Abstract
In response to the threat of DNA damage, cells exhibit a dramatic and multi-factorial response spanning from transcriptional changes to protein modifications, collectively known as the DNA damage response (DDR). Here, we review the literature surrounding the transcriptional response to DNA damage. We review differences in observed transcriptional responses as a function of cell cycle stage and emphasize the importance of experimental design in these transcriptional response studies. We additionally consider topics including structural challenges in the transcriptional response to DNA damage as well as the connection between transcription and protein abundance.
Introduction
DNA is vulnerable to damage from a variety of endogenous and exogenous sources, ranging from metabolic side products to sunlight [1]. Each damaging agent is capable of producing a different type of lesion: ionizing radiation and reactive oxygen species can produce single and double-strand breaks, UV light can cause the formation of pyrimidine dimers, and DNA replication errors can result in mismatch lesions, insertions, and deletions [1–3]. Though DNA damage is mostly considered to be unplanned and undesired, cells can also employ DNA damage in a controlled fashion to facilitate DNA replication and meiotic recombination. Thus, to ensure maintenance of genetic integrity for cellular and organism survival, cells have developed a response mechanism to repair damaged DNA, termed the DNA damage response (DDR). In general, the DNA damage response comprises the variety of intra and inter-cellular processes that occur following the detection of DNA damage, ultimately culminating in the choice to utilize one of several DNA repair modules of distinct but overlapping function, and occasionally resulting in cell death [1]. Following detection of DNA damage, a robust signaling cascade must occur, rapidly leading to protein modifications, activation of cell cycle checkpoints, and chromatin remodeling; more slowly, changes in cellular transcriptional programs occur. The result is a cell that is poised to repair the lesioned DNA before resuming the cell cycle [1,2,4]. At the cellular level, repair failure can lead to apoptosis or senescence. At the level of the organism, consequences of deficient DNA damage repair include the development of detrimental diseases such as cancer, neurological defects, infertility, and immune deficiencies [1,2].
Decades of research have revealed much regarding the mechanisms of the DDR, and the specific details of an elicited response depend heavily upon several factors: the type of DNA damage detected and, importantly, the position of the cell in the mitotic cell cycle. The DDR is a modular system, equipped with the tools to repair the diverse repertoire of DNA lesions. Small lesions, such as nucleotide mismatches, are repaired by the mismatch repair (MMR) module. Base excision repair (BER) is responsible for the repair of chemically-altered bases or single-strand breaks. Bulky or other helix-distorting lesions, such as pyrimidine dimers, are repaired by nucleotide excision repair (NER). Finally, double-strand breaks (DSB) may be repaired accurately by homologous recombination repair (HRR) when possible, or by the more error-prone nonhomologous end-joining (NHEJ) pathway [1–3]. Although some proteins are module-specific, a recent survey of the conserved DNA damage network found many interactions between module components, [5] highlighting the dense interconnectedness of DDR pathways. These major repair modules are reviewed in references [1–3].
As mentioned above, the cell’s position in the mitotic cell cycle is also extremely important; specific types of DNA damage and DDR choice can be cell cycle-specific. For example, nucleotide mismatches are associated with DNA replication, and replication fork collapse can result in the accumulation of single-stranded DNA. Conversely, specific types of damage repair can only occur during specific cell cycle phases: the double-strand break homologous repair pathway requires the presence of a sister chromatid– a condition only met during the S and G2 phases [6,7].
Many studies have considered the mechanisms of DNA repair, especially as these mechanisms pertain to cell survival following a DNA damage insult. This review specifically covers the transcriptional changes that take place inside cells as they respond to DNA damage, the machinery regulating that response, and interactions between transcriptional changes and the cell’s position in the cell cycle. We first briefly review the important background topic of cell cycle checkpoints and then discuss conserved transcriptional programs induced specifically in response to replication stress versus DNA damage experienced outside of G1/S phase. Next, we discuss the importance of experimental approach in attempts to study these transcriptional responses. Then, we consider the relationship between transcriptional programs and protein abundance, followed by a brief discussion regarding conflicts between transcription and DNA replication. Finally, we consider future directions.
Calling the Shots: DNA Damage-Relevant Transcription Throughout the Cell Cycle– Goals and Foul Plays
Many genes are under control of the cell cycle and exhibit periodic transcriptional patterns (Supplemental Table 1) [8–14]. Control of general cell cycle-regulated transcription has been thoroughly reviewed elsewhere [8,9]. Here, we highlight snapshots of the transcriptional program that appear to play a role in the response to genotoxic assaults.
G1/S Transcription
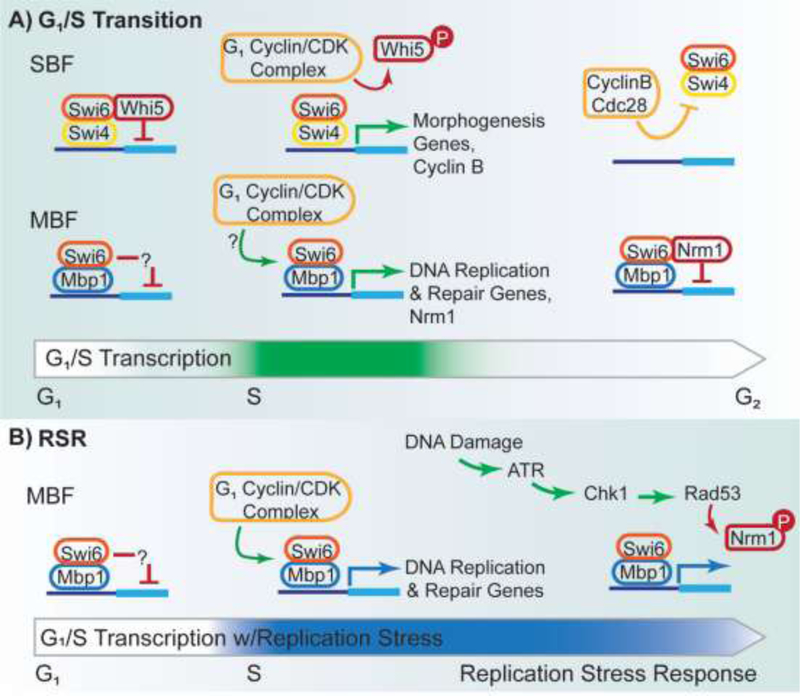
In S. cerevisiae, G1 transcription is mediated by the heteromeric transcription factors SBF (Swi4/6 cell cycle box binding factor) and MBF (MluI cell cycle box binding factor). SBF transcription is inhibited by Whi5, while MBF acts as a transcriptional repressor before S-phase [8,15]. Upon commitment, G1 cyclin-CDK complexes inactivate Whi5 by phosphorylation [16], permitting SBF to initiate G1 transcription. A positive feedback loop reinforces continued Whi5 phosphorylation, producing a strong wave of G1 transcription that peaks at the S-phase transition [8,17–19]. MBF transcriptional activation is also dependent upon G1 cyclin-CDK complexes. However, the exact mechanism remains unknown [9]. It is unlikely to be controlled by alteration of DNA binding levels, as it is present at target genes throughout the cell cycle. Instead, evidence points toward chromatin remodeling by the INO80 complex at target genes [20]. Transcription is normally shut down in S-phase via a negative feedback loop involving Cyclin B/Cdc28-mediated dissociation of SBF from promoters [8]. MBF transcription is tuned down via a negative feedback loop involving its transcriptional co-repressor Nrm1 and the repressor protein Yox1 [15,21].
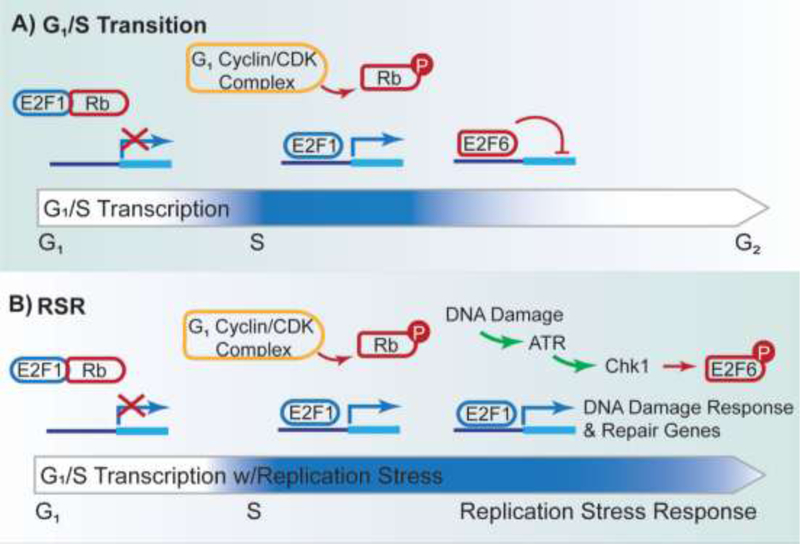
Despite a lack of sequence homology, there is striking functional conservation for the regulation of the G1/S transition in higher-order eukaryotic cells [8,22]. Pocket proteins, including the well-known retinoblastoma protein (Rb), as well as p103 and p107, play a role analogous to the role of Whi5 in S. cerevisiae, inhibiting G1 transcription by sequestering E2F transcription factors. G1 cyclin-CDK complexes phosphorylate pocket proteins, liberating the E2F1–3 transcriptional activators to trigger the G1/S transcriptional wave [8,23]. After cells progress into S-phase, transcription is downregulated via a combinatorial approach. First, the transcriptional E2F1–3 transcriptional activators are thought to be deactivated via a negative feedback loop. In addition, transcriptional activity is limited by the transcriptional repressors E2F6–8, analogous to the regulatory role played by yeast Nrm1 [8,24].
The Intra-S-phase checkpoint
The intra-S-phase checkpoint assesses the integrity of DNA replication and is responsible for the detection of DNA replication stress, recognizable by slowed, stalled, or collapsed replication forks. Structural blocks, such as those caused by DNA damage, or nucleotide supply deficiencies may lead to replication fork collapse. Uncoupling of DNA helicase and polymerase at stalled forks exposes single-stranded DNA, which binds Replication Protein A (RPA) [25]. The ATR/ATRIP kinase complex is recruited by RPA and, in turn, recruits activator proteins. Importantly, ATR activates the S-phase checkpoint via phosphorylation of the checkpoint protein kinase Chk1 [25–27]. Activated Chk1 perturbs the cell cycle by limiting cyclin-dependent kinase activity [28]. Collectively, this ATR-mediated response is known as the replication stress response (RSR).
Global Transcriptional Inhibition in Response to DNA Damage
It has been well-established in yeast and mammalian systems that DNA damage results in the repression of RNA synthesis and ribosomal genes. Most notable is the regulation of the RNA polymerase. In yeast, it has been found that the environmental stress response (discussed below) involves repression of genes involved in the production and translation of mRNA. [10,29,30] In human cells, it has been shown that p53 represses the expression of ribosomal genes [31] and that DNA damage limits the activity of RNA polymerase II by a combination of post-translational modifications and controlled degradation [32]. As transcription and translation are necessarily tied to the cell cycle, it is conceivable that cells experiencing stress exert global reductions in transcription to promote exit from the cell cycle. Conceivably, cells must overcome global transcriptional limitations in order to produce a targeted response to DNA damage. Such a system has recently been demonstrated in human cells where p53-mediated reduction of Myc levels was shown to reduce global transcriptional reduction, yet leave the expression of p53 direct targets intact [33].
Conserved Transcriptional Responses During Replication Stress
The first studies revealing that genes could be induced in response to DNA damage were performed in Escherichia coli. Using a random-integration LacZ reporter approach, researchers succeeded in delineating a subset of genes whose expression increased in response to DNA damage [34]. Subsequent studies found that, in addition to post-transcriptional regulation, transcriptional control is used to ensure that essential DNA repair proteins are present in bacterial cells that have suffered DNA damage [35]. In yeast and mammalian cells, early experimental approaches to identify DNA damage inducible transcripts successfully employed a differential hybridization approach [36,37]. Subsequently, the development of microarray [38–40] and chromatin immunoprecipitation (ChIP) technology permitted genome-wide surveys of transcriptional landscapes in S. cerevisiae in response to different challenges, including genotoxic stress [29,30,41–56].
In yeast, activation of the RSR produces activated checkpoint kinase Rad53. It has been demonstrated that Rad53-mediated inhibition of Nrm1 results in prolonged G1 transcription, specifically of MBF targets. [50,57–59] MBF targets are enriched for genes involved in DNA replication and repair as well as nucleotide synthesis [60,61]. Accordingly, prolonged MBF G1 transcription was associated with increased resistance to hydroxyurea (HU)-induced replication stress in fission yeast [15]. Consistent with these findings, Jaehnig et. al found that Rad53-dependent genes induced following treatment with the alkylating agent methyl methane sulfonate (MMS) were enriched for G1 transcripts [53]. Interestingly, G2/M transcripts were downregulated, consistent with reports of downregulation of this subset of genes in fission yeast [50]. De novo protein synthesis does not appear to be required for survival during yeast replication stress. However, cells treated with cycloheximide following HU exhibited much longer DNA replication times than cells treated with HU alone, prompting investigators to conclude that protein synthesis may be required for resumption of normal DNA synthesis rates following replication stress [62]. In addition, the transcriptional repressor Crt1 is phosphorylated by activated Rad53/Dun1, liberating cells to transcribe the ribonucleotide reductase (RNR) genes, which catalyze the rate-limiting step in maintenance of the deoxyribonucleotide triphosphate pool [63,64].
In mammalian cells, a similar circuit has been demonstrated. Relief of E2F6-mediated repression of G1 transcription plays an important role in limiting genome instability [28,65]. E2F6 depletion was important for the regulation of genes involved in DNA replication and the response to DNA damage, among others [28]. In contrast to yeast, de novo protein synthesis is important in limiting DNA damage during replication stress. Specifically, cells unable to maintain G1 transcription due to E2F6 overexpression were not only unable to appropriately arrest replication forks in response to RS, but also demonstrated decreased recruitment of the stabilization proteins Rad51, FANCD2, and Cdc7 to chromatin in HU-treated cells. In addition, relief of E2F6 transcriptional repression was sufficient to promote cell recovery from replication stress, even in the context of checkpoint deficiency induced by Chk1 drug blockade. Intriguingly, maintenance of E2F expression in the context of Chk1 drug blockade rescued the shortened DNA replication track length defect of these cells following replication stress, indicating that E2F expression is sufficient to permit DNA replication to resume following replication stress, even in the context of checkpoint deficiency [65]. To our knowledge, such experiments have not yet been conducted in S. cerevisiae. It will be interesting to see whether MBF-specific transcription in yeast can produce similar findings. The Rfx family of genes is closely related to S. cerevisiae Crt1. Similar to yeast, it has been shown that Rfx1 binds to the promoter of the RNR2 gene, but is released in the context of HU treatment [66].
The G1 and G2 Checkpoints
Each time a cell re-enters the cell cycle, there are ample opportunities for the introduction of new mutations, making the “restriction point” in late G1, also known as the G1/S checkpoint when the cell commits to cell cycle entry, a crucial decision point for the cell [4,67–69]. Upon detection of DNA damage, the ATM/ATR kinases activate Chk1/Chk2, which trigger the degradation of Cdc25 phosphatase and, in multicellular organisms, activate p53. The destabilization of Cdc25 occurs rapidly through post-translational modifications, while the activation of p53 occurs somewhat more slowly, requiring transcriptional activation. Ultimately, cell entry into S-phase is prevented [4,69,70]. The G2/M checkpoint is activated in the event that any unrepaired DNA damage has been detected. The ATM/ATR kinases again activate Chk1/Chk2 to mediate Cdc25 phosphatase degradation, preventing the activation of Cyclin B/Cdk1 complex needed for the G2/M transition. In mammalian cells, p53 and BRCA1 play a role in mediating a sustained G2/M checkpoint response [69]. Activation of any checkpoint can result in a delay in cell cycle progression [4,67–69]. These ATM/ATR-mediated responses govern the transcriptional programs described below.
Transcriptional Responses to DNA Damage Outside of S-Phase
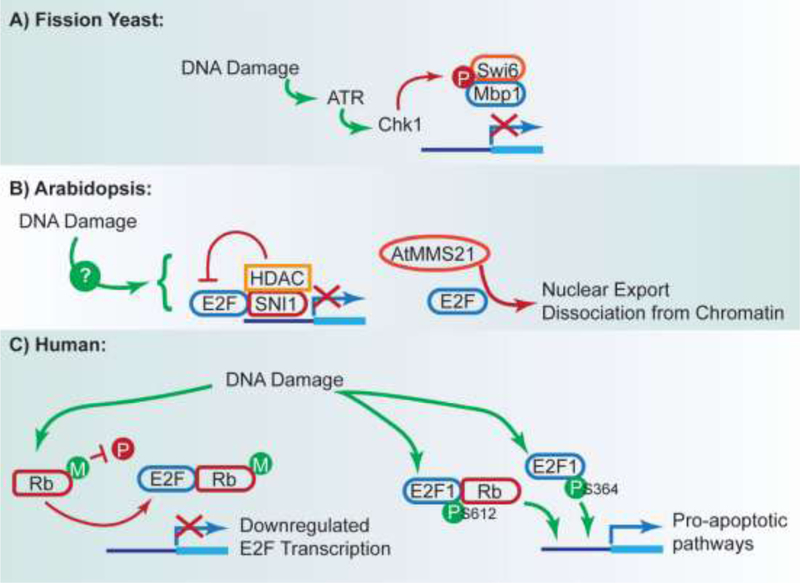
In Schizosaccharomyces pombe, the replication stress checkpoint and DNA damage checkpoint are controlled by two distinct kinases, Cds1 and Chk1, respectively. While activation of the replication stress checkpoint by Cds1 results in prolonged MBF target gene expression, as described above, activation of the DNA damage checkpoint results in direct phosphorylation of MBF by Chk1. MBF phosphorylation releases it from chromatin and leads to a concomitant decrease in MBF target gene expression, as measured by ChIP and qPCR using a subset of well-known MBF target genes [71]. The extent to which the full panel of MBF target genes is affected has yet to be specifically queried. Aflatoxin B is thought to alkylate DNA and other biological molecules as well as produce apurinic sites and precursors to reactive oxygen species [72]. Thus, aflatoxin likely elicits a combination of DDRs. In a study using a low dose of aflatoxin, cell populations demonstrated delayed progression through S-phase, and approximately half differentially-expressed genes were cell cycle-regulated genes. Notably, the differentially expressed cell cycle gene set included repression of S-phase histone genes and late M-phase specific genes [45]. This finding supports damage-dependent control of multiple cell cycle checkpoints. Similar suppression of histone genes has been observed in human cells in response to ionizing radiation [73,74], restriction-induced double-strand breaks, and p53 stabilization by nutilin [74].
Notably, a recent study in Arabidopsis thaliana examined the role of SNI1, a subunit of the Structural Maintenance of Chromosome (SMC) 5/6 complex with some sequence homology to mouse Rb. It was found that sni1 mutant strains exhibited an over-activated, but deficient, homologous repair pathway, leading to increased amounts of DNA damage [75]. Further work demonstrated that SNI1 also suppressed E2F transcription by recruitment of histone deacetylase to E2F targets. Interestingly, the root growth defect and endoreplication defects of the sni1 mutant were suppressed by loss of E2F function, though this strain still demonstrated some sensitivity to DNA-damaging agents [76]. Loss of RBR (Retinoblastoma Related) in Arabidopsis has been shown to result in an E2F-dependent hypersensitive DDR, with excessive cell death [77]. In addition, recent work has indicated that AtMMS21, a DNA damage protein that is an E3 ubiquitin ligase, can negatively regulate the activity of E2F [78]. Though it remains to be directly demonstrated that these events occur in response to DNA damage, it has been demonstrated that AtMMS21 is required for the DDR in Arabidopsis [79]. Taken together, these studies demonstrate the adoption of multiple mechanisms to regulate E2F transcription and that failure to do so may lead to DNA damage.
In mammalian cells, multiple species of modified E2F following DNA damage have been reported, with somewhat conflicting conclusions regarding the downstream transcriptional effects. However, the consensus is that the E2F transcriptional program resulting from these DNA damage-specific species is distinct from the normal cell cycle transcriptional program.
Direct phosphorylation of E2F1 at serine 612 results in Rb binding, creating a complex that, surprisingly, participates in repression of cell cycle control genes and activation of proapoptotic pathways [80]. In an independent study, researchers concluded that an Rb-free population of E2F1 and an pRb-E2F1-ser364 variant produce a net positive effect on the transcription of proapoptotic genes [81]. In Drosophila, loss of E2F1 function was shown to result in apoptosis block, even though these mutants exhibited ‘normal’ apoptotic transcriptional programs. It was discovered that dysregulation of mitochondrial genes led to poor mitochondrial function, explaining the lack of apoptosis [82].
Rb itself has also proven to be a target of the DNA damage response. In mammalian cells, treatment with etoposide, a topoisomerase inhibitor known to produce double-strand breaks, resulted in the methylation of Rb at K810, which overlaps known CDK consensus sites. Rb methylation was shown to antagonize phosphorylation of Rb and was important for efficient cell cycle blockade following etoposide treatment [83]. In a related study, methylation of a nearby site, K860, was shown to influence Rb binding to the transcriptional repressor L3MBTL1, though no consistent effect on E2F binding was observed [84]. In both studies, methylation reduced transcriptional induction of E2F targets. To our knowledge, a similar circuit with Whi5 is not seen in yeast, possibly due to the finding that the pocket protein Whi5 regulates SBF transcription, while NRM1 regulates MBF targets [15].
More recently, a repressor function has been reported for E2F7 and E2F8 in mammalian cells. Loss of E2F7/8 resulted in increased expression of E2F1 and concomitant increased apoptosis, as well as worsened tumorigenesis in mouse models of melanoma [85]. Conversely, loss of E2F7 function at lower, non-lethal doses of DNA damage was shown to result in p53-independent upregulation of DNA damage repair genes, as well as improved DNA repair. It was proposed that E2F7-mediated regulation may be important to prevent inappropriate repair activities once DNA has been restored, which can lead to genomic instability, supporting a role for the E2F7 factor as a tumor suppressor [86]. Another study that analyzed the appearance of nascent transcripts in response to DNA damage found coordinated, p53-dependent, suppression of G2/M transcripts and E2F targets in gene set enrichment analysis [74]. CHiP seq data and motif analysis seem to support that p53 is directly responsible for approximately half of the ionizing radiation-induced transcriptional response, while repression events, mostly surrounding M-phase genes, are indirectly mediated by p53 [87]. This result is consistent with findings from a large network motif analysis study in which p53 motifs are not found upstream of genes downregulated in response to p53 stabilization [88]. As E2F7 is a direct p53 target, E2F7 may account for some transcriptional repression.
Structural Challenges
Replication Stress versus Transcription
The processes of DNA replication and G1-S transcription create a conflict for resources, as both make use of the same DNA template. In the context of compromised DNA replication — for example, replication in highly-transcribed regions of the genome, or increased replication due to an oncogene — the conflict between transcription and replication is exacerbated [89].
The process of DNA replication is highly regulated and organized to ensure that the genome is duplicated exactly once per cell cycle. Only during G1 does a cell begin to prepare for replication. Origins of replication, located in excess throughout the genome, are licensed by the formation of pre-replication complexes (pre-RC), consisting of the origin recognition complex, Cdc6, Cdt1, and the mini-chromosome maintenance complex (MMC). Many more origins are licensed than are fired during S-phase. Cyclin dependent kinase (CDK) and DBF4-dependent kinase activity at the G1-S transition activate MCM to recruit additional components, thus permitting them to begin DNA replication. Firing of an origin creates two replication forks which move away from one another as they replicate strands of DNA. Replication is complete when replisomes converge [90,91].
Multiple regulatory mechanisms ensure singular duplication of the genome and reduce conflicts that may produce replication stress. As replisomes traverse other origins of replication, they must dis-assemble pre-replicative complexes, thus preventing that origin from firing later. In addition, origin licensing and firing are temporally restricted: licensing occurs only during G1 and firing occurs in a scheduled manner throughout S-phase, though the fine details are still unknown to researchers [91,92].
Collisions of the replicative forks with physical barriers, such as transcription factors, may stall fork progress, and may even lead to DNA damage. In Escherichia coli, these collisions are largely avoided by co-orientation of DNA replication and transcription of the most abundant E. coli transcripts. Occasionally, a faster-moving DNA polymerase may catch up to a slower-moving RNA polymerase on a chromosome. In vitro reconstitution experiments indicate that the RNA-DNA conflict may be resolved either by the DNA polymerase either slowing down or displacing the RNA polymerase. Head-on collisions between DNA polymerases appear to have lethal consequences in E. coli [93]. Similar co-orientation of replication and transcription of ribosomal genes has been observed in yeast. [94] In human cells, the situation is not as well-defined; there is currently no evidence to support the co-orientation of DNA replication and transcription [95]. In fact, it has been demonstrated in B-lymphoblasts that the increased breakage at regions known as “chromosomal fragile sites” is due to a spatio-temporal overlap of transcription and DNA replication of long genes (i.e. those which take longer than one cell cycle to transcribe). Importantly, it has been proposed that oncogene-induced DNA damage may occur through replication stress. Oncogenes may increase replication stress in a number of ways, from interfering with the replication process itself to increasing the frequency with which cells must replicate their DNA. Interestingly, over-expression of the oncogene Cyclin E has been shown to induce transcription-dependent replication stress by increasing replication initiation [96]. Furthermore, the Myc oncogene has also been shown to increase replication stress by increasing the number of active origins during S-phase [97]. Together, these findings underscore the importance of coordinated replication and transcription.
The Role of the Chromatin Environment
It is becoming apparent that diseases such as cancer are the result not only of accumulated mutations due to DNA repair failure, but also of epigenetic alterations. The role of chromatin modifications and the histone code in promoting repair activity by DNA repair enzymes has been reviewed elsewhere [98]. In this section, we will discuss the interface between transcription and the chromatin environment in the context of DNA damage— both in promoting DNA damage repair and the potential long-term effects on the transcriptome following DNA damage.
Following UV-induced DNA damage, it has been demonstrated that waves of RNA polymerase II are released into transcriptionally active genes, thus permitting transcription-coupled NER of actively-transcribed genes [99]. It is proposed that waves of transcriptional progression through active genes may also create an open chromatin environment, thus promoting further DNA damage repair. Though this hypothesis remains to be directly tested, it is supported by the observation that transcriptionally active regions of the genome in tumor samples demonstrated lower mutation prevalence than inactive regions [99].
DDR activity on damaged DNA has been demonstrated to produce rapid loss of transcription at the site of damage. In yeast, transcriptional inhibition following double-strand break is mediated by strand resection. [100] In mammalian cells, however, the response is more complex, involving the integration of ATM signaling, histone 2A ubiquitination, histone deacetylation [101], and even changes in the DNA structure, itself [102]. While originally thought that damage-associated chromatin changes reverted following repair, recent evidence indicates that chromatin modifications following DNA damage are heritable, including retention of the repressive histone 3 lysine 9 (H3K9) methylation or recovery of the transcriptionally-active H3K4 signal, as observed in GFP HR reporter systems. Notably, final methylation status at repair sites could be permanently altered by depletion of BER activity following HR. Drastic differences in expression were observed in clones with the same number of methylated CpG sites surrounding the repaired lesion indicate that specific CpG methylation governs the transcriptional activity of the repaired gene [103]. Together, these observations support a model in which methylation status is determined by repair activities, as well as by modifications caused by BER proteins and transcriptional remodeling.
Recent intriguing work has taken advantage of chromosome conformation capture-derivative technique (HiC), which enables 3D mapping of chromosomal conformations, to investigate DSB repair pathway choice. Using a cell line in which DSBs were introduced by restriction enzyme cleavage, it was discovered that DSBs clustered with one another in the nucleus in a manner that was associated with transcriptional activity. HRR was the preferred repair mechanism for actively transcribed regions, while NHEJ was preferentially used in silenced genomic regions. In G1 cells, DSB clustering was associated with delayed repair; inhibition of transcription alone did not consistently alter clustering, possibly indicating that this phenomenon is due to secondary structures or RNA:DNA hybrids at transcribed regions. The nuclear cytoskeleton, however, was necessary for clustering, indicating that there may be active governance in repair pathway choice. Delayed repair may sequester DSBs to avoid deleterious NHEJ and promote HRR at a more desirable cell cycle point [95]. Given that cell cycle position governs the sensitivity of many standard chemotherapeutic agents [104], it will be essential to thoroughly investigate the potential implications of this study.
Global Models of the Transcriptional Response
Large collections of genome-wide expression data have made possible the analysis of transcriptional activity from a global perspective. One such study used genetic, biochemical and ChIP-chip data from S. cerevisiae to construct a transcriptional network containing known direct transcriptional regulatory motifs [105]. Conditional gene expression data were then used to highlight active regulatory pathways that were specific to a given condition, resulting in condition-specific sub-networks. Contrary to the longstanding belief that similar transcriptional motifs are used with some constancy across conditions, this dynamic transcriptional network demonstrated that the relative occurrence of transcriptional motifs varied across the surveyed conditions. For DNA damage, stress response, and diauxic shift sub-networks (these conditions were termed ‘exogenous states’), researchers observed that each active transcription factor regulated a large number of target genes; each target gene was, in turn, regulated by only a few transcription factors. Networks generated for ‘endogenous states’ (cell cycle and sporulation) showed the opposite behavior. Biologically, the larger regulatory network of each transcription factor in exogenous states can be interpreted as evidence that each active transcription factor has greater overall influence by regulating more genes at once. The smaller number of regulatory interactions for any given target gene can be interpreted as evidence that the target genes are regulated by simpler combinations of upstream transcription factors. In addition, the average path length (the shortest path between any two nodes) was shorter in the exogenous states, suggesting that signals might spread more rapidly through these networks in the setting of genotoxic stress. Master regulatory transcription factors, which govern activity of large subsets of genes for each condition, may explain condition-dependent lethality [105]. Workman et al. found that the MMS sensitivity of transcription factor knockouts was highly correlated with the number of genes regulated by that transcription factor [46]. Other studies have demonstrated that transcriptionally responsive gene sets are generally not enriched for genes that affect sensitivity to DNA-damaging agents (though individual cases where deletion of a transcriptionally responsive DNA repair gene does result in DNA damage sensitivity [49]). That transcription factor deletions result in sensitivity while independent gene deletion does not support a scenario in which master regulators produce a wide response that is important to limit damage sensitivity.
By integrating transcriptional, epigenetic, and post-translational modification data, we have begun to create a picture of the global changes that occur in cells in response to DNA damage. Integration of multiple types of data has improved the sensitivity of a study to detect general responses to damage. For example, a study integrating mass-spectrometry data and global expression changes in response to an siRNA screen produced a picture of transcription-coupled NER. Importantly, all known components of transcription-coupled NER could only be identified when integrating all data types [106]. This highlights the observation that the biases inherent to any one experimental approach may be overcome by utilizing multiple data types; increasing numbers of studies are integrating multiple data types to study the DNA damage response.
In human cells, it has been demonstrated that p53 directly governs ~50% of the transcriptional response to ionizing radiation in an ATM-dependent manner. Interestingly, while p53 bound many regions of the genome by CHip-seq, its ability to influence transcription was determined by binding-affinity and distance to the transcriptional start site. While induced genes were directly governed, repressed genes were indirectly governed [87]. This is consistent with a more recent study in which downregulation was shown to be attributable to a p53-dependent decrease in Myc levels [33]. Interestingly, while p53-regulated genes were enriched for apoptosis following damage, there was a temporary rise in NFkB-regulated genes, which were associated with anti-apoptotic signals [87]. As the authors note, it is interesting to speculate whether the rise of NFkB target genes occurred in a subpopulation of cells, or whether represents more general transcriptional response of any cell responding to damage that would present a limited window in which a cell must repair damage before succumbing to apoptosis. The NFkB wave likely influences cell survival in p53-deficient cells and presents a tempting therapeutic target.
The Importance of Experimental Approach
Damage Matters
Many investigators have studied the effects of methyl methane-sulfonate (MMS), an alkylating agent capable of altering not only DNA but also RNA and proteins [107]. Thus, cells responding to MMS treatment are responding to the onslaught of alkylated proteins and RNA, in addition to lesioned DNA. Such a broad response is supported by the body of work surrounding what is known as the environmental stress response (ESR). Jelinsky et. al first demonstrated that a diverse set of genes was differentially expressed in response to MMS [29]. Further work by the Samson group demonstrated that some of these same genes could be induced in response to stress, independent of MMS treatment [41]. Gasch et. al thoroughly and clearly delineated the ESR as a stereotyped transcriptional program of ~900 genes employed by cells to combat environmental stressors. The ESR is a graded response that positions the cell to maintain essential homeostatic functions such as carbohydrate metabolism, cellular osmolarity, and cellular redox potential, thus promoting survival [10]. In fact, in unsynchronized cell populations, the ESR was strongly detected in MMS-treated yeast cells [30]. Similarly, ionizing radiation is known to produce free radicals whose damage capacity is not limited to DNA [108]. Accordingly, the ESR signal was also strongly detected, though transiently, in irradiated cells [30]. Studies using genetic models of replication stress [49] and radiomimetic chemicals that specifically damage DNA [47] have demonstrated that, in these cases, the DNA damage-specific transcriptional response is significantly attenuated (hundreds of differentially regulated genes) compared to the damage induced by the nonspecific damaging agents (thousands of genes). In addition, the ESR signal is largely absent, indicating that the choice of DNA-damaging agent is critical.
Cell Cycle Position
Many investigations have been performed in asynchronous starting cell populations. It has been shown that treatment with 0.02% MMS or 170 Gray of ionizing radiation both induce 100% cell cycle arrest at S-phase and G2/M phase, respectively, by 90 minutes post-treatment [30], indicating that cell cycle stage is of great importance in th study of the DDR. The most deleterious type of alkylation-induced DNA lesion, O6-methylguanine (O6MeG) may be directly repaired throughout the cell cycle by o-6-methylguanine-DNA methyltransferase (MGMT). Recent evidence has demonstrated lower abundance of MGMT during S-phase in human cells [109]. Whether decreased MGMT abundance correlates with decreased repair activity remains to be determined. During S-phase, MMR activity on unrepaired O6MeG produces faulty O6MeG-T pairings. Subsequent rounds of S-phase and associated MMR activity continue this cycle to eventually produce a double-strand break [110]. In contrast, double-strand breaks, such as those induced by ionizing radiation, may be detected and repaired by one of two pathways: homologous repair or non-homologous end-joining. Repair pathway choice depends on Cdk1 activity and the presence of a sister chromatid [7,111]. It is thus conceivable that transcriptional response may correlate with cell cycle arrest position, and this association is supported by multiple reports. While most cell-cycle-regulated genes did not overtly correlate with cell cycle arrest point, Gasch et. al found that a small number of G1-specific genes were induced in response to MMS, consistent with later reports [57,59,112] of sustained G1 expression in response to replication stress. In addition, some G2-specific genes were expressed in response to irradiation [30]. DNA damage-specific transcriptional signals have been detected [30,41] and have been proposed to differentiate cytotoxicity and genotoxicity [43]. Finally, Chu and colleagues found a ~3-fold increase in the number of differentially-expressed genes due to HU treatment in a synchronized versus asynchronous population comparison. Cell cycle-specific transcriptional responses may thus be more robustly identified in synchronized populations [50]. Regardless, it is clear that DDR transcriptional datasets are not all equal and should be interpreted with consideration of factors such as off-target cell stress and cell cycle position.
The next level: Transcription to protein abundance
Researchers have traditionally used mRNA abundance as a proxy for protein abundance, partly due to the difficulty of assessing the abundance of thousands of proteins at once. As our ability to detect proteins has improved, however, it is becoming clearer that changes in mRNA levels cannot explain the entire picture. In yeast, for example, imaging analyses under MMS treatment have identified a subset of genes exhibiting differential protein abundance, though this subset is substantially smaller than previously-identified transcriptional responders. This study confirmed a correlation of mRNA induction with increased protein abundance; transcriptional upregulation accounted for ~60% of protein abundance changes in response to MMS treatment [113]. Conversely, there was almost no concordance (6% in Mazumder, et al.) between transcriptional repression and protein abundance, as determined by single-cell imaging [113,114]. The correlation between mRNA levels and protein abundance can vary widely across conditions and organisms, typically trending towards lower correlation values in multicellular organisms compared to single-celled organisms such as yeast [115]. Such discoveries beg the question- What is driving the remainder of observed differences in protein levels?
With regard to downregulated genes, poor correlation between transcriptional repression and protein abundance (by mass spectrometry) has previously been observed in response to NaCl treatment, prompting the suggestion that transcriptional repression might serve the purpose of freeing up resources for the production of needed, induced transcripts in the setting of stress, rather than reducing levels of unneeded proteins [116]. Other factors may contribute to these observations, including different protein half-lives, as well as difficulty in detecting proteins at or near background cutoffs in mass spectrometry and image analysis. Anecdotal scenarios for correlation between transcriptional repression and protein reduction exist. For example, E2F7 was observed by mRNA analysis and western blot to repress E2F1 and CDC25 in response to doxorubicin treatment [117]; p53-dependent repression of PLK1 in response to adriamycin treatment has also been observed by mRNA analysis and western blot [118]. The extent to which protein turnover and detection issues may contribute at a global level to previously observed poor correlation for downregulated genes remains to be determined.
One interesting new area concerns the role of tRNAs. In yeast, the methyltransferase Trm9 has been demonstrated to produce modifications on the wobble position of the arginine (UCU) and glutamic acid (UUC) tRNAs. Importantly, these modifications were needed to enhance transcript levels of RNR1 and 3 in a codon-dependent manner, and trm9Δ cells were sensitive to damaging agents [119]. tRNA modifications have also been discovered to play a role in the mammalian cells. Wobble position modifications mediated by AlkBh8, homolog of Trm9, were found to be important in stop codon reprogramming, which is necessary in the production of selenoproteins. AlkBh8-deficient MEFs were deficient in synthesis of selenoproteins involved in detection of reactive oxygen species and demonstrated more double-strand breaks by comet assay [120].
The role of other post-transcriptional regulation mechanisms, including RNA splicing, RNA turnover, miRNAs, codon reprogramming, codon bias, RNA structures, and RNA binding proteins are all active areas of research, and have been reviewed elsewhere [121–127]. What is clear is that mRNA abundance alone cannot explain the full breadth of protein level variations seen in the DNA damage response. More research in these areas will be required to determine how the integration of transcription and translation produces a coordinated DDR cell state.
Perspective and Future Directions
Several aspects of the transcriptional response to DNA damage remain particularly unclear. The contribution of transcription to cell survival following DNA damage remains contested, though evidence seems to indicate that disruption of the transcriptional program does contribute to survival, while singular disruption of responsive genes may not. While the entire class of DNA damage repair genes may not be upregulated in response to DNA damage, there is strong evidence for the upregulation of specific pathways such as base excision repair [128]. In addition, systematic analyses of tumor transcriptional landscapes have demonstrated disruption of DNA damage repair genes, also at the level of specific pathways [129]. Recent evidence has also indicated that one of the pathways by which PARP inhibitors function is by limiting homologous repair factor availability via transcriptional regulation [130]. While the role of transcription in the DNA damage response is still being elucidated, it is becoming more and more clear that this axis of DDR regulation is important. In addition, the role of the cell cycle should be considered. More studies need to be performed in synchronized cell systems to fully illuminate this topic. The use of multiple DNA damaging agents that produce similar types of DNA damage will help to distinguish true responses to DNA damage from off-target responses related to cell stress and potentially other factors. Finally, an emerging topic in mammalian DDR systems is the contribution of the circadian clock, which is regulated by a transcription-translation feedback loop [131–134]. Elucidating the contribution of these different factors will be critical as we move into the age of targeted cancer treatments.
We apologize to any researchers whose work was not cited due to space constraints.
Supplementary Material
Figure 1: Yeast G1/S Transcription and Replication Stress Response.
A) Transcription by SBF and MBF is inhibited prior to S-phase. SBF is inhibited by Whi5; MBF acts as a transcriptional repressor by an unknown mechanism. Upon entrance to S-phase, G1 cyclin/CDK complex promotes transcription by SBF and MBF. The cyclin/CDK complex promotes Whi5 dissociation from SBF by phosphorylation. At the end of S-phase, SBF and MBF transcription are downregulated by negative feedback loops. The Cyclin B/Cdc28 complex promotes SBF dissociation from chromatin, and Nrm1 acts as a transcriptional co-repressor for MBF. B) Replication stress results in activated Rad53, which phosphorylates Nrm1, interfering with its interaction with MBF. This results in continued MBF transcription as repair occurs.
Figure 2: Human G1/S Transition and Replication Stress Response.
A) Prior to S-phase, E2F transcription is inhibited by Rb binding. Upon entrance to S-phase, G1 cyclin/CDK complex phosphorylates Rb, releasing E2F1 to permit transcription. E2F6 then inhibits transcription of G1/S targets. B) Upon replication stress, ATR activates Chk1 to phosphorylate E2F6, promoting the transcription of E2F1 targets.
Figure 3: Summary of Transcriptional Responses to DNA Damage.
A) In fission yeast, MBF transcription is phosphorylated by Chk1 in response to DNA damage, leading to decreased transcription. B) Studies in Arabidopsis indicate that E2F transcription is regulated by a variety of pathways. SNI1 has been demonstrated to decrease E2F transcription by recruiting HDAC. In addition, AtMMS21 reduces E2F chromatin binding and nuclear translocation. C) In humans, DNA damage results in diversification of E2F species. Rb methylation interferes with its phosphorylation, resulting in continued sequestration of E2F and reduced E2F transcription. Conversely, specific phosphorylated E2F1 species, with or without Rb are induced by DNA damage and upregulate pro-apoptotic pathways.
Funding:
The authors gratefully acknowledge the following funding sources: NCI 5T32CA67754-22; NIEHS R01ES014811
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40: 179–204. doi: 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461: 1071–1078. doi: 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481: 287–294. doi: 10.1038/nature10760 [DOI] [PubMed] [Google Scholar]
- 4.Bartek J, Lukas J. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol. 2001;13: 738–747. doi: 10.1016/s0955-0674(00)00280-5 [DOI] [PubMed] [Google Scholar]
- 5.Pearl LH, Schierz AC, Ward SE, Al-Lazikani B, Pearl FMG. Therapeutic opportunities within the DNA damage response. Nat Rev Cancer. 2015;15: 166–180. doi: 10.1038/nrc3891 [DOI] [PubMed] [Google Scholar]
- 6.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9: 297–308. doi: 10.1038/nrm2351 [DOI] [PubMed] [Google Scholar]
- 7.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18: 134–147. doi: 10.1038/cr.2007.111 [DOI] [PubMed] [Google Scholar]
- 8.Bertoli C, Skotheim JM, de Bruin RAM. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 2013;14: 518–528. doi: 10.1038/nrm3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haase SB, Wittenberg C. Topology and control of the cell-cycle-regulated transcriptional circuitry. Genetics. 2014;196: 65–90. doi: 10.1534/genetics.113.152595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11: 4241–4257. doi: 10.1091/mbc.11.12.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pramila T, Wu W, Miles S, Noble WS, Breeden LL. The Forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev. 2006;20: 2266–2278. doi: 10.1101/gad.1450606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rowicka M, Kudlicki A, Tu BP, Otwinowski Z. High-resolution timing of cell cycle-regulated gene expression. Proc Natl Acad Sci U S A. 2007;104: 16892–16897. doi: 10.1073/pnas.0706022104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Chen S, Wang S, Soares F, Fischer M, Meng F, et al. Transcriptional landscape of the human cell cycle. Proc Natl Acad Sci U S A. 2017;114: 3473–3478. doi: 10.1073/pnas.1617636114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Granovskaia MV, Jensen LJ, Ritchie ME, Toedling J, Ning Y, Bork P, et al. High-resolution transcription atlas of the mitotic cell cycle in budding yeast. Genome Biol. 2010;11: R24. doi: 10.1186/gb-2010-11-3-r24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Bruin RAM, Kalashnikova TI, Chahwan C, McDonald WH, Wohlschlegel J, Yates J 3rd, et al. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol Cell. 2006;23: 483–496. doi: 10.1016/j.molcel.2006.06.025 [DOI] [PubMed] [Google Scholar]
- 16.Costanzo M, Nishikawa JL, Tang X, Millman JS, Schub O, Breitkreuz K, et al. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117: 899–913. doi: 10.1016/j.cell.2004.05.024 [DOI] [PubMed] [Google Scholar]
- 17.Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993;12: 1955–1968. Available: https://www.ncbi.nlm.nih.gov/pubmed/8387915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stuart D, Wittenberg C. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 1995;9: 2780–2794. Available: https://www.ncbi.nlm.nih.gov/pubmed/7590253 [DOI] [PubMed] [Google Scholar]
- 19.Dirick L, Böhm T, Nasmyth K. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. Wiley Online Library; 1995;14: 4803–4813. Available: https://onlinelibrary.wiley.com/doi/abs/10.1002/j.1460-2075.1995.tb00162.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knezevic I, González-Medina A, Gaspa L, Hidalgo E, Ayté J. The INO80 complex activates the transcription of S-phase genes in a cell cycle-regulated manner. FEBS J. 2018;285: 3870–3881. doi: 10.1111/febs.14640 [DOI] [PubMed] [Google Scholar]
- 21.Gómez-Escoda B, Ivanova T, Calvo IA, Alves-Rodrigues I, Hidalgo E, Ayté J. Yox1 links MBF-dependent transcription to completion of DNA synthesis. EMBO Rep. EMBO Press; 2011;12: 84–89. doi: 10.1038/embor.2010.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rb Cooper K., whi it’s not just for metazoans anymore. Oncogene. 2006;25: 5228–5232. doi: 10.1038/sj.onc.1209630 [DOI] [PubMed] [Google Scholar]
- 23.Henley SA, Dick FA. The retinoblastoma family of proteins and their regulatory functions in the mammalian cell division cycle. Cell Div. 2012;7: 10. doi: 10.1186/1747-1028-7-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giangrande PH, Zhu W, Schlisio S, Sun X, Mori S, Gaubatz S, et al. A role for E2F6 in distinguishing G1/S- and G2/M-specific transcription. Genes Dev. 2004;18: 2941–2951. doi: 10.1101/gad.1239304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol Cell. 2017;66: 801–817. doi: 10.1016/j.molcel.2017.05.015 [DOI] [PubMed] [Google Scholar]
- 26.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9: 616–627. doi: 10.1038/nrm2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osborn AJ, Elledge SJ, Zou L. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 2002;12: 509–516. Available: https://www.ncbi.nlm.nih.gov/pubmed/12446112 [DOI] [PubMed] [Google Scholar]
- 28.Bertoli C, Klier S, McGowan C, Wittenberg C, de Bruin RAM. Chk1 inhibits E2F6 repressor function in response to replication stress to maintain cell-cycle transcription. Curr Biol. 2013;23: 1629–1637. doi: 10.1016/j.cub.2013.06.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jelinsky SA, Samson LD. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc Natl Acad Sci U S A. 1999;96: 1486–1491. Available: https://www.ncbi.nlm.nih.gov/pubmed/9990050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gasch AP, Huang M, Metzner S, Botstein D, Elledge SJ, Brown PO. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell. 2001;12: 2987–3003. doi: 10.1091/mbc.12.10.2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhai W, Comai L. Repression of RNA Polymerase I Transcription by the Tumor Suppressor p53. Mol Cell Biol. American Society for Microbiology Journals; 2000;20: 5930–5938. doi: 10.1128/MCB.20.16.5930-5938.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heine GF, Horwitz AA, Parvin JD. Multiple mechanisms contribute to inhibit transcription in response to DNA damage. J Biol Chem. 2008;283: 9555–9561. doi: 10.1074/jbc.M707700200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Porter JR, Fisher BE, Baranello L, Liu JC, Kambach DM, Nie Z, et al. Global Inhibition with Specific Activation: How p53 and MYC Redistribute the Transcriptome in the DNA Double-Strand Break Response. Mol Cell. 2017;67: 1013–1025.e9. doi: 10.1016/j.molcel.2017.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kenyon CJ, Walker GC. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci U S A. 1980;77: 2819–2823. Available: https://www.ncbi.nlm.nih.gov/pubmed/6771759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutton MD, Walker GC. Managing DNA polymerases: coordinating DNA replication, DNA repair, and DNA recombination. Proc Natl Acad Sci U S A. 2001;98: 8342–8349. doi: 10.1073/pnas.111036998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McClanahan T, McEntee K. Specific transcripts are elevated in Saccharomyces cerevisiae in response to DNA damage. Mol Cell Biol. 1984;4: 2356–2363. Available: https://www.ncbi.nlm.nih.gov/pubmed/6440006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fornace AJ Jr, Alamo I Jr, Hollander MC. DNA damage-inducible transcripts in mammalian cells. Proc Natl Acad Sci U S A. 1988;85: 8800–8804. Available: https://www.ncbi.nlm.nih.gov/pubmed/3194391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270: 467–470. Available: https://www.ncbi.nlm.nih.gov/pubmed/7569999 [DOI] [PubMed] [Google Scholar]
- 39.Shalon D, Smith SJ, Brown PO. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 1996;6: 639–645. Available: https://www.ncbi.nlm.nih.gov/pubmed/8796352 [DOI] [PubMed] [Google Scholar]
- 40.Kafatos FC, Jones CW, Efstratiadis A. Determination of nucleic acid sequence homologies and relative concentrations by a dot hybridization procedure. Nucleic Acids Res. Oxford University Press; 1979;7: 1541–1552. doi: 10.1093/nar/7.6.1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jelinsky SA, Estep P, Church GM, Samson LD. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol Cell Biol. 2000;20: 8157–8167. Available: https://www.ncbi.nlm.nih.gov/pubmed/11027285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keller-Seitz MU, Certa U, Sengstag C, Würgler FE, Sun M, Fasullo M. Transcriptional response of yeast to aflatoxin B1: recombinational repair involving RAD51 and RAD1. Mol Biol Cell. 2004;15: 4321–4336. doi: 10.1091/mbc.e04-05-0375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caba E, Dickinson DA, Warnes GR, Aubrecht J. Differentiating mechanisms of toxicity using global gene expression analysis in Saccharomyces cerevisiae. Mutat Res. 2005;575: 34–46. doi: 10.1016/j.mrfmmm.2005.02.005 [DOI] [PubMed] [Google Scholar]
- 44.Mercier G, Berthault N, Touleimat N, Képès F, Fourel G, Gilson E, et al. A haploid-specific transcriptional response to irradiation in Saccharomyces cerevisiae. Nucleic Acids Res. 2005;33: 6635–6643. doi: 10.1093/nar/gki959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo Y, Breeden LL, Fan W, Zhao LP, Eaton DL, Zarbl H. Analysis of cellular responses to aflatoxin B(1) in yeast expressing human cytochrome P450 1A2 using cDNA microarrays. Mutat Res. 2006;593: 121–142. doi: 10.1016/j.mrfmmm.2005.07.001 [DOI] [PubMed] [Google Scholar]
- 46.Workman CT, Mak HC, McCuine S, Tagne B, Agarwal M, Ozier O, et al. A systems approach to mapping DNA damage response pathways. Science. 2006;312: 1054–1059. doi: 10.1126/science.1122088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fry RC, DeMott MS, Cosgrove JP, Begley TJ, Samson LD, Dedon PC. The DNA-damage signature in Saccharomyces cerevisiae is associated with single-strand breaks in DNA. BMC Genomics. 2006;7: 313. doi: 10.1186/1471-2164-7-313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beyer A, Workman C, Hollunder J, Radke D, Möller U, Wilhelm T, et al. Integrated assessment and prediction of transcription factor binding. PLoS Comput Biol. 2006;2: e70. doi: 10.1371/journal.pcbi.0020070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rusyn I, Fry RC, Begley TJ, Klapacz J, Svensson JP, Ambrose M, et al. Transcriptional networks in S. cerevisiae linked to an accumulation of base excision repair intermediates. PLoS One. 2007;2: e1252. doi: 10.1371/journal.pone.0001252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chu Z, Li J, Eshaghi M, Peng X, Karuturi RKM, Liu J, et al. Modulation of Cell Cycle–specific Gene Expressions at the Onset of S Phase Arrest Contributes to the Robust DNA Replication Checkpoint Response in Fission Yeas. MBoC. American Society for Cell Biology (mboc); 2007;18: 1756–1767. doi: 10.1091/mbc.e06-10-0928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fu Y, Zhu Y, Zhang K, Yeung M, Durocher D, Xiao W. Rad6-Rad18 mediates a eukaryotic SOS response by ubiquitinating the 9-1-1 checkpoint clamp. Cell. 2008;133: 601–611. doi: 10.1016/j.cell.2008.02.050 [DOI] [PubMed] [Google Scholar]
- 52.Shalem O, Dahan O, Levo M, Martinez MR, Furman I, Segal E, et al. Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation. Mol Syst Biol. 2008;4: 223. doi: 10.1038/msb.2008.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jaehnig EJ, Kuo D, Hombauer H, Ideker TG, Kolodner RD. Checkpoint kinases regulate a global network of transcription factors in response to DNA damage. Cell Rep. 2013;4: 174–188. doi: 10.1016/j.celrep.2013.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsang CK, Liu Y, Thomas J, Zhang Y, Zheng XFS. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat Commun. 2014;5: 3446. doi: 10.1038/ncomms4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lindgren E, Hägg S, Giordano F, Björkegren J, Ström L. Inactivation of the budding yeast cohesin loader Scc2 alters gene expression both globally and in response to a single DNA double strand break. Cell Cycle. 2014;13: 3645–3658. doi: 10.4161/15384101.2014.964108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Z, Humphryes N, Van Eijk P, Waters R. UV induced ubiquitination of the yeast Rad4– Rad23 complex promotes survival by regulating cellular dNTP pools. Nucleic acids. academic.oup.com; 2015; Available: https://academic.oup.com/nar/article-abstract/43/15/7360/2414360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Travesa A, Kuo D, de Bruin RAM, Kalashnikova TI, Guaderrama M, Thai K, et al. DNA replication stress differentially regulates G1/S genes via Rad53-dependent inactivation of Nrm1. EMBO J. 2012;31: 1811–1822. doi: 10.1038/emboj.2012.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bastos de Oliveira FM, Harris MR, Brazauskas P, de Bruin RAM, Smolka MB. Linking DNA replication checkpoint to MBF cell-cycle transcription reveals a distinct class of G1/S genes. EMBO J. 2012;31: 1798–1810. doi: 10.1038/emboj.2012.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dutta C, Patel PK, Rosebrock A, Oliva A, Leatherwood J, Rhind N. The DNA replication checkpoint directly regulates MBF-dependent G1/S transcription. Mol Cell Biol. 2008;28: 5977–5985. doi: 10.1128/MCB.00596-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wittenberg C, Reed SI. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene. 2005;24: 2746–2755. doi: 10.1038/sj.onc.1208606 [DOI] [PubMed] [Google Scholar]
- 61.Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409: 533–538. doi: 10.1038/35054095 [DOI] [PubMed] [Google Scholar]
- 62.Tercero JA, Longhese MP, Diffley JFX. A central role for DNA replication forks in checkpoint activation and response. Mol Cell. 2003;11: 1323–1336. Available: https://www.ncbi.nlm.nih.gov/pubmed/12769855 [DOI] [PubMed] [Google Scholar]
- 63.Huang M, Zhou Z, Elledge SJ. The DNA Replication and Damage Checkpoint Pathways Induce Transcription by Inhibition of the Crt1 Repressor. Cell. 1998;94: 595–605. doi: 10.1016/S0092-8674(00)81601-3 [DOI] [PubMed] [Google Scholar]
- 64.Zaim J, Speina E, Kierzek AM. Identification of New Genes Regulated by the Crt1 Transcription Factor, an Effector of the DNA Damage Checkpoint Pathway in Saccharomyces cerevisiae. J Biol Chem. 2005;280: 28–37. doi: 10.1074/jbc.M404669200 [DOI] [PubMed] [Google Scholar]
- 65.Bertoli C, Herlihy AE, Pennycook BR, Kriston-Vizi J, de Bruin RAM. Sustained E2F-Dependent Transcription Is a Key Mechanism to Prevent Replication-Stress-Induced DNA Damage. Cell Rep. 2016;15: 1412–1422. doi: 10.1016/j.celrep.2016.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lubelsky Y, Reuven N, Shaul Y. Autorepression of rfx1 gene expression: functional conservation from yeast to humans in response to DNA replication arrest. Mol Cell Biol. 2005;25: 10665–10673. doi: 10.1128/MCB.25.23.10665-10673.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408: 433–439. doi: 10.1038/35044005 [DOI] [PubMed] [Google Scholar]
- 68.Bartek J, Lukas J. Pathways governing G1/S transition and their response to DNA damage. FEBS Lett. 2001;490: 117–122. Available: https://www.ncbi.nlm.nih.gov/pubmed/11223026 [DOI] [PubMed] [Google Scholar]
- 69.Lukas J, Lukas C, Bartek J. Mammalian cell cycle checkpoints: signalling pathways and their organization in space and time. DNA Repair. 2004;3: 997–1007. doi: 10.1016/j.dnarep.2004.03.006 [DOI] [PubMed] [Google Scholar]
- 70.Mailand N, Falck J, Lukas C, Syljuâsen RG, Welcker M, Bartek J, et al. Rapid destruction of human Cdc25A in response to DNA damage. Science. 2000;288: 1425–1429. Available: https://www.ncbi.nlm.nih.gov/pubmed/10827953 [DOI] [PubMed] [Google Scholar]
- 71.Ivanova T, Alves-Rodrigues I, Gómez-Escoda B, Dutta C, DeCaprio JA, Rhind N, et al. The DNA damage and the DNA replication checkpoints converge at the MBF transcription factor. Mol Biol Cell. 2013;24: 3350–3357. doi: 10.1091/mbc.E13-05-0257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bedard LL, Massey TE. Aflatoxin B1-induced DNA damage and its repair. Cancer Lett. 2006;241: 174–183. doi: 10.1016/j.canlet.2005.11.018 [DOI] [PubMed] [Google Scholar]
- 73.Su C, Gao G, Schneider S, Helt C, Weiss C, O’Reilly MA, et al. DNA damage induces downregulation of histone gene expression through the G1 checkpoint pathway. EMBO J. 2004;23: 1133–1143. doi: 10.1038/sj.emboj.7600120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Venkata Narayanan I, Paulsen MT, Bedi K, Berg N, Ljungman EA, Francia S, et al. Transcriptional and post-transcriptional regulation of the ionizing radiation response by ATM and p53. Sci Rep. 2017;7: 43598. doi: 10.1038/srep43598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yan S, Wang W, Marqués J, Mohan R, Saleh A, Durrant WE, et al. Salicylic acid activates DNA damage responses to potentiate plant immunity. Mol Cell. 2013;52: 602–610. doi: 10.1016/j.molcel.2013.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang L, Chen H, Wang C, Hu Z, Yan S. Negative regulator of E2F transcription factors links cell cycle checkpoint and DNA damage repair. Proc Natl Acad Sci U S A. 2018;115: E3837–E3845. doi: 10.1073/pnas.1720094115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Horvath BM, Kourova H, Nagy S, Nemeth E, Magyar Z, Papdi C, et al. Arabidopsis RETINOBLASTOMA RELATED directly regulates DNA damage responses through functions beyond cell cycle control. EMBO J. 2017;36: 1261–1278. doi: 10.15252/embj.201694561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu Y, Lai J, Yu M, Wang F, Zhang J, Jiang J, et al. The Arabidopsis SUMO E3 Ligase AtMMS21 Dissociates the E2Fa/DPa Complex in Cell Cycle Regulation. Plant Cell. 2016;28: 2225–2237. doi: 10.1105/tpc.16.00439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu P, Yuan D, Liu M, Li C, Liu Y, Zhang S, et al. AtMMS21, an SMC5/6 complex subunit, is involved in stem cell niche maintenance and DNA damage responses in Arabidopsis roots. Plant Physiol. 2013;161: 1755–1768. doi: 10.1104/pp.112.208942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Inoue Y, Kitagawa M, Taya Y. Phosphorylation of pRB at Ser612 by Chk1/2 leads to a complex between pRB and E2F-1 after DNA damage. EMBO J. EMBO Press; 2007;26: 2083–2093. doi: 10.1038/sj.emboj.7601652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Carnevale J, Palander O, Seifried LA, Dick FA. DNA damage signals through differentially modified E2F1 molecules to induce apoptosis. Mol Cell Biol. 2012;32: 900–912. doi: 10.1128/MCB.06286-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ambrus AM, Islam ABMMK, Holmes KB, Moon NS, Lopez-Bigas N, Benevolenskaya EV, et al. Loss of dE2F compromises mitochondrial function. Dev Cell. 2013;27: 438–451. doi: 10.1016/j.devcel.2013.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Carr SM, Munro S, Kessler B, Oppermann U, La Thangue NB. Interplay between lysine methylation and Cdk phosphorylation in growth control by the retinoblastoma protein. EMBO J. 2011;30: 317–327. doi: 10.1038/emboj.2010.311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Saddic LA, West LE, Aslanian A, Yates JR 3rd, Rubin SM, Gozani O, et al. Methylation of the retinoblastoma tumor suppressor by SMYD2. J Biol Chem. 2010;285: 37733–37740. doi: 10.1074/jbc.M110.137612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thurlings I, Martínez-López LM, Westendorp B, Zijp M, Kuiper R, Tooten P, et al. Synergistic functions of E2F7 and E2F8 are critical to suppress stress-induced skin cancer. Oncogene. 2017;36: 829–839. doi: 10.1038/onc.2016.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mitxelena J, Apraiz A, Vallejo-Rodríguez J, García-Santisteban I, Fullaondo A, Alvarez-Fernández M, et al. An E2F7-dependent transcriptional program modulates DNA damage repair and genomic stability. Nucleic Acids Res. 2018;46: 4546–4559. doi: 10.1093/nar/gky218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rashi-Elkeles S, Warnatz H-J, Elkon R, Kupershtein A, Chobod Y, Paz A, et al. Parallel profiling of the transcriptome, cistrome, and epigenome in the cellular response to ionizing radiation. Sci Signal. 2014;7: rs3. doi: 10.1126/scisignal.2005032 [DOI] [PubMed] [Google Scholar]
- 88.Verfaillie A, Imrichová H, Van de Sande B, Standaert L, Christiaens V, Hulselmans G, et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol. Public Library of Science; 2014;10: e1003731 Available: http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1003731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Macheret M, Halazonetis TD. DNA replication stress as a hallmark of cancer. Annu Rev Pathol. 2015;10: 425–448. doi: 10.1146/annurev-pathol-012414-040424 [DOI] [PubMed] [Google Scholar]
- 90.Sarni D, Kerem B. Oncogene-Induced Replication Stress Drives Genome Instability and Tumorigenesis. Int J Mol Sci. Multidisciplinary Digital Publishing Institute; 2017;18: 1339. doi: 10.3390/ijms18071339 [DOI] [Google Scholar]
- 91.Kotsantis P, Petermann E, Boulton SJ. Mechanisms of Oncogene-Induced Replication Stress: Jigsaw Falling into Place. Cancer Discov. 2018;8: 537–555. doi: 10.1158/2159-8290.CD-17-1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11: 208–219. doi: 10.1038/nrm2852 [DOI] [PubMed] [Google Scholar]
- 93.Brewer BJ. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell. 1988;53: 679–686. Available: https://www.ncbi.nlm.nih.gov/pubmed/3286014 [DOI] [PubMed] [Google Scholar]
- 94.Brewer BJ, Fangman WL. A replication fork barrier at the 3’ end of yeast ribosomal RNA genes. Cell. 1988;55: 637–643. Available: https://www.ncbi.nlm.nih.gov/pubmed/3052854 [DOI] [PubMed] [Google Scholar]
- 95.Necşulea A, Guillet C, Cadoret J-C, Prioleau M-N, Duret L. The Relationship between DNA Replication and Human Genome Organization. Mol Biol Evol. Oxford University Press; 2009;26: 729–741. doi: 10.1093/molbev/msn303 [DOI] [PubMed] [Google Scholar]
- 96.Jones RM, Mortusewicz O, Afzal I, Lorvellec M, García P, Helleday T, et al. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene. 2013;32: 3744–3753. doi: 10.1038/onc.2012.387 [DOI] [PubMed] [Google Scholar]
- 97.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448: 445–451. doi: 10.1038/nature05953 [DOI] [PubMed] [Google Scholar]
- 98.Liu F, Wang L, Perna F, Nimer SD. Beyond transcription factors: how oncogenic signalling reshapes the epigenetic landscape. Nat Rev Cancer. 2016;16: 359–372. doi: 10.1038/nrc.2016.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lavigne MD, Konstantopoulos D, Ntakou-Zamplara KZ, Liakos A, Fousteri M. Global unleashing of transcription elongation waves in response to genotoxic stress restricts somatic mutation rate. Nat Commun. 2017;8: 2076. doi: 10.1038/s41467-017-02145-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Manfrini N, Clerici M, Wery M, Colombo CV, Descrimes M, Morillon A, et al. Resection is responsible for loss of transcription around a double-strand break in Saccharomyces cerevisiae. Elife. 2015;4. doi: 10.7554/eLife.08942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141: 970–981. doi: 10.1016/j.cell.2010.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fleming AM, Ding Y, Burrows CJ. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc Natl Acad Sci U S A. 2017;114: 2604–2609. doi: 10.1073/pnas.1619809114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Russo G, Landi R, Pezone A, Morano A, Zuchegna C, Romano A, et al. DNA damage and Repair Modify DNA methylation and Chromatin Domain of the Targeted Locus: Mechanism of allele methylation polymorphism. Sci Rep. 2016;6: 33222. doi: 10.1038/srep33222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shah MA, Schwartz GK. Cell cycle-mediated drug resistance: an emerging concept in cancer therapy. Clin Cancer Res. 2001;7: 2168–2181. Available: https://www.ncbi.nlm.nih.gov/pubmed/11489790 [PubMed] [Google Scholar]
- 105.Luscombe NM, Babu MM, Yu H, Snyder M, Teichmann SA, Gerstein M. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431: 308–312. doi: 10.1038/nature02782 [DOI] [PubMed] [Google Scholar]
- 106.Boeing S, Williamson L, Encheva V, Gori I, Saunders RE, Instrell R, et al. Multiomic Analysis of the UV-Induced DNA Damage Response. Cell Rep. 2016;15: 1597–1610. doi: 10.1016/j.celrep.2016.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Roberts JJ, Pascoe JM, Plant JE, Sturrock JE, Crathorn AR. Quantitative aspects of the repair of alkylated DNA in cultured mammalian cells: I. The effect on HeLa and Chinese hamster cell survival of alkylation of cellular macromolecules. Chem Biol Interact. 1971;3: 29–47. doi: 10.1016/0009-2797(71)90024-X [DOI] [PubMed] [Google Scholar]
- 108.Riley PA. Free radicals in biology: oxidative stress and the effects of ionizing radiation. Int J Radiat Biol. 1994;65: 27–33. Available: https://www.ncbi.nlm.nih.gov/pubmed/7905906 [DOI] [PubMed] [Google Scholar]
- 109.Mostofa AGM, Punganuru SR, Madala HR, Srivenugopal KS. S-phase Specific Downregulation of Human O6-Methylguanine DNA Methyltransferase (MGMT) and its Serendipitous Interactions with PCNA and p21cip1 Proteins in Glioma Cells [Internet]. Neoplasia. 2018. pp. 305–323. doi: 10.1016/j.neo.2018.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Christmann M, Roos WP, Kaina B. DNA Methylation Damage: Formation, Repair and Biological Consequences In: Vijayalaxmi Obe G, editors. Chromosomal Alterations: Methods, Results and Importance in Human Health. Berlin, Heidelberg: Springer Berlin Heidelberg; 2007. pp. 99–121. doi: 10.1007/978-3-540-71414-9_7 [DOI] [Google Scholar]
- 111.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431: 1011–1017. doi: 10.1038/nature02964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.de Bruin RAM, Kalashnikova TI, Aslanian A, Wohlschlegel J, Chahwan C, Yates JR 3rd, et al. DNA replication checkpoint promotes G1-S transcription by inactivating the MBF repressor Nrm1. Proc Natl Acad Sci U S A. 2008;105: 11230–11235. doi: 10.1073/pnas.0801106105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mazumder A, Pesudo LQ, McRee S, Bathe M, Samson LD. Genome-wide single-cell-level screen for protein abundance and localization changes in response to DNA damage in S. cerevisiae. Nucleic Acids Res. 2013;41: 9310–9324. doi: 10.1093/nar/gkt715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tkach JM, Yimit A, Lee AY, Riffle M, Costanzo M, Jaschob D, et al. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat Cell Biol. 2012;14: 966–976. doi: 10.1038/ncb2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5: 1512–1526. doi: 10.1039/b908315d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee MV, Topper SE, Hubler SL, Hose J, Wenger CD, Coon JJ, et al. A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol Syst Biol. 2011;7: 514. doi: 10.1038/msb.2011.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Carvajal LA, Hamard P-J, Tonnessen C, Manfredi JJ. E2F7, a novel target, is up-regulated by p53 and mediates DNA damage-dependent transcriptional repression. Genes Dev. 2012;26: 1533–1545. doi: 10.1101/gad.184911.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jackson MW, Agarwal MK, Yang J, Bruss P, Uchiumi T, Agarwal ML, et al. p130/p107/p105Rb-dependent transcriptional repression during DNA-damage-induced cell-cycle exit at G2. J Cell Sci. 2005;118: 1821–1832. doi: 10.1242/jcs.02307 [DOI] [PubMed] [Google Scholar]
- 119.Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, et al. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol Cell. 2007;28: 860–870. doi: 10.1016/j.molcel.2007.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Endres L, Begley U, Clark R, Gu C, Dziergowska A, Małkiewicz A, et al. Alkbh8 Regulates Selenocysteine-Protein Expression to Protect against Reactive Oxygen Species Damage. PLoS One. 2015;10: e0131335. doi: 10.1371/journal.pone.0131335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Boucas J, Riabinska A, Jokic M, Herter-Sprie GS, Chen S, Höpker K, et al. Posttranscriptional regulation of gene expression—adding another layer of complexity to the DNA damage response. Front Genet. 2012;3. doi: 10.3389/fgene.2012.00159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McKay BC. Post-transcriptional regulation of DNA damage-responsive gene expression. Antioxid Redox Signal. 2014;20: 640–654. doi: 10.1089/ars.2013.5523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shkreta L, Chabot B. The RNA Splicing Response to DNA Damage [Internet]. Biomolecules. 2015. pp. 2935–2977. doi: 10.3390/biom5042935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Naro C, Bielli P, Pagliarini V, Sette C. The interplay between DNA damage response and RNA processing: the unexpected role of splicing factors as gatekeepers of genome stability. Front Genet. 2015;6: 142. doi: 10.3389/fgene.2015.00142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wickramasinghe VO, Venkitaraman AR. RNA Processing and Genome Stability: Cause and Consequence. Mol Cell. 2016;61: 496–505. doi: 10.1016/j.molcel.2016.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang X, Lu X. Posttranscriptional regulation of miRNAs in the DNA damage response. RNA Biol. 2011;8: 960–963. doi: 10.4161/rna.8.6.17337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Truitt ML, Ruggero D. New frontiers in translational control of the cancer genome. Nat Rev Cancer. 2016;16: 288–304. doi: 10.1038/nrc.2016.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fry RC, Begley TJ, Samson LD. Genome-wide responses to DNA-damaging agents. Annu Rev Microbiol. 2005;59: 357–377. doi: 10.1146/annurev.micro.59.031805.133658 [DOI] [PubMed] [Google Scholar]
- 129.Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD, et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018;23: 239–254.e6. doi: 10.1016/j.celrep.2018.03.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schiewer MJ, Mandigo AC, Gordon N, Huang F, Gaur S, de Leeuw R, et al. PARP-1 regulates DNA repair factor availability. EMBO Mol Med. 2018;10. doi: 10.15252/emmm.201708816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gaddameedhi S, Reardon JT, Ye R, Ozturk N, Sancar A. Effect of circadian clock mutations on DNA damage response in mammalian cells. Cell Cycle. 2012;11: 3481–3491. doi: 10.4161/cc.21771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hogenesch JB. It’s all in a day’s work: Regulation of DNA excision repair by the circadian clock. Proc Natl Acad Sci U S A. 2009;106: 2481–2482. doi: 10.1073/pnas.0813323106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sancar A, Lindsey-Boltz LA, Kang T-H, Reardon JT, Lee JH, Ozturk N. Circadian clock control of the cellular response to DNA damage. FEBS Lett. 2010;584: 2618–2625. doi: 10.1016/j.febslet.2010.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fang M, Ohman Strickland PA, Kang H-G, Zarbl H. Uncoupling genotoxic stress responses from circadian control increases susceptibility to mammary carcinogenesis. Oncotarget. 2017;8: 32752–32768. doi: 10.18632/oncotarget.15678 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.