Abstract
Ogura-type cytoplasmic male sterility (Ogura-CMS) has been widely used in the hybrid breeding industry for cruciferous vegetables. Turnip (Brassica rapa ssp. rapifera) is one of the most important local cruciferous vegetables in China, cultivated for its fleshy root as a flat disc. Here, morphological characteristics of an Ogura-CMS line ‘BY10-2A’ and its maintainer fertile (MF) line ‘BY10-2B’ of turnip were investigated. Ogura-CMS turnip showed a reduction in the size of the fleshy root, and had distinct defects in microspore development and tapetum degeneration during the transition from microspore mother cells to tetrads. Defective microspore production and premature tapetum degeneration during microgametogenesis resulted in short filaments and withered white anthers, leading to complete male sterility of the Ogura-CMS line. Additionally, the mechanism regulating Ogura-CMS in turnip was investigated using inflorescence transcriptome analyses of the Ogura-CMS and MF lines. The de novo assembly resulted in a total of 84,132 unigenes. Among them, 5,117 differentially expressed genes (DEGs) were identified, including 1,339 up- and 3,778 down-regulated genes in the Ogura-CMS line compared to the MF line. A number of functionally known members involved in anther development and microspore formation were addressed in our DEG pool, particularly genes regulating tapetum programmed cell death (PCD), and associated with pollen wall formation. Additionally, 185 novel genes were proposed to function in male organ development based on GO analyses, of which 26 DEGs were genotype-specifically expressed. Our research provides a comprehensive foundation for understanding anther development and the CMS mechanism in turnip.
Introduction
As an important and valuable resource, male-sterile varieties are extensively exploited in crop hybrid breeding. Cytoplasmic male-sterility (CMS) is a category of male-sterility resulted from a genomic conflict between the mitochondrial and nuclear genomes, and has been extensively utilized [1]. Various types of CMS have been developed and adopted in plant breeding [2].
It has been proposed that normal microsporogenesis needs appropriate timing of tapetum degeneration and specific gene expression [3]. In CMS system, this elaborate process is complex because of the mitochondrial retrograde signaling pathway and the interaction of nuclear and organelle genomes [4–7]. Considerable variations in morphological phenotype of anther development, particularly of microspore and tapetum behaviors, arise with different nuclear backgrounds and/or cytoplasmic genotype [8]. Mostly, for example, CMS causes the premature degradation of the tapetal cells [9]. However, CMS sorghum and CMS-T maize show a persistent tapetum which likely inhibits nutrient delivery, resulting in the failure of microspore development [8,10].
Ogura-type CMS was first discovered in Japanese radish (Raphanus sativus) and is now widely applied in the breeding of Brassicaceae crops, such as Brassica napus, B. juncea, and B. oleracea, providing a classic model to probe the role of nuclear-cytoplasmic genome interactions [11]. Although interactions of specific mitochondrial gene orf138 with different nuclear backgrounds have been reported to be responsible for Ogura-CMS, diverse floral behaviors attributed to the same cytoplasm in different species have not been fully investigated [11]. For example, anther morphology in Ogura-CMS B. napus is normal, whereas pollen development is impaired and sensitive to temperature [12–16]. However, Ogura-CMS Chinese cabbage (B. rapa ssp. pekinensis) shows reduced plant height and delayed flowering, has shorter filaments, and produces few and infertile pollen grains in indehiscent anthers [17]. All these differences suggest the presence of various regulatory mechanisms and/or multiple regulatory pathways in Brassica spp.
Recently, numerous candidate genes involved in CMS have been discovered in different species, such as onion (Allium cepa), cabbage (B. olerace var. capitata), rice (Oryza sativa), and pepper (Capsicum annuum) [7,9,18,19]. In addition, participation of miRNAs and non-coding RNAs is becoming increasingly evident in retrograde regulation of CMS [20–23]. Despite previous extensive work, no specific retrograde pathway has been reported for CMS to date and the regulatory mechanism of CMS is still largely unknown.
Exploring the molecular mechanisms underlying CMS is of great importance for improving seed yield in many crop species, especially in crucifers. As a Brassica root crop, turnip (Brassica rapa ssp. rapifera) has been important for human consumption for thousands of years [24]. In this study, the morphological characteristics of an Ogura-CMS line ‘BY10-2A’ and its maintainer fertile (MF) line ‘BY10-2B’ of turnip were investigated, and a detailed RNA sequencing (RNA-Seq) analysis for inflorescences in turnip was conducted. These data provide a comprehensive view on the dynamic gene expression networks and their potential roles in controlling anther development. Using pairwise comparisons, we identified 5,117 DEGs, which might respond to the mutation of the mitochondrial ORF138 locus. Among them, 185 novel genes were proposed to function in male organ development based on GO analyses. These findings provide a comprehensive insight into the regulatory networks responsible for Ogura-CMS tapetum abnormality and pollen abortion in turnip, and demonstrate that cytoplasmic retrograde regulation is probably a principal molecular mechanism for CMS in turnip.
Materials and methods
Plant materials and growth conditions
Previously, the Ogura-CMS line ‘BY10-2A’ of B. rapa ssp. rapifera was developed by inter-specific hybridization between B. rapa ssp. chinensis as the Ogura-CMS cytoplasm donor and fertile B. rapa L. ssp. rapifera, followed by 10 recurrent generations of back-crossing. The Ogura-CMS line and its maintainer fertile (MF) line ‘BY10-2B’ were cultivated in the experimental farm of Wenzhou Vocational College of Science and Technology, Wenzhou, Zhejiang, China.
Plant morphological analysis and floret structure observation
Plants were observed and photographed at 32, 48, 110, and 180 days after germination. The length and diameter of fleshy roots were measured at 110 and 180 days after germination. A week after the first anthesis, florets of both the Ogura-CMS and MF lines were collected. The floret structures were observed under a Leica MZ16FA stereoscopic microscope (Leica Microsystems, Wetzlar, Germany).
Floral buds, floral organs, anthers and pollen grains morphological analysis
Ogura-CMS and MF floral buds at different anther developmental stages, and various floral organs were fixed with 2.5% glutaraldehyde in phosphate buffer (pH 7.0) overnight, and post-fixed with 1% OsO4 in phosphate buffer for 1 h. Subsequently, the specimens were dehydrated in a graded ethanol series (50%, 70%, 80%, 90%, 95%, 2×100%). For scanning electron microscopy, the dehydrated specimens were coated with gold-palladium in an Eiko Model IB5 ion coater (Eiko Engineering Company, Ibaraki, Japan), and photographed in a Hitachi Model TM-1000 scanning electron microscope (Tokyo, Japan) [http://dx.doi.org/10.17504/protocols.io.zz4f78w]. For semi-thin section analyses, the dehydrated specimens were embedded in Spurr resin. Semi-thin sections (1 μm) were sliced under a LKB 11800 PYRAMITOME ultramicrotome (Stockholm, Sweden) and stained with 0.5% toluidine blue. Images of anther cross-sections were obtained with a Leica DMLB fluorescence microscope (Leica Microsystems, Wetzlar, Germany) [http://dx.doi.org/10.17504/protocols.io.zz5f786]. For transmission electron microscopy (TEM), ultrathin sections (70 nm) were obtained and stained with uranyl acetate followed by alkaline lead citrate, and photographed with a Hitachi Model H-7650 transmission electron microscope (Tokyo, Japan) [http://dx.doi.org/10.17504/protocols.io.zz6f79e].
RNA sample collection and total RNA isolation
After flowering, all floral buds of an inflorescence from the Ogura-CMS and MF lines of B. rapa ssp. rapifera were collected. In each case, samples were harvested and pooled from ten individual plants with transcriptome profiles representing ‘f’ difference, then immediately frozen in liquid nitrogen and stored at -70°C until RNA isolation. For biological repetitions, RNA was extracted from three samples using the EASYspin Plant RNA kit (Aidlab Biotechnologies Corporation, Bejing, China). RNA quality and quantity were characterized on a 1% agarose gel, and determined with a NanoPhotometer spectrophotometer (IMPLEN, CA, USA) and a Qubit RNA Assay Kit in Qubit2.0 Flurometer (Life Technologies, CA, USA). RNA integrity was assessed using the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
Illumina sequencing and de novo transcriptome assembly
A total of 3 μg RNA per sample was used for library preparation using a NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA). The library preparations were then sequenced on an Illumina Hiseq 2000 platform by the Biomarker Biotechnology Corporation (Beijing, China). Clean reads were filtered from the raw reads by removing reads containing adapter, poly-N and low-quality reads. All the downstream analyses were based on clean data with quality determined by Q20, Q30, GC-content and sequence duplication levels. Then de novo transcriptome assembly was accomplished using the Trinity platform [25] with min-kmer-cov set to 2 by default and all other parameters set to default. The RNA-Seq data were uploaded to the Sequence Read Archive of the National Center for Biotechnology Information (NCBI) (DOI: https://www.ncbi.nlm.nih.gov/sra/PRJNA505114; accession number: PRJNA505114).
Annotation of differentially expressed genes
Differentially expressed genes (DEGs) were screened out using the DESeq (2010) R Package. Genes with a Benjamini-Hochberg false discovery rate (FDR) < 0.05 and a log2 fold change (FC) ≥ 1 or ≤ -1 in each pairwise comparison were assigned as differential expressed. DEG sequences were blast in the NCBI non-redundant protein (Nr) database, Swiss-Prot database and orthologous groups of genes (eggNOG) database, and also aligned to the Clusters of Orthologous Group (COG), Clusters of Protein homology database (KOG), and Homologous protein family (Pfam) database to predict and classify functions [26,27]. Gene ontology (GO) enrichment analysis of DEGs was performed by the topGO (2007) R packages based on a Kolmogorov-Smirnov test. Pathway analysis of DEGs was carried out to detect the important pathways, based on the database of Kyoto Encyclopedia of Genes and Genomes (KEGG). Statistical enrichment of DEGs in KEGG pathways was identified by KOBAS software using a hypergeometric test [28].
Real-time reverse transcription polymerase chain reaction (RT-PCR) validation
Sixteen DEGs were randomly selected for validation using real-time RT-PCR. Residual RNA samples for DEG analysis were transcribed into cDNA with a HiScrip II 1st Strand cDNA Synthesis Kit (Vazyme Biotech, Nanjing, China) as the template. ACT7 was used as the normalization control [29]. Real-time RT-PCR was performed on an Applied Biosystems 7500 Real-time PCR System (ThermoFisher, MA, USA) and the relative expression levels were analyzed. Three technical repeats were performed. All primers used were listed in S1 Table.
Results
The Ogura-CMS turnip displays complete male sterility
Despite 10 generations of back-crossings, Ogura-CMS plants showed a reduction in fleshy root size compared with its MF line (Fig 1A–1C). Furthermore, during the reproductive growth phase, the Ogura-CMS line was distinguishable from the MF line by its short filaments and withered white anthers (Fig 1D and 1E). The floral buds from the Ogura-CMS line showed the same developmental pattern as those of its MF line (Fig 2A and 2C). However, compared with the yellow and plump anthers of the MF line (Fig 2B), no pollen was observed in the mature Ogura-CMS anthers (Fig 2D). Other than pollen absence and stamen abnormality, floral organs presented normal morphologies (S1 Fig), including female gametophytes as pollinating the MF pollen grains on the Ogura-CMS stigma led to normal silique growth and full seed set (S2 Fig).
Fig 1. Morphological features of fleshy roots and flowers in the Ogura-CMS line and its maintainer fertile (MF) line of turnip.
(A) A MF plant and its fleshy roots at 110 days after germination. (B) An Ogura-CMS plant and its fleshy roots at 110 days after germination. (C) The length and diameter of fleshy roots at 110 days after germination. The values are the mean ± SD (standard deviation). Asterisks indicate statistical significance versus the length of MF fleshy root (**P<0.01); crosses indicate statistical significance versus the diameter of MF fleshy root (++P<0.01). Statistical significance is determined by a one-way ANOVA test. (D) A MF floret at anthesis stage with normal floral organs. (E) An Ogura-CMS floret at anthesis stage with short filaments and withered white anthers. Bars = 5 cm in (A, B), 2 mm in (D, E).
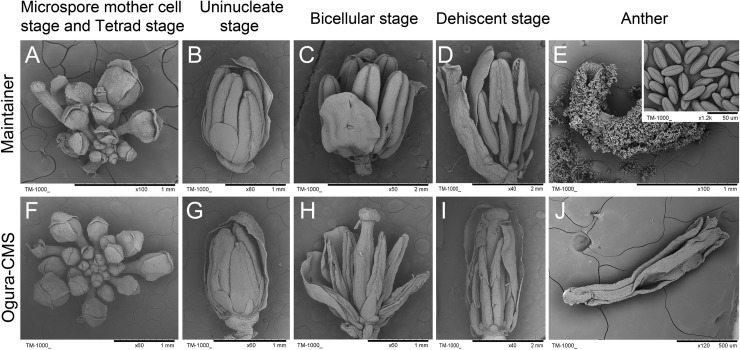
Fig 2. Scanning electron microscopy observation of flower development in the Ogura-CMS line and its maintainer fertile (MF) line of turnip.
(A-D) Floral bud morphology in the MF line from the microspore mother cell stage to the mature pollen stage. (E) A MF dehiscent anther with normal oval pollen grains (inset shows the mature pollen grains). (F-I) Floral bud morphology in the Ogura-CMS line from the microspore mother cell stage to the mature pollen stage. (A, F) Microspore mother cell stage and tetrad stage. (B, G) Uninucleate stage. An Ogura-CMS floral bud exhibits the same morphology as a MF bud. (C, H) Bicellular stage. The withered anthers of the Ogura-CMS line are evident, compared with the lump anthers of the MF line. (D, I) Dehiscent stage. The MF anthers split open along the stomium, whereas the collapse of the Ogura-CMS anthers is evident. (J) An Ogura-CMS anther without any pollen grains. Bars = 1 mm in (A, B, E-H), 2 mm in (C, D, I), 500 μm in (J).
Aberrant anther development occurs during transition from microspore mother cells to tetrads
To determine the precise stage at which Ogura-CMS anther shrinkage begins, semi-thin sections of anthers from various developmental stages were prepared and further analyzed by microscopy. No differences in microspore development were observed inside the anther locules during the process of meiotic division up to the tetrad stage (Fig 3A, 3B, 3F and 3G). Each Ogura-CMS tetrad (Fig 3G) contained four microspores, similar to those of the MF line (Fig 3B), indicating that meiosis is normal in the Ogura-CMS line. Both Ogura-CMS and MF tetrads were surrounded by a tapetum, a middle layer, an endothecium, and an epidermis from the inside out at the tetrad stage (Fig 3B and 3G). However, it was noticeable that the Ogura-CMS tapetum (Fig 3G) swelled at the center of the locule, and the cytoplasm was distinguishably clear from that of the MF line (Fig 3B). After the tetrad stage, the MF middle layer degenerated, then disappeared by the uninucleate stage, and the MF anthers released microspores that developed into mature pollen grains (Fig 3C–3E). However, the collapse of the Ogura-CMS microspores started at the uninucleate microspore stage and was accompanied by extensive degeneration (Fig 3H). Moreover, the middle layer persisted at this point, together with significantly enlarged tapetum, crushing the free microspores to the locule center. At later stages of development, the collapse of the microspore was even more remarkable due to the degeneration of its entire contents (Fig 3I). Eventually, defective microspore development and early clearing of tapetal cytoplasm led to shrunken anthers with collapsed locules and remnants of pollen grains adhered to the inner face of the epidermis (Fig 3J).
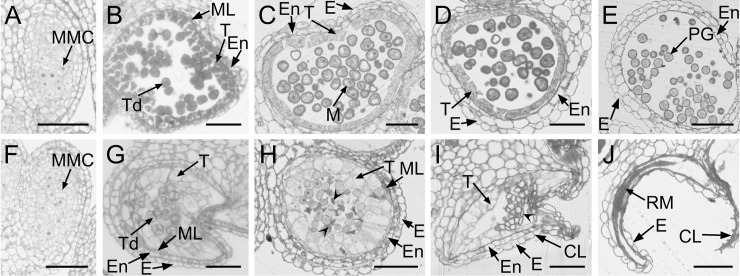
Fig 3. Anther and microspore development in the Ogura-CMS line and its maintainer fertile (MF) line of turnip.
(A-E) Semi-thin sections of the MF anthers. (F-J) Semi-thin sections of the Ogura-CMS anthers. (A, F) Microspore mother cell stage. (B, G) Tetrad stage. The young microspores are surrounded by a callose wall, a tapetum, a middle layer, an endothecium, and an epidermis from the inside out at the tetrad stage. The tapetum in (G) swells at the center of the locule. (C, H) Uninucleate microspore stage. The middle layer persisted in (J). The aborted microspores indicated by arrowheads in (J) was surrounded by a swollen tapetal layer. (D, I) Bicellular stage. The collapse of anther locule is obvious with the aborted microspores indicated by arrowhead in (I). (E, J) Dehiscent stage. Endothecium layer is absent in the surrounding walls and remnants of the aborted microspores adhere to the inner face of the epidermis in (J). CL, collapsed locule; E, epidermis; En, endothecium; M, microspore; ML, middle layer; MMC, microspore mother cell; PG, pollen grain; RM, remnants of microspores; T, tapetum; Td, tetrads. Bars = 50 μm.
Abnormal Ogura-CMS microspore development was further confirmed by TEM. Ogura-CMS microspores underwent similar development to those of MF from the microspore mother cell stage to the tetrad stage (Fig 4A, 4B, 4F and 4G) and, at the uninucleate stage, were distinguishable from MF microspores (Fig 4C and 4H). At the uninucleate stage, the MF microspores had almost complete basic intine and exine structure (Fig 4C). Exine was comprised of inner nexine and outer sexine. Sexine further possessed a three-dimensional structure composed of baculae and a roof-like tectum, whereas the bilayer nexine, consisting of nexine I and nexine II, was laid down on the intine layer (Fig 4K). Subsequently, bicellular microspores were generated concurrently with the size increase of exine, and the mature exine structure was visually completed at this point (Fig 4D and 4L). Finally, the mature pollen grain was complete with tryphine (Fig 4E and 4M). However, after dissolution of the callose wall, free Ogura-CMS microspores were deformed with a thin and incomplete exine layer (Fig 4H). The nexine I was the last layer overlying the microspore plasma membrane but not the intine layer (Fig 4N). By the bicellular stage, with an empty body, pollen grains were prepared with incomplete-developed layers of exine and tryphine (Fig 4O), which developed into remnants at the dehiscent stage (Fig 4J).
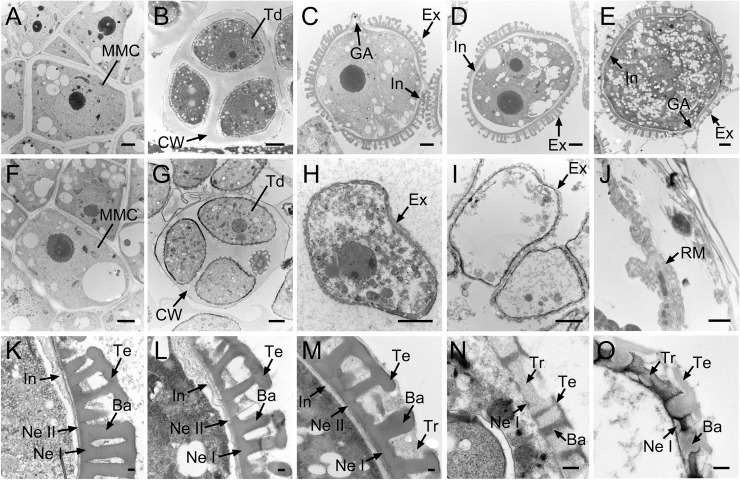
Fig 4. Transmission electron microscopy observation of microspore development in the Ogura-CMS line and its maintainer fertile (MF) line of turnip.
(A-E) Images of microspore development in the MF line from the microspore mother cell stage to the mature pollen stage. (F-J) Images of microspore development in the Ogura-CMS line from the microspore mother cell stage to the mature pollen stage. (A, F) Microspore mother cell stage. (B, G) Tetrad stage, showing four young microspores surrounded by the callose wall. (C, H) Uninucleate microspore stage, showing the intine and the germinal apertures commenced in (C). (D, I) Bicellular stage, showing the degenerated microspores in (I). (E, J) Mature pollen stage, showing the mature pollen grain in the MF line (E) and the remnants of microspores in the Ogura-CMS line (J). (K-M) Magnified images of pollen wall in (C-E), showing the multilayered structure. (N, O) Magnified images of pollen wall in (H, I), showing the incomplete-developed exine layer and the absence of inine layer. Ba, baculum; CW, callose wall; Ex, exine; GA, germinal aperture; In, intine; M, microspore; MMC, microspore mother cell; Ne I, nexine I; Ne II, nexine II; PG, pollen grain; RM, remnants of microspores; Td, tetrads; Te, tectum; Tr, tryphine. Bars = 2 μm in (A-J), 0.2 μm in (K-O).
Coordinated with microspore development, visible changes occurred to the surrounding walls in the anther locules and were also observed when using TEM. No defects on Ogura-CMS surrounding walls were detected at the microspore mother cell stage (Fig 5F) compared with those of the MF line (Fig 5A). Anther primordia which were enclosed in an epidermis, differentiated inwardly into the endothecium, middle layer, tapetum, and microspore mother cells. At the tetrad stage, MF tapetal cells became mature and vacuolated, with a heterogeneous density in the cytoplasm (Fig 5B). However, the anther pattern was significantly altered in Ogura-CMS tapetum at this stage. Ogura-CMS tapetal cells enlarged and swelled to expand to the center of the locules, with larger vacuoles and a clearing cytoplasm (Fig 5G). When the callose wall totally dissolved and free individual microspores released into the anther locules, a large number of elaioplasts emerged in the MF tapetum, and remnants of the middle layer were absent in the MF anther (Fig 5C), but still clearly present in the Ogura-CMS anther (Fig 5H). In addition, Ogura-CMS tapetal cells were full of shedding materials such as tapetosomes and significantly swelled, crushing the free microspores to the locule center (Fig 5H). This premature degradation of the tapetum was obvious, which fulfilled the programmed cell death ahead of schedule. All that remained of the Ogura-CMS tapetum was an almost empty shell at the bicellular stage (Fig 5I), but integral tapetal cells with a large amount of elaioplasts were still observed in the MF anther (Fig 5D). Moreover, the endothecium also appeared abnormal in the Ogura-CMS anther, devoid of any content in the cytoplasm at the bicellular stage (Fig 5I), and had disappeared completely at the mature pollen stage (Fig 5J).
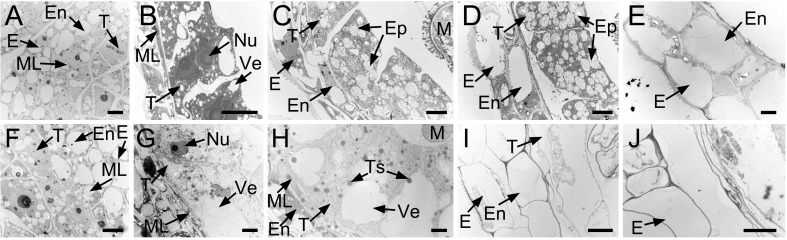
Fig 5. Transmission electron microscopy observation of tapetum development in the Ogura-CMS line and its maintainer fertile (MF) line of turnip.
(A, F) Microspore mother cell stage. Micropores mother cells are surrounded by the tapetum, middle layer, endothecium, and epidermis from the inside out. (B, G) Tetrad stage, showing four distinctive surrounding walls and vacuolated tapetums. The tapetal cells in (G) swell to expand at the center of the locule, with larger vacuoles and a clearing cytoplasm. (C, H) Uninucleate microspore stage. Middle layer disappears and elaioplasts emerge in (C), whereas middle layer persists and tapetosomes were ubiquitous in (H). (D, I) Bicellular stage. Premature degradation of the tapetum occurs in (I), compared with integral tapetal cells with a large amount of elaioplasts in (D). (E, J) Mature pollen stage, showing the absence of the endothecium in (J). E, epidermis; En, endothecium; Ep, elaioplast; M, microspore; ML, middle layer; Nu, nuclei; T, tapetum; Ts, tapetosome; Ve, vacuole. Bars = 5 μm.
Overall, the Ogura-CMS anthers showed two distinct defects that occurred during the transition from microspore mother cells to tetrads: the failure of microspore development and the swollen tapetum layer. Defective microspore production and premature tapetum degeneration during microgametogenesis led to complete male sterility of the Ogura-CMS line.
RNA-Seq analysis on inflorescences of the Ogura-CMS line and its maintainer fertile line in turnip
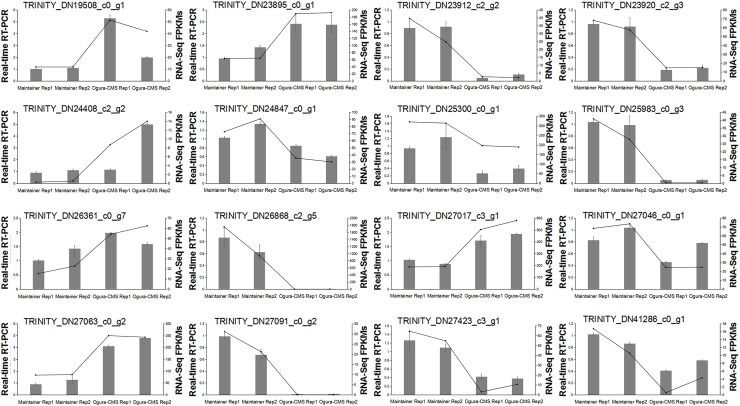
To explore the molecular basis for the morphological differences in anther and microspore development described above, RNA-Seq analyses were conducted to generate transcriptome profiles of the whole inflorescences from the Ogura-CMS and MF lines. RNA-Seq analysis was performed with three biological replicates for each. After removing low-quality reads, an average of 24.2 × 106 clean reads per library were generated (S2 Table). The de novo assembly resulted in a total of 84,132 unigenes (S3 Fig and S3 Table), which were proposed to be expressed during floral bud development in turnip. To verify the quality of the RNA-Seq data, real-time RT-PCR analysis was conducted on 16 randomly selected genes (Fig 6). The strong correlation between the RNA-Seq and real-time RT-PCR results indicated high reliability of our transcriptomic profiling data.
Fig 6. Experimental validation of the quality of the RNA-Seq data by real-time RT-PCR.
The columns indicate the relative RNA levels of selected differentially expressed genes (DEGs) identified between the Ogura-CMS inflorescences and its maintainer inflorescences. The lines show the FPKM expression data of RNA-Seq.
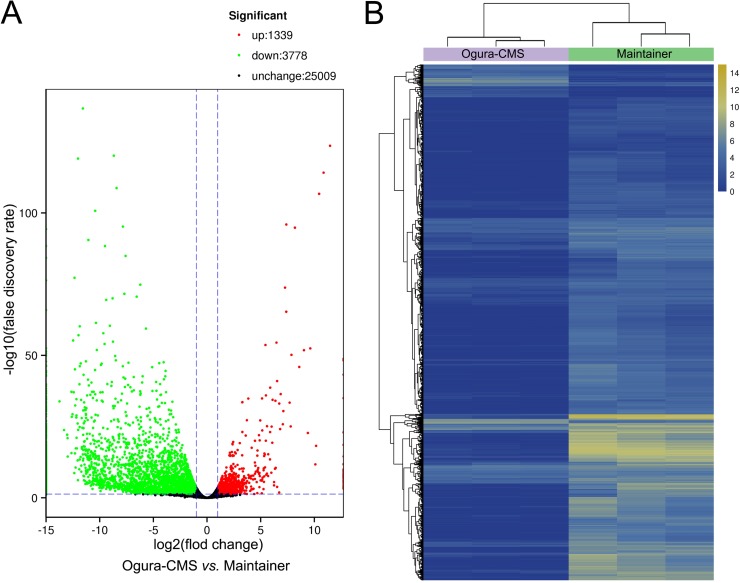
To determine whether the genes regulating pollen and tapetum development are defective or expressed abnormally, differential expression of genes during thereproductive development of the MF and CMS lines was analyzed. The reads were mapped to unigenes and quantified to show gene expression abundance by Fragments Per Kilobase of transcript per Million mapped reads (FPKM) [30]. The FPKM expression data were tested by correlation analysis to evaluate sampling between biological replicates, and all correlation coefficients were ≥ 0.82. Using the DESeq (2010) R Package with a FDR < 0.05 and a log2 FC ≥ 1 or ≤ -1, pairwise comparisons of Ogura-CMS versus MF showed that 5,117 genes were significantly differentially expressed, of which 1,339 genes were significantly up-regulated and 3,778 genes significantly down-regulated in the Ogura-CMS line relative to the MF line (Fig 7). Representative genes for the up- and down-regulated DEGs are listed in Tables 1 and 2, respectively, according to their functional categories. Referencing to the chromosome of Chinese cabbage (Brassica rapa ssp. pekinensis), the DEGs are widely distributed in all chromosomes (S4 Fig).
Fig 7. Expression profiles of differentially expressed genes (DEGs) in the Ogura-CMS and its maintainer fertile (MF) inflorescences of turnip.
(A) Volcano plot showing significantly DEGs with log2 fold change (FC) ≥ 1 or ≤ -1 (Benjamini-Hochberg false discovery rate < 0.05). (B) A hierarchical clustering graph based on the expression values of all significantly DEGs identified in (A).
Table 1. Functional categories of representative genes significantly up-regulated in inflorescences of the Ogura-CMS line relative to its maintainer fertile (MF) line.
| Gene ID | log2 fold change (Ogura-CMS line/MF line) | Description |
|---|---|---|
| Transcription factors | ||
| TRINITY_DN24247_c4_g1 | 4.989846 | Transcription factor MYB39 |
| TRINITY_DN22677_c1_g9 | 3.653598 | Probable WRKY transcription factor 71 |
| TRINITY_DN25088_c3_g1 | 3.295980 | NAC transcription factor 29 |
| TRINITY_DN22677_c1_g2 | 3.196358 | Probable WRKY transcription factor 71 |
| TRINITY_DN23536_c1_g7 | 3.049643 | Homeobox-leucine zipper protein ATHB-21 |
| Carbohydrate transport and metabolism | ||
| TRINITY_DN24721_c0_g1 | 5.481076 | β-glucosidase 27 |
| TRINITY_DN22492_c0_g5 | 5.408367 | Peroxidase 15 |
| TRINITY_DN24408_c2_g2 | 4.629681 | Probable xyloglucan endotransglucosylase/hydrolase protein 18 |
| TRINITY_DN22492_c0_g7 | 4.542994 | Peroxidase |
| TRINITY_DN22492_c0_g3 | 3.409620 | Peroxidase 49 |
| Lipid transport and metabolism | ||
| TRINITY_DN25317_c1_g5 | 1.323946 | Fatty acyl-CoA reductase 1 |
| TRINITY_DN24410_c0_g2 | 1.435586 | Probable acyl-activating enzyme 16 |
| TRINITY_DN25317_c1_g16 | 1.800464 | Fatty acyl-CoA reductase 1 |
| Plant hormone signal transduction | ||
| TRINITY_DN22137_c0_g2 | 2.385851 | Auxin-induced protein X15 |
| TRINITY_DN24146_c0_g1 | 2.268715 | Indole-3-acetic acid-amido synthetase GH3.5 |
| TRINITY_DN24009_c3_g3 | 1.789990 | Indole-3-acetic acid-induced protein ARG7 |
| TRINITY_DN26266_c0_g2 | 1.751535 | Serine/threonine-protein kinase SRK2J |
| TRINITY_DN23900_c1_g2 | 1.737397 | Auxin-responsive protein IAA12 |
Table 2. Functional categories of representative genes significantly down-regulated in inflorescences of the Ogura-CMS line relative to its maintainer fertile (MF) line.
| Gene ID | log2 fold change (Ogura-CMS line/MF line) | Description |
|---|---|---|
| Transcription factors | ||
| TRINITY_DN25469_c0_g5 | -8.944461 | Zinc finger protein ZAT2 |
| TRINITY_DN24004_c2_g7 | -8.817322 | NAC transcription factor 25 |
| TRINITY_DN23291_c0_g9 | -6.858249 | Transcription factor GAMYB |
| TRINITY_DN25812_c2_g1 | -6.813564 | MADS-box transcription factor 16 |
| TRINITY_DN22189_c1_g8 | -6.333698 | Probable WRKY transcription factor 31 |
| Carbohydrate transport and metabolism | ||
| TRINITY_DN27564_c2_g4 | -12.994995 | Exopolygalacturonase clone GBGA483 |
| TRINITY_DN22409_c1_g6 | -12.479006 | Probable pectate lyase 4 |
| TRINITY_DN26448_c4_g4 | -11.859968 | Pectinesterase 21 |
| TRINITY_DN23204_c0_g1 | -11.382811 | Pectinesterase PPME1 |
| TRINITY_DN27059_c3_g2 | -11.281507 | Xyloglucan endotransglucosylase/hydrolase protein 3 |
| Lipid transport and metabolism | ||
| TRINITY_DN24821_c1_g3 | -9.749978 | Delta(8)-fatty-acid desaturase 2 |
| TRINITY_DN26166_c1_g2 | -8.847981 | Dehydrodolichyl diphosphate synthase 8 |
| TRINITY_DN18964_c0_g2 | -8.041336 | Probable lipid phosphate phosphatase 4 |
| TRINITY_DN22477_c0_g1 | -7.414755 | Long chain acyl-CoA synthetase 5 |
| TRINITY_DN26830_c1_g1 | -7.196643 | Phosphoinositide phospholipase C 6 |
| Plant hormone signal transduction | ||
| TRINITY_DN22372_c0_g1 | -9.961644 | Indole-3-acetic acid-amido synthetase GH3.17 |
| TRINITY_DN17602_c0_g1 | -6.559842 | 4-substituted benzoates-glutamate ligase GH3.12 |
| TRINITY_DN22992_c3_g4 | -5.265387 | Auxin-induced protein 15A |
| TRINITY_DN27694_c2_g5 | -2.799438 | ABSCISIC ACID-INSENSITIVE 5-like protein 1 |
Functional annotation of differentially expressed genes during reproductive development of the Ogura-CMS line and its maintainer fertile line of turnip
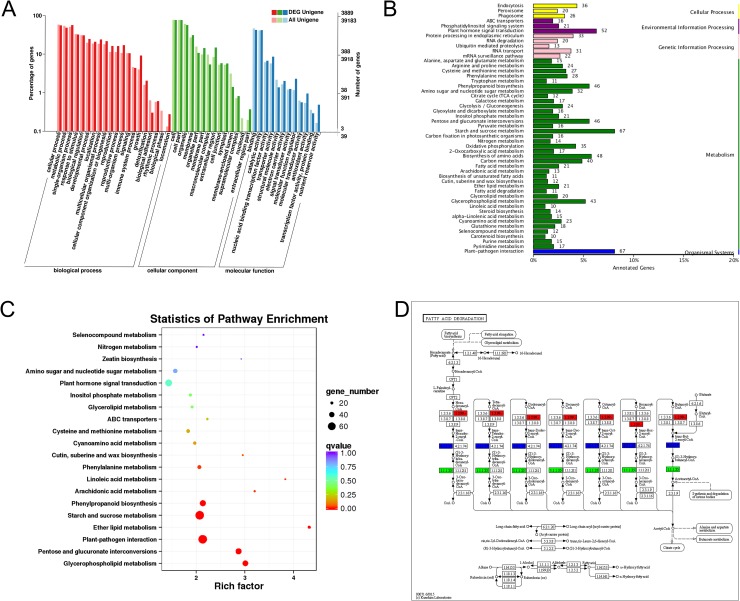
To perform annotation analysis of the DEGs, eight public databases including COG, GO, KEGG, Swiss-prot database, KOG, Pfam, eggNOG, and Nr were searched. In total, 4,864 DEGs were found and annotated in detail in at least one of these databases (S4 Table). To forecast functional classifications of annotated DEGs, the GO and KEGG pathway analyses were performed to provide a clue. We utilized all DEGs for GO analysis and found that 76% of DEGs (3,889 out of 5,117) have at least one GO term assigned and were categorized into 46 functional groups (Fig 8A). Among these, the top three dominant categories were involved in cellular, metabolic, and single-organism processes (Fig 8A). The KEGG pathway analysis manifested that 50 pathways were significantly enriched, particularly metabolic pathways, plant-pathogen interaction, and plant hormone signal transduction (Fig 8B). The DEGs were found to be mostly enriched in ether lipid metabolism (Fig 8C). Furthermore, many genes involved in fatty acid metabolism which is essential for the assembly of exine and tryphine [3,31] were dysregulated (Fig 8D).
Fig 8. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses.
(A) GO annotations of all unigenes and differential expressed genes (DEGs) in the Ogura-CMS and its maintainer fertile (MF) inflorescences of turnip. The results are summarized in three main categories: biological process, cellular component and molecular function. The y-axis on the right indicates the number of genes in a category. The x-axis on the left indicates the percentage of a specific category of genes in that main category. (B) KEGG pathway annotations of DEGs. (C) KEGG pathway enrichment analysis of DEGs with top 20 enrichment scores. (D) KEGG pathway annotations of the fatty acid degradation pathway. Red marked nodes are associated with up-regulated genes; green nodes are associated with down-regulated genes; blue nodes are associated with both up-regulated and down-regulated genes.
In all annotated DEGs, 1,289 genes were significantly up-regulated and 3,575 genes were significantly down-regulated in the Ogura-CMS relative to MF inflorescences (S4 Table). Among these, 610 DEGs were specifically expressed in the MF inflorescences and only 31 DEGs in the Ogura-CMS inflorescences (S5 and S6 Tables), implying that there is a considerable scope for further research to discover novel CMS-associated genes in turnip. In addition, several genes that were classified as function unknown but specifically expressed in the MF inflorescences, such as TRINITY_DN22922_c0_g2, TRINITY_DN37525_c0_g1, and TRINITY_DN23102_c0_g1, could be good candidates for CMS-related genes.
Genes related to anther development and microspore formation
Based on genetic and transcriptomic studies, it has been long assumed that Arabidopsis pollen development involves precise spatial and temporal cooperation of the tapetum and the gametophyte itself, and relies on the functions of numerous genes and their dynamic regulatory network [32]. We compared the expressive alteration of homologs of Arabidopsis genes previously reported to be associated with anther and pollen development, to unravel whether cytoplasmic retro-regulated counterparts of those genes from the nucleus. In addition, some functionally known genes involved in this unique process in species other than Arabidopsis were also compared. As the innermost layer surrounding the sporogenous cells in the anther, the tapetum provides not only energy, but also nutrients, metabolites, and sporopollenin precursors for microspore development [33]. Coordinated with the defective tapetum, homologs of some extensively demonstrated genes and enzymes associated with tapetum development in Arabidopsis (Table 3) and other species (Table 4) exhibited altered expression in Ogura-CMS. For example, AMS is a basic helix-loop-helix (bHLH) transcription factor, one of the master regulators for tapetum and microspore development in Arabidopsis [3]. Expression of a counterpart of AMS (TRINITY_DN27860_c1_g1) was down-regulated in Ogura-CMS. Some turnip homologs of AMS-dependent genes including QRT3 (TRINITY_DN22468_c2_g3), CYP98A8 (TRINITY_DN26854_c1_g2), CHS (TRINITY_DN27063_c0_g1), EXL6 (TRINITY_DN24807_c1_g1, and TRINITY_DN26730_c0_g3), and PAB5 (TRINITY_DN22996_c2_g9, TRINITY_DN22996_c2_g5, and TRINITY_DN22996_c2_g10), showed reduced expression, but some genes did not, such as CYP703A2, CYP704B1, KCS7, LAP5 and LTP12 (Table 3). It was noteworthy that counterparts of two AMS-dependent genes, TKPR1 (TRINITY_DN23571_c0_g2, TRINITY_DN25196_c0_g1, and TRINITY_DN23571_c0_g1) and A6 (TRINITY_DN25127_c0_g3), suggested to be directly regulated by AMS in Arabidopsis, had increased expression in Ogura-CMS (Table 3). In addition, AMS regulatory pathway oriented analyses showed that turnip homologs of ATA20 (TRINITY_DN25943_c1_g3 and TRINITY_DN25943_c1_g1), bHLH89 (TRINITY_DN24940_c2_g5), and bHLH91 (TRINITY_DN22984_c1_g2 and TRINITY_DN22984_c1_g1) were down-regulated (Table 3), but DYT1, TDF1, MS188, and MS1 were not in the pool of DEGs.
Table 3. List of known anther and microspore development-involved genes in Arabidopsis and turnip.
| Gene ID | log2 fold change (Ogura-CMS line/ maintainer line) | Up/down-regulation (Ogura-CMS line/ maintainer line) | Arabidopsis | References | ||
|---|---|---|---|---|---|---|
| Gene name | Locus | Description | ||||
| Anther development/Tapetum development and degeneration | ||||||
| TRINITY_DN26683_c5_g2 | 1.652018 | up | RBOHE | AT1G19230 | NADPH oxidase | [34] |
| TRINITY_DN27860_c1_g1 | -1.816880 | down | AMS | AT2G16910 | bHLH transcription factor | [33,35] |
| TRINITY_DN25433_c1_g3 | 1.150095 | up | BAM1 | AT5G65700 | Leucine-rich repeat receptor-like serine/threonine-protein kinase | [36] |
| TRINITY_DN24940_c2_g5 | -3.073631 | down | bHLH89 | AT1G06170 | bHLH transcription factor | [37] |
| TRINITY_DN22984_c1_g2 | -2.032122 | down | bHLH91 | AT2G31210 | bHLH transcription factor | [37] |
| TRINITY_DN22984_c1_g1 | -2.529790 | down | bHLH91 | AT2G31210 | bHLH transcription factor | [37] |
| TRINITY_DN25943_c1_g3 | -9.043751 | down | ATA20 | AT3G15400 | Anther 20 | [38] |
| TRINITY_DN25943_c1_g1 | -9.481635 | down | ATA20 | AT3G15400 | Anther 20 | [38] |
| TRINITY_DN26731_c0_g2 | -1.383573 | down | GPAT6 | AT2G38110 | Glycerol-3-phosphate 2-O-acyltransferase | [39] |
| TRINITY_DN23446_c2_g8 | -2.186224 | down | GPAT6 | AT2G38110 | Glycerol-3-phosphate 2-O-acyltransferase | [39] |
| TRINITY_DN23672_c1_g1 | -1.126727 | down | GPAT6 | AT2G38110 | Glycerol-3-phosphate 2-O-acyltransferase | [39] |
| Anther dehiscence | ||||||
| TRINITY_DN24200_c1_g2 | -3.476626 | down | ADPG1 | AT3G57510 | Polygalacturonase | [40] |
| TRINITY_DN24097_c1_g2 | 2.667609 | up | ADPG2 | AT2G41850 | Polygalacturonase | [40] |
| TRINITY_DN24097_c1_g3 | * | up | ADPG2 | AT2G41850 | Polygalacturonase | [40] |
| Pollen development | ||||||
| TRINITY_DN25738_c0_g3 | -1.398578 | down | ASKβ | AT3G61160 | Shaggy-like protein kinase | [41] |
| TRINITY_DN25738_c0_g5 | -8.831513 | down | ASKβ | AT3G61160 | Shaggy-like protein kinase | [41] |
| TRINITY_DN25738_c0_g2 | # | down | ASKβ | AT3G61160 | Shaggy-like protein kinase | [41] |
| TRINITY_DN23175_c0_g2 | -6.511601 | down | ASKβ | AT3G61160 | Shaggy-like protein kinase | [41] |
| TRINITY_DN25619_c0_g5 | -2.895881 | down | AtMGT4/AtMRS2-3 | AT3G19640 | Magnesium transporter | [42] |
| TRINITY_DN25619_c0_g7 | -6.739006 | down | AtMGT9/AtMRS2-2 | AT5G64560 | Magnesium transporter | [43] |
| TRINITY_DN23465_c1_g2 | * | up | Exportin-1 | AT5G17020 | Exportin protein family protein | [44] |
| TRINITY_DN24192_c2_g4 | # | down | WRKY34 | AT4G26440 | WRKY transcription factor | [45] |
| TRINITY_DN24036_c0_g3 | -1.197370 | down | WRKY2 | AT5G56270 | WRKY transcription factor | [45] |
| TRINITY_DN26176_c2_g4 | -1.741099 | down | WRKY2 | AT5G56270 | WRKY transcription factor | [45] |
| TRINITY_DN27988_c0_g1 | -8.710511 | down | SCP/LBD27 | AT3G47870 | LBD transcription factor | [46,47] |
| TRINITY_DN23313_c2_g1 | -1.780221 | down | MYB32 | AT4G34990 | R2R3 MYB transcription factor | [48] |
| TRINITY_DN23983_c0_g5 | -1.147885 | down | MYB32 | AT4G34990 | R2R3 MYB transcription factor | [48] |
| Pollen wall development | ||||||
| TRINITY_DN23912_c2_g2 | -4.032420 | down | CalS5 | AT2G13680 | Callose synthase | [49] |
| TRINITY_DN24310_c1_g1 | -6.224270 | down | CalS5 | AT2G13680 | Callose synthase | [49] |
| TRINITY_DN24929_c2_g3 | # | down | ROCK1 | AT5G65000 | Nucleotide sugar transporter | [50] |
| TRINITY_DN23139_c0_g6 | -5.759143 | down | AtbZIP34 | AT2G42380 | bZIP transcription factor | [51] |
| TRINITY_DN24551_c0_g3 | 2.504648 | up | CDM1/AtTTP | AT1G68200 | CCCH-type zinc finger protein | [52,53] |
| TRINITY_DN24329_c2_g4 | -6.144312 | down | QRT1 | AT5G55590 | Pectinesterase | [54,55] |
| TRINITY_DN24097_c1_g4 | -4.462741 | down | QRT2 | AT3G07970 | Polygalacturonase | [55] |
| TRINITY_DN22468_c2_g3 | -4.018069 | down | QRT3 | AT4G20050 | Polygalacturonase | [56] |
| TRINITY_DN22996_c2_g9 | -5.452547 | down | PAB5 | AT1G71770 | Polyadenylate-binding protein | [35] |
| TRINITY_DN22996_c2_g5 | -4.602617 | down | PAB5 | AT1G71770 | Polyadenylate-binding protein | [35] |
| TRINITY_DN22996_c2_g10 | -5.410000 | down | PAB5 | AT1G71770 | Polyadenylate-binding protein | [35] |
| TRINITY_DN24807_c1_g1 | -8.742684 | down | EXL6 | AT1G75930 | GDSL esterase/lipase | [33] |
| TRINITY_DN26730_c0_g3 | -6.884745 | down | EXL6 | AT1G75930 | GDSL esterase/lipase | [33] |
| TRINITY_DN27063_c0_g1 | -7.907580 | down | CHS | AT4G00040 | Chalcone synthase | [35] |
| TRINITY_DN26854_c1_g2 | -3.366574 | down | CYP98A8 | AT1G74540 | Cytochrome P450 | [33] |
| Pollen exine development | ||||||
| TRINITY_DN23571_c0_g2 | 1.261739 | up | TKPR1 | AT4G35420 | Tetraketide α-pyrone reductase 1 | [57] |
| TRINITY_DN25196_c0_g1 | 2.205715 | up | TKPR1 | AT4G35420 | Tetraketide α-pyrone reductase 1 | [57] |
| TRINITY_DN23571_c0_g1 | 1.101840 | up | TKPR1 | AT4G35420 | Tetraketide α-pyrone reductase 1 | [57] |
| TRINITY_DN22625_c4_g1 | -1.977965 | down | AtAPY6 | AT2G02970 | ATP-diphosphohydrolase | [58] |
| TRINITY_DN24453_c1_g2 | 3.497293 | up | AtACBP4 | AT3G054420 | Acyl-CoA-binding protein | [59,60] |
| Pollen coat formation | ||||||
| TRINITY_DN27258_c1_g1 | -1.035989 | down | FLP1/CER3 | AT5G57800 | Aldehyde decarboxylase | [61] |
| TRINITY_DN25053_c4_g1 | -6.822741 | down | ABCG9 | AT4G27420 | ATP binding cassette transporter | [62] |
| TRINITY_DN27312_c0_g2 | -4.364649 | down | ABCG31 | AT2G29940 | ATP binding cassette transporter | [62] |
| TRINITY_DN27312_c0_g1 | -3.151138 | down | ABCG31 | AT2G29940 | ATP binding cassette transporter | [62] |
| TRINITY_DN22659_c0_g1 | -11.567435 | down | GRP17 | AT5G07530 | Oleosin-like protein | [63] |
| TRINITY_DN23861_c0_g2 | -10.827264 | down | GRP17 | AT5G07530 | Oleosin-like protein | [63] |
| TRINITY_DN22389_c1_g4 | # | down | GRP17 | AT5G07530 | Oleosin-like protein | [63] |
| Pollen intine development | ||||||
| TRINITY_DN25796_c0_g4 | -11.203487 | down | FLA3 | AT2G24450 | Fasciclin-like arabinogalactan protein | [64] |
| TRINITY_DN21829_c0_g1 | -9.421529 | down | AGP6 | AT5G14380 | Arabinogalactan protein | [65] |
| TRINITY_DN23900_c2_g6 | # | down | AGP11 | AT3G01700 | Arabinogalactan protein | [65] |
| TRINITY_DN23900_c2_g1 | -4.547789 | down | AGP11 | AT3G01700 | Arabinogalactan protein | [65] |
| TRINITY_DN23148_c0_g1 | -2.519718 | down | RPG1 | AT3G02230 | Reversibly glycosylated polypeptide | [66] |
| TRINITY_DN27324_c0_g2 | -1.867440 | down | RPG1 | AT3G02230 | Reversibly glycosylated polypeptide | [66] |
| TRINITY_DN27324_c0_g1 | -1.279723 | down | RPG2 | AT5G15650 | Reversibly glycosylated polypeptide | [66] |
| TRINITY_DN23148_c0_g5 | -2.965583 | down | RPG2 | AT5G15650 | Reversibly glycosylated polypeptide | [66] |
| TRINITY_DN26054_c1_g12 | 1.360432 | up | ABCG1 | AT2G39350 | ATP binding cassette transporter | [67] |
| TRINITY_DN24546_c0_g2 | 1.303258 | up | ABCG1 | AT2G39350 | ATP binding cassette transporter | [67] |
* specifically expressed genes in the Ogura-CMS inflorescences of turnip
# specifically expressed genes in inflorescences of the maintainer fertile (MF) line of turnip.
Table 4. List of known anther and microspore development-related genes in turnip and species other than Arabidopsis.
| Gene ID | log2 fold change (Ogura-CMS line/ maintainer line) | Up/down-regulation (Ogura-CMS line/ maintainer line) | Species | Gene name | Description | References |
|---|---|---|---|---|---|---|
| Anther development/Tapetum development and degeneration | ||||||
| TRINITY_DN23291_c0_g9 | -6.858249 | down | Oryza sativa | GAMYB | MYB transcription factor | [68,69] |
| TRINITY_DN24913_c2_g6 | -3.344908 | down | Oryza sativa | GAMYB | MYB transcription factor | [68,69] |
| TRINITY_DN24913_c2_g4 | -5.085345 | down | Oryza sativa | GAMYB | MYB transcription factor | [68,69] |
| Pollen development | ||||||
| TRINITY_DN25127_c0_g3 | 1.157526 | up | Brassica napus/Oryza sativa | A6/Osg1 | Similar to β-1,3-glucanase | [70,71] |
| TRINITY_DN25738_c0_g3 | -1.398578 | down | Brassica rapa | BrASK2 | Shaggy-like protein kinase | [72] |
| TRINITY_DN25738_c0_g5 | -8.831513 | down | Brassica rapa | BrASK2 | Shaggy-like protein kinase | [72] |
| TRINITY_DN25738_c0_g2 | # | down | Brassica rapa | BrASK2 | Shaggy-like protein kinase | [72] |
| TRINITY_DN23175_c0_g2 | -6.511601 | down | Brassica rapa | BrASK2 | Shaggy-like protein kinase | [72] |
| Pollen wall development | ||||||
| TRINITY_DN27860_c1_g1 | -1.816880 | down | Oryza sativa | TDR | bHLH transcription factor | [73] |
| TRINITY_DN24360_c3_g3 | -11.780927 | down | Brassica campestris | BcMF9 | Polygalacturonase 3 (PGA3) | [74] |
| TRINITY_DN26838_c0_g1 | -11.405028 | down | Brassica campestris | BcMF9 | Polygalacturonase 3 (PGA3) | [74] |
| Pollen intine development | ||||||
| TRINITY_DN27040_c1_g9 | -9.032317 | down | Brassica campestris | BcPLL9 | Pectatelyase-like 9 | [75] |
| TRINITY_DN27040_c1_g15 | # | down | Brassica campestris | BcPLL9 | Pectatelyase-like 9 | [75] |
| TRINITY_DN27040_c1_g4 | -8.186261 | down | Brassica campestris | BcPLL9 | Pectatelyase-like 9 | [75] |
| TRINITY_DN27040_c1_g6 | -9.846965 | down | Brassica campestris | BcPLL9 | Pectatelyase-like 9 | [75] |
| TRINITY_DN23900_c2_g6 | # | down | Brassica campestris | BcMF8 | Arabinogalactan protein | [76,77] |
| TRINITY_DN23900_c2_g1 | -4.547789 | down | Brassica campestris | BcMF8 | Arabinogalactan protein | [76,77] |
| TRINITY_DN21829_c0_g1 | -9.421529 | down | Brassica campestris | BcMF18 | Arabinogalactan protein | [78] |
# specifically expressed genes in inflorescences of the maintainer fertile (MF) line of turnip.
Based on our cytological results in pollen wall development of turnip, a dramatically altered expression of numerous counterparts of known genes associated with pollen wall formation in CMS florets (Tables 3 and 4) also seemed to be responsible for obvious defects in male gametophyte development. Accordingly, we suggest a somewhat similar regulatory network underlying microspore development in Arabidopsis and turnip.
Apart from the functionally known genes presented in Tables 3 and 4, 185 other novel DEGs were addressed to participate in male organ development in our GO analyses (S7 Table). Among these DEGs, 26 were specifically expressed in the MF inflorescences and none in the Ogura-CMS inflorescences (Table 5). Of the down-regulated genes, two were predicted to encode uncharacterized protein and four were functionally unknown, implying that they could be good candidates for further research to discover novel anther and microspore development-associated genes in turnip.
Table 5. Novel genotype-specific expressed genes with proposed function in male organ development based on GO analyses.
| Gene ID | Maintainer line of turnip | Ogura-CMS line of turnip | Regulated | Proposed function | ||||
|---|---|---|---|---|---|---|---|---|
| Rep1_FPKM | Rep2_FPKM | Rep3_FPKM | Rep1_FPKM | Rep2_FPKM | Rep3_FPKM | |||
| TRINITY_DN19577_c0_g2 | 1.25 | 1.08 | 1.12 | 0 | 0 | 0 | down | Protein GAMETE EXPRESSED 2 |
| TRINITY_DN23051_c0_g11 | 1.67 | 2.06 | 1.41 | 0 | 0.04 | 0 | down | Zinc finger protein ZAT3 |
| TRINITY_DN23051_c0_g7 | 1.67 | 2.06 | 1.41 | 0 | 0.04 | 0 | down | Zinc finger protein ZAT3 |
| TRINITY_DN21111_c0_g1 | 23.61 | 14.06 | 28.67 | 0 | 0 | 0 | down | MADS-box transcription factor 56 |
| TRINITY_DN27168_c1_g1 | 21.32 | 11.07 | 30.7 | 0 | 0 | 0 | down | MADS-box transcription factor |
| TRINITY_DN26124_c2_g12 | 4.78 | 2.73 | 5.97 | 0 | 0 | 0 | down | Transcription factor bHLH84 |
| TRINITY_DN24536_c1_g10 | 13.25 | 5.04 | 49.07 | 0 | 0 | 0 | down | Copper transporter 1-like |
| TRINITY_DN21496_c0_g1 | 4.45 | 9.51 | 3.35 | 0 | 0 | 0 | down | Nicotianamine synthase |
| TRINITY_DN26355_c0_g14 | 75.57 | 24.19 | 181.55 | 0 | 0 | 0 | down | ADP-ribosylation factor GTPase-activating protein AGD10 |
| TRINITY_DN26355_c0_g13 | 61.77 | 24.55 | 145.28 | 0 | 0 | 0.17 | down | ADP-ribosylation factor GTPase-activating protein AGD10 |
| TRINITY_DN25568_c0_g4 | 0.68 | 1.2 | 0 | 0 | 0 | 0 | down | Receptor-like protein 12 |
| TRINITY_DN23305_c1_g3 | 14.47 | 5.96 | 22.17 | 0 | 0 | 0.1 | down | E3 ubiquitin-protein ligase ORTHRUS 2 |
| TRINITY_DN24462_c0_g13 | 23.71 | 15.5 | 22.11 | 0 | 0 | 0 | down | Indole-3-acetic acid-amido synthetase GH3.9 |
| TRINITY_DN22388_c0_g1 | 191.36 | 144.98 | 23.21 | 0 | 0 | 0 | down | Non-specific lipid-transfer protein-like protein |
| TRINITY_DN22388_c0_g4 | 113.92 | 90.76 | 14.87 | 0 | 0 | 0.25 | down | Non-specific lipid-transfer protein-like protein |
| TRINITY_DN22388_c0_g5 | 34.33 | 26.18 | 18.52 | 0 | 0 | 0 | down | Non-specific lipid-transfer protein-like protein |
| TRINITY_DN26904_c2_g5 | 29.98 | 23.43 | 32.42 | 0 | 0 | 0 | down | Probable methyltransferase PMT17 |
| TRINITY_DN23083_c4_g4 | 37.56 | 23.91 | 15.94 | 0 | 0 | 0 | down | Probable methyltransferase PMT17 |
| TRINITY_DN26904_c2_g3 | 51.31 | 36.79 | 4.6 | 0 | 0 | 0 | down | Probable methyltransferase PMT17 |
| TRINITY_DN4311_c0_g1 | 2.76 | 1.4 | 1.71 | 0 | 0 | 0 | down | Probable serine protease EDA2 |
| TRINITY_DN24972_c0_g2 | 20.13 | 17.06 | 0.99 | 0 | 0 | 0 | down | Uncharacterized protein |
| TRINITY_DN27618_c0_g1 | 78.27 | 95.23 | 75.59 | 0 | 0 | 0.69 | down | Uncharacterized protein |
| TRINITY_DN22737_c1_g7 | 6.71 | 5.23 | 22.32 | 0 | 0 | 0 | down | Function unkown |
| TRINITY_DN24700_c2_g1 | 35.39 | 23.24 | 13.14 | 0 | 0 | 0 | down | Function unkown |
| TRINITY_DN22388_c0_g7 | 101.23 | 70.46 | 98.04 | 0 | 0 | 0 | down | Function unkown |
| TRINITY_DN24377_c2_g5 | 6.15 | 8.72 | 19.02 | 0 | 0 | 0 | down | Function unkown |
Overall, these results indicated that mutation of cytoplasmic ORF138 retro-regulated genes from the nucleus, and interactions between them led to male sterility in Ogura-CMS turnip.
Discussion
Morphological characteristics of Ogura-CMS turnip
Ogura-CMS originated from Japanese radish and has been extensively applied in hybrid breeding of crop species [1]. The morphological changes occurring during anther development, particularly in microspore and tapetum behavior, show considerable variation with different nuclear backgrounds and/or cytoplasmic genotype [8]. Here, we emphasized the value of detecting variation in apparently uniform turnip. In Ogura-CMS flowers of turnip, the length of the stamens was greatly reduced, with shortened filaments and thin withered white anthers (Figs 1 and 2). It has been proposed that filament elongation concerns the polar auxin transport from pollen grains/anther to filaments, and hormone-biosynthetic enzymes are also essential for stamen development, including filament elongation [79–81]. When RNA-Seq results were classified by function, we assumed that altered expression of the auxin-, jasmonate-, and gibberellin-related genes (Fig 8 and S4 Table) may be the reason for short Ogura-CMS filaments.
Further investigation showed that microspores exhibited aberrant development in Ogura-CMS anthers and this was initially observed at the uninucleate stage, leading to remnants of the aborted microspores with incomplete-developed layers of exine and tryphine (Fig 4). Defects in Ogura-CMS pollen wall formation were consistent with the observation that apart from the abortion of microspore development, the tapetum in Ogura-CMS anthers was swollen and abnormally vacuolated, initially at the tetrad stage (Figs 3 and 5). Tapetal organelles, such as elaioplasts and tapetosomes, were abnormally formed. In Brassicaceae plants, such as Arabidopsis, elaioplasts and tapetosomes are the carriers in the traffic of lipid molecules from the tapetum to the microspore [82]. Proper lipid accumulation in these organelles is critical for normal tapetum development and pollen wall formation, as disruptions of lipid metabolism-related genes affect tapetum morphology and pollen wall patterning [31]. These results showed that defective microspore production and early tapetum degeneration during microgametogenesis led to complete male sterility of the Ogura-CMS line.
It was also noteworthy that in sterile anthers, the middle layer persisted for a long time (Fig 5). The question is still poorly documented for that if other non-tapetal anther layers, such as the middle layer, endothecium, and epidermis, are also essential for microspore development. Previously, a delay in PCD of the middle layer was reported to induce male sterility in kiwifruit (Actinidia deliciosa) [83]. The absence of well-defined genes in which mutation leads to delayed degeneration of the middle layer has enriched our knowledge on microspore development. For instance, Oryza sative tdr mutant shows a delay in middle layer degeneration as well as tapetum PCD [73]. In our RNA-seq data, the turnip homologue (TRINITY_DN27860_c1_g1) of this bHLH transcription factor-encoding gene showed a significant decrease in transcript level in the Ogura-CMS line relative to the MF line (Table 4). Mutation of a leucine-rich repeat receptor protein kinase EXS/EMS1 in Arabidopsis causes abnormal persistence of the middle layer [84,85]. It is not clear what genes regulate the middle layer degeneration. Although non-tapetal anther layers argue against an essential role in pollen development, the molecular basis and effort of non-tapetal anther layers in microspore development still need further exploration in the future.
Morphology of all other floral organs was normal, suggesting that genes involved in floral organ identity were normal. Apart from affecting stamen development, several negative effects other than heterosis are resulted from the introduction of Ogura-CMS from radish, via B. napus into B. rapa [17]. It has been reported that the CMS mitochondrial signaling can also cause aberrations in other aspects including vegetative growth [2]. Though 10 generations of back-crossing, protoplast fusion eliminated the negative effects on crown diameter and plant height of Ogura-CMS plants, the fleshy root still had reduced size (Fig 1 and S2 Fig).
Cytoplasmic retro-regulates nuclear gene expression
Based on findings on Arabidopsis microsporogenesis [86], the early stage of this process in the Ogura-CMS line should be normal (Fig 3). Instead, genes associated with tapetal development and meiotic tapetal function are defective in the Ogura-CMS line. Moreover, the expression of genes associated with microspore development could be altered. It has been reported in CMS rice, carrots, wheat, and B. napus that mitochondrial mutations affect nuclear gene expression, through a communication pathway from mitochondria to the nucleus called the mitochondrial retrograde signaling pathway [4–7]. In addition, variation in anther behavior among Brassica species suggests the presence of different regulatory mechanisms and/or multiple regulatory pathways [17]. Therefore, we explored the differential gene expression patterns of transcripts between the Ogura-CMS and its MF inflorescences in turnip. RNA-Seq identified prodigious DEGs (S4 Table), supporting an extensive involvement of mRNAs in anther development. The complex transcript network was simplified by modularization. A lower number of dysfunctional DEGs in the Ogura-CMS inflorescences may also reflect the underdevelopment of stamens. Overall, these results indicate that the regulatory network of anther development is altered in Ogura-CMS turnip, the nuclear genes involved in this complex process are retro-regulated by mitochondrial orf138, and mitochondrial-to-nucleus signaling in Ogura-CMS plants perhaps causes aberrant male sporophyte and gametophyte development.
Genes involved in tapetum development and microspore formation show dysregulated expression
Pollen development involves delicate and coordinated spatio-temporal regulation of numerous biosynthetic genes by specific transcriptional regulators, and requires coordinated activity of different cell types and tissues of both gametophytic and sporophytic origin [3,32,87,88]. It is estimated that about 14,000 genes give a positive expression signal during microspore development, of which only a few have been shown to be pollen-specific with a defined requirement at different stages of pollen development [32]. In the CMS system, it may be more complicated because of the presence of the mitochondrial retrograde signaling pathway and the interaction between the nucleus and mitochondria [89]. We performed a transcriptomic comparison between the expression of some well-known anther and tapetum development-associated genes and their homologs in turnip. The results showed that cytoplasmic retro-regulated counterparts of some genes from the nucleus (Tables 3 and 4), such as GPAT6, which is essential for tapetum development, when disrupted causes defective exine deposition and partial pollen degradation [39]; RBOHE, encoding a NADPH oxidase and required for tapetal PCD, if interfered results in aborted male gametophytes [34]; and BAM1, a Leu-rich repeat receptor-like kinase encoding gene, is an important regulator for anther development [36].
During early tapetum development in Arabidopsis, EXS/EMS1 is a trigger to the signaling pathway for tapetal fate determination [84,85]. Here, the counterparts of EXS/EMS1 were not distinguishable in turnip (S3 and S4 Tables), indicating that retrograde signaling did not affect gene expression at the early stage of tapetum development and function, consistent with normal early tapetum behavior we observed (Fig 5). Later, core genetic networks involving several transcription factors regulate tapetum and microspore development, including a bHLH transcription factor Arabidopsis AMS/rice TDR, one of the master regulators [3]. AMS triggers at least 23 pollen wall development-related genes, such as A6, TKPR1 [33]. Mutation in AMS causes tapetal hypertrophy extending into the locule and subsequently leads to abnormality of microspore development [33,35]. The turnip ortholog of AMS was found to be down-regulated in the Ogura-CMS inflorescence (Table 3). Some AMS-dependent genes were also dysregulated in the Ogura-CMS inflorescence, but others, such as CYP703A2, CYP703B1, WBC27, TEK, and LAP5, were not. In addition, a transcriptional regulatory pathway involving AMS with DYT1-TDF1-AMS-MS188-MS1 has been proposed for tapetal and microspore development in Arabidopsis [3]. DYT1 is a bHLH transcriptional factor and TDF1 is a putative R2R3 MYB transcriptional factor, both of which are responsible for tapetum development and microspore formation [90,91]. However, the turnip orthologs of DYT1 and TDF1 were absent in our RNA-Seq data. Instead, the counterparts of the other two Arabidopsis bHLH transcriptional factors, bHLH89 and bHLH91 [37], and another R2R3 MYB transcription factor, MYB32 [48], had decreased expression in the Ogura-CMS inflorescence. It has been suggested that BrbHLH89 might replace DYT1 function and transcription factor genes BrTDF1, BrAMS, BrMS188, and BrMS1 in Chinese cabbage, however, it exhibited delayed or extended expression in the Ogura-CMS line [17]. Therefore, we suggest that the function of AMS may be retained but the regulatory AMS pathway may have some alteration between turnip and Arabidopsis, or other Brassica spp.
Recently, based on research on the inflorescence transcriptome of Ogura-CMS cabbage, 22 DEGs were assigned to pollen development [18]. Among these DEGs, some functionally known key members involving pollen development, such as BoLAP5, BoLAP6, BoACOS5, BoMS2, BoCYP703A2, and BoCYP704B1, were found to be down-regulated in Ogura-CMS anthers compared to the fertile line. In addition, some new members, such as BobHLH1, BoMYB1, and BoMF2, were also identified [92–94]. However, some of these well-defined genes were not part of the pool of our DEGs. These differences in gene expression may be the cause of different anther behaviors that Ogura-CMS cabbage has an abnormally fat tapetum with a delayed tapetum PCD, compared to our Ogura-CMS turnip which exhibited premature tapetum degeneration during microgametogenesis. These results provide new clues for a better understanding of different Ogura-CMS Brassica spp regulatory mechanisms.
Apart from those well-defined genes, 185 other novel DEGs were predicted to be involved in male organ development based on GO analyses, of which 26 DEGs were genotype-specific (S7 Table and Table 5), implying that they are good candidates for further research on novel anther and microspore development-associated genes in turnip. It is interesting that some members were previously reported to function in other developmental processes, not just anther development. For example, putative indole-3-acetic acid-amido synthetase GH3.9, a member of the GH3 family of auxin-responsive genes, was previously reported to contribute to primary root length [95]; GAMETE EXPRESSED 2 encodes a trans-membrance domain containing protein which is targeted by DUO1 in Arabidopsis [96]. These genes may be endowed with new putative roles in male organ development.
Genes related to pollen wall development
The indispensable biological function of pollen is somehow reflected by its unique surrounding wall. Pollen wall synthesis is regulated by both the sporophytic tapetum and the microspore [97]. As the innermost layer of the pollen wall, intine not only plays an important role in aperture configuration and pollen stability, but also functions in pollen tube growth into the stigma [77,87,98,99]. Intine comprises of pectin, cellulose, hemicellulose, hydrolytic enzymes and hydrophobic proteins, and is controlled by the gametophyte [97,100]. Several genes and proteins have been identified in intine formation [66,74,75,97,101–107]. Here, we found that the expression of homologs of RGP1, RGP2, and PLL9 in the Ogura-CMS inflorescences of turnip was reduced (Tables 3 and 4), disruptions of which contribute to the premature degradation of developing microspores [66,75]. Arabinogalactan proteins (AGPs) are extensively glycosylated hydroxyproline-rich glycoproteins and are also proved to be indispensable for intine formation [78,108]. Currently, two classical AGP-encoding genes AGP6 and AGP11 and, their B. campestris ortholog BcMF18 and BcMF8, and a fasciclin-like AGP (FLA) gene FLA3, have been isolated and identified [77,78,109,110]. The orthologs of all these AGP-encoding genes were found in turnip and showed down-regulated expression in the Ogura-CMS inflorescences (Tables 3 and 4). In addition, counterparts of another two FLA genes, FLA5 (TRINITY_DN22476_c2_g15 and TRINITY_DN22476_c2_g16) and FLA14 (TRINITY_DN23986_c3_g2, TRINITY_DN23986_c3_g5, and TRINITY_DN23986_c3_g6), showed down-regulation in the Ogura-CMS inflorescences (S4 Table). Similarly, in Chinese cabbage, BrFLA14 was highly expressed in fertile buds and could be associated with pollen development [72], implying that this FLA gene could be important for intine formation in turnip.
In addition to the extensively reviewed genes and enzymes associated with pollen intine formation summarized above, the expression of some genes acting at different steps of exine formation was also dysregulated (Table 3). Exine formation requires proper pollen wall pattern determination and sporopollenin deposition. Pollen wall pattern determination is dependent on the formation and dissolution of the callose wall, which provides a structural basis for the primexine deposition [3]. Previous attempts to explore the roles of callose wall-related genes have indicated that overexpression of β-1,3-glucanase in the tapetum results in defective exine pattern formation [111]. Brassica napus and Arabidopsis A6 and its rice ortholog Osg1 encode β-1,3-glucanase that are essential for timely callose degradation [70,71]. As for the abnormality of tapetum, their counterpart in turnip (TRINITY_DN25127_c0_g3) was found to be up-regulated in the Ogura-CMS inflorescences (Table 4). In addition, the counterparts of CalS5 (TRINITY_DN23912_c2_g2 and TRINITY_DN24310_c1_g1), in which mutation causes abnormal exine wall formation [49], had a decreased expression in the Ogura-CMS inflorescences (Table 3). Although the appearance of the Ogura-CMS callose wall at the tetrad stage was normal, the dysregulated expression of homologous genes of A6/Osg1 and CalS5 was suggested to be positively correlated with efficient microspore development. Sporopollenin deposition is dependent on the synthesis, secretion and translocation of sporopollenin precursors, which is closely associated with fatty acid metabolism [3,31]. In our RNA-Seq data, many genes involved in fatty acid metabolism pathway were dysregulated in the Ogura-CMS inflorescences (Fig 8D), indicating that they might also be responsible for exine defects. Notably, a number of functionally known key members of the complex biochemical biosynthesis of sporopollenin precursors in the tapetum were not in the pool of DEGs. ACOS5, a medium- and long-chain fatty acyl CoA-synthetase [112], are condensed with fatty acyl ACP reductase MS2 [113], cytochrome P450 family members CYP703As and CYP704Bs [114–116], flavonoid synthases LAP5 (PKSA) and LAP6 (PKSB) [117], together with tetraketide α-pyrone reductases TKPR1 and TKPR2 [57], yield precursors of sporopollenin building units. Disruption of these genes drastically hampers biosynthesis of sporopollenin precursors in the tapetum. Among all these genes, only the counterparts of TKPR1 were found up-regulated in our RNA-Seq results. We suggest that the reduced expression of ATP-binding cassette subfamily G (ABCG) transporters contributes to the transport of sporopollenin precursors across anther tissues [67]. Here, we only found ABCG1 had decreased expressed counterparts in the Ogura-CMS inflorescences (Table 3), which functions redundantly with ABCG16 in exine layer formation. These results suggest that the synthesis and secretion of sporopollenin precursors may be little affected by mitochondrial retrograde signaling. In addition, RNA-Seq analyses showed that a large number of lipid transfer proteins (LTPs)-encoding genes were dysregulated (S4 Table). Presently, no experimental evidence was obtained for the mechanism by which LTPs sporopollenin precursors are translocated, but LTPs are likely candidates for their delivery from the tapetum to the microspores [100]. Overall, these results indicated that during the process of sporopollenin deposition in Ogura-CMS turnip, the focal point of the effect of mitochondrial orf138 may be sporopollenin precursor translocation.
Genes related to pollen coat formation, which is also closely associated with fatty acid metabolism, are also indispensable in Ougra-CMS turnip. FLP1/CER3, is a member of the ECERIFERUM family, which appears to participate in the synthesis of wax, components of tryphine and sporopollenin of exine in Arabidopsis [61]. In the flp-1 mutant, excessive tryphine fills the cavity of the exine leading to a smooth surface that is sensitive to acetolysis. We found the homolog of FLP1/CER3 (TRINITY_DN27258_c1_g1) was down-regulated (Table 3). In addition, the expression of the counterparts of ABCG9 (TRINITY_DN25053_c4_g1), ABCG31 (TRINITY_DN27312_c0_g2 and TRINITY_DN27312_c0_g1) and GRP17 (TRINITY_DN22659_c0_g1, TRINITY_DN23861_c0_g2, and TRINITY_DN22389_c1_g4), which contribute to the accumulation of pollen coat [62,63], was also decreased (Table 3). The altered expression of genes associated with the pollen coat may be related to male sterility in Ogura-CMS turnip.
Conclusions
In this study, detailed morphological characteristics of an Ogura-CMS line and its MF line of turnip were described. We employed high-throughput sequencing approaches to identify candidate nuclear genes that responded to mitochondrial retrograde signaling, and focused on the anther and microspore development-associated genes. Comparison of the gene expression between CMS and MF lines supported the notion that CMS is attributed to the mitochondrial retrograde signaling pathway and the interaction of nuclear and organelle genomes. Furthermore, our results suggest that different regulatory mechanisms and/or multiple regulatory pathways may exist in Brassica spp.
Supporting information
(A-D) Scanning electron microscopy observation of floral organ morphology in the MF line. (E-H) Scanning electron microscopy observation of floral organs in the Ogura-CMS line showing normal morphology that are similar to those of the MF line. (A, E) The outer surface of a petal. (B, F) The outer surface of a sepal with numerous stomata (indicated by arrows). (C, G) The finger-like papillae of a mature stigma with a smooth surface. Arrows indicate the stomata on the style outer epidermis. (D, H) The outer surface of a nectary. Bars = 50 μm in (A, E), 100 μm in (B, F), 300 μm in (C, G), 200 μm in (D, H).
(JPG)
(A) External morphology of seedlings at 32 days after germination. (B) The fleshy roots thickening at 48 days after germination. (C) Morphological features of flowering plants at 180 days after germination. (D) The length and diameter of fleshy roots at 180 days after germination. The values are the mean ± SD (standard deviation). (E) Normal silique growth and seed set on an Ogura-CMS plant and a MF plant pollinated with MF pollen grains as the male donor. Bars = 5 cm.
(JPG)
(JPG)
From the outside in, the first circle of the Circos plot is a chromosome map of the Chinese cabbage genome. The homologous genes of all DEGs of turnip in Chinese cabbage are showed in the second circle. All DEGs are marked in the third circle.
(JPG)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank Dr. Heng Dong (Hangzhou Normal University, China) for his comments on this manuscript.
Abbreviations
- ABCG
ATP-binding cassette subfamily G
- AGP
arabinogalactan proteins
- bHLH
basic helix-loop-helix
- CMS
cytoplasmic male sterility
- COG
Clusters of Orthologous Group
- DEG
differentially expressed gene
- eggNOG
orthologous groups of genes
- FC
fold change
- FDR
false discovery rate
- FLA
fasciclin-like AGP
- FPKM
Fragments Per Kilobase of transcript per Million mapped reads
- GO
Gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KOG
Clusters of Protein homology database
- LTP
lipid transfer protein
- MF
maintainer fertile
- NCBI
National Center for Biotechnology Information
- Nr
NCBI non-redundant protein
- PCD
programmed cell death
- Pfam
Homologous protein family
- PPI
predicted protein-protein interaction
- RNA-seq
RNA sequencing
- RT-PCR
reverse transcription polymerase chain reaction
- STRING
Search Tool for the Retrieval of Interacting Genes/Proteins
- TEM
transmission electron microscopy.
Data Availability
The RNA-Seq data were uploaded to the Sequence Read Archive of the National Center for Biotechnology Information (NCBI) (DOI: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA505114; accession number: PRJNA505114).
Funding Statement
This work was jointly supported by funds from the Special Project on Science and Technology innovation of Seed and Seedling of Wenzhou [N20160008] (SL), and the National Natural Science Foundation of China [31501764] (SL).
References
- 1.Touzet P, Meyer EH. Cytoplasmic male sterility and mitochondrial metabolism in plants. Mitochondrion. 2014;19: 166–171. 10.1016/j.mito.2014.04.009 [DOI] [PubMed] [Google Scholar]
- 2.Carlsson J, Glimelius K. Cytoplasmic male-sterility and nuclear encoded fertility restoration In: Kempken F, editor. Plant Mitochondria. New York: Springer; 2011. pp. 469–491. [Google Scholar]
- 3.Lou Y, Zhu J, Yang Z. Molecular cell biology of pollen walls In: Nick P, Opatrný Z, editors. Applied Plant Cell Biology; Berlin, Heidelberg: Springer; 2014. pp. 179–205. [Google Scholar]
- 4.Murai K, Takumi S, Koga H, Ogihara Y. Pistillody, homeotic transformation of stamens into pistil-like structures, caused by nuclear-cytoplasm interaction in wheat. The Plant Journal. 2002;29: 169–181. [DOI] [PubMed] [Google Scholar]
- 5.Linke B, Nothnagel T, Börner T. Flower development in carrot CMS plants: mitochondria affect the expression of MADS box genes homologous to GLOBOSA and DEFICIENS. The Plant Journal. 2003;34: 27–37. [DOI] [PubMed] [Google Scholar]
- 6.Teixeira RT, Farbos I, Glimelius K. Expression levels of meristem identity and homeotic genes are modified by nuclear-mitochondrial interactions in alloplasmic male-sterile lines of Brassica napus. The Plant Journal. 2005;42: 731–742. 10.1111/j.1365-313X.2005.02407.x [DOI] [PubMed] [Google Scholar]
- 7.Fujii S, Komatsu S, Toriyama K. Retrograde regulation of nuclear gene expression in CW-CMS of rice. Plant Molecular Biology. 2007;63: 405–417. 10.1007/s11103-006-9097-8 [DOI] [PubMed] [Google Scholar]
- 8.Colhoun CW, Steer MW. Microsporogenesis and the mechanism of cytoplasmic male sterility in maize. Annals of Botany. 1981;48: 417–424. [Google Scholar]
- 9.Yuan Q, Song C, Gao L, Zhang H, Yang C, Sheng J, et al. Transcriptome de novo assembly and analysis of differentially expressed genes related to cytoplasmic male sterility in onion. Plant Physiology and Biochemistry. 2018;125: 35–44. 10.1016/j.plaphy.2018.01.015 [DOI] [PubMed] [Google Scholar]
- 10.Overman MA, Warmke HE. Cytoplasmic male sterility in sorghum: II. Tapetal behavior in fertile and sterile anthers. Journal of Heredity. 1972;63: 227–234. [Google Scholar]
- 11.Yamagishi H, Bhat SR. Cytoplasmic male sterility in Brassicaceae crops. Breeding Science. 2014;64: 38–47. 10.1270/jsbbs.64.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polowick PL, Sawhney VK. A scanning electron microscopic study on the influence of temperature on the expression of cytoplasmic male sterility in Brassica napus. Canadian Journal of Botany. 1987;65: 807–814. [Google Scholar]
- 13.Polowick PL, Sawhney VK. High temperature induced male and female sterility in canola (Brassica napus L.). Annals of Botany. 1988;62: 83–86. [Google Scholar]
- 14.Polowick PL, Sawhney VK. Microsporogenesis in a normal line and in the ogu cytoplasmic male-sterile line of Brassica napus. I. The influence of high temperatures. Sexual Plant Reproduction. 1991;4: 22–27. [Google Scholar]
- 15.Gourret JP, Delourme R, Renard M. Expression of ogu cytoplasmic male sterility in cybrids of Brassica napus. Theoretical and Applied Genetics. 1992;83: 549–556. 10.1007/BF00226898 [DOI] [PubMed] [Google Scholar]
- 16.González-Melendi P, Uyttewaal M, Morcillo CN, Mora JRH, Fajardo S, Budar F, et al. A light and electron microscopy analysis of the events leading to male sterility in Ogu-INRA CMS of rapeseed (Brassica napus). Journal of Experimental Botany. 2008;59: 827–838. 10.1093/jxb/erm365 [DOI] [PubMed] [Google Scholar]
- 17.Dong X, Kim WK, Lim YP, Kim YK, Hur Y. Ogura-CMS in Chinese cabbage (Brassica rapa ssp. pekinensis) causes delayed expression of many nuclear genes. Plant Science. 2013;199: 7–17. 10.1016/j.plantsci.2012.11.001 [DOI] [PubMed] [Google Scholar]
- 18.Xing M, Sun C, Li H, Hu S, Lei L, Kang J. Integrated analysis of transcriptome and proteome changes related to the Ogura cytoplasmic male sterility in cabbage. PLoS ONE. 2018;13: e0193462 10.1371/journal.pone.0193462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu Y, Liao L, Jin X, Mao D, Liu R. Analysis of the meiotic transcriptome reveals the genes related to the regulation of pollen abortion in cytoplasmic male-sterile pepper (Capsicum annuum L.). Gene. 2018;641: 8–17. 10.1016/j.gene.2017.10.022 [DOI] [PubMed] [Google Scholar]
- 20.Yang J, Liu X, Xu B, Zhao N, Yang X, Zhang M. Identification of miRNAs and their targets using high-throughput sequencing and degradome analysis in cytoplasmic male-sterile and its maintainer fertile lines of Brassica juncea. BMC Genomics. 2013;14: 9 10.1186/1471-2164-14-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei X, Zhang X, Yao Q, Yuan Y, Li X, Wei F, et al. The miRNAs and their regulatory networks responsible for pollen abortion in Ogura-CMS Chinese cabbage revealed by high-throughput sequencing of miRNAs, degradomes, and transcriptomes. Frontiers in Plant Science. 2015;6: 894 10.3389/fpls.2015.00894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding X, Li J, Zhang H, He T, Han S, Li Y, et al. Identification of miRNAs and their targets by high-throughput sequencing and degradome analysis in cytoplasmic male-sterile line NJCMS1A and its maintainer NJCMS1B of soybean. BMC Genomics. 2016;17: 24 10.1186/s12864-015-2352-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mishra A, Bohra A. Non-coding RNAs and plant male sterility: current knowledge and future prospects. Plant Cell Reports. 2018;37: 177–191. 10.1007/s00299-018-2248-y [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Schonhof I, Krumbein A, Gutezeit B, Li L, Stützel H, et al. Water supply and growing season influence glucosinolate concentration and composition in turnip root (Brassica rapa ssp. rapifera L.). Journal of Plant Nutrition and Soil Science. 2010;171: 255–265. [Google Scholar]
- 25.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology. 2011;29: 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Research. 2000;28: 33–36. 10.1093/nar/28.1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Research. 2014;42: 222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21: 3787–3793. 10.1093/bioinformatics/bti430 [DOI] [PubMed] [Google Scholar]
- 29.Wang C, Cui HM, Huang TH, Liu TK, Hou XL, Li Y. Identification and validation of reference genes for RT-qPCR analysis in non-heading Chinese cabbage flowers. Frontiers in Plant Science. 2016;7: 811 10.3389/fpls.2016.00811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Baren MJV, et al. Transcript assembly and abundance estimation from RNA-Seq reveals thousands of new transcripts and switching among isoforms. Nature Biotechnology. 2010;28: 511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang J, Zhang Z, Cao J. Pollen wall development: the associated enzymes and metabolic pathways. Plant Biology. 2013;15: 249–263. 10.1111/j.1438-8677.2012.00706.x [DOI] [PubMed] [Google Scholar]
- 32.Twell D. Male gametophyte development In: Pua EC, Davey MR, editors. Plant Developmental Biology-Biotechnological Perspectives. Berlin, Heidelberg: Springer; 2010. pp. 225–244. [Google Scholar]
- 33.Xu J, Ding Z, Vizcay-Barrena G, Shi J, Liang W, Yuan Z, et al. ABORTED MICROSPORES acts as a master regulator of pollen wall formation in Arabidopsis. The Plant Cell. 2014;26: 1544–1556. 10.1105/tpc.114.122986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie HT, Wan ZY, Li S, Zhang Y. Spatiotemporal production of reactive oxygen species by NADPH oxidase is critical for tapetal programmed cell death and pollen development in Arabidopsis. The Plant Cell. 2014;26: 2007–2023. 10.1105/tpc.114.125427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J, Yang C, Yuan Z, Zhang D, Gondwe MY, Ding Z, et al. The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. The Plant Cell. 2010;22: 91–107. 10.1105/tpc.109.071803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hord CLH, Chen C, DeYoung BJ, Clark SE, Ma H. The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. The Plant Cell. 2006;18: 1667–1680. 10.1105/tpc.105.036871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu E, You C, Wang S, Cui J, Niu B, Wang Y, et al. The DYT1-interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for Arabidopsis anther development and transcriptome. The Plant Journal. 2015;83: 976–990. 10.1111/tpj.12942 [DOI] [PubMed] [Google Scholar]
- 38.Rubinelli P, Hu Y, Ma H. Identification, sequence analysis and expression studies of novel anther-specific genes of Arabidopsis thaliana. Plant Molecular Biology. 1998;37: 607–619. [DOI] [PubMed] [Google Scholar]
- 39.Li XC, Zhu J, Yang J, Zhang GR, Xing WF, Zhang S, et al. Glycerol-3-Phosphate Acyltransferase 6 (GPAT6) is important for tapetum development in Arabidopsis and plays multiple roles in plant fertility. Molecular Plant. 2012;5: 131–142. 10.1093/mp/ssr057 [DOI] [PubMed] [Google Scholar]
- 40.Ogawa M, Kay P, Wilson S, Swain SM. ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. The Plant Cell. 2009;21: 216–233. 10.1105/tpc.108.063768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong X, Nou IS, Yi H, Hur Y. Suppression of ASKβ (AtSK32), a clade III Arabidopsis GSK3, leads to the pollen defect during late pollen development. Molecules and Cells. 2015;38: 506–517. 10.14348/molcells.2015.2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Huang Y, Tan H, Yang X, Tian L, Luan S, et al. An endoplasmic reticulum magnesium transporter is essential for pollen development in Arabidopsis. Plant Science. 2015;231: 212–220. 10.1016/j.plantsci.2014.12.008 [DOI] [PubMed] [Google Scholar]
- 43.Chen J, Li L, Liu Z, Yuan Y, Guo L, Mao D, et al. Magnesium transporter AtMGT9 is essential for pollen development in Arabidopsis. Cell Research. 2009;19: 887–898. 10.1038/cr.2009.58 [DOI] [PubMed] [Google Scholar]
- 44.Blanvillain R, Boavida LC, McCormick S, Ow DW. EXPORTIN1 genes are essential for development and function of the gametophytes in Arabidopsis thaliana. Genetics. 2008;180: 1493–1500. 10.1534/genetics.108.094896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guan Y, Meng X, Khanna R, LaMontagne E, Liu Y, Zhang S. Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLoS Genetics. 2014;10: e1004384 10.1371/journal.pgen.1004384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oh SA, Park KS, Twell D, Park SK. The SIDECAR POLLEN gene encodes a microspore-specific LOB/AS2 domain protein required for the correct timing and orientation of asymmetric cell division. The Plant Journal. 2010;64: 839–850. 10.1111/j.1365-313X.2010.04374.x [DOI] [PubMed] [Google Scholar]
- 47.Kim MJ, Kim M, Lee MR, Park SK, Kim J. LATERAL ORGAN BOUNDARIES DOMAIN (LBD) 10 interacts with SIDECAR POLLEN/LBD27 to control pollen development in Arabidopsis. The Plant Journal. 2015;81: 794–809. 10.1111/tpj.12767 [DOI] [PubMed] [Google Scholar]
- 48.Preston J, Wheeler J, Heazlewood J, Li SF, Parish RW. AtMYB32 is required for normal pollen development in Arabidopsis thaliana. The Plant Journal. 2004;40: 979–995. 10.1111/j.1365-313X.2004.02280.x [DOI] [PubMed] [Google Scholar]
- 49.Dong X, Hong Z, Sivaramakrishnan M, Mahfouz M, Verma DPS. Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. The Plant Journal. 2005;42: 315–328. 10.1111/j.1365-313X.2005.02379.x [DOI] [PubMed] [Google Scholar]
- 50.Niemann MCE, Bartrina I, Ashikov A, Weber H, Novák O, Spíchal L, et al. Arabidopsis ROCK1 transports UDP-GlcNAc/UDP-GalNAc and regulates ER protein quality control and cytokinin activity. Proceedings of the National Academy of Sciences. 2015;112: 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gibalová A, Reňák D, Matczuk K, Dupl’áková N, Cháb D, Twell D, et al. AtbZIP34 is required for Arabidopsis pollen wall patterning and the control of several metabolic pathways in developing pollen. Plant Molecular Biology. 2009;70: 581–601. 10.1007/s11103-009-9493-y [DOI] [PubMed] [Google Scholar]
- 52.Lu P, Chai M, Yang J, Ning G, Wang G, Ma H. The Arabidopsis CALLOSE DEFECTIVE MICROSPORE1 gene is required for male fertility through regulating callose metabolism during microsporogenesis. Plant Physiology. 2014;164: 1893–1904. 10.1104/pp.113.233387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi ZH, Zhang C, Xu XF, Zhu J, Zhou Q, Ma LJ, et al. Overexpression of AtTTP affects ARF17 expression and leads to male sterility in Arabidopsis. PLoS ONE. 2015;10: e0117317 10.1371/journal.pone.0117317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rhee SY, Somerville CR. Tetrad pollen formation in quartet mutants of Arabidopsis thaliana is associated with persistence of pectic polysaccharides of the pollen mother cell wall. The Plant Journal. 1998;15: 79–88. [DOI] [PubMed] [Google Scholar]
- 55.Francis KE, Lam SY, Copenhaver GP. Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiology. 2006;142: 1004–1013. 10.1104/pp.106.085274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rhee SY, Osborne E, Poindexter PD, Somerville CR. Microspore separation in the quartet 3 mutants of Arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiology. 2003;133: 1170–1180. 10.1104/pp.103.028266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grienenberger E, Kim SS, Lallemand B, Geoffroy P, Heintz D, Souza C de A, et al. Analysis of TETRAKETIDE α-PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. The Plant Cell. 2010;22: 4067–4083. 10.1105/tpc.110.080036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang J, Wu J, Romanovicz D, Clark G, Roux SJ. Co-regulation of exine wall patterning, pollen fertility and anther dehiscence by Arabidopsis apyrases 6 and 7. Plant Physiology and Biochemistry. 2013;69: 62–73. 10.1016/j.plaphy.2013.04.022 [DOI] [PubMed] [Google Scholar]
- 59.Leung KC, Li HY, Mishra G, Chye ML. ACBP4 and ACBP5, novel Arabidopsis acyl-CoA-binding proteins with kelch motifs that bind oleoyl-CoA. Plant Molecular Biology. 2004;55: 297–309. 10.1007/s11103-004-0642-z [DOI] [PubMed] [Google Scholar]
- 60.Hsiao AS, Yeung EC, Ye ZW, Chye ML. The Arabidopsis cytosolic acyl-CoA-binding proteins play combinatory roles in pollen development. Plant and Cell Physiology. 2015;56: 322–333. 10.1093/pcp/pcu163 [DOI] [PubMed] [Google Scholar]
- 61.Ariizumi T, Hatakeyama K, Hinata K, Sato S, Kato T, Tabata S, et al. A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Molecular Biology. 2003;53: 107–116. 10.1023/B:PLAN.0000009269.97773.70 [DOI] [PubMed] [Google Scholar]
- 62.Choi H, Ohyama K, Kim YY, Jin JY, Lee SB, Yamaoka Y, et al. The role of Arabidopsis ABCG9 and ABCG31 ATP binding cassette transporters in pollen fitness and the deposition of steryl glycosides on the pollen coat. The Plant Cell. 2014;26: 310–324. 10.1105/tpc.113.118935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mayfield JA, Preuss D. Rapid initiation of Arabidopsis pollination requires the oleosin-domain protein GRP17. Nature Cell Biology. 2000;2: 128–130. 10.1038/35000084 [DOI] [PubMed] [Google Scholar]
- 64.Li J, Yu M, Geng L-L, Zhao J. The fasciclin-like arabinogalactan protein gene, FLA3, is involved in microspore development of Arabidopsis: Arabidopsis FLA3 is involved in microspore development. The Plant Journal. 2010;64: 482–497. 10.1111/j.1365-313X.2010.04344.x [DOI] [PubMed] [Google Scholar]
- 65.Levitin B, Richter D, Markovich I, Zik M. Arabinogalactan proteins 6 and 11 are required for stamen and pollen function in Arabidopsis. The Plant Journal. 2008;56: 351–363. 10.1111/j.1365-313X.2008.03607.x [DOI] [PubMed] [Google Scholar]
- 66.Drakakaki G, Zabotina O, Delgado I, Robert S, Keegstra K, Raikhel N. Arabidopsis Reversibly Glycosylated Polypeptides 1 and 2 are essential for pollen development. Plant Physiology. 2006;142: 1480–1492. 10.1104/pp.106.086363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yadav V, Molina I, Ranathunge K, Castillo IQ, Rothstein SJ, Reed JW. ABCG transporters are required for suberin and pollen wall extracellular barriers in Arabidopsis. The Plant Cell. 2014;26: 3569–3588. 10.1105/tpc.114.129049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aya K, Ueguchi-Tanaka M, Kondo M, Hamada K, Yano K, Nishimura M, et al. Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. The Plant Cell. 2009;21: 1453–1472. 10.1105/tpc.108.062935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aya K, Hiwatashi Y, Kojima M, Sakakibara H, Ueguchi-Tanaka M, Hasebe M, et al. The Gibberellin perception system evolved to regulate a pre-existing GAMYB-mediated system during land plant evolution. Nature Communications. 2011;2: 544 10.1038/ncomms1552 [DOI] [PubMed] [Google Scholar]
- 70.Hird DL, Worrall D, Hodge R, Smartt S, Paul W, Scott R. The anther‐specific protein encoded by the Brassica napus and Arabidopsis thaliana A6 gene displays similarity to β‐1,3‐glucanases. The Plant Journal. 1993;4: 1023–1033. [DOI] [PubMed] [Google Scholar]
- 71.Wan L, Zha W, Cheng X, Liu C, Lv L, Liu C, et al. A rice β-1,3-glucanase gene Osg1 is required for callose degradation in pollen development. Planta. 2011;233: 309–323. 10.1007/s00425-010-1301-z [DOI] [PubMed] [Google Scholar]
- 72.Dong X, Feng H, Xu M, Lee J, Kim YK, Lim YP, et al. Comprehensive analysis of genic male sterility-related genes in Brassica rapa using a newly developed Br300K oligomeric chip. PLoS ONE. 2013;8: e72178 10.1371/journal.pone.0072178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li N, Zhang DS, Liu HS, Yin CS, Li X, Liang W, et al. The rice Tapetum Degeneration Retardation gene is required for tapetum degradation and anther development. The Plant Cell. 2006;18: 2999–3014. 10.1105/tpc.106.044107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang L, Ye Y, Zhang Y, Zhang A, Liu T, Cao J. BcMF9, a novel polygalacturonase gene, is required for both Brassica campestris intine and exine formation. Annals of Botany. 2009;104: 1339–1351. 10.1093/aob/mcp244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jiang J, Yao L, Yu Y, Liang Y, Jiang J, Ye N, et al. PECTATE LYASE-LIKE 9 from Brassica campestris is associated with intine formation. Plant Science. 2014;229: 66–75. 10.1016/j.plantsci.2014.08.008 [DOI] [PubMed] [Google Scholar]
- 76.Huang L, Cao JS, Zhang AH, Ye YQ. Characterization of a putative pollen-specific arabinogalactan protein gene, BcMF8, from Brassica campestris ssp. chinensis. Molecular Biology Reports. 2008;35: 631–639. 10.1007/s11033-007-9133-z [DOI] [PubMed] [Google Scholar]
- 77.Lin S, Dong H, Zhang F, Qiu L, Wang F, Cao J, et al. BcMF8, a putative arabinogalactan protein-encoding gene, contributes to pollen wall development, aperture formation and pollen tube growth in Brassica campestris. Annals of Botany. 2014;113: 777–788. 10.1093/aob/mct315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin S, Yue X, Miao Y, Yu Y, Dong H, Huang L, et al. The distinct functions of two classical arabinogalactan proteins BcMF8 and BcMF18 during pollen wall development in Brassica campestris. The Plant Journal. 2018;94: 60–76. 10.1111/tpj.13842 [DOI] [PubMed] [Google Scholar]
- 79.Feng X., Ni WM, Elge S, Mueller-Roeber B, Xu ZH, Xue HW. Auxin flow in anther filaments is critical for pollen grain development through regulating pollen mitosis. Plant Molecular Biology. 2006;61: 215–226. 10.1007/s11103-006-0005-z [DOI] [PubMed] [Google Scholar]
- 80.Cecchetti V, Altamura MM, Falasca G, Costantino P, Cardarelli M. Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. The Plant Cell. 2008;20: 1760–1774. 10.1105/tpc.107.057570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song S, Qi T, Huang H, Xie D. Regulation of stamen development by coordinated actions of jasmonate, auxin, and gibberellin in Arabidopsis. Molecular Plant. 2013;6: 1065–1073. 10.1093/mp/sst054 [DOI] [PubMed] [Google Scholar]
- 82.Quilichini TD, Douglas CJ, Samuels AL. New views of tapetum ultrastructure and pollen exine development in Arabidopsis thaliana. Annals of Botany. 2014;114: 1189–1201. 10.1093/aob/mcu042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Falasca G, D’Angeli S, Biasi R, Fattorini L, Matteucci M, Canini A, et al. Tapetum and middle layer control male fertility in Actinidia deliciosa. Annals of Botany. 2013;112: 1045–1055. 10.1093/aob/mct173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Canales C, Bhatt AM, Scott R, Dickinson H. EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Current Biology. 2002;12: 1718–1727. [DOI] [PubMed] [Google Scholar]
- 85.Zhao DZ, Wang GF, Speal B, Ma H. The EXCESS MICROSPOROCYTES1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes & Development. 2002;16: 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chang F, Wang Y, Wang S, Ma H. Molecular control of microsporogenesis in Arabidopsis. Current Opinion in Plant Biology. 2011;14: 66–73. 10.1016/j.pbi.2010.11.001 [DOI] [PubMed] [Google Scholar]
- 87.Shi J, Cui M, Yang L, Kim YJ, Zhang D. Genetic and biochemical mechanisms of pollen wall development. Trends in Plant Science. 2015;20: 741–753. 10.1016/j.tplants.2015.07.010 [DOI] [PubMed] [Google Scholar]
- 88.Hafidh S, Fíla J, Honys D. Male gametophyte development and function in angiosperms: a general concept. Plant Reproduction. 2016;29: 31–51. 10.1007/s00497-015-0272-4 [DOI] [PubMed] [Google Scholar]
- 89.Yurina NP, Odintsova MS. Signal transduction pathways of plant mitochondria: retrograde regulation. Russian Journal of Plant Physiology. 2010;57: 7–19. [Google Scholar]
- 90.Zhang W, Sun Y, Timofejeva L, Chen C, Grossniklaus U, Ma H. Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development. 2006;133: 3085–3095. 10.1242/dev.02463 [DOI] [PubMed] [Google Scholar]
- 91.Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, et al. Defective in Tapetal Development and Function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. The Plant Journal. 2008;55: 266–277. 10.1111/j.1365-313X.2008.03500.x [DOI] [PubMed] [Google Scholar]
- 92.Liu HX, Kang JG, Xie JM, Jian YC, Ding Y. Cloning and expression of an OguCMS-related anther-preferential transcription factor in Brassica oleracea. Acta Horticulturae Sinica. 2010;37: 1953–1960. [Google Scholar]
- 93.Zhang L, Kang Z, Liu H, Kang J. Cloning and expression of an Ogura cytoplasmic male sterile (OguCMS)-related MYB transcription factor in Brassica oleracea var. capitata. Journal of Agricultural Biotechnology. 2012;20: 627–635. [Google Scholar]
- 94.Kang J, Guo Y, Chen Y, Li H, Zhang L, Liu H. Upregulation of the AT-hook DNA binding gene BoMF2 in OguCMS anthers of Brassica oleracea suggests that it encodes a transcriptional regulatory factor for anther development. Molecular Biology Reports. 2014;41: 2005–2014. 10.1007/s11033-014-3048-2 [DOI] [PubMed] [Google Scholar]
- 95.Khan S, Stone JM. Arabidopsis thaliana GH3.9 influences primary root growth. Planta. 2007;226: 21–34. 10.1007/s00425-006-0462-2 [DOI] [PubMed] [Google Scholar]
- 96.Borg M, Brownfield L, Khatab H, Sidorova A, Lingaya M, Twell D. The R2R3 MYB transcription factor DUO1 activates a male germline-specific regulon essential for sperm cell differentiation in Arabidopsis. The Plant Cell. 2011;23: 534–549. 10.1105/tpc.110.081059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schnurr JA, Storey KK, Jung HJG, Somers DA, Gronwald JW. UDP-sugar pyrophosphorylase is essential for pollen development in Arabidopsis. Planta. 2006;224: 520–532. 10.1007/s00425-006-0240-1 [DOI] [PubMed] [Google Scholar]
- 98.Marquez J, Seoane‐Camba JA, Suarez‐Cervera M. The role of the intine and cytoplasm in the activation and germination processes of Poaceae pollen grains. Grana. 1997;36: 328–342. [Google Scholar]
- 99.Hesse M. Pollen wall stratification and pollination. Plant Systematics and Evolution. 2000;222: 1–17. [Google Scholar]
- 100.Ariizumi T, Toriyama K. Genetic regulation of sporopollenin synthesis and pollen exine development. Annual Review of Plant Biology. 2011;62: 1.1–1.24. [DOI] [PubMed] [Google Scholar]
- 101.Fei H, Sawhney VK. Ultrastructural characterization of male sterile33 (ms33) mutant in Arabidopsis affected in pollen desiccation and maturation. Canadian Journal of Botany. 2001;79: 118–129. [Google Scholar]
- 102.Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, et al. Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proceedings of the National Academy of Sciences. 2007;104: 15566–15571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huang L, Cao J, Zhang A, Ye Y, Zhang Y, Liu T. The polygalacturonase gene BcMF2 from Brassica campestris is associated with intine development. Journal of Experimental Botany. 2009;60: 301–313. 10.1093/jxb/ern295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Moon S, Kim SR, Zhao G, Yi J, Yoo Y, Jin P, et al. Rice GLYCOSYLTRANSFERASE1 encodes a glycosyltransferase essential for pollen wall formation. Plant Physiology. 2013;161: 663–675. 10.1104/pp.112.210948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ueda K, Yoshimura F, Miyao A, Hirochika H, Nonomura K-I, Wabiko H. COLLAPSED ABNORMAL POLLEN1 gene encoding the arabinokinase-like protein is involved in pollen development in rice. Plant Physiology. 2013;162: 858–871. 10.1104/pp.113.216523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jiang J, Yao L, Yu Y, Lv M, Miao Y, Cao J. PECTATE LYASE-LIKE 10 is associated with pollen wall development in Brassica campestris. J Integr Plant Biol. 2014;56: 1095–1105. 10.1111/jipb.12209 [DOI] [PubMed] [Google Scholar]
- 107.Xu D, Shi J, Rautengarten C, Yang L, Qian X, Uzair M, et al. Defective Pollen Wall 2 (DPW2) encodes an acyl transferase required for rice pollen development. Plant Physiology. 2017;173: 240–255. 10.1104/pp.16.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Showalter AM. Arabinogalactan-proteins: structure, expression and function. Cellular & Molecular Life Sciences. 2001;58: 1399–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Coimbra S, Costa M, Jones B, Mendes MA, Pereira LG. Pollen grain development is compromised in Arabidopsis agp6 agp11 null mutants. Journal of Experimental Botany. 2009;60: 3133–3142. 10.1093/jxb/erp148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li J, Yu M, Geng LL, Zhao J. The fasciclin-like arabinogalactan protein gene, FLA3, is involved in microspore development of Arabidopsis. The Plant Journal. 2010;64: 482–497. 10.1111/j.1365-313X.2010.04344.x [DOI] [PubMed] [Google Scholar]
- 111.Tsuchiya T, Toriyama K, Yoshikawa M, Ejiri S, Hinata K. Tapetum-specific expression of the gene for an endo-beta-1,3-glucanase causes male sterility in transgenic tobacco. Plant & Cell Physiology. 1995;36: 487–494. [DOI] [PubMed] [Google Scholar]
- 112.de Azevedo Souza C, Kim SS, Koch S, Kienow L, Schneider K, McKim SM, et al. A novel fatty Acyl-CoA Synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. The Plant Cell. 2009;21: 507–525. 10.1105/tpc.108.062513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen W, Yu XH, Zhang K, Shi J, De Oliveira S, Schreiber L, et al. Male Sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiology. 2011;157: 842–853. 10.1104/pp.111.181693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Morant M, Jorgensen K, Schaller H, Pinot F, Moller BL, Werck-Reichhart D, et al. CYP703 is an ancient Cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. The Plant Cell. 2007;19: 1473–1487. 10.1105/tpc.106.045948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dobritsa AA, Shrestha J, Morant M, Pinot F, Matsuno M, Swanson R, et al. CYP704B1 is a long-chain fatty acid ω-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiology. 2009;151: 574–589. 10.1104/pp.109.144469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li H, Pinot F, Sauveplane V, Werck-Reichhart D, Diehl P, Schreiber L, et al. Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. The Plant Cell. 2010;22: 173–190. 10.1105/tpc.109.070326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dobritsa AA, Lei Z, Nishikawa S, Urbanczyk-Wochniak E, Huhman DV, Preuss D, et al. LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiology. 2010;153: 937–955. 10.1104/pp.110.157446 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A-D) Scanning electron microscopy observation of floral organ morphology in the MF line. (E-H) Scanning electron microscopy observation of floral organs in the Ogura-CMS line showing normal morphology that are similar to those of the MF line. (A, E) The outer surface of a petal. (B, F) The outer surface of a sepal with numerous stomata (indicated by arrows). (C, G) The finger-like papillae of a mature stigma with a smooth surface. Arrows indicate the stomata on the style outer epidermis. (D, H) The outer surface of a nectary. Bars = 50 μm in (A, E), 100 μm in (B, F), 300 μm in (C, G), 200 μm in (D, H).
(JPG)
(A) External morphology of seedlings at 32 days after germination. (B) The fleshy roots thickening at 48 days after germination. (C) Morphological features of flowering plants at 180 days after germination. (D) The length and diameter of fleshy roots at 180 days after germination. The values are the mean ± SD (standard deviation). (E) Normal silique growth and seed set on an Ogura-CMS plant and a MF plant pollinated with MF pollen grains as the male donor. Bars = 5 cm.
(JPG)
(JPG)
From the outside in, the first circle of the Circos plot is a chromosome map of the Chinese cabbage genome. The homologous genes of all DEGs of turnip in Chinese cabbage are showed in the second circle. All DEGs are marked in the third circle.
(JPG)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
The RNA-Seq data were uploaded to the Sequence Read Archive of the National Center for Biotechnology Information (NCBI) (DOI: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA505114; accession number: PRJNA505114).