Abstract
Bioorthogonal strategies are continuing to pave the way for new analytical tools in biology. Although a significant amount of progress has been made in developing covalent reaction based bioorthogonal strategies, balanced reactivity, and stability are often difficult to achieve from these systems. Alternatively, despite being kinetically beneficial, the development of noncovalent approaches that utilize fully synthetic and stable components remains challenging due to the lack of selectivity in conventional noncovalent interactions in the living cellular environment. Herein, we introduce a bioorthogonal assembly strategy based on a synthetic host–guest system featuring Cucurbit[7]uril (CB[7]) and adamantylamine (ADA). We demonstrate that highly selective and ultrastable host–guest interaction between CB[7] and ADA provides a noncovalent mechanism for assembling labeling agents, such as fluorophores and DNA, in cells and tissues for bioorthogonal imaging of molecular targets. Additionally, by combining with covalent reaction, we show that this CB[7]–ADA based noncovalent interaction enables simultaneous bioorthogonal labeling and multiplexed imaging in cells as well as tissue sections. Finally, we show that interaction between CB[7] and ADA fulfills the demands of specificity and stability that is required for assembling molecules in the complexities of a living cell. We demonstrate this by sensitive detection of metastatic cancer-associated cell surface protein marker as well as by showing the distribution and dynamics of F-actin in living cells.
Developing strategies to selectively assemble bioorthogonal molecular components within the complexities of cells and tissues is of great interest in biology.1,2 It continues to drive advancements in various domains of fundamental and medical research, including protein modification,3−5 sensitive assay development,6 therapeutic targeting,7 and cell surface engineering.8 Additionally, such bioorthogonal strategies have become the key components in applications that require molecular tagging or labeling, such as imaging.9−14 To date, a variety of covalent approaches were developed for this purpose. Notable examples include the Staudinger ligation reaction,15 Cu(I) catalyzed or strain promoted azide–alkyne cycloaddition reaction (“click” chemistry),16,17 and inverse electron demand 1,2,4,5-tetrazine (Tz) ligation.18−20
Noncovalent interactions offer an alternative approach for bioorthogonal assembly with kinetic benefit. The association between noncovalent affinity pairs is typically diffusion controlled and proceeds at a significantly higher rate (kon ∼ 109–1010 M–1 s–1) as compared to their covalent counterparts (kon ∼ 1–104 M–1 s–1).21 Such fast kinetics offers important advantage to achieve efficient labeling when using low nanomolar (nM) concentration of a small and rapidly clearing probe, which is often the case in vivo. While kinetically beneficial, the use of noncovalent recognition motifs in biological labeling and imaging experiments is majorly restricted to naturally occurring biotin–streptavidin system.22 Biotin–streptavidin based systems suffer from few intrinsic drawbacks, including immunogenicity, background, and specificity issues from the endogenous presence of biotin. Additionally, large molecular weight of streptavidin (∼66 kDa) limits the potential application of this system in studies that require rapid diffusion (e.g., pretargeted nuclear imaging and therapy) or intracellular labeling in live cells. On the other hand, the use of synthetic recognition motifs, while a promising avenue, poses major challenges due to the chemical complexities of the living system and lack of selectivity in conventional noncovalent interactions. Recently, we reported a nanoparticle labeling approach for cancer cell detection via utilization of a β-cyclodextrin (β-CD) based synthetic binding pair.23 Although this method provided increased sensitivity in magnetic resonance-based detection, the method is restricted to multivalent nanoparticle based probes due to low affinity (Kd ∼ 10–4 M) and selectivity issues with β-CD based binding pairs.
In recent years, Cucurbit[n]uril (CB[n]), a family of synthetic macrocyclic hosts, has gained considerable attention due to their attractive molecular recognition property.24−32 It has led to the development of new studies in materials fabrication,33−39 bio-sensing,40−42 protein immobilization and separation,43−52 and drug delivery.53−60 Of this CB[n] family, the heptameric member CB[7] is specifically interesting because of their well-defined structure, superior water solubility, and biocompatibility, coupled with their ability to form highly selective and ultrastable host–guest inclusion complexes in aqueous medium.24,25,28 These remarkable attributes have motivated us to investigate whether CB[7] based synthetic host–guest platform could be used for fast and efficient noncovalent labeling of cells and tissue samples. Importantly, we envisioned that the high affinity and selectivity of the CB[7] host–guest system would be well suited for targeting molecules inside living cells. While this manuscript was under review, Kimoon Kim, Kyeng Min Park, and co-workers reported an elegant example of CB[7] based system for bioimaging application.61
Herein, we report a CB[7] based noncovalent tag for bioorthogonal labeling of cells and tissue samples. By attaching CB[7] with primary targeting agents (e.g., antibodies) and employing ADA conjugated fluorophore, we show that host–guest interactions between CB[7] and ADA provides an in situ noncovalent coupling mechanism for bioorthogonal imaging in cells. The CB[7]–ADA system displays exceptional serum stability and maintains high coupling efficiency even after incubation of the components at 37 °C for 16 h. We demonstrate the versatility of this labeling platform by employing the CB[7]–ADA interaction to achieve high-density DNA labeling for PAINT (points accumulation for imaging in nanoscale topography) based super-resolution imaging. Importantly, we show that high density labeling via CB[7]–ADA interaction helps to improve imaging resolution for visualizing structures that were impossible to resolve using biotin–streptavidin based labeling platform. We establish the translation of this noncovalent labeling platform to image target molecules in the complexities of tissue samples by using the Drosophila melanogaster model system. Additionally, we show that the CB[7]–ADA based noncovalent system provides a completely orthogonal labeling platform against the existing covalent system (e.g., Tz ligation) and thus their combination could be used for simultaneous labeling of multiple biomolecules within a single biological system. Finally, we establish the utility of CB[7]–ADA system to enable specific labeling of molecules in a live cell environment by imaging metastatic cancer-associated cell surface protein marker and showing distribution and dynamics of small molecule targeted F-actin in living cell.
Experimental Section
Antibody Modification with CB[7]
Secondary antibodies were purchased from commercial sources and passed through a Zeba spin column (Thermo Fisher Scientific) pre-equilibrated with PBS (pH 7.4) containing 10% 1 M NaHCO3. Antibodies were then incubated with tetrazine-NHS (Tz-NHS) at room temperature (RT) for 2 h. The conjugated product was purified using Zeba spin column pre-equilibrated with PBS (pH 7.4). CB[7]–PEG-TCO was synthesized by following the protocol mentioned in the Supporting Information. Afterward strain promoted cycloaddition reaction between antibody-Tz and CB[7]–PEG-TCO in PBS (pH 7.4) resulted in the formation of CB[7] conjugated antibodies. The conjugated product was characterized by matrix-assisted laser desorption ionization (MALDI) mass spectrometry.
Phalloidin Conjugation with CB[7]
Phalloidin amine was purchased from commercial source and dissolved in dry dimethylformamide (DMF). Then it was reacted with Tz-NHS in the presence of triethyl amine at RT for 3 h for formation of Tz conjugated phalloidin. Conjugated product was purified by reversed phase high-performance liquid chromatography (HPLC) using water/acetonitrile containing 0.1% trifluoroacetic acid (TFA) as eluent. The tetrazine-conjugated phalloidin was characterized by high-resolution mass spectrometry (HRMS). The purified product was reacted with CB[7]–PEG-TCO at room temperature in water for 12 h to obtain CB[7] conjugated phalloidin.
ADA Conjugation with Fluorophores (ADA Imager)
Commercially available NHS ester derivatives of fluorophores (Cy5 and Rhodamine) were conjugated with maleimide via amine-NHS ester coupling. The conjugated product was purified by reversed phase HPLC using water/acetonitrile containing 0.1% TFA as eluent. Maleimide conjugated fluorophores were characterized by HRMS. ADA was modified to a thiol derivative on one end for coupling with maleimide-modified fluorophores. A thiol-ene coupling reaction has been carried out between thiol modified ADA and maleimide conjugated fluorophores in PBS medium at RT for 12 h. Then ADA imagers were purified by HPLC using water/acetonitrile containing 0.1% TFA as eluent and characterized by HRMS. The imagers were dissolved in PBS and used in host–guest based imaging.
ADA Conjugation with DNA
ADA was conjugated with DNA through thiol–maleimide coupling conjugation reactions. At first, amine modified DNA was conjugated with NHS ester of maleimide to synthesize maleimide conjugated DNA. Conjugated product was purified using 0.1 M TEAA buffer (pH 7.4) as eluent and characterized by MALDI-MS. After that, DNA–maleimide was reacted with thiol modified ADA to get ADA conjugated DNA. We purified the conjugated product by using reversed phase HPLC and employing 0.1 M TEAA buffer (pH 7.4) as eluent. The conjugated product was characterized using MALDI-MS.
Optical Setup for Host–Guest Based Imaging in Structured Illumination Method
The fluorescence images were acquired using an inverted Zeiss ELYRA PS1 microscope in structured illumination mode. Two lasers (561 nm (200 mW) and 642 nm (150 mW)) have been used for respective excitation of fluorophores. 10% Laser power from the objective top was used for structured illumination imaging. Imaging was performed using a Zeiss oil-immersion objective (alpha Plan-apochromat DIC 63×/1.40 Oil DIC M27, numerical aperture (NA) 1.40 oil). Fluorescence light was spectrally filtered with emission filters (MBS-561+EF BP 570–650/LP 750 for laser line 561 and MBS-642+EF LP 655 for laser line 642) and imaged using a PCO edge sCMOS camera (quantum yield >70%). Structured illumination 2D and 3D images were processed using structured illumination analysis package for Zen software (Zeiss). Additional softwares have been used for color adjustment (ImageJ) and data analysis (Origin 9.0).
Immunostaining Protocol for CB[7]–ADA Interaction Based Imaging
Approximately 25 000 cells/well were plated in 8-well chamber slides and allowed to grow up to 60% confluence. Culture media was removed and cells were washed using PBS (pH 7.4). The cells were immediately fixed using suitable fixation methods (4% PFA for 15 min or chilled methanol for 7 min at −20 °C or 0.1% glutaraldehyde and 3% PFA for 10 min at RT). Afterward, cells were permeabilized using 0.25% triton X-100 for 10 min (this step is skipped for methanol fixation) and blocked using 3% BSA and 0.1% triton X-100 in PBS for 2 h at RT. Cells were then stained with 10 μg mL–1 primary antibody for 24 h at 4 °C. After removing excess primary antibody by washing with PBS, the cells were stained with 10 μg mL–1 CB[7] conjugated secondary antibodies for 2 h at RT. Excess secondary antibodies were removed by washing with PBS and ADA conjugated fluorophores were incubated for 30 min. Cells were subsequently used for host–guest based imaging after removing excess unbound fluorophores by washing with PBS.
DNA–PAINT Imaging Using Host–Guest Interaction Based Labeling
HeLa cells were used for the DNA–PAINT imaging. Methanol fixed cells were treated with primary and CB[7] conjugated secondary antibodies. Subsequently, ADA conjugated DNA docking strand was incubated with CB[7] tagged cells for 30 min. Gold nanoparticle (drift marker) was deposited on the imaging well by centrifuging the nanoparticle solution at 450 rcf for 3 min. Atto 655 conjugated DNA (1 nM) in PBS containing 500 mM NaCl was incubated with the cells for DNA–PAINT imaging. Single molecule blinking was recorded using a 642 nm excitation laser line. 50% Laser power has been used for image acquisition using a Zeiss oil-immersion TIRF objective (alpha Plan-apochromat DIC 100×/1.46 Oil DIC M27, NA 1.46). Fluorescence light was spectrally filtered with emission filter (MBS-642+EF LP 655 for laser line 642) and imaged on an electron-multiplying charge-coupled device (EMCCD) camera (Andor iXon DU897, quantum yield >90%, 512 × 512 pixels). EMCCD camera gain was kept at 50 during image acquisition. Exposure time of 100 ms was used for image acquisition. A total 10 000 frames were reconstructed to a super resolution image using Zen software package (ZEN 2.0).
Small Molecule Based Labeling of F-Actin
For labeling F-actin, fixed (4% PFA) and permeabilized cells were incubated with 2 μM solution of CB[7] conjugated phalloidin for 12 h at 4 °C. Excess phalloidin was removed and host–guest based imaging was performed after addition of ADA conjugated fluorophores and subsequent washing with PBS.
Multiplexed Imaging Protocol in Cells and Tissues
Cells or thoracic muscle tissues from adult flies were treated with primary antibodies against microtubule (or mitochondria). Then CB[7] conjugated secondary antibody (or CB[7] conjugated phalloidin) were treated for labeling of CB[7] in cells. The orthogonal pair, TCO conjugated secondary antibody was also added together with CB[7] secondary antibody. After removal of excess antibodies, ADA–fluorophores and Tz–fluorophores were added to the cells (or tissues). Structured illumination images were recorded after removal of excess unbound fluorophores.
Phalloidin Delivery and Host–Guest Based Live-Cell Microscopy
HeLa cells were used for the live-cell microscopy experiment. Approximately 25 000 cells/well were plated in 8-well chamber slides and allowed to grow up to 60% confluence over 18 h. Cell culture media was then removed and the cells were washed using DPBS (pH 7.4). Two μM concentration of phalloidin-CB[7] conjugate was assembled with HMD conjugated AuNP at a concentration ratio of 10:1. The assembly was subsequently added to the cells. Cells were then maintained at 37 °C with 5% CO2 in a humidified atmosphere for 3 h. After 3 h incubation, cells were washed twice with DPBS. The cells were immediately incubated with 100 nM solution of Rhodamine conjugated ADA imager and kept inside the microscope at 37 °C and 5% CO2 atmosphere for host–guest interaction-based imaging. Structured illumination images were recorded for hours with every 1 min interval.
Results and Discussion
Host–Guest Interaction Mediated Bioorthogonal Labeling in Fixed Cell
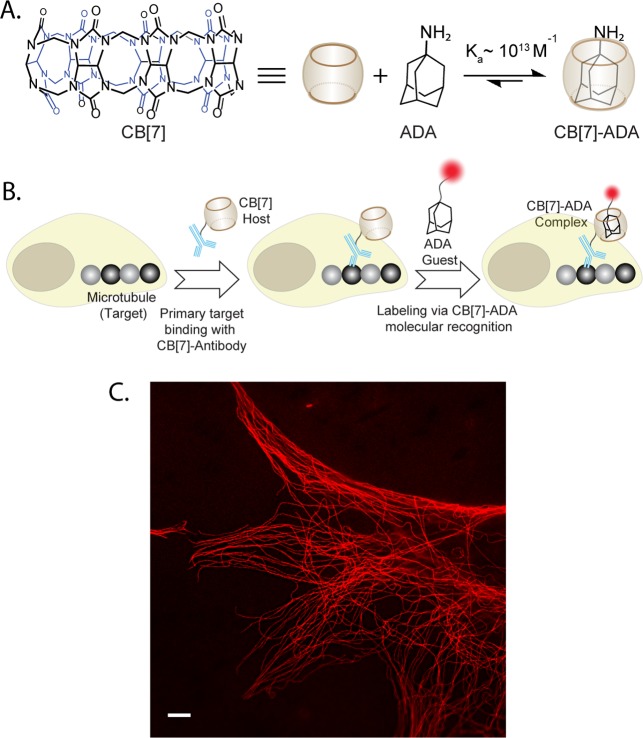
To probe the feasibility of the CB[7] based host–guest interaction as a tool for bioorthogonal labeling, we first used an antibody based targeting platform and fluorophore as an imaging agent. We selected ADA as the guest component for this study and harnessed its ability to form highly selective and ultrastable monovalent host–guest inclusion complex with CB[7] with a Kd of ∼10–13 M (Figure 1A).24 The microtubule structures in fixed mouse embryonic fibroblast (MEF) cells were targeted using CB[7] conjugated antibodies (CB[7]–Ab) and subsequently host–guest interactions between CB[7] and ADA were used as the in situ coupling mechanism between fluorophores and antibodies (Figure 1B). Antibodies were covalently conjugated with CB[7] through utilization of photochemically functionalized CB[7] (Scheme S1). Successful antibody conjugation was verified by MALDI-MS analysis (Figure S5). To evaluate the effectiveness of the labeling, Structured Illumination Microscopy (SIM) images were acquired after coupling CB[7] labeled microtubules with ADA conjugated Cy5 dye (ADA–Cy5) through CB[7]–ADA host–guest interaction. Figure 1C shows SIM image from MEF cells that was acquired with a 642 nm excitation laser. Notably, the CB[7]–ADA interaction based labeling was highly selective to the microtubule structure with minimal nonspecific binding. In addition, the labeling was stable, as multiple washing cycles did not result in loss of signal intensity (Figure S26). Overall, these results demonstrate that highly selective and ultrastable host–guest interactions between CB[7] and ADA offers a novel noncovalent coupling platform for bioorthogonal labeling of target molecules in cellular complexities.
Figure 1.
Host–guest interaction mediated bioorthogonal labeling in cells. (A) Structure of CB[7] and ADA. (B) Schematic showing the strategy for noncovalent labeling. Microtubules were tagged with CB[7] using CB[7]–antibody. Fluorophore conjugated ADA finds its high affinity host through molecular recognition, resulting in specific fluorescent labeling of microtubules. (C) SIM image of microtubules from CB[7]–ADA labeling platform. Scale bar: 5 μm.
In Situ Labeling Kinetics and Stability of Host–Guest Bioorthogonal Platform
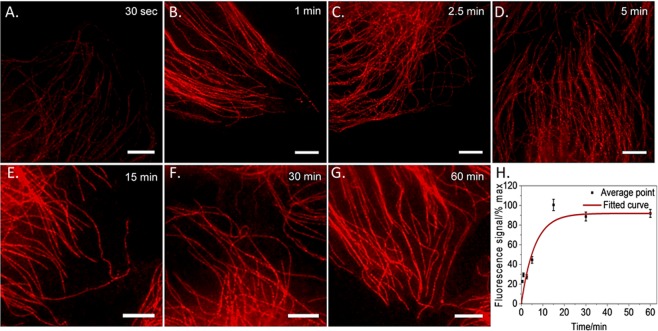
We characterized in situ binding kinetics of CB[7]–ADA based labeling via time–lapse fluorescence imaging. After targeting microtubules with CB[7]–Ab, cells were incubated with ADA–Cy5 for different time duration. Figure 2A–G shows time–lapse fluorescence images that confirms fast CB[7]–ADA mediated fluorophore binding with fluorescence signal reaching saturation in less than 15 min. Notably, it is clearly evident from Figure 2A that incubation for only 60 s with 250 nM ADA–Cy5 was sufficient to clearly visualize microtubule structure, indicating fast and efficient target labeling with ADA–Cy5. We also observed that increasing imager concentration to 1 μM resulted in efficient labeling even with 5 s of incubation time (Figure S27). Next, we tested whether exceptionally high association rate between CB[7] and ADA would allow efficient labeling and no-wash imaging at low nM imager concentration. Three targets, including microtubules, mitochondria, and actin, were imaged using CB[7]–ADA mediated interaction. We observed that the use of 5–10 nM ADA imager provides sufficient probe accumulation at the target site, enabling high-contrast and real-time imaging of target biomolecules without employing washing step (Figures S28 and S29). Taken together, these results shows that extremely fast, highly selective, and ultrastable host–guest interactions between CB[7] and ADA offers a novel noncovalent coupling platform for bioorthogonal labeling of target molecules at low nanomolar probe concentration.
Figure 2.
In situ binding kinetics of CB[7]–ADA based labeling platform. (A–G) Structured illumination imaging of microtubule after incubation of the CB[7] targeted cells with 250 nM ADA–Cy5 for different time duration. (H) Plot of fluorescence intensity vs time indicates saturation of labeling arises within 15 min of incubation with ADA–Cy5 imager. Scale bar 2 μm (A–G).
Coupling platforms that provide high reactivity while maintaining stability are the most ideally suited for biological studies.62 We subsequently conducted a physiologically relevant stability experiment with CB[7]–ADA system and compared it with a frequently used Tz based covalent system. Reactive reagents were incubated in serum-supplemented culture media at 37 °C. Microtubule labeling efficiency was evaluated after 16 h of incubation via fluorescence imaging. Figures S30 and S31 show that CB[7]–ADA was able to maintain high-efficiency labeling while Tz ligation-based reagents showed significant deterioration, clearly indicating the higher stability of the synthetic host–guest system under biological conditions.
pH is an important consideration in labeling experiment as it varies between different intracellular organelles. For example, pH of lysosome is ∼4.5 whereas pH of mitochondria is ∼8. To understand the effect of pH, we performed CB[7]–ADA host–guest interaction based labeling experiment under various pH conditions ranging from 4.5 to 9.2. Fluorescence microscopy images clearly showed the specific and efficient labeling of microtubules under all different pH conditions (Figure S32). Besides pH, high labeling efficiency was observed when CB[7]–ADA interaction mediated coupling was performed in various cell culture media (Figure S33). These results indicate the efficacy of CB[7]–ADA interaction under stringent labeling conditions.
CB[7]-ADA Based DNA Labeling and Super-Resolution DNA–PAINT Imaging
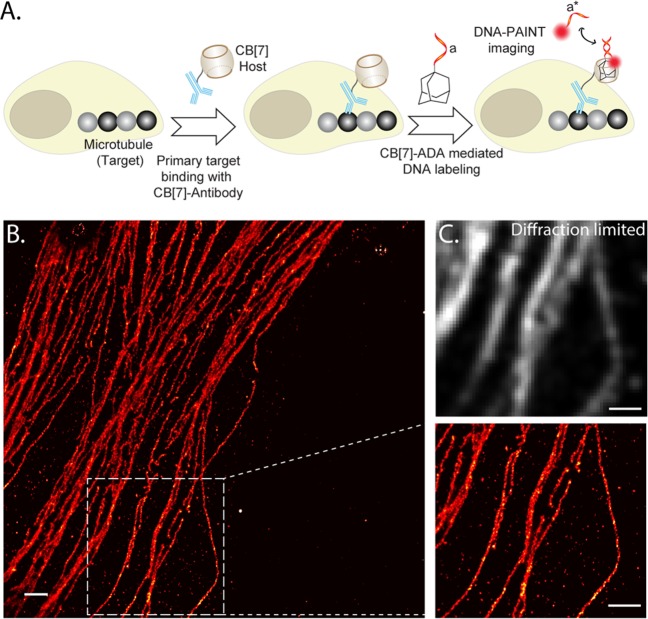
In recent years, labeling biomolecules with DNA has gained considerable interest due to its utility in various emerging technologies, including immuno-PCR and super-resolution imaging.63 To demonstrate the versatility of the CB[7]–ADA based labeling platform, we choose DNA as a second labeling agent. A 11 NT DNA sequence was selected for this labeling study with a goal to perform DNA-PAINT based super-resolution imaging of biomolecules. To label biomolecules with DNA via CB[7]–ADA based interaction, we synthesized ADA functionalized DNA strand (ADA–DNA) via thiol–maleimide coupling (Scheme S8). Similar to the case of fluorophore, we targeted microtubule using CB[7]–Ab and subsequently incubated the cells with ADA–DNA for specific DNA immobilization on microtubule via CB[7]–ADA interactions (Figure 3A). To test super-resolution imaging capability, we carried out single-molecule imaging using complementary ATTO655-conjugated DNA strand. Figure 3A shows the super-resolved image that was reconstructed from DNA–PAINT imaging. A significant increase in resolution as compared to the diffraction-limited image could be easily observed by visualizing a dense microtubule region (Figure 3B). To highlight the labeling quality, we measured the cross-sectional profile of localization of a single microtubule filament. Importantly, as shown in the magnified view (Figure S34), we were able to resolve the hollow tubular structure of the filament having diameter of ∼42 nm, while previously demonstrated DNA–PAINT imaging using biotin–streptavidin mediated DNA labeling was unable to resolve this hollow tubular structure.62 This underlines the importance of the high labeling density and efficiency of CB[7]–ADA based coupling method, as both of these parameters significantly contribute toward achieving improved spatial resolution.
Figure 3.
Host–guest interactions mediated DNA labeling and DNA–PAINT super-resolution imaging. (A) Schematic showing the strategy for noncovalent DNA labeling via CB[7]–ADA interaction. The installed DNA tag was used as docking sites for DNA–PAINT based imaging. (B) Super-resolved image of microtubules using CB[7]–ADA interaction mediated labeling and DNA–PAINT imaging. Magnified image of the boxed region clearly shows the distribution of closely spaced microtubules. (C) Diffraction limited image of the same magnified region shows indistinguishable microtubules. Scale bar: 1 μm (B) and 500 nm (magnified view and part C).
Host–Guest Labeling in Tissue
After successful demonstration of the CB[7]–ADA based noncovalent labeling in cells, we progressed to study the utility of this labeling method in tissue samples. The vast diversity and complexity existing within tissue specimens presents a significant challenge for noncovalent labeling tools to operate with high selectivity. For evaluating the CB[7]–ADA based labeling method, we have chosen ovary tissues of Drosophila melanogaster model and specifically labeled the actin cytoskeleton. Phalloidin, a bicyclic heptapeptide, was used as a highly specific small molecule binder for F-actin. Approximately 30 μm thick ovary tissue sections were dissected, fixed and stained with CB[7] conjugated phalloidin derivative by overnight incubation. Excess CB[7]–phalloidin was removed by washing and ADA–Cy5 imager was added before mounting on microscopic slide. SIM image (Figure 4) clearly shows the fluorescently labeled actin network in the complex ovary tissue, which is wrapped around by a layer of surrounding muscle tissue. Importantly, a negligible amount of nonspecifically bound probes were observed from the SIM image. The specificity in actin staining demonstrates the ability of ADA imager to selectively find its CB[7] binding partner even in the complexities of tissues. Additionally, primary target labeling using phalloidin probe establishes the small molecule imaging potential of CB[7]–ADA based labeling platform.
Figure 4.
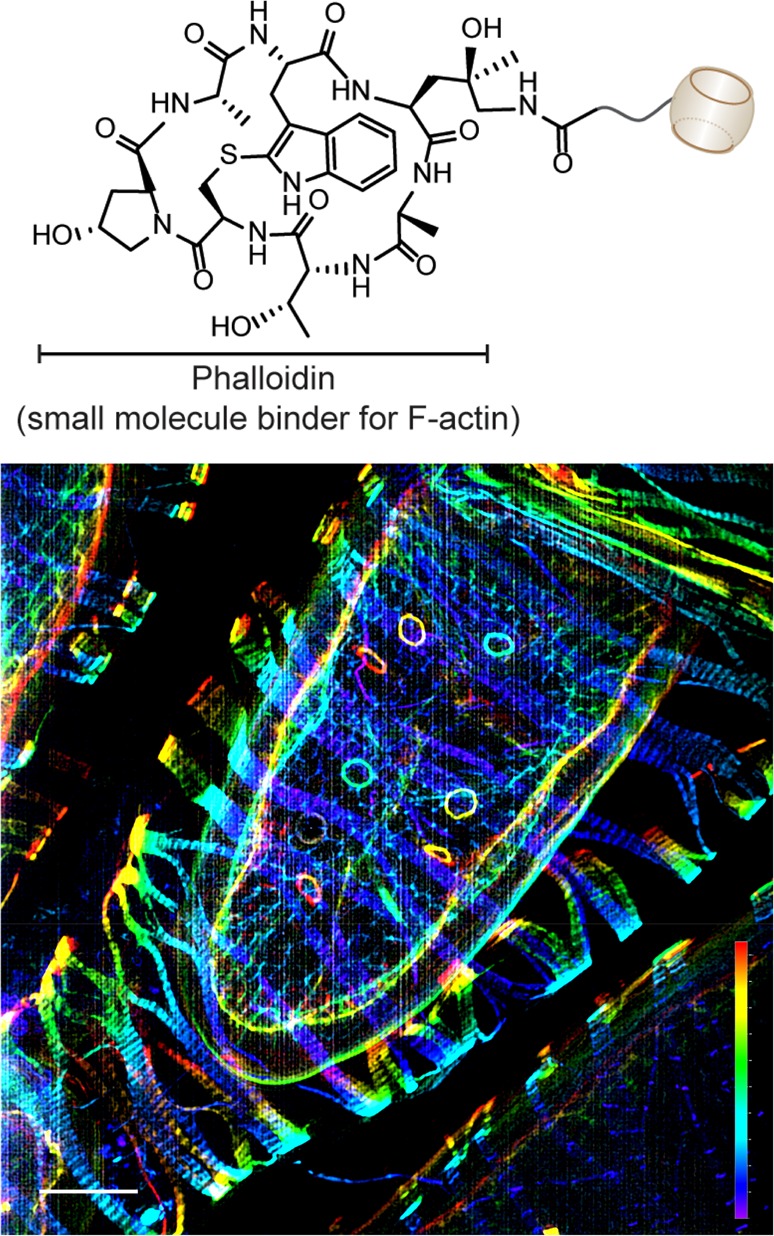
CB[7]–ADA interaction mediated labeling and imaging of actin filaments in the ovary tissue and the muscle sheath enveloping the egg chamber of Drosophila model system. Actin filaments were targeted using CB[7] conjugated phalloidin derivative. Scale bar: 10 μm (xy) and 17.3 μm (z depth).
Multiplexed Bioorthogonal Labeling in Cells and Tissue
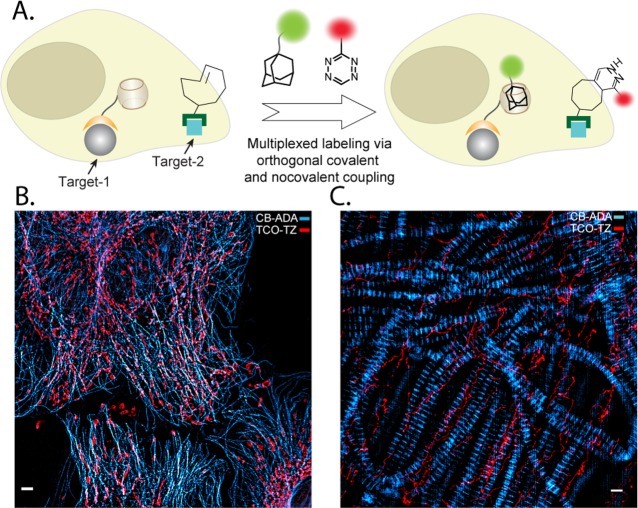
Modern biochemical studies often require simultaneous tracking of multiple biomolecules within a single system. These studies demand applications of two bioorthogonal pairs that are mutually orthogonal. Although a variety of covalent bioorthogonal reaction exists, often time cross-reactivity between several bioorthogonal covalent reagents limits their potential application in multiplexed experiments.17 The CB[7]–ADA based labeling method presents a new opportunity for multiplexed labeling as this noncovalent interaction based bioorthogonal labeling approach is orthogonal to the existing covalent approaches. To demonstrate proof-of-principle validation, we used Tz–TCO ligation as a covalent component along with CB[7]–ADA based noncovalent interaction for simultaneous labeling and imaging of multiple target molecules in cells and tissue sections (Figure 5A). In cells, microtubules were stained using CB[7] conjugated antibodies and mitochondria were stained using TCO conjugated antibodies. The corresponding imager strands, selected for dual-color imaging, are ADA conjugated Rhodamine and Tz conjugated Cy5. We acquired dual-color SIM images after incubating the cells with dye modified binding partners for 15 min. Figure 5B shows that microtubule network has strong fluorescence signal from Rhodamine, whereas mitochondrial compartment has strong signal from Cy5. This suggests that ADA–Rhodamine and Tz–Cy5 are able to assemble selectively with CB[7] and TCO, respectively. Similar experiments performed in Drosophila muscle tissue with CB[7]–ADA targeting actin and Tz–TCO targeting microtubule also showed specific dual color labeling (Figure 5C). Additionally, we show that integrating more number of orthogonal labeling pairs can effectively increase the multiplexing power. To demonstrate this, we performed three color multiplexed imaging using one noncovalent (CB[7]–ADA) and two covalent pairs (Tz–TCO and azide–dibenzocyclooctyne (DBCO)). We targeted three intracellular proteins by these three orthogonal pairs: CB[7]–antibody to microtubule, TCO–antibody to mitochondria, and DBCO–phalloidin to actin filaments. ADA–rhodamine, Tetrazine–Cy5, and azide–bodipy were used as the recognition pairs for target visualization. As shown in Figure S35, multicolour imaging performed via SIM indicated specific labeling of all three targeted intracellular proteins with negligible cross reactivity. These experiments demonstrate that CB[7]–ADA based noncovalent interaction along with covalent bioorthogonal reaction provide excellent mutual orthogonality to enable simultaneous labeling and multiplexed imaging in biological complexities.
Figure 5.
Multiplexed labeling. (A) Schematic showing the use of CB[7]–ADA and TCO–Tz orthogonal labeling platforms for simultaneous labeling and multiplexed imaging. (B) Multiplexed SIM imaging of microtubules and mitochondria. Microtubules and mitochondria were labeled using CB[7] and TCO conjugated antibodies, respectively. Multiplexed images were captured after addition of ADA–Rhodamine and Tz–Cy5. (C) Multiplexed imaging of actin and microtubule in thoracic muscle tissue of Drosophila using orthogonal labeling pairs. Actin was targeted using phalloidin–CB[7] and microtubule was targeted using antibody–TCO conjugate. Scale bars: 5 μm (parts B and C).
To achieve multiplexed imaging by only applying CB[7]–ADA based interaction, we devised a sequential multiplexing approach. In this case targets were sequentially labeled (via CB[7]–ADA interaction), imaged, and bleached to finally achieve a multiplexed readout. For this purpose, initially we targeted mitochondria using CB[7] conjugated antibody and imaged using ADA–Cy5 fluorophore. Then the mitochondria labeled fluorophores were photobleached by illuminating high intensity laser for 30 s. Next, microtubules were labeled and imaged using antibody–CB[7] followed by ADA–Cy5 incubation. Finally, actin was labeled by using phalloidin–CB[7] and ADA–Cy5 after photobleaching of microtubule labeled fluorophores (Figure S36). This sequential labeling approach by using only a single dye demonstrate the effectiveness of CB[7]–ADA based labeling approach for higher order multiplexed imaging.
Live Cell Bioorthogonal Imaging via Host–Guest Mediated Interaction
We next investigated the compatibility of CB[7]–ADA interaction for labeling in live cell environment by imaging a vascular adhesion protein (VCAM) that localizes on the cell membrane. VCAM was selected as our potential target given its clinical importance as an important marker for determining ovarian cancer metastasis and its therapeutic response. Bioorthogonal noncovalent labeling of VCAM expressing SVEC (Saphenous Vein Endothelial Cell) cell was carried out by using CB[7]–Ab followed by incubation with ADA–Cy5 imager. The SIM image that was captured from live cells showed fluorescence from cell membrane, indicating specific staining of VCAM via CB[7]–ADA mediated interaction (Figure 6A,B). To support the specificity in the labeling process, two control experiments were performed, where we first omitted the primary antibody from the labeling scheme and second cells were treated with ADA–Cy5 alone. Both of these control experiments resulted in the loss of fluorescence from cell membrane as observed via fluorescence microscopy (Figures S37 and S38). We further used Fluorescence Activated Cell Shorting (FACS) to quantitatively analyze the live cell labeling data (Figure S39). Strong fluorescence response in the presence of CB[7] targeting conjugate and ADA imager agent as compared to the controls clearly demonstrate the specificity of the ADA imaging agent for CB[7]-modified antibody in the presence of live cells and serum environment.
Figure 6.
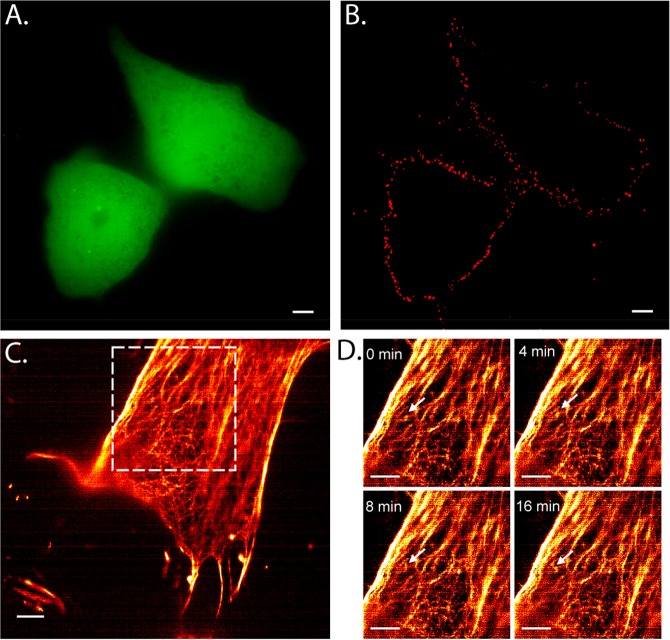
Live cell imaging using CB[7]–ADA interaction. (A) RFP (pseudo-colored green) transfected SVEC cells. (B) Live cell imaging of VCAM in SVEC cell surface using CB[7]–ADA interaction. Cells were imaged after exciting with a 642 nm laser line (corresponding to Cy5 excitation). Cells were treated with primary (against VCAM) and CB[7] modified secondary antibodies. Imaging was carried out after incubation with ADA–Cy5 imager. (C) Intracellular labeling of actin filaments in live HeLa cells using phalloidin–CB[7] and ADA–Rhodamine imager. (D) Actin dynamics were indicated in the arrowed region from the boxed region in part C. Scale bars: 5 μm (A–C) and 2 μm (D).
The demand for precise control over selectivity is amplified when it is necessary to perform coupling experiment in highly complex chemical environments, such as those found in intracellular environment of a living cell. To evaluate the potential of CB[7]–ADA based bioorthogonal chemistry for live intracellular labeling, we imaged F-actin using phalloidin–CB[7] derivative in live HeLa cells. The labeling of F-actin in live cell was carried out by gold nanoparticle assisted delivery of phalloidin–CB[7] conjugates inside live cell (Figure S40). While previous studies indicated the cell permeable nature of CB[7],41,42,60,63−65 nanoparticle assistance was necessary for this study presumably due to cell impermeable nature of phalloidin or the larger size of the resultant phalloidin–CB[7] complex. Gold nanoparticle surface was functionalized with hexamethylene diamine (HMD) ligand through a biocompatible tetraethylene glycol linker. Noncovalent interaction between HMD and CB[7] drives phalloidin–CB[7] to enter intracellular environment and interact with F-actin. Two μM phalloidin–CB[7] labeled the actin fibers sufficiently with the aid of 200 nM gold nanoparticles. 100 nM ADA–Rhodamine was used to acquire SIM images of F-actin in living cells. It should be noted that HMD guest binds with CB[7] with an affinity of ∼10–7 M (Kd) whereas ADA possess an affinity of ∼10–13 M (Kd) toward CB[7]. Therefore, the initial complex between HMD–CB[7] will be disassembled by presentation of the orthogonal guest molecule ADA, which has relatively high affinity for CB[7]. This guest exchange process leads to the formation of more stable CB[7]–ADA complex, leading to efficient target labeling. SIM image, shown in Figure 6C, clearly visualizes the high-density actin networks, reflecting the ability of ADA to specifically find its binding partner in a living intracellular environment. To study the dynamic nature of actin, time-lapse SIM images were recorded over 36 min with 1-min interval (Figure 6D and Supporting Movie 1). 0, 4, 8, and 16 min time point images show distribution of actin filaments over the cell, whereas the arrowed region in each frame shows the dynamic nature of actin filaments (Figure 6D). This study illustrates that the interaction between CB[7] and ADA satisfies the demands of specificity and stability that is required for assembling molecules in the complexities of a living cell and serum environment.
Conclusions
In conclusion, we have introduced a fully synthetic and noncovalent CB[7] based tag for fast and efficient bioorthogonal assembly in cells and tissues. Benefiting from the extremely high association rate via noncovalent interactions, the CB[7] tag allows efficient labeling at low nM probe concentration and enables microscopic investigation without excess probe washing. The CB[7] tag is highly stable in biological media under physiologically relevant temperature. It is capable of maintaining strong selectivity and affinity toward ADA even in the complexities of a living intracellular environment, and this feature can be used to perform live-cell imaging. CB[7]–ADA based labeling not only works well in biological systems but also works well with existing covalent bioorthogonal chemistries. This mutual orthogonality provides ability to perform multiplexed labeling. CB[7] tag provides a synthetic alternative to the biotin–streptavidin system with features that are often more appropriate for biological studies, such as pretargeted in vivo imaging, where efficient labeling is desirable using small and rapidly clearing probe. We expect that this versatile noncovalent platform will find application in various studies, including cell surface engineering, nanoparticle targeting, and creating macroscale cellular assembly.
Acknowledgments
This work is supported by a Wellcome Trust-DBT India Alliance Intermediate Fellowship (Grant IA/I/16/1/502368), a DBT Innovative Young Biotechnologist Award (IYBA) (Grant BT/09/IYBA/2015/15), a SERB Early Career Research Award (Grant ECR/2016/002052), a DAE-BRNS Young Scientist’s Research Award (YSRA) (Grant 37(1)/20/01/2017-BRNS), and a JNCASR Startup Fund to S.S.A. R.S. acknowledges support from CSIR for Graduate Research Fellowship. M.P. acknowledges support from UGC for a Graduate Research Fellowship. We acknowledge MBGU (JNCASR) for helping with the FACS analysis.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.8b01851.
Author Contributions
R.S. and S.S.A. conceived the study, designed and performed the experiments, analyzed and interpreted the data, and wrote the manuscript. N.D.S. prepared cells for the imaging experiment and reviewed the manuscript. M.P. prepared the tetrazine derivative. D.J. and M.S.I. helped with the cell culture experiments. S.R. and S.V. helped with the tissue imaging experiments. S.S.A. supervised the overall study.
The authors declare the following competing financial interest(s): A patent has been filed based on this work.
Supplementary Material
References
- McKay C. S.; Finn M. G. Chem. Biol. 2014, 21, 1075–1101. 10.1016/j.chembiol.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sletten E. M.; Bertozzi C. R. Angew. Chem., Int. Ed. 2009, 48, 6974–6998. 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd A. K.; Valls Cuevas J. M.; Devaraj N. K. J. Am. Chem. Soc. 2015, 137, 4884–4887. 10.1021/jacs.5b00040. [DOI] [PubMed] [Google Scholar]
- Li J.; Yu J.; Zhao J.; Wang J.; Zheng S.; Lin S.; Chen L.; Yang M.; Jia S.; Zhang X.; Chen P. R. Nat. Chem. 2014, 6, 352. 10.1038/nchem.1887. [DOI] [PubMed] [Google Scholar]
- Tian Y.; Jacinto M. P.; Zeng Y.; Yu Z.; Qu J.; Liu W. R.; Lin Q. J. Am. Chem. Soc. 2017, 139, 6078–6081. 10.1021/jacs.7b02615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budin G.; Chung H. J.; Lee H.; Weissleder R. Angew. Chem., Int. Ed. 2012, 51, 7752–7755. 10.1002/anie.201202982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H.; Lee S.; Na J. H.; Kim S. H.; Hahn S. K.; Choi K.; Kwon I. C.; Jeong S. Y.; Kim K. Angew. Chem., Int. Ed. 2012, 51, 11836–11840. 10.1002/anie.201206703. [DOI] [PubMed] [Google Scholar]
- Pulsipher A.; Dutta D.; Luo W.; Yousaf M. N. Angew. Chem., Int. Ed. 2014, 53, 9487–9492. 10.1002/anie.201404099. [DOI] [PubMed] [Google Scholar]
- Lang K.; Davis L.; Wallace S.; Mahesh M.; Cox D. J.; Blackman M. L.; Fox J. M.; Chin J. W. J. Am. Chem. Soc. 2012, 134, 10317–10320. 10.1021/ja302832g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y.; Jiang X.; Yuan R.; Zhou Y.; Ji C.; Yang L.; Chen H.; Wang Q. Anal. Chem. 2017, 89, 538–543. 10.1021/acs.analchem.6b04141. [DOI] [PubMed] [Google Scholar]
- Wu H.; Cisneros B. T.; Cole C. M.; Devaraj N. K. J. Am. Chem. Soc. 2014, 136, 17942–17945. 10.1021/ja510839r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Qian L.; Li L.; Bernhammer J. C.; Huynh H. V.; Lee J. S.; Yao S. Q. Angew. Chem., Int. Ed. 2016, 55, 3254–3254. 10.1002/anie.201600314. [DOI] [PubMed] [Google Scholar]
- Huang L.-L.; Liu K.; Zhang Q.; Xu J.; Zhao D.; Zhu H.; Xie H.-Y. Anal. Chem. 2017, 89, 11620–11627. 10.1021/acs.analchem.7b03043. [DOI] [PubMed] [Google Scholar]
- Cheng M.; Zhang W.; Yuan J.; Luo W.; Li N.; Lin S.; Yang Y.; Fang X.; Chen P. R. Chem. Commun. 2014, 50, 14724–14727. 10.1039/C4CC02804J. [DOI] [PubMed] [Google Scholar]
- van Berkel S. S.; van Eldijk M. B.; van Hest J. C. M. Angew. Chem., Int. Ed. 2011, 50, 8806–8827. 10.1002/anie.201008102. [DOI] [PubMed] [Google Scholar]
- Jiang H.; López-Aguilar A.; Meng L.; Gao Z.; Liu Y.; Tian X.; Yu G.; Ovryn B.; Moremen K. W.; Wu P. Angew. Chem., Int. Ed. 2018, 57, 967–971. 10.1002/anie.201706535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson D. M.; Nazarova L. A.; Prescher J. A. ACS Chem. Biol. 2014, 9, 592–605. 10.1021/cb400828a. [DOI] [PubMed] [Google Scholar]
- Patterson D. M.; Nazarova L. A.; Xie B.; Kamber D. N.; Prescher J. A. J. Am. Chem. Soc. 2012, 134, 18638–18643. 10.1021/ja3060436. [DOI] [PubMed] [Google Scholar]
- Seitchik J. L.; Peeler J. C.; Taylor M. T.; Blackman M. L.; Rhoads T. W.; Cooley R. B.; Refakis C.; Fox J. M.; Mehl R. A. J. Am. Chem. Soc. 2012, 134, 2898–2901. 10.1021/ja2109745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez A.; Dzijak R.; Dračínský M.; Rampmaier R.; Siegl S. J.; Vrabel M. Angew. Chem., Int. Ed. 2017, 56, 1334–1337. 10.1002/anie.201610491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Casado A.; Dam H. H.; Yilmaz M. D.; Florea D.; Jonkheijm P.; Huskens J. J. Am. Chem. Soc. 2011, 133, 10849–10857. 10.1021/ja2016125. [DOI] [PubMed] [Google Scholar]
- Liu W.; Samanta S. K.; Smith B. D.; Isaacs L. Chem. Soc. Rev. 2017, 46, 2391–2403. 10.1039/C7CS00011A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agasti S. S.; Liong M.; Tassa C.; Chung H. J.; Shaw S. Y.; Lee H.; Weissleder R. Angew. Chem., Int. Ed. 2012, 51, 450–454. 10.1002/anie.201105670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow S. J.; Kasera S.; Rowland M. J.; del Barrio J.; Scherman O. A. Chem. Rev. 2015, 115, 12320–12406. 10.1021/acs.chemrev.5b00341. [DOI] [PubMed] [Google Scholar]
- Assaf K. I.; Nau W. M. Chem. Soc. Rev. 2015, 44, 394–418. 10.1039/C4CS00273C. [DOI] [PubMed] [Google Scholar]
- Lazar A. I.; Biedermann F.; Mustafina K. R.; Assaf K. I.; Hennig A.; Nau W. M. J. Am. Chem. Soc. 2016, 138, 13022–13029. 10.1021/jacs.6b07655. [DOI] [PubMed] [Google Scholar]
- Ma X.; Zhao Y. Chem. Rev. 2015, 115, 7794–7839. 10.1021/cr500392w. [DOI] [PubMed] [Google Scholar]
- Stoffelen C.; Voskuhl J.; Jonkheijm P.; Huskens J. Angew. Chem., Int. Ed. 2014, 53, 3400–3404. 10.1002/anie.201310829. [DOI] [PubMed] [Google Scholar]
- Nguyen H. D.; Dang D. T.; van Dongen J. L. J.; Brunsveld L. Angew. Chem., Int. Ed. 2010, 49, 895–898. 10.1002/anie.200904413. [DOI] [PubMed] [Google Scholar]
- Yan B.; Yesilbag Tonga G.; Hou S.; Fedick P. W.; Yeh Y.-C.; Alfonso F. S.; Mizuhara T.; Vachet R. W.; Rotello V. M. Anal. Chem. 2014, 86, 6710–6714. 10.1021/ac501682y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Yun G.; Khan S.; Kim J.; Murray J.; Lee Y. M.; Kim W. J.; Lee G.; Kim S.; Shetty D.; Kang J. H.; Kim J. Y.; Park K. M.; Kim K. Mater. Horiz. 2017, 4, 450–455. 10.1039/C7MH00038C. [DOI] [Google Scholar]
- Kang Y.; Tang X.; Yu H.; Cai Z.; Huang Z.; Wang D.; Xu J.-F.; Zhang X. Chem. Sci. 2017, 8, 8357–8361. 10.1039/C7SC04125J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Yu Y.; Gao J.; Wang Z.; Zhang X. Angew. Chem., Int. Ed. 2010, 49, 6576–6579. 10.1002/anie.201002415. [DOI] [PubMed] [Google Scholar]
- Rauwald U.; Scherman O. A. Angew. Chem. 2008, 120, 4014–4017. 10.1002/ange.200705591. [DOI] [PubMed] [Google Scholar]
- An Q.; Brinkmann J.; Huskens J.; Krabbenborg S.; de Boer J.; Jonkheijm P. Angew. Chem., Int. Ed. 2012, 51, 12233–12237. 10.1002/anie.201205651. [DOI] [PubMed] [Google Scholar]
- Sankaran S.; de Ruiter M.; Cornelissen J. J. L. M.; Jonkheijm P. Bioconjugate Chem. 2015, 26, 1972–1980. 10.1021/acs.bioconjchem.5b00419. [DOI] [PubMed] [Google Scholar]
- Neirynck P.; Brinkmann J.; An Q.; van der Schaft D. W. J.; Milroy L.-G.; Jonkheijm P.; Brunsveld L. Chem. Commun. 2013, 49, 3679–3681. 10.1039/c3cc37592g. [DOI] [PubMed] [Google Scholar]
- Sankaran S.; Cavatorta E.; Huskens J.; Jonkheijm P. Langmuir 2017, 33, 8813–8820. 10.1021/acs.langmuir.7b00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaf K. I.; Alnajjar M. A.; Nau W. M. Chem. Commun. 2018, 54, 1734–1737. 10.1039/C7CC09519H. [DOI] [PubMed] [Google Scholar]
- Hennig A.; Bakirci H.; Nau W. M. Nat. Methods 2007, 4, 629. 10.1038/nmeth1064. [DOI] [PubMed] [Google Scholar]
- Bockus A. T.; Smith L. C.; Grice A. G.; Ali O. A.; Young C. C.; Mobley W.; Leek A.; Roberts J. L.; Vinciguerra B.; Isaacs L.; Urbach A. R. J. Am. Chem. Soc. 2016, 138, 16549–16552. 10.1021/jacs.6b11140. [DOI] [PubMed] [Google Scholar]
- Li M.; Lee A.; Kim K. L.; Murray J.; Shrinidhi A.; Sung G.; Park K. M.; Kim K. Angew. Chem., Int. Ed. 2018, 57, 2120–2125. 10.1002/anie.201711629. [DOI] [PubMed] [Google Scholar]
- Lee D.-W.; Park K. M.; Banerjee M.; Ha S. H.; Lee T.; Suh K.; Paul S.; Jung H.; Kim J.; Selvapalam N.; Ryu S. H.; Kim K. Nat. Chem. 2011, 3, 154. 10.1038/nchem.928. [DOI] [PubMed] [Google Scholar]
- Van Dun S.; Ottmann C.; Milroy L.-G.; Brunsveld L. J. Am. Chem. Soc. 2017, 139, 13960–13968. 10.1021/jacs.7b01979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vink P. J.; Briels J. M.; Schrader T.; Milroy L. G.; Brunsveld L.; Ottmann C. Angew. Chem., Int. Ed. 2017, 56, 8998–9002. 10.1002/anie.201701807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goor O. J. G. M.; Bosmans R. P. G.; Brunsveld L.; Dankers P. Y. W. J. Polym. Sci., Part A: Polym. Chem. 2017, 55, 3607–3616. 10.1002/pola.28743. [DOI] [Google Scholar]
- Murray J.; Sim J.; Oh K.; Sung G.; Lee A.; Shrinidhi A.; Thirunarayanan A.; Shetty D.; Kim K. Angew. Chem., Int. Ed. 2017, 56, 2395–2398. 10.1002/anie.201611894. [DOI] [PubMed] [Google Scholar]
- Logsdon L. A.; Urbach A. R. J. Am. Chem. Soc. 2013, 135, 11414–11416. 10.1021/ja406032x. [DOI] [PubMed] [Google Scholar]
- Li W.; Bockus A. T.; Vinciguerra B.; Isaacs L.; Urbach A. R. Chem. Commun. 2016, 52, 8537–8540. 10.1039/C6CC03193E. [DOI] [PubMed] [Google Scholar]
- Lee J. W.; Shin M. H.; Mobley W.; Urbach A. R.; Kim H. I. J. Am. Chem. Soc. 2015, 137, 15322–15329. 10.1021/jacs.5b10648. [DOI] [PubMed] [Google Scholar]
- Ali O. A.; Olson E. M.; Urbach A. R. Supramol. Chem. 2013, 25, 863–868. 10.1080/10610278.2013.810338. [DOI] [Google Scholar]
- Logsdon L. A.; Schardon C. L.; Ramalingam V.; Kwee S. K.; Urbach A. R. J. Am. Chem. Soc. 2011, 133, 17087–17092. 10.1021/ja207825y. [DOI] [PubMed] [Google Scholar]
- Tonga G. Y.; Jeong Y.; Duncan B.; Mizuhara T.; Mout R.; Das R.; Kim S. T.; Yeh Y.-C.; Yan B.; Hou S.; Rotello V. M. Nat. Chem. 2015, 7, 597. 10.1038/nchem.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta S. K.; Quigley J.; Vinciguerra B.; Briken V.; Isaacs L. J. Am. Chem. Soc. 2017, 139, 9066–9074. 10.1021/jacs.7b05154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathinte Meethal S.; Sasmal R.; Pahwa M.; C S.; Das Saha N.; Agasti S. S. Langmuir 2018, 34, 693–699. 10.1021/acs.langmuir.7b03554. [DOI] [PubMed] [Google Scholar]
- Park K. M.; Baek K.; Ko Y. H.; Shrinidhi A.; Murray J.; Jang W. H.; Kim K. H.; Lee J. S.; Yoo J.; Kim S.; Kim K. Angew. Chem., Int. Ed. 2018, 57, 3132–3136. 10.1002/anie.201713059. [DOI] [PubMed] [Google Scholar]
- Samanta S. K.; Moncelet D.; Briken V.; Isaacs L. J. Am. Chem. Soc. 2016, 138, 14488–14496. 10.1021/jacs.6b09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X.; Isaacs L. Angew. Chem., Int. Ed. 2016, 55, 8076–8080. 10.1002/anie.201602671. [DOI] [PubMed] [Google Scholar]
- Zhou X.; Su X.; Pathak P.; Vik R.; Vinciguerra B.; Isaacs L.; Jayawickramarajah J. J. Am. Chem. Soc. 2017, 139, 13916–13921. 10.1021/jacs.7b07977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L.; Hettiarachchi G.; Briken V.; Isaacs L. Angew. Chem., Int. Ed. 2013, 52, 12033–12037. 10.1002/anie.201305061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. L.; Sung G.; Sim J.; Murray J.; Li M.; Lee A.; Shrinidhi A.; Park K. M.; Kim K. Nat. Commun. 2018, 9, 1712. 10.1038/s41467-018-04161-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrey H. E.; Judkins J. C.; am Ende C. W.; Ballard T. E.; Fang Y.; Riccardi K.; Di L.; Guilmette E. R.; Schwartz J. W.; Fox J. M.; Johnson D. S. J. Am. Chem. Soc. 2015, 137, 11461–11475. 10.1021/jacs.5b06847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann R.; Avendano M. S.; Woehrstein J. B.; Dai M.; Shih W. M.; Yin P. Nat. Methods 2014, 11, 313–318. 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettiarachchi G.; Nguyen D.; Wu J.; Lucas D.; Ma D.; Isaacs L.; Briken V. PLoS One 2010, 5, e10514. 10.1371/journal.pone.0010514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes-Navajas P.; Gonzalez-Bejar M.; Scaiano J. C.; Garcia H. Photochem. Photobiol. Sci. 2009, 8, 1743–1747. 10.1039/b9pp00041k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.