain why the zif/268gene, which contains several SREs in its promoter (Changelian et al., 1989; Christy et al., 1989), is activated even with moderate LTP-inducing stimuli that may not increase the nuclear calcium concentration (Cole et al., 1989; Wisden et al., 1990; Worley et al., 1993). The molecular mechanisms controlled by nuclear calcium may involve CaM kinase IV. CaM kinase IV is localized to the nucleus of neuronal cells (Jensen et al., 1991) and can phosphorylate CREB, a mediator of calcium-activated transcription that interacts with the CRE, on serine 133 (Sheng et al., 1991; Matthews et al., 1994; Sun et al., 1994). This residue is critical for CREB to function as a transcriptional activator (Gonzales and Montminy, 1989; Sheng et al., 1991; Matthews et al., 1994; Sun et al., 1994). In addition, expression of a constitutively active form of this CaM kinase IV is sufficient to activate CRE/CREB-dependent transcription (Matthews et al., 1994; Sun et al., 1994). Further studies are required to investigate the expression, subcellular localization, and function of CaM kinase IV in PC12 and AtT20 cells.
The SRE integrates two calcium-signaling pathways
The SRE is a second site of transcriptional regulation by calcium-signaling pathways. The mechanisms by which calcium signals activate the SRE are cell type-dependent. In PC12 cells, calcium-activated transcription via the c-fos SRE requires the TCF site, whereas this response in AtT20 cells and primary hippocampal neurons occurs via a TCF-independent SRF-linked mechanism. These cell type differences may be also relevant for gene regulation in the brain. A TCF-dependent pathway may be functional in one area of the brain, whereas in a different area the SRF-linked pathway may be operating. As a consequence, genes that contain SRF binding sites without adjacent TCF binding motif in their promoter will respond to calcium signals only in those cell types in which the SRF-linked signaling pathway is functional. One example is the β-actin gene that contains in its promoter a binding site for SRF and an adjacent inverted Ets motif, which when compared with the c-fos SRE, less efficiently forms a ternary complex (Treisman et al., 1992). Indeed, in PC12 cells in which the TCF-dependent mechanism targets the c-fos SRE, β-actin expression is not induced by KCl-induced membrane depolarization (Bartel et al., 1989). In contrast, in hippocampal neurons in which the c-fos SRE is activated by an SRF-linked mechanism, transcription of β-actin is stimulated by calcium signals (Bading et al., 1995). Thus, the cell type is an important determinant for the functional significance of a particular calcium-signaling pathway and provides a means for differential gene expression by calcium signals.
The nature of the calcium-signaling pathway targeting the TCF site remains unclear. In cortical neurons, calcium signals generated by NMDA receptor activation can stimulate transcription through a MAP kinase signaling cascade targeting the TCF Elk-1 (Xia et al., 1996). However, our analysis of the TCF-dependent pathway in PC12 cells demonstrated that neither expression of RasN17 nor treatment of the cells with MAP kinase kinase 1 inhibitor PD 98059 significantly reduced the TCF-dependent calcium response, indicating that the Ras/MAP kinases (ERKs) pathway is not critical. Moreover, we find that, in primary hippocampal neurons, NMDA receptor-regulated gene expression is mainly independent of the TCF site and requires the SRF binding site (see below). In the experiments described by Xia et al. (1996), a c-fos-based reporter gene was used in which the binding site for TCF Elk-1 was inserted in close proximity to the c-fosTATA box, whereas in our experiment the TCF site remained in its natural position (nucleotide −320) in the intact c-fospromoter. It is possible that MAP kinases/ERK, which are known to be activated in response to NMDA receptor activation (Bading and Greenberg, 1991), can stimulate transcription through TCF Elk-1 if the TCF binding site is close to the start site of transcription. If, however, the TCF site is several hundred base pairs upstream of the TATA box (as in the constructs used in this study), transcriptional activation through this site may be mediated by a mechanism that functions independently of the Ras/MAP kinases (ERKs) signaling cascade. Thus, differences in the promoter constructs used for the transcriptional analysis and cell type differences may explain this discrepancy.
The TCF-independent SRF-linked pathway of calcium-activated transcription in AtT20 cells and primary hippocampal neurons resembles the mechanism by which serum stimulation or activation of heterotrimeric G-proteins induces c-fos expression. Signals generated by these stimuli are transduced to the nucleus by a mechanism involving members of the rho family of GTPases (Hill et al., 1995). This raises the possibility that calcium signals also stimulate transcription by a rho-dependent mechanism. Our finding that the TCF-independent SRF-linked pathway is insensitive to PD 98059 is consistent with this hypothesis, because a rho-mediated mechanism of transcriptional activation may function independently of MAP kinase kinase 1 activation. Although our experiments demonstrate that, in the context of the entire c-fos gene, the SRF binding site is not sufficient to mediate c-fos induction in PC12 cells upon KCl stimulation, two other studies indicate that a TCF-independent SRF-linked pathway may be also functional in PC12 cells (Misra et al., 1994; Miranti et al., 1995). In these studies it was shown that the SRF binding site, when placed close to the c-fos TATA box, is capable of mediating calcium-dependent transcriptional activation via a mechanism that involves CaM kinases (Misra et al., 1994; Miranti et al., 1995). These results also illustrate that the promoter context in which a particular transcriptional control element is analyzed is an important, often unappreciated, determinant of the signaling mechanism that is functionally significant.
Conclusion
Calcium controls gene expression by multiple mechanistically distinct pathways activating transcription via the CRE and the SRE. The SRE is bifunctional and integrates two signaling mechanisms. The significance of these pathways for the regulation of transcription depends on the spatial properties of the calcium signals (Hardingham et al., 1997) and the cell type. Given the differences in the composition and arrangements of calcium response elements in transcriptional control regions, these findings constitute the basic principles of calcium-activated transcription that may underlie differential gene expression by electrical activity in neurons.
Footnotes
This work was supported by the Medical Research Council, Glaxo Wellcome, Imperial Cancer Research Fund, and the Howard Hughes Medical Institute. We thank Bill Wisden for helpful discussions and comments on this manuscript.
Correspondence should be addressed to Dr. Hilmar Bading, Division of Neurobiology, Medical Research Council Laboratory of Molecular Biology, Hills Road, Cambridge CB2 2QH, England.
Dr. Hill’s present address: Ludwig Institute for Cancer Research, 91 Riding House Street, London W1P 8BT, England.
REFERENCES
- 1.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. Greene and Wiley; New York: 1987. [Google Scholar]
- 2.Bading H, Greenberg ME. Stimulation of protein tyrosine phosphorylation by NMDA receptor activation. Science. 1991;253:912–914. doi: 10.1126/science.1715095. [DOI] [PubMed] [Google Scholar]
- 3.Bading H, Ginty DD, Greenberg ME. Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science. 1993;260:181–186. doi: 10.1126/science.8097060. [DOI] [PubMed] [Google Scholar]
- 4.Bading H, Segal MM, Sucher NJ, Dudek H, Lipton SA, Greenberg ME. N-methyl-d-aspartate receptors are critical for mediating the effects of glutamate on intracellular calcium concentration and immediate early gene expression in cultured hippocampal neurons. Neuroscience. 1995;64:653–664. doi: 10.1016/0306-4522(94)00462-e. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DF, Sheng M, Lau LF, Greenberg ME. Growth factors and membrane depolarization activate distinct programs of early response gene expression: dissociation of fos and jun induction. Genes Dev. 1989;3:304–313. doi: 10.1101/gad.3.3.304. [DOI] [PubMed] [Google Scholar]
- 6.Changelian PS, Feng P, King TC, Milbrandt J. Structure of the NGFI-A gene and detection of upstream sequences responsible for its transcriptional induction by nerve growth factor. Proc Natl Acad Sci USA. 1989;86:377–381. doi: 10.1073/pnas.86.1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christy B, Nathans D. Functional serum response elements upstream of the growth factor-inducible gene zif 268. Mol Cell Biol. 1989;9:4889–4895. doi: 10.1128/mcb.9.11.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole AJ, Saffen DW, Baraban JM, Worley PF. Rapid increase of an immediate early gene messenger RNA in hippocampal neurons by synaptic NMDA receptor activation. Nature. 1989;340:474–476. doi: 10.1038/340474a0. [DOI] [PubMed] [Google Scholar]
- 9.Dalton S, Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68:597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- 10.Darnell JE, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional regulation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 11.Feig LA, Cooper GM. Inhibition of NIH 3T3 cell proliferation by a mutant ras protein with preferential affinity for GDP. Mol Cell Biol. 1988;8:3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu X-Y, Zhang J-J. Transcription factor p91 interacts with the epidermal growth factor receptor and mediates activation of the c-fos gene promoter. Cell. 1993;74:1135–1145. doi: 10.1016/0092-8674(93)90734-8. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh A, Greenberg ME. Calcium signaling in neurons: molecular mechanisms and cellular consequences. Science. 1995;268:239–247. doi: 10.1126/science.7716515. [DOI] [PubMed] [Google Scholar]
- 14.Gille H, Sharrocks AD, Shaw PE. Phosphorylation of p62TCF by MAP kinase stimulates ternary complex formation at c-fos promoter. Nature. 1992;358:414–417. doi: 10.1038/358414a0. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 16.Graham R, Gilman M. Distinct protein targets for signals acting at the c-fos serum response element. Science. 1991;251:189–192. doi: 10.1126/science.1898992. [DOI] [PubMed] [Google Scholar]
- 17.Hardingham GE, Chawla S, Johnson CM, Bading H. Distinct functions of nuclear and cytoplasmic calcium in the control of gene expression. Nature. 1997;385:260–265. doi: 10.1038/385260a0. [DOI] [PubMed] [Google Scholar]
- 18.Hayes TE, Kichen AM, Cochran BH. Inducible binding of a factor to the c-fos regulatory region. Proc Natl Acad Sci USA. 1987;84:1272–1276. doi: 10.1073/pnas.84.5.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill CS, Treisman R. Differential activation of c-fos promoter elements by serum, lysophosphatidic acid, G-proteins, and polypeptide growth factors. EMBO J. 1995;14:5037–5047. doi: 10.1002/j.1460-2075.1995.tb00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill CS, Marais R, John S, Wynne J, Dalton S, Treisman R. Functional analysis of a growth factor-responsive transcription factor complex. Cell. 1993;73:395–406. doi: 10.1016/0092-8674(93)90238-l. [DOI] [PubMed] [Google Scholar]
- 21.Hill CS, Wynne J, Treisman R. Serum-regulated transcription by serum response factor (SRF): a novel role for the DNA-binding domain. EMBO J. 1994;13:5421–5432. doi: 10.1002/j.1460-2075.1994.tb06877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill CS, Wynne J, Treisman R. The Rho family GTPase RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 23.Hipskind RA, Rao VN, Mueller CG, Reddy ES, Nordheim A. Ets-related protein Elk-1 is homologous to the c-fos regulatory factor p62TCF. Nature. 1991;354:531–534. doi: 10.1038/354531a0. [DOI] [PubMed] [Google Scholar]
- 24.Ihle JN, Witthuhn BA, Quelle FW, Yamamoto K, Thierfelder WE, Kreider B, Silvennoinen O. Signaling by the cytokine receptor superfamily: JAKs and STATs. Trends Biochem Sci. 1994;19:222–227. doi: 10.1016/0968-0004(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 25.Impey S, Mark M, Villacres EC, Poser S, Chavkin C, Storm DR. Induction of CRE-mediated gene expression by stimuli that generate long-lasting LTP in area CA1 of the hippocampus. Neuron. 1996;16:973–982. doi: 10.1016/s0896-6273(00)80120-8. [DOI] [PubMed] [Google Scholar]
- 26.Janknecht R, Ernst WH, Pingoud V, Nordheim A. Activation of TCF Elk-1 by MAP kinases. EMBO J. 1993;12:5097–5104. doi: 10.1002/j.1460-2075.1993.tb06204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen KF, Ohmstede C-A, Fisher RS, Sayhoun N. Nuclear and axonal localization of Ca2+/calmodulin-dependent protein kinase type Gr in rat cerebellar cortex. Proc Natl Acad Sci USA. 1991;88:2850–2853. doi: 10.1073/pnas.88.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lerea LS, McNamara JO. Ionotropic glutamate receptor subtypes activate c-fos transcription by distinct calcium-requiring intracellular signaling pathways. Neuron. 1993;10:31–41. doi: 10.1016/0896-6273(93)90239-n. [DOI] [PubMed] [Google Scholar]
- 29.Lerea LS, Butler LS, McNamara JO. NMDA and non-NMDA receptor-mediated increase of c-fos mRNA in dentate gyrus neurons involved calcium influx via different routes. J Neurosci. 1992;12:2973–2981. doi: 10.1523/JNEUROSCI.12-08-02973.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- 31.Marshall CJ. MAP kinase kinase kinase, MAP kinase kinase, and MAP kinase. Curr Opin Genet Dev. 1994;4:82–89. doi: 10.1016/0959-437x(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 32.Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 33.Matthews RP, Guthrie CR, Wailes LM, Zhao X, Means AR, McKnight GS. Calcium/calmodulin-dependent protein kinase types II and IV differentially regulate CREB-dependent gene expression. Mol Cell Biol. 1994;14:6107–6116. doi: 10.1128/mcb.14.9.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miranti CK, Ginty DD, Huang G, Chatila T, Greenberg ME. Calcium activates serum response factor-dependent transcription by a ras- and Elk-1-independent mechanism that involves a Ca2+/calmodulin-dependent kinase. Mol Cell Biol. 1995;15:3672–3684. doi: 10.1128/mcb.15.7.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Misra RP, Bonni A, Miranti CK, Rivera VM, Sheng M, Greenberg ME. L-type voltage-sensitive calcium channel activation stimulates gene expression by a serum response factor-dependent pathway. J Biol Chem. 1994;269:25483–25493. [PubMed] [Google Scholar]
- 36.Morgan JL, Curran T. Stimulus–transcription coupling in the nervous system: involvement of the inducible proto-oncogenes fos and jun. Annu Rev Neurosci. 1991;14:421–451. doi: 10.1146/annurev.ne.14.030191.002225. [DOI] [PubMed] [Google Scholar]
- 37.Norman C, Runswick M, Pollock R, Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988;55:989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- 38.Prywes R, Roeder RG. Purification of the c-fos enhancer-binding protein. Mol Cell Biol. 1987;7:3482–3489. doi: 10.1128/mcb.7.10.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robertson LM, Kerppola TK, Vendrell M, Luk D, Smeyne RJ, Bocchiaro C, Morgan JI, Curran T. Regulation of c-fos expression in transgenic mice requires multiple interdependent transcription control elements. Neuron. 1995;14:241–252. doi: 10.1016/0896-6273(95)90282-1. [DOI] [PubMed] [Google Scholar]
- 40.Rosen LB, Ginty DD, Weber MJ, Greenberg ME. Membrane depolarization and calcium influx stimulate MEK and MAP kinase via activation of Ras. Neuron. 1994;12:1207–1221. doi: 10.1016/0896-6273(94)90438-3. [DOI] [PubMed] [Google Scholar]
- 41.Rusanescu G, Qi H, Thomas SM, Brugge JS, Halegoua S. Calcium influx induces neurite growth through a Src-Ras signaling cassette. Neuron. 1995;15:1415–1425. doi: 10.1016/0896-6273(95)90019-5. [DOI] [PubMed] [Google Scholar]
- 42.Sadowski HB, Shuai K, Darnell JE, Jr, Gilman MZ. A common nuclear signal transduction pathway activated by growth factor and cytokine receptors. Science. 1993;261:1739–1743. doi: 10.1126/science.8397445. [DOI] [PubMed] [Google Scholar]
- 43.Schröter H, Shaw PE, Nordheim A. Purification of intercalator-released p67, a polypeptide that interacts specifically with the c-fos serum response element. Nucleic Acids Res. 1987;15:10145–10157. doi: 10.1093/nar/15.24.10145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaw P, Schröter H, Nordheim A. The ability of a ternary complex to form over the serum response element correlates with serum inducibility of the c-fos promoter. Cell. 1989;56:563–572. doi: 10.1016/0092-8674(89)90579-5. [DOI] [PubMed] [Google Scholar]
- 45.Sheng M, Greenberg ME. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron. 1990;4:477–485. doi: 10.1016/0896-6273(90)90106-p. [DOI] [PubMed] [Google Scholar]
- 46.Sheng M, Dougan ST, McFadden G, Greenberg ME. Calcium and growth factor pathways of c-fos transcriptional activation require distinct upstream regulatory sequences. Mol Cell Biol. 1988;8:2787–2796. doi: 10.1128/mcb.8.7.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheng M, McFadden G, Greenberg ME. Membrane depolarization and calcium induce c-fos transcription via phosphorylation of transcription factor CREB. Neuron. 1990;4:571–582. doi: 10.1016/0896-6273(90)90115-v. [DOI] [PubMed] [Google Scholar]
- 48.Sheng M, Thompson MA, Greenberg ME. CREB: a Ca2+-regulated transcription factor phosphorylated by calmodulin-dependent kinases. Science. 1991;252:1427–1430. doi: 10.1126/science.1646483. [DOI] [PubMed] [Google Scholar]
- 49.Sun P, Enslen H, Myung PS, Maurer RA. Differential activation of CREB by Ca2+/calmodulin-dependent protein kinases type II and type IV involves phosphorylation of a site that negatively regulates activity. Genes Dev. 1994;8:2527–2539. doi: 10.1101/gad.8.21.2527. [DOI] [PubMed] [Google Scholar]
- 50.Thomas SM, DeMarco M, D’Arcangelo G, Halegoua S, Brugge JS. Ras is essential for nerve growth factor- and phorbol ester-induced tyrosine phosphorylation of MAP kinases. Cell. 1992;68:1031–1040. doi: 10.1016/0092-8674(92)90075-n. [DOI] [PubMed] [Google Scholar]
- 51.Treisman R. Transient accumulation of c-fos RNA following serum stimulation requires a conserved 5′ element and c-fos 3′ sequences. Cell. 1985;42:889–902. doi: 10.1016/0092-8674(85)90285-5. [DOI] [PubMed] [Google Scholar]
- 52.Treisman R. Identification and purification of a polypeptide that binds to the c-fos serum response element. EMBO J. 1987;6:2711–2717. doi: 10.1002/j.1460-2075.1987.tb02564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Treisman R. Ternary complex factors: growth factor-regulated transcriptional activators. Curr Opin Genet Dev. 1994;4:96–101. doi: 10.1016/0959-437x(94)90097-3. [DOI] [PubMed] [Google Scholar]
- 54.Treisman R, Marais R, Wynne J. Spatial flexibility in ternary complexes between SRF and its accessory proteins. EMBO J. 1992;11:4631–4640. doi: 10.1002/j.1460-2075.1992.tb05565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagner BJ, Hayes TE, Hoban CJ, Cochran BH. The SIF binding element confers sis/PDGF inducibility onto the c-fos promoter. EMBO J. 1990;9:4477–4484. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wisden W, Errington ML, Williams S, Dunnett SB, Waters C, Hitchcock D, Evan G, Bliss TVP, Hunt SP. Differential expression of immediate early genes in the hippocampus and spinal cord. Neuron. 1990;4:603–614. doi: 10.1016/0896-6273(90)90118-y. [DOI] [PubMed] [Google Scholar]
- 57.Wood KW, Sarnecki C, Roberts TM, Blenis J. Ras mediates nerve growth factor receptor modulation of three signal-transducing protein kinases: MAP kinase, Raf-1, and RSK. Cell. 1992;68:1041–1050. doi: 10.1016/0092-8674(92)90076-o. [DOI] [PubMed] [Google Scholar]
- 58.Worley PF, Bhat RV, Baraban JM, Erickson CA, McNaughton BL, Barnes CA. Thresholds for synaptic activation of transcription factors in hippocampus: correlation with long-term enhancement. J Neurosci. 1993;13:4776–4786. doi: 10.1523/JNEUROSCI.13-11-04776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xia Z, Dudek H, Miranti CK, Greenberg ME. Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism. J Neurosci. 1996;16:5425–5436. doi: 10.1523/JNEUROSCI.16-17-05425.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng W, Rampe D, Triggle DJ. Pharmacological, radioligand binding, and electrophysiological characteristics of FPL 64176, a novel nondihydropyridine Ca2+ channel activator, in cardiac and vascular preparations. Mol Pharmacol. 1991;40:734–741. [PubMed] [Google Scholar]
- 61.Zhong Z, Wen Z, Darnell JE., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]