Abstract
Of numerous genes regulated by retinoic acid (RA), CYP26A1 is the most inducible gene by RA. In this study, we have used a shortened construct form, E4, of the CYP26A1 gene promoter, in a promoter-less vector with either luciferase or red fluorescent protein (RFP) as the reporter gene and have tested its responses to retinoids in transfected HepG2 and HEK293T cells. The promoter responded linearly to a wide concentration range of RA in cells cotransfected with retinoic acid receptors. It also responded quantitatively to retinol and other retinoids. An isolated clonal line of HEK293T cells permanently transfected with the promoter driving the expression of RFP responded to both RA and retinol, and the responses could be measured by fluorescence microscopy and flow cytometry. The promoter was used to assess the retinoid activity of 3 novel synthetic retinoid analogues, as well as of the intact serum samples of rats. Among the synthetic retinoid analogues tested, EC23 is more potent than RA at lower concentrations and was more stable than RA. The retinoid activities could be measured in control rat serum samples and were increased in the serum of RA-treated rats. This system offers a biologically-based alternative to massbased retinoid analysis.
Keywords: CYP26A1 Gene promoter, Retinol, Retinoic acid, Red fluorescent protein, Luciferase assay
1. Introduction
Retinoic acid (RA), a major active metabolite of vitamin A, functions as a pan-agonist ligand of nuclear retinoic acid receptors (RARα, β, and γ), which partner with retinoid X receptors (RXRα, β, and γ) and bind specifically to retinoic acid response elements (RARE) in target genes [1–4]. The canonical RARE consists of a core of two hexameric motifs of PuG(G/T)TCA(X)nPuG(G/T)TCA, generally oriented as a direct repeat (DR) spaced by two or five nucleotides (called DR2 or DR5). Tissue levels of RA are regulated both through synthesis by a series of dehydrogenases from retinol and oxidative catabolism by CYP26 enzymes [5]. RA is not only the specific substrate for CYP26 enzymes but also is the key regulator for these genes. CYP26A1, as a member of the CYP26 gene subfamily [6], has been shown to be highly inducible by RA during embryonic development as well as in adult tissues including the liver. Of the numerous genes regulated by RA only a few possess a perfect functional retinoic acid response element, known as DR5, present in the promoter region upstream of the transcription start site [7]. CYP26A1 is one of these genes with at least 3 DR5 elements in addition one DR half site present within 2 kb upstream of the transcription start site [8–10]. Of the three functional DR5 elements, one is located in the proximal region and the other two together with one functional DR half site are present in the distal region within 2 kb upstream of the promoter region of the transcription start site [9, 10].
Previously we cloned the full-length (FL) promoter of the human CYP26A1 gene into pGL3basic-luc with luciferase as the reporter gene [9]. By a deletion study, we eliminated the intermediary region lacking the RA response elements and brought together the distal region close to the proximal region to make a construct (termed E4) which was shown to exhibit a stronger response to RA when co-transfected with retinoic acid receptors [10]. In the current study, we have used this promoter construct in the pGL3-basic vector with either luciferase or mCherry red fluorescent protein (RFP) as the reporter gene to evaluate the promoter response to retinoids in human hepatoma HepG2 cells and human embryonic kidney HEK293T cells when the cells are cotransfected with retinoid receptors. The promoter responded in a roughly linear manner to logarithmic concentrations of RA and could potentially be used to estimate the retinoid activities of novel synthetic retinoids as well as rat serum samples.
2. Material and Methods
2.1. Materials
Human hepatoma HepG2 cells, and human embryonic kidney HEK293T cells were both obtained from the American Type Culture Collection (ATCC, Manassas, VA). HepG2 cells were maintained in Eagle’s Minimum Essential Medium (EMEM) and HEK293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM), each supplemented with 10% heat inactivated fetal bovine serum (FBS) and 0.5% penicillin-streptomycin at 37 ºC in a 5% CO2-air incubator. The cells were plated and used at 60 to 70% confluency.
All-trans (AT)Retinol and all-trans- retinoic acid (ATRA) were purchased from Sigma–Aldrich Chemical Company and prepared as stock solutions at 1 and 10 mM in ethanol. Am580, a specific ligand for RARα, was kindly provided by Dr. K. Shudo and Y. Kadeshika. Plasmid vectors including pGEMT-Easy, pGL3-Basic-luc and pRLTK, were purchased from Promega (Madison, WI) and pcDNA3.1 plasmid vector for protein expression was from Invitrogen (Carsbad, CA). MMLV Reverse transcriptase, oligo dT nucleotide, T4 polynucleotide kinase, and T4 DNA ligase were all from Promega, High Fidelity Taq DNA polymerase, Lipofectamine 2000 transfection reagent and Trizol reagent were from Invitrogen. Reagents for RT-qPCR was purchased from Thermo-Fisher (https://www.thermofisher.com), plasmid DNA Purification Kits were from Qiagen (Valencia, CA), and DNA gel extraction kit was from Omega Bio-Tek (Thermo Scientific). All the restriction enzymes were obtained from New England Biolab (Ipswich, MA). The full-length (FL) promoter of CYP26A1 gene was cloned from a human CYP26A1 genomic clone containing the extended 5′- and 3′-flanking regions in FOSMID vector (obtained from Children’s Hospital Oakland-BACPAC Resources, Oakland, CA) and the deleted CYP26A1 promoter clone in pGL3-Basic-luc vector were reported previously [9]. For preparation of all other reagents in our laboratory, we followed the protocols reported in [11].
2.3. Synthetic retinoids
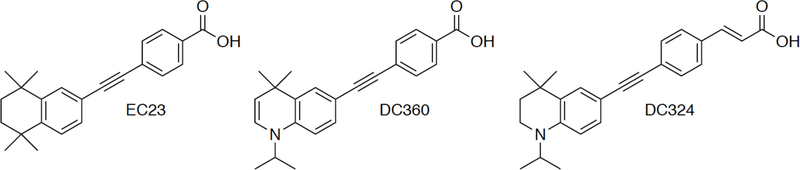
EC23, DC324 and DC360 (Figure 1) were synthesized according to the published procedures [12–15]. EC23 and DC360 are synthetic analogues of ATRA, and DC324 is a nonactive analogue that exhibits poor binding affinity for the retinoic acid receptors due to the compound’s extended structure.
Figure 1. Structure of Synthetic Retinoids.
2.4. Animal experiment
Procedures for animal use were approved by The Pennsylvania State University’s Animal Care and Use Committee. Adult female rats were orally treated once with either canola oil, as the vehicle, or ATRA at 1 mg, 2 mg or 5 mg/kg body weight for 6 h and then euthanized by CO2 inhalation for blood withdrawal. Serum samples were prepared by centrifugation and stored frozen at −20oC until use. For determination of total retinoid activity, serum samples were added directly to DMEM media without FBS at 20% dilution (v/v), and incubated, with cells; both retinol and RA added directly to 20% FBS-medium were used as standards.
2.5. RNA extraction and analysis
Total RNA was extracted from the cells using Trizol reagent and then dissolved in DEPCtreated autoclaved water for analysis. The RNA samples were quantified by Nano-Drop spectrophotometry and then reverse transcribed using MMLV reverse transcriptase and oligo dT from Promega in a 25 µl reaction mixture following the procedure recommended by the manufacturer. The first stranded cDNA was first diluted and then analyzed by realtime PCR for CYP26A1 and RARβ mRNA transcripts with 18 S ribosomal RNA as an internal control using primer pairs (Table 1). The PCR program was set to run first at 94 °C for 10 min for activation of the polymerase and then 40 cycles of 20 s at 94°C for melting, 30 s at 60°C for annealing, 30 s at 72°C for extension.
Table 1.
List of primers for RT-PCR analysis and cloning.
| Gene name | Analysis | Primer Pairs |
|---|---|---|
| CYP26A1 | RT-PCR | S: GCTGCCTCTCTAACCTGCAC A: TGCTTTAGTGCCTGCATGTC |
| RARβ | RT-PCR | S: GTACCACTATGGGGTCAGCG A: AATTTGTCCCAGAGGCCCAG |
| 18S Ribosomal RNA | RT-PCR | S: CGCGGTTCTATTTTGTTGGT A: AGTCGGCATCGTTTATGGTC |
| RARα | Cloning | S: atgctagcGCCACCATGGCCAGCAACAGCAGC A: ttaagcttCACGGGGAGTGGGTGGC |
| RARβ | Cloning | S: atgctagcGCCACCATGACCACCAGCGGCCACG A: ttaagcttATTGCACGAGTGGTGACTG |
| RARγ | Cloning | S: atgctagcGCCACCATGGCCACCAATAAGGAGC A: ttaagcttCAGGCTGGGGACTTCAGGC |
| RXRα | Cloning | S: gccaccatggACACCAAACATTTCCTG A: tattcgaaCTAAGTCATTTGGTGCGGCG |
| RARβ Gene promoter | Cloning | F: CCAATGCACATTCCAACACT R: GATCCCAAGTTCTCCTTCCA |
| mCherry | Cloning and RT-PCR | S: aaccatggTGAGCAAGGGCGAGG A: aatctagaTTACTTGTACAGCTCGTCCATG |
Lower case letters indicate the restriction sites for cloning
S: Sense; A: Antisense; F: Forward; R: Reverse
2.6. Cloning the human retinoid receptors, human RARβ promoter and mCherry open reading frame
Human RARα, RARβ, RARγ and RXRα were cloned from poly A+ RNA isolated from human liver samples [16]. Poly A+ RNA was first reverse transcribed using MMLV reverse transcriptase and oligo dT from Promega in a 25 µl reaction mixture. Following dilution to 150 µl with deionized water, 5 µl was used to amplify the open reading frame of retinoid receptors with specific primer pairs for RARα, RARβ, RARγ, and RXRα (Table 1) using High Fidelity Taq DNA polymerase from Invitrogen following the manufacturer protocol for 30 cycles. The PCR product was run on agarose gel by electrophoresis and the DNA band was cut and extracted from the gel using DNA gel extraction kit. The DNA was first cloned into pGEMT-Easy vector and then subjected to DNA sequencing for confirmation. The isolated plasmid containing the receptor DNA was double digested with Bam HI/XbaI and subcloned into the pcDNA3.1(+) expression vector. The human retinoid expression vectors were first transformed into JM109 bacteria for screening and amplification and then purified using Plasmid Midi Kit from Qiagen. The DNA expression plasmid was subjected to sequencing for confirmation in the Nucleic Acid Facility, Pennsylvania State University. For construction of pcDNA3.1-hRAR-hRXRα expression vectors, the pcDNA3.1-RAR was first double digested with StuI or SmaI/BstBI to remove the DNA reading frame for neomycin and replaced with ORF for RXRα. The plasmid DNA was transformed into bacteria, amplified and then purified as described above.
An about 2.0 kb DNA fragment of the human RARβ gene was cloned by PCR from human liver genomic DNA using a specific primer pair (Table 1) and High Fidelity Taq DNA polymerase as described above [17]. The DNA fragment including the full length RARβ promoter and partial 5’-UTR of the RARβ mRNA was cloned in pGEM-T Easy plasmid vector by TA cloning, double digested with NdeI/SacII, blunt-ended and then sub-cloned into the Sma-I site of the pGL3-Basic-Luciferase.
A DNA fragment containing the mCherry ORF was first amplified by PCR from a commercial plasmid vector using 5’- aaccatggTGAGCAAGGGCGAGG-3’ as the forward and 5’- aatctagaTTACTTGTACAGCTCGTCCATG-3’ as the reverse primers (lower case letters indicate the restriction sites for cloning) and the cloned into the pGEM-T Easy vector by TA-cloning. An isolated clone was subjected to DNA sequencing for confirmation and then double digested with NcoI/XbaI to obtain the mCherry fragment for subcloning into pGL3Basic plasmid vector. For construction of pGL3-Basic-hCYP26A1P-E4-mCherry clone containing the human CYP26A1 promoter, the pGL3-Basic- hCYP26A1P-E4 Luciferase [9, 10] was first double digested with NcoI/XbaI to remove luciferase DNA fragment and then replaced with mCherry ORF using T4-DNA ligase.
2.7. Transfection, luciferase assay and RFP analysis
HepG2 cells grown in EMEM and HEK293T cells in DMEM, each with 10% fetal bovine serum in T-75 flask, were trypsinized and transferred into 12- or 24-well plates 1–2 day before transfection. When the cells were about 60 to 70% confluent, the pGL3-BasichCYP26A1-E4-luc plasmid constructs, together with the expression vectors, were added into the same medium containing 3% fetal bovine serum with no antibiotics using Lipofectamine 2000 following the protocol recommended for transient transfection. pRLTK plasmid containing the Renilla-Luc gene was cotransfected to provide an internal control for transfection efficiency. After overnight transfection the cells were incubated with fullgrowth medium containing 10% fetal bovine serum and 0–1 µM RA, or retinol in ethanol at the final concentration of 0.001% ethanol. After incubation at 37 °C for up to 24 h the cells were washed with PBS and then lysed to assay for firefly-Luc and Renilla-luc activities using the DRL luciferase assay system from Promega in Luminator 20. Promoter activity was defined as the ratio of firefly-luciferase to Renilla-luciferase activity. Each reported activity is an average of 3 wells with standard error of the mean.
For RFP analysis by fluorescence microscopy, the pGL3-Basic-hCYP26A1-E4-mCherry plasmid construct together with the retinoid expression vectors and p-YFP vector as an internal control were transiently transfected and then cells were treated with RA as described above. Following treatment with RA the cells were observed using an Olympus IX70 inverted fluorescence microscope equipped with an Olympus DP72 camera and cellSens standard 1.14 image acquisition software. At least 3 regions of the monolayer cells in each well were evaluated and recorded. The RFP fluorescence microscopy data were analyzed, quantified, and normalized against YFP fluorescence as an internal control using NIH ImageJ 1.49d.
To make a stable cell line for expression of RFP, HEK293T cells were co-transfected with pGL3-basic-hCYP26A1-E4 promoter-mCherry vector together with human RARα and human RXRα expression vectors as described above. The cells were then transferred by trypsinization to a new plate with fresh DMEM medium containing 10% FBS and 300 µg/ml G418 as the selection marker. After 2 weeks, the live cells were grown in new plates and incubated with 1µM RA for 24 h. The cells expressing RFP were first cloned by limited dilution, then sorted into individual wells by a Beckman Coulter MoFlo Astrios EQ Cell Sorter based on RA induced RFP expression. The individual cells were grown in 96 well plates for 2 weeks and then transferred into 12-well plates with full growth medium containing G418. The resulting monoclonal lines were assessed for expression of RFP after treatment with RA under a fluorescence microscope.
2.8. Statistical analysis
All data shown are the mean ± standard deviation or standard error of the mean of at least 3 repeats in each experiment. The data were analyzed by one-way ANOVA followed by Fisher’s least significant difference test using Prism6 Statistical Software (GraphPad). A p value <0.05 was considered statistically significant.
3. Results
3.1. CYP26A1 gene is highly responsive to retinoic acid in the HepG2 hepatoma cells.
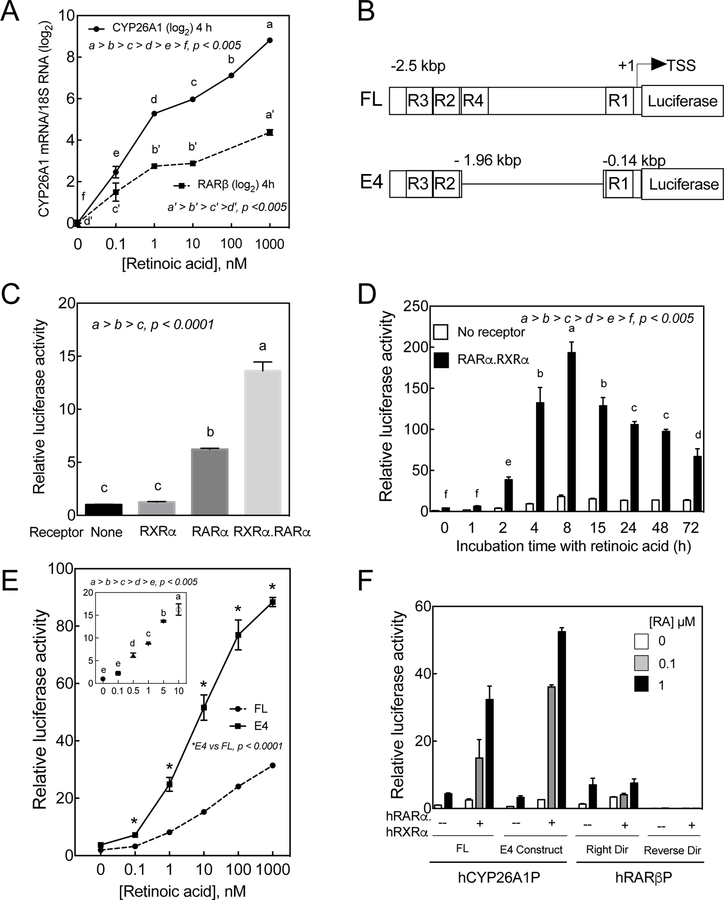
The endogenous mRNA level of CYP26A1 was nearly undetectable in HepG2 cells incubated in full growth medium for 4 h; however, upon addition of RA at concentrations as low as 0.1 nM, the expression level was increased by 5 fold, and further induced proportionally by all doses of RA (Figure 2A). As a comparison, the mRNA expression of RARβ, another potentially RA inducible gene, was also responsive to RA dose dependently although the response was of a much lower magnitude compared to the CYP26A1 gene (Figure 2A).
Figure 2. CYP26A1 gene is highly responsive to RA in HepG2 cells.
A. HepG2 cells were incubated with 0 to 1 µM RA for 4 h and then subjected to total RNA extraction and analysis for CYP26A1 and RARβ mRNA expression using 18S ribosomal RNA as the internal control. B. Schematic representation of the full-length* (FL) and deleted forms (E4) of hCYP26A1 gene in pGL3-basic vector containing luciferase as the reported gene. R1, R2, R3 and R4 represent retinoic acid response elements (RARE) C. HepG2 cells cotransfected with E4 construct together with an expression vector containing both RARα and RXRα were more responsive to RA (1 nM) than with the expression vector containing only RARα. D. Kinetic response to RA (1 µM) of HepG2 cells cotransfected with the E4 promoter construct of hCYP26A1 gene without or with the RARα.RXRα expression vector. E. Full-length or E4 constructs of hCYP26A1 gene responded dose dependently to RA in HepG2 cells cotransfected with RARα.RXRα expression vector. F. Both FL and E4 promoter constructs of hCYP26A1 gene responded significantly better to RA than FL promoter of RARβ promoter in HepG2 cells cotransfected with RARα.RXRα expression vector.
3.2. The promoter of the CYP26A1 gene is highly responsive to retinoic acid in HepG2 cells cotransfected with retinoic acid receptor alpha
Since the expression of CYP26A1 is highly responsive to RA, we initiated this study to evaluate the use of the CYP26A1 promoter as a tool to estimate vitamin A bioactivity levels, including retinoic acid in biological system. Previously, we have cloned the full-length (FL) promoter of CYP26A1 gene extending from a few nucleotides to 2.5 kb upstream of the ATG coding region into pGL3-basic-luc plasmid vector containing the coding region of firefly luciferase as a reporter gene [9] (Figure 2B). By a deletion we made a construct (E4 construct; Figure 2B) [9], which was shown to be highly responsive to RA in HepG2 cells cotransfected with RA receptors [10].
Retinoid X receptors act as partners with RARs to activate the retinoid responsive genes. Previously, we have shown that combination of RARα and RXRα are more effective than RARα alone on the promoter activity of the CYP26A1 gene in HepG2 cells in response to RA [10]. Both RARα and RXRα are expressed at much higher levels in the liver, particularly in hepatocytes, in comparison to the other retinoid receptors. Therefore, we first cloned these receptors individually from human liver and then placed both genes into one expression vector as described in Materials and Methods. This dual expression clone was cotransfected with the CYP26A1 promoter into HepG2 cells. Whereas RXRα alone was shown to have no significant effect on the promoter activity of CYP26A1 gene in HepG2 cells in response to RA, it increased the effectiveness of RARα by more than 2 fold (Figure 2C).
To evaluate the kinetic response of the CYP26A1 promoter to RA, HepG2 cells were cotransfected with the E4 promoter construct together with or without RARα.RXRα expression vector for 24 h after which the cells were incubated with 1 µM RA for 1 to 72 h. The cells were then washed and lysed in order to assess the resultant luciferase activity. The promoter activity rapidly increased approximately 40 times within 2 h, reached a maximum of 200 times after 8 h and then started to gradually decline after 15 to 72 h (Figure 2D). The receptors had a dramatic effect on the promoter response to RA.
Similar to the mRNA expression of CYP26A1 gene, the promoter activity responded proportionally to RA. HepG2 cells cotransfected with the promoter of CYP26A1 gene together with RAR.RXR expression vector were incubated with 0 to 1 µM RA for 4 h and then lysed to assay for luciferase activity. The promoter responded significantly to as low a concentration as 1 nM RA and proportionally further to higher concentrations up to 1µM (Figure 2E). Results from further experiments showed that the CYP26A1 promoter responded linearly to RA within the log concentration ranges between 0.1 to 10 nM (Figure 2E insert). At all treatment concentrations tested, the E4 promoter was shown to respond significantly higher than the FL promoter construct (Figure 2E).
3.3. mCherry RFP may be used as a reporter for the promoter of the CYP26A1 gene
To directly observe the promoter response to retinoids, we have cloned mCherry ORF and then subcloned into the luciferase frame site of the pGL3-basic-luc vector driven by the hCYP26A1 E4 promoter. This vector, together with RARα.RXRα expression vector and pCMV-YFP expression vector as the internal control were cotransfected into HepG2 cells after which the cells were incubated with RA for 24 h. The expression of RFP in response to RA treatment was evaluated by fluorescence microscopy as compared to the expression of YFP as an internal control for transfection. Expression of RFP was almost none in the transfected cells treated with vehicle (Figure S1). However, treatment of the cells with as low as 0.1 nM RA resulted in quantifiable expression of RPF in the cells (Figure S1). A gradual increase in the expression of RFP was observed in the cells treated with up to 10 nM RA (Figure S1), similar to that of luciferase used as the reporter gene (see Figure 2E).
3.4. CYP26A1 promoter is much more highly responsive to retinoic acid as compared to the promoter of RARβ gene
To compare the activity of the promoter of CYP26A1 gene in response to RA with that of RARβ gene, as another potential RA responsive gene, we have amplified from human liver a fragment of the RARβ promoter including the upstream proximal region of the gene and cloned into pGL3-basic-luc vector containing luciferase as the reporter gene as described in Materials and Methods. We then cotransfected this vector in the absence or presence of the RARα.RXRα expression vector into HepG2 cells and compared the results with those of FL and E4 promoter constructs of CYP26A1. In the absence of the receptors, the RARβ promoter responded to RA in a similar manner to those of the CYP26A1 promoter (Figure 2F). However, cotransfection of the receptors did not have any significant effect on the RARβ promoter, in contrast to those of the CYP26A1 promoters, which responded 10 to 50 times more strongly to RA. (Figure 2F).
3.5. RARα was shown to be more effective on the CYP26A1 promoter in response to retinoic acid than the other retinoid receptors
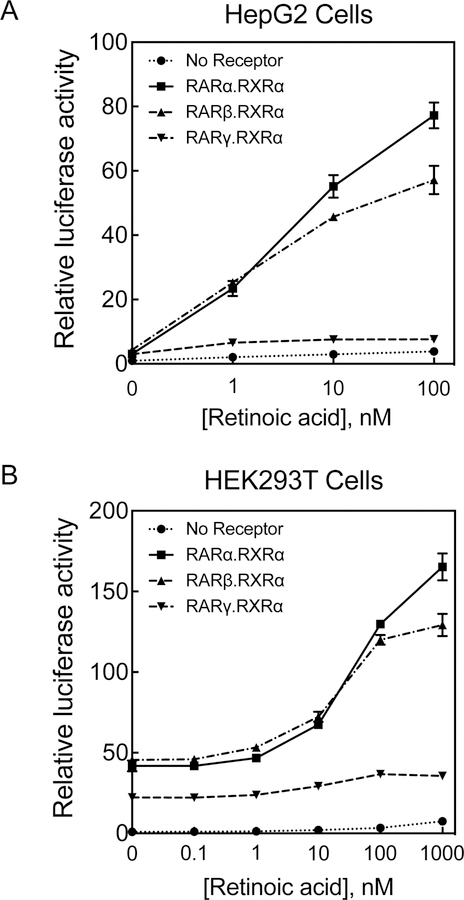
To compare the effectiveness of RARα with that of other RA receptors including hRARβ and hRARγ, on the promoter of CYP26A1 gene in response to RA we cloned each with hRXRα into pcDNA3.1 expression vectors using analogous procedure to that used for RARα (see Material and Methods). The E4 promoter construct of the CYP26A1 gene together with the individual receptors were cotransfected into the cells, after which the cells were incubated with RA at different concentrations for 24 h. The cells were then washed and lysed for luciferase activity assays. At low concentrations of RA, both RARα and RARβ had a similar effect in both HepG2 cells (Figure 3A) as well as HEK293T cells, the human embryonic kidney cells (Figure 3B). However, at higher concentrations, RARα was shown to have a stronger effect than RARβ on the promoter of CYP26A1 gene in response to RA (Figure 3A and 3B). Both RARα and RARβ had a stronger effect than RARγ at all the RA concentrations examined (Figure 3). Apparently, the CYP26A1 promoter was less responsive to RA at low concentrations in HEK293T cells (Figure 3B) than in HepG2 cells (Figure 3A). This may be due to the moderate background displayed by HEK293T cells in response to the cotransfected receptors without RA (Figure 3). This background appears absent in HepG2 cells. Similar results were observed when the CYP26A1 promoter with RFP as the reporter gene was used for cotransfection with either of the receptors in both HepG2 cells (Figure S2a) and HEK293T cells (Figure S2b).
Figure 3. RARα makes CYP26A1 promoter respond to RA better than either RARβ or RARγ in both HepG2 cells and HEK293T cells.
Mono-layered cells were cotransfected with CYP26A1 promoter E4 construct in pGL3-basic-luc together with either hRARα.hRXRα, hRARβ.hRXRα or hRARγ.hRXRα and then treated with different concentrations of RA for 24h. The cells were used for luciferase assay in HepG2 cells (A) and HEK293T cells (B).
3.6. CYP26A1 promoter responds to retinol and to different retinoid analogues
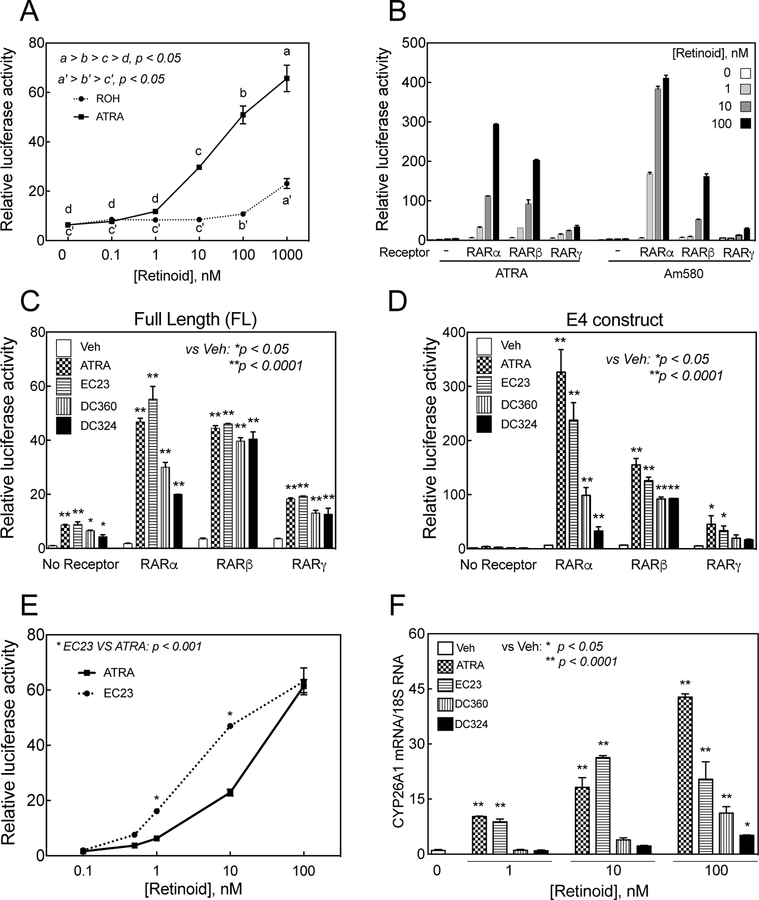
To determine whether the CYP26A1 promoter responds to retinol, HepG2 cells cotransfected with the E4 promoter construct of the human CYP26A1 gene together with the hRARα.hRXRα expression vector, were incubated with either retinol or RA at different concentrations for 24 h and then washed and lysed for luciferase activity. The results showed that the promoter did not respond significantly to retinol treatment at or below a concentration of 10 nM (Figure 4A). However, at 100 nM retinol the relative luciferase activity increased significantly (p<0.05) to a level similar to that obtained by 1 nM RA. At 100 nM, the response to RA was more than 10 times of that to retinol at the same concentration (Figure 4A).
Figure 4. CYP26A1 promoter responds to retinol as well as to different retinoid analogues in HepG2 cells.
A. HepG2 cells cotransfected with E4 promoter construct in pGL3-basic -luc vector together with hRARα.hRXRα expression vector were treated with either RA or retinol at different concentrations for 24 h and then lysed for luciferase assay. The promoter responded to retinol but at higher concentrations than that of RA. B. Cells were cotransfected with CYP26A1 E4 promoter construct in pGL3-basic-luc vector together with RARα, RARβ, or RARγ and then treated with either RA or Am580, an RARα specific ligand, at different concentrations for 24 h after which the cells were collected for luciferase assay. To measure the activity of the synthetic retinoid analogues, HepG2 cells grown in 24-well plates were cotransfected in triplicate with pGL3-Basic-luc plasmid containing full-length human CYP26A1 promoter (C) or E4 promoter (D) without or with 0.1 µg retinoid receptors and then treated with either ethanol as vehicle or retinoid compounds including RA at 0.1 µM in EMEM medium for 24 h after which the cells were collected for luciferase assay. E. HepG2 cells cotransfected with human CYP26A1 E4 promoter together with hRARα.hRXRα were then incubated with either RA or EC23 at different concentrations for 24 h after which the cells were collected for luciferase activity assay. F. Newly synthesized retinoid analogues induced CYP26A1 mRNA expression in HepG2 cells. HepG2 human hepatoma cells were grown in 12-well plates and treated with either ethanol as the vehicle control or retinoid compounds including RA at 3 different concentrations in triplicate for 4 h and then collected for RNA analysis by RT-PCR. The relative expression of CYP26A1/18S ribosomal RNA level, as the control, was set as 1 in HepG2 cells treated with the vehicle in bar graphs.
To assess the promoter responses to the synthetic retinoids, we first examined Am580, a synthetic retinoid known to be a specific ligand for RARα. HepG2 cells were first cotransfected with the CYP26A1 promoter together with RA receptors and then incubated with retinoids including RA. The cells were washed and then assayed for luciferase activity. In the presence of RARα, Am580 was shown to have a stronger effect than RA at all the concentrations used (Figure 4B). Whereas in the presence of RARβ the promoter responded modestly to both Am580 and to RA, in the presence of RARγ the promoter was much less affected by both ligands (Figure 4B).
Whereas ATRA is a pan-agonist for RARs, 9-cis RA is an active ligand for both RARs and RXRs. Here we have examined the effect of 9-cis RA on the promoter activity of CYP26A1 gene in HepG2 cells cotransfected with RARs together with RXRα and compared the results with those using ATRA. 9-cis RA at 100 nM concentration activated the CYP26A1 promoter to the same extent as ATRA in HepG2 cells cotransfected withe any of RARs, each in combination with RXRα (Figure S3). Combination of 9-cis RA with ATRA did not further activate the promoter of the CYP26A1 gene. Similar to ATRA, 9-cis RA more strongly activated in cells cotransfected with either RARα or RARβ as compared to RARγ, in combination with RXRa (Figure S3).
In a separate experiment, we tested the CYP26A1 promoter responses to three novel synthetic retinoids, namely, EC23, DC360 and DC324. When full-length promoter was used, all three synthetic retinoids significantly increased the promoter activity of CYP26A1 gene through all three individual RARs (Figure 4C). However, when the E4 promoter was used all three synthetic retinoids had a significant effect on increasing the promoter activity of the CYP26A1 gene through RARα and RARβ, but not through RARγ (Figure 3D). Only EC23 significantly increased the promoter activity through RARγ (p<0.05)( Figure 4D). Through RARβ all three retinoids increased the promoter activity of the CYP26A1 gene to the same extent when either promoter was used (Figure 4C and D). The E4 promoter was more responsive to retinoid than the full-length promoter when the promoter constructs were cotransfected with the individual RARs (Figure 4C and D).
In another separate experiment, HepG2 cells cotransfected with the E4 promoter together with the hRARa.hRXRa expression vector were treated with either EC23 or RA over concentrations from 0 to 100 nM for 24 h, after which the cells were collected for luciferase activity. At 10 nM concentrations or lower, EC23 was more effective than RA at inducing of the CYP26A1 promoter (Figure 4E). At 100 nM, however, EC23 was equally effective to RA at inducing the CYP26A1 promoter.
3.7. Synthetic retinoid analogues induced the expression of CYP26A1 and RARβ genes in HepG2 cells
HepG2 cells were treated with different retinoids including RA at 0, 1, 10, and 100 nM concentrations for 4 h, followed by analysis of the relative mRNA levels of CYP26A1 and RARβ, two genes known to be responsive to retinoid treatment in vivo. Among three synthetic retinoids, EC23 increased the expression of CYP26A1 mRNA in HepG2 cells similar to that of RA at 1 and 10 nM concentrations (p<0001 vs. vehicle, Figure 4F). However, the effect of EC23 was significantly lower than that of RA at100 nM (p<0.0001), although it increased the level of CYP26A1 mRNA significantly compared to vehicle-treated control cells (p<0.0001). Neither DC360 nor DC324 were significantly effective at low concentrations of 1 and 10 nM (p>0.05) (Figure 4F); however, both compounds at 100 nM increased CYP26A1 mRNA significantly (p<0.05), although their effect was significantly lower than that of RA at the same concentration (p<0.0001). A similar pattern of results, but at lower magnitude, was observed when RARβ mRNA was analyzed in HepG2 cells (Figure S4). The effect of EC23 was comparable to that of RA at all concentrations, however, DC360, but not DC324, increased RARβ RNA levels at the concentration of 100 nM (p<0.001), similar to the increase by RA (Figure S4).
3.8. HEK293T cells permanently transfected with CYP26A1 promoter respond quantitatively to retinoic acid
In order to observe the quantitative response to the presence of RA or other retinoids without repeated transfections, we constructed a permanently transfected cell line containing the E4 promoter of the CYP26A1 gene together with individual RARα and RXRα expression vectors. For this, HEK293T cells were cotransfected with the pGL3-basicmCherry vector cloned with the E4 promoter construct of the human CYP26A1 gene together with human RARα and human RXRα expression vectors as described in Material and Methods. After transfection, the cells were trypsinized and grown in full growth medium containing G418 for selection. After two weeks, the majority of the cells expressed RFP following treatment with RA. We isolated individual colonies for expression of RFP, first by limited dilution, and then by individual cell sorting using flow cytometry. Initially, we isolated 14 different individual cells in 96-well plates and propagated them to select 4 different colonies. Of these 4 colonies from individual cells, we selected colony #1A1 for further processing and analysis for two main reasons: first, the cells showed a low RFP background when grown in full growth media without added RA, and secondly, they expressed significant RFP in response to RA treatment.
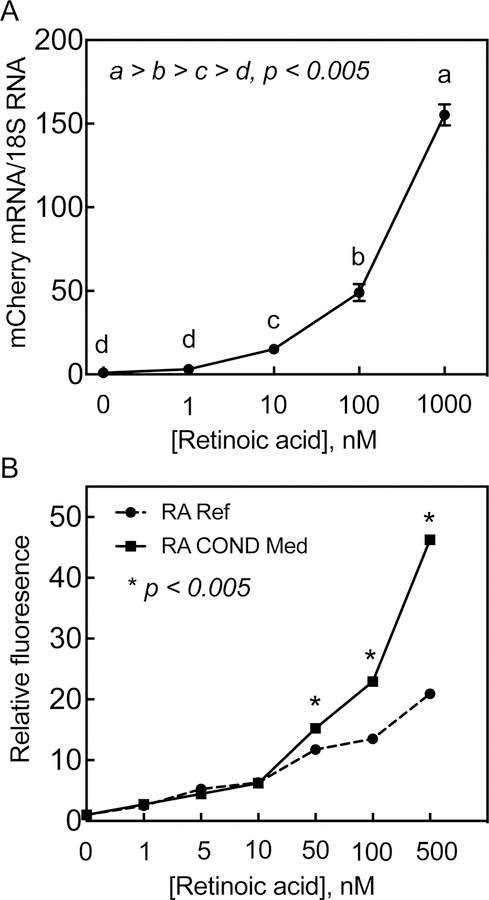
The expression of the reporter mCherry mRNA increased proportionally in HEK293T 1A1cells in response to RA added at different concentrations for 24 h. The expression of mCherry mRNA (using primers for the mCherry coding region as well as for 18S rRNA) was nearly undetectable in 1A1 cells incubated with vehicle, however, it was induced 3-fold, though not significantly, in the same cells in response to RA as low as 1 nM, and further increased 15-, 50-, and 150-fold at concentrations of 10, 100, and 1000 nM, respectively, as compared to vehicle-treated control cells (Figure 5A).
Figure 5. Red fluorescent protein (RFP) is expressed quantitatively in HEK293T 1A1 cells in response to RA.
A. HEK293T 1A1 cells grown in 12-well plates were incubated with RA at different concentrations for 24 h after which the cells were washed and collected for total RNA isolation. The total RNA samples were then subjected to RT-PCR analysis for expression of mCherry RNA in triplicate using 18S ribosomal RNA expression as the internal control. B. HEK293T 1A1 cells were grown in 12-well plates then incubated 24 h with either RA at different concentrations or 24-h RA treated conditioned media from HepG2 cells. For conditioned media preparation, HepG2 cells were incubated RA at different concentrations in EMEM full growth media for 24 h. The media were collected and then incubated at 50% with HEK293T 1A1 cells without addition of any RA. For reference standard, the HEK293T 1A1 cells were incubated with 50% HepG2 cell 24-h vehicle-treated conditioned media with added RA at different concentrations for 24 h. The HEK293T 1A1 cells in duplicate wells were then trypsinized and collected for flow cytometry for the cells expressing RFP.
RFP expressed in the HEK293T 1A1 cell line could be measured by flow cytometry. For this, cell monolayer grown in 12-well plates were incubated for 24 h with either RA-treated conditioned media from HepG2 cells or an RA reference standard at concentrations from 1 to 500 nM and then prepared for flow cytometry according to the procedure described in the Material and Methods section. RFP expression levels exhibited a linear correlation in response to RA treatment (R2 = 0.96, p<0.0001) (Figure 5B). The conditioned media was as effective as the RA reference standard at the lower concentration levels, however, they were more effective than the reference standards at the higher concentrations of RA. For example, RFP expression induced in the cells by the conditioned medium was about 2 times of that induced by the RA reference standard at 500 nM concentration (Figure 5B).
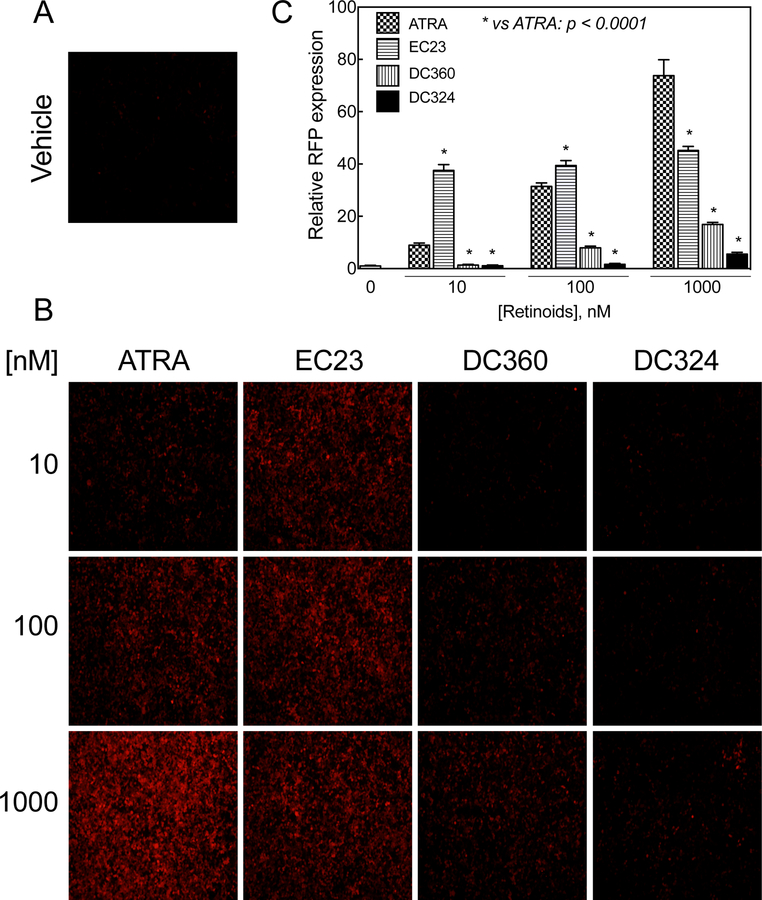
3.9. HEK293T 1A1 cells may be used to determine the retinoid activity of synthetic retinoid analogues
Monolayer HEK293T 1A1 cells in 12-well plates were incubated with either synthetic retinoid analogues or RA, each at 3 different concentrations of 10, 100, and 1000 nM for 24h and then analyzed by fluorescence microscopy for the expression of RFP. The RFP expressing cells were counted and the relative fluorescence intensity levels were calculated based on the expressed RFP in cells treated with vehicle (Figure 6A). The cells were significantly more responsive regarding RFP to EC23 than to RA at 10 and 100 nM (Figure 6B and C). At 1000 nM, however, EC23 was significantly less effective than RA, probably indicating saturation of EC23 as a potent ligand for retinoic acid receptors. In fact, the cells responded to RA by the expression of RFP dose-dependently. Two other retinoid analogues, DC360 and DC324, were significantly less effective than RA in inducing expression of RFP in these cells (Figure 6B and C). Whereas DC360 and DC324 had no effect at 10 nM, DC360, at or above 100 nM induced RFP expression significantly as compared to the vehicle control, whereas DC324 was effective only at 1000 nM. However, both were less effective than RA (Figure 6B and C) (p<0.0001).
Figure 6. HEK293T 1A1 cells response to the newly synthesized retinoid analogues.
A and B. Monolayer HEK293T 1A1 cells in 12-well plates were incubated with either newly synthesized retinoid analogues or RA, each at different concentrations for 24h and then observed under a microscope for expression of RFP. C. The RFP expressing cells were counted using the NIH ImageJ program and the relative fluorescence intensity levels were calculated based on the expression of RFP in vehicle-treated cells. Values represent the mean of 3 different regions ± SD.
In a separate experiment, monolayer HEK293T 1A1 cells in 12-well plates were treated with either EC23 or RA at concentrations from 0 to 100 nM for 24 h, after which the cells were analyzed by fluorescence microscopy for RFP (Figure 7A). The RFP expressing cells were counted and the relative fluorescence intensity levels were calculated compared to RFP background in vehicle-treated cells (Figure 7B). EC23 was more effective (p<0.0001) than RA in inducing RFP expression in these cells at concentrations from 0.5 to 100 nM (Figure 7A and B).
Figure 7. EC23 was shown to have more retinoid activity than RA at low concentrations.
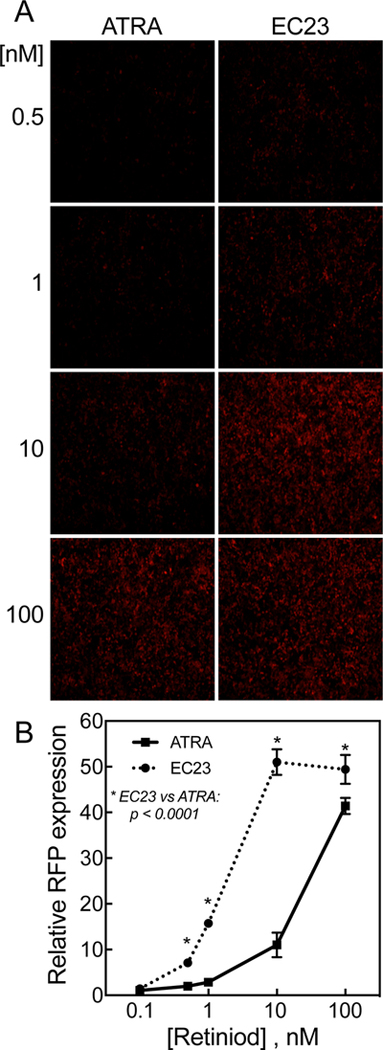
A. Monolayer HEK293T 1A1 cells in 12-well plates were incubated with either EC23 or RA, each at different concentrations for 24h and then observed under microscope for RFP expression. B. The RFP expressing cells were counted using NIH ImageJ program and the relative fluorescence intensity levels were calculated based on the expressed RFP in cells treated with vehicle. The values represent the mean of 3 different regions ± SD.
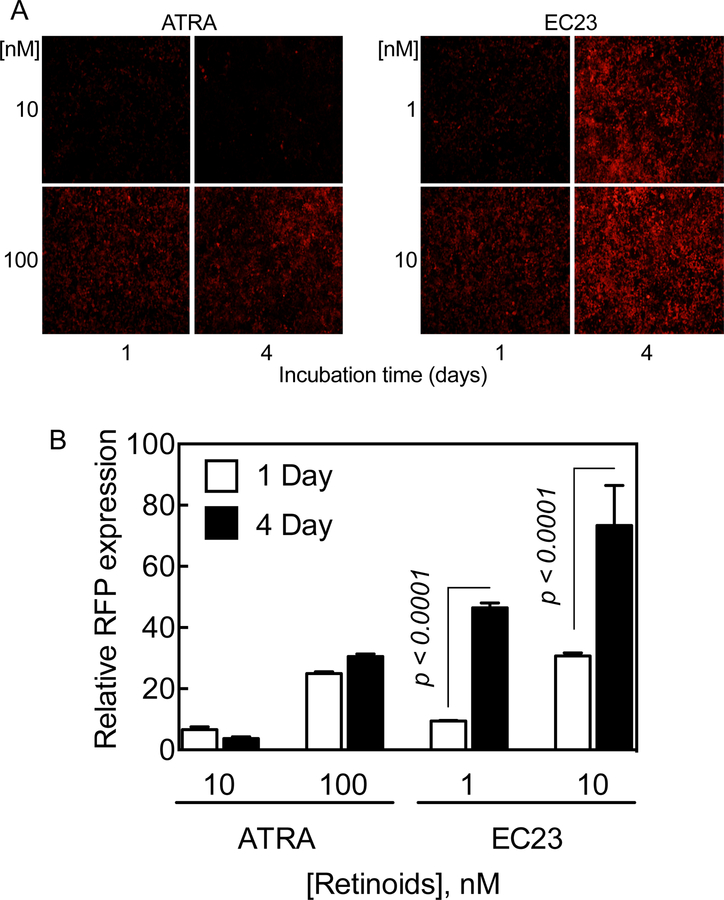
These synthetic retinoid analogues, compared to RA, were shown to exhibit a longer-lasting effect on RFP expression in HEK293T 1A1 cells, measured after treatment with different concentrations for either 1 or 4 days. The RFP signals in the cells treated with EC23, compared to RA, were higher in the 4-day treatment than in the 24-hour treatment (Figure 8). Likewise, RFP signals were doubled in cells treated with either DC360 or DC324 for 4 days as compared to those in the cells treated for 24 h (data not shown).
Figure 8. Synthetic retinoid analogue, EC23, has apparently more extended retinoid activity than RA in expression of RFP in HEK293T 1A1 cells.
A. Monolayer HEK293T 1A1 cells grown in 12-well plates were incubated with either EC23 at 1 and 10 nM or RA at 10 and 100 nM concentrations for either 1 or 4 days and RFP expression was observed by fluorescence microscopy, and, B. measured using NIH ImageJ. The values represent the mean of 3 different sections ± SD.
3.10. CYP26A1 promoter may be used to determine the retinoid activity levels in serum
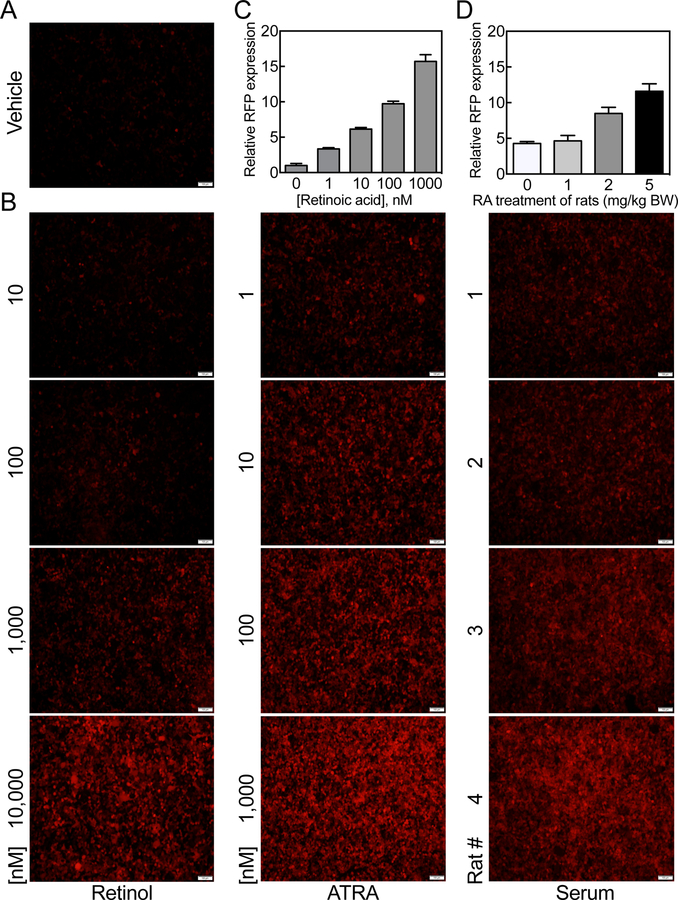
For analysis of RFP expression resulting from treatment of cells with serum of animals treated in vivo with RA, we incubated HEK293T 1A1 cells directly with rat serum at a 20% dilution in DMEM media without FBS for 24 h and then observed cells by fluorescence microscopy; either exogenous retinol or RA was included in separate wells as reference standards, added in DMEM media with 20% FBS. As in the previous experiments, there was no significant expression of RFP in 1A1 cells treated with vehicle (Figure 9A). Whereas 1A1 cells responded effectively to RA in the expression of RFP at a concentration as low as 1 nM, retinol did not have a significant effect on the expression of RFP at a concentration of 100 nM (Figure 9B and C). As compared to the RA standards, the control serum sample from the vehicle-treated rat was equivalent to ~2.5 nM at the 20% serum dilution used (Figure 9B, C and D). Then, serum samples from rat treated with 1, 2 and 5 mg of RA/kg BW were tested. Serum (after 1 mg RA/kg) increased the RFP expression slightly (Figure 9B and D), however, serum from rats treated with 2 and 5 mg RA/kg BW elevated the RA concentrations in the serum to about 30 nM and 180 nM, respectively, as determined by RFP expression in HEK293T 1A1 cells (Figure 9B and D).
Figure 9. Total retinoid activity may be estimated in intact rat serum by HEK293T 1A1 cells.
To prepare serum for testing, adult female rats were treated orally with either oil as the vehicle control or RA at 1, 2, and 5 mg/kg body weight for 6 h, at which time blood was collected and serum prepared by centrifugation and stored at −20oC until analysis. Total serum retinol concentrations as determined by UPLC were all in the normal ranges of 0.98 for the control rat and 0.77, 0.74, and 1.74 µM for rats treated with RA at 1, 2, and 5 mg/kg body weight, respectively. A and B. Cells were incubated with intact rat serum samples at 20% in DMEM media without added FBS for 24 h, and then observed by fluorescence microscopy for expression of RFP using retinol and RA at different concentrations in DMEM media with added 20% FBS as the standards. C and D. RFP expressing cells were counted using NIH ImageJ program and the relative fluorescence intensity levels were calculated based on the expressed RFP in cells treated with vehicle. The retinoid activity levels of the serum samples tested were calculated based on the RA standard. The values represent the mean of 3 different regions ± SD.
4. Discussion
In this study we have used a short form of the CYP26A1 gene promoter (E4 construct) in a pGL3 promoter-less vector with either luciferase or mCherry as the reporter gene to evaluate the promoter response to retinoids in HepG2 cells as well as HEK293T cells when the cells are cotransfected with an RARα.RXRα expression vector. We found that: 1) the E4 promoter construct responds to RA within 2 h and significantly at a concentration as low as 0.5 nM. In the logarithmic dose response study, which encompassed up to 1 µM of RA concentration the promoter response was nearly linear in the range of 0.1 to 10 nM. 2) Similar to the gene itself, the CYP26A1 promoter responded to RA at a much higher extent than the promoter of the RARβ gene, which possesses two functional DR5 elements in the proximal region upstream of the transcription start site. In fact, CYP26A1 is a more useful tool for measuring the intracellular RA activity than RARβ because of a greater dynamic range of responses as compared to RARβ response, which is more attenuated. 3) RARα and RARβ had a greater effect on the promoter responses to RA than RARγ in both HepG2 cells and HEK293T cells partly because of the specificity for RARγ receptor for DR2 [18] rather than DR5, 4) In the transient transfection experiments, the CYP26A1 promoter response to RA treatment was much higher in HepG2 cells than in HEK293T cells. For example, the CYP26A1 promoter response was about 20 times higher in HepG2 cells treated with 1 nM RA, whereas no significant response was observed from the same promoter at the same concentration of RA in HEK293T cells. A similar trend has been observed for the expression of CYP26A1 mRNA in these 2 cell lines [9, 10]. Whereas the expression of CYP26A1 is almost undetectable in HepG2 cells treated with vehicle, significant expression of the CYP26A1 gene was observed in vehicle-treated HEK293T cells [9, 10]. This may be due to relaxation of the chromatin in the region where the CYP26A1 gene is located [9]. In fact, we previously found no significant increase in binding of histone H3 to the promoter region in the vicinity of the transcription start site in HEK293T cells treated with RA as compared with vehicle-treated cells [9]. 5) The CYP26A1 promoter responded to RAR specific ligands in HepG2 cells. The promoter responded comparably to Am580, an RARα specific ligand as well as 9-cis-RA, which binds to both RAR and RXR receptors. However, the promoter did not respond to the RXR specific ligand significantly, unless it was used in combination with RAR specific ligands (data not shown). Nuclear receptors such as RARs, TRs, and VDR are considered non-permissive receptors, which are enhanced by their own agonists but are not affected by RXR ligands [19]. This is in contrast to the permissive action of RXR heterodimers, which can be affected by either RXR ligands and/or the ligands of the nuclear receptors such as PPARs, LXR, PXR, FXR and CAR [19]. 6) The CYP26A1 promoter responded to retinol in both HepG2 cells and HEK293T cells, however, the promoter was enhanced by retinol at higher treatment concentrations compared to that by RA. It is not known whether retinol acts as a potential ligand for retinoic acid receptors, or if it needs to be oxidized first for formation of RA. All-trans-retinol may act as a functional ligand for retinoic acid receptors [20]. 7) The CYP26A1 promoter was used to evaluate the retinoid activities of three synthetic retinoids, namely, EC23, DC360 and DC324. EC23 was shown to have higher activity than the other two retinoids, DC360 and DC324. As compared to RA, this retinoid, EC23, was more effective than RA at low concentrations based on the data from the promoter activity as well as on the CYP26A1 gene expression in HepG2 cells.
Based on the quantitative responses we observed from the CYP26A1 promoter to RA as well as other retinoids, we decided to produce a monoclonal cell line possessing the CYP26A1 promoter with a reporter gene, which can be directly measured in the cells treated with retinoids without repeated transfection. The CYP26A1 promoter construct in pGL3-basic vector with mCherry open reading frame as the reporter gene was used together with RARs to transfect HEK293T cells, which grow rapidly and are easily trypsinized into individual cells for flow cytometry. A cell line clone, 1A1, which expresses permanently the RFP protein, the fluorescent signal from mCherry, upon treatment with retinoids, was isolated and characterized for further analysis. The 1A1 clone was found to have a low background level of RFP without retinoid treatment and to be resistant to high concentrations of retinoids while maintaining high expression of RFP. Additionally, the 1A1 clone was found to have a quantitative response to RA by expression of mCherry mRNA as determined by RT-PCR and by RFP protein expression as analyzed by fluorescence microscopy and flow cytometry. The cells responded quantitatively to retinoids in the expression of RFP and the responses were comparable to those measured for luciferase activities in HepG2 cells. Among three synthetic retinoids we examined, EC23 was found to exhibit stronger retinoid activity than RA at low concentrations; similar to the values measured by luciferase activity in HepG2 cells. Moreover, the synthetic retinoids were shown to be more stable than RA as tested in HEK293T 1A1 cells for RFP expression.
The HEK293T 1A1 cells may be used to estimate the activities of retinoids in biological systems. For example, the cells were used to estimate the retinoid activities of the preconditioned media from HepG2 cells treated with RA by the expression of RFP, which was easily measured by flow cytometry. The cells could also be used to estimate the retinoid activities by direct incubation of serum samples from rats treated without or with RA. We found that the RFP expression levels increased in the cells treated with serum samples of the rats treated with RA as compared to treatment with vehicle.
In summary, a short version promoter of the CYP26A1 gene driving the expression of either luciferase or RFP in a promoter-less vector was used to test its response to RA in HepG2 cells and HEK293T cells cotransfected with RARs. The promoter responded nearly linearly to a wide range of the log concentrations of RA and was sensitive enough to detect RA activity as low as 0.5 nM. The promoter responded better to RA when the cells were cotransfected with RARα.RXRα as compared to the other RARs. It responded to retinol and was used to estimate the retinoid activities of three synthetic retinoids. A stable cell line of HEK293T cells was established to express RFP under the CYP26A1 E4 promoter to respond to RA as well as to retinol. The cell line could be observed live for expression of RFP using fluorescence microscopy or trypsinized easily into individual cells for quantification of RFP expression using flow cytometry. This system was used to estimate the retinoid activities of synthetic retinoids as well as biological specimens such as serum.
Supplementary Material
Figure S1. Red fluorescent protein (RFP) as the reporter gene driven by CYP26A1 promoter responds to RA dose-dependently in HepG2 cells. Monolayer cells grown in 12-well plates were cotransfected with human E4 promoter construct of CYP26A1 gene pGL3-basic-mCherry together with hRARα.hRXRα expression vector and yellow fluorescent protein expression vector (pCMV-YFP), as the internal control for transfection and then incubated with RA at different concentrations for 6 h at 37oC. The live cells in 3 sections of each well were observed by microscopy for relative expression of RFP and YFP.
Figure S2a. RARα makes CYP26A1 promoter respond to RA better than either RARβ or RARγ in HepG2 cells. Mono-layer HepG2 cells were cotransfected with the CYP26A1 promoter construct E4 in pGL3-basic-mCherry, together with either hRARα.hRXRα, hRARβ.hRXRα or hRARγ.hRXRα and then treated with different concentrations of RA for 24 h. Live cells were assessed for RFP expression by fluorescence microscopy. Representative images are shown.
Figure S2b. RARα makes CYP26A1 promoter respond to RA better than either RARβ or RARγ in HEK293T cells. Mono-layered HEK293T cells were cotransfected with CYP26A1 promoter E4 construct in pGL3-basic-mCherry together with either hRARα.hRXRα, hRARβ.hRXRα or hRARγ.hRXRα and then treated with different concentrations of RA for 24 h. Live cells were assessed for RFP expression by fluorescence microscopy. Representative images are shown.
Figure S3. CYP26A1 promoter responds to 9-Cis RA at the same extent as compared to ATRA. HepG2 cells were cotransfected with CYP26A1 E4 promoter construct in pGL3basic-luc vector together with RARα, RARβ, or RARγ and then treated with either ATRA or 9-Cis RA or both, at 100 nM for 24 h after which the cells were collected for luciferase assay.
Figure S4. Retinoid analogues induce RARβ mRNA expression in HepG2 cells. HepG2 cells were grown in 12-well plates, treated with either ethanol as the vehicle control or retinoid compounds including ATRA at 3 different concentrations in triplicate for 4 h, after which total RNA was analyzed by RT-PCR. In the bar graphs the expression of RARβ relative to18S ribosomal RNA, used as the control, was set to 1.0 in vehicle-treated cells.
Highlights.
CYP26A1 promoter is highly responsive to retinoic acid (RA) in culture cells
RARα is most effective on the CYP26A1 promoter in response to RA
CYP26A1 promoter responds to retinol and to retinoid analogues
HEK293T cells stably transfected with CYP26A1 promoter respond quantitatively to RA
These cells were used to determine the retinoid activity of synthetics and rat serum
Acknowledgements
Supported by NIH grants DK-41479 and HD-066982 and funds from the Pennsylvania State University.
Abbreviations used are:
- RA
retinoic acid
- ATRA
all-trans RA
- DR
direct repeat
- RARE
RA response element
- RAR
RA receptor
- RXR
retinoid X receptor
- CYP
cytochrome 450
- RFP
red fluorescent protein
- RT-PCR
real-time polymerase chain reaction
- UPLC
ultra performance liquid chromatography
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Carrier M, Rochette-Egly C, Control of Gene Expression by Nuclear Retinoic Acid Receptors, The Retinoids, John Wiley & Sons, Inc; 2015, pp. 91–116. [Google Scholar]
- [2].Urban S, Ye T, Davidson I, How the RAR–RXR Heterodimer Recognizes the Genome, The Retinoids, John Wiley & Sons, Inc; 2015, pp. 151–163. [Google Scholar]
- [3].Wei L-N, Retinoic Acid Receptor Coregulators in Epigenetic Regulation of Target Genes, The Retinoids, John Wiley & Sons, Inc; 2015, pp. 117–130. [Google Scholar]
- [4].Mendoza-Parra M-A, Bourguet W, de Lera AR, Gronemeyer H, Retinoid ReceptorSelective Modulators, The Retinoids, John Wiley & Sons, Inc; 2015, pp. 165–192. [Google Scholar]
- [5].Napoli JL, Physiological insights into all-trans-retinoic acid biosynthesis, Biochim Biophys Acta, 1821 (2012) 152–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ross AC, Zolfaghari R, Cytochrome P450s in the Regulation of Cellular Retinoic Acid Metabolism, Annual Review of Nutrition, 31 (2011) 65–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Balmer JE, Blomhoff R, Gene expression regulation by retinoic acid, Journal of Lipid Research, 43 (2002) 1773–1808. [DOI] [PubMed] [Google Scholar]
- [8].Loudig O, Maclean GA, Dore NL, Luu L, Petkovich M, Transcriptional co-operativity between distant retinoic acid response elements in regulation of Cyp26A1 inducibility, Biochemical Journal, 392 (2005) 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhang Y, Zolfaghari R, Ross AC, Multiple retinoic acid response elements cooperate to enhance the inducibility of CYP26A1 gene expression in liver, Gene, 464 (2010) 32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zolfaghari R, Ross AC, Hepatocyte Nuclear Factor 4α (HNF4α) in Coordination With Retinoic Acid Receptors Increases all-trans-Retinoic Acid-Dependent CYP26A1 Gene Expression in HepG2 Human Hepatocytes, Journal of Cellular Biochemistry, 115 (2014) 1740–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sambrook J, Russell DW, Molecular cloning: a laboratory manual, Third ed. ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2001. [Google Scholar]
- [12].Chisholm DR, Lamb R, Pallett T, Affleck V, Holden C, Marrison J, O’Toole P, Steffen A, Nelson AK, Mahler C, Girkin J, Valentine R, Marder TB, Whiting A, Ambler CA, Small Molecule Photosensitisers for Photodynamic Therapy Applications, (Unpublished results).
- [13].Chisholm DR, Tomlinson C, Zhou G-L, Holden C, Affleck V, Lamb R, Newling K, Ashton P, Valentine R, Redfern C, Erostyak J, Makkai G, Ambler CA, Whiting A, Pohl E, (Unpublished results). [DOI] [PubMed]
- [14].Christie VB, Barnard JH, Batsanov AS, Bridgens CE, Cartmell EB, Collings JC, Maltman DJ, Redfern CPF, Marder TB, Przyborski S, Whiting A, Synthesis and evaluation of synthetic retinoid derivatives as inducers of stem cell differentiation, Organic & Biomolecular Chemistry, 6 (2008) 3497–3507. [DOI] [PubMed] [Google Scholar]
- [15].Chisholm DR, Tomlinson CWE, Zhou G-L, Holden C, Affleck V, Lamb R, Newling K, Ashton P, Valentine R, Redfern C, Erostyák J, Makkai G, Ambler CA, Whiting A, Pohl E, Fluorescent Retinoic Acid Analogues as Probes for Biochemical and Intracellular Characterization of Retinoid Signaling Pathways, ACS Chemical Biology, (2019). [DOI] [PubMed]
- [16].Zolfaghari R, Ross AC, Cloning, gene organization and identification of an alternative splicing process in lecithin:retinol acyltransferase cDNA from human liver, Gene, 341 (2004) 181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].de Thé H, Vivanco-Ruiz MM, Tiollais P, Stunnenberg H, Dejean A, Identification of a retinoic acid responsive element in the retinoic acid receptor beta gene, Nature, 343 (1990) 177–180. [DOI] [PubMed] [Google Scholar]
- [18].Ganti KP, Mukherji A, Surjit M, Li M, Chambon P, Similarities and differences in the transcriptional control of expression of the mouse TSLP gene in skin epidermis and intestinal epithelium, Proceedings of the National Academy of Sciences, 114 (2017) E951E960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gilardi F, Desvergne B, RXRs: Collegial Partners, in: Asson-Batres MA, Rochette-Egly C (Eds.) The Biochemistry of Retinoic Acid Receptors I: Structure, Activation, and Function at the Molecular Level, Springer; Netherlands, Dordrecht, 2014, pp. 75–102. [Google Scholar]
- [20].Repa JJ, Hanson KK, Clagett-Dame M, All-trans-retinol is a ligand for the retinoic acid receptors, Proceedings of the National Academy of Sciences, 90 (1993) 7293–7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Red fluorescent protein (RFP) as the reporter gene driven by CYP26A1 promoter responds to RA dose-dependently in HepG2 cells. Monolayer cells grown in 12-well plates were cotransfected with human E4 promoter construct of CYP26A1 gene pGL3-basic-mCherry together with hRARα.hRXRα expression vector and yellow fluorescent protein expression vector (pCMV-YFP), as the internal control for transfection and then incubated with RA at different concentrations for 6 h at 37oC. The live cells in 3 sections of each well were observed by microscopy for relative expression of RFP and YFP.
Figure S2a. RARα makes CYP26A1 promoter respond to RA better than either RARβ or RARγ in HepG2 cells. Mono-layer HepG2 cells were cotransfected with the CYP26A1 promoter construct E4 in pGL3-basic-mCherry, together with either hRARα.hRXRα, hRARβ.hRXRα or hRARγ.hRXRα and then treated with different concentrations of RA for 24 h. Live cells were assessed for RFP expression by fluorescence microscopy. Representative images are shown.
Figure S2b. RARα makes CYP26A1 promoter respond to RA better than either RARβ or RARγ in HEK293T cells. Mono-layered HEK293T cells were cotransfected with CYP26A1 promoter E4 construct in pGL3-basic-mCherry together with either hRARα.hRXRα, hRARβ.hRXRα or hRARγ.hRXRα and then treated with different concentrations of RA for 24 h. Live cells were assessed for RFP expression by fluorescence microscopy. Representative images are shown.
Figure S3. CYP26A1 promoter responds to 9-Cis RA at the same extent as compared to ATRA. HepG2 cells were cotransfected with CYP26A1 E4 promoter construct in pGL3basic-luc vector together with RARα, RARβ, or RARγ and then treated with either ATRA or 9-Cis RA or both, at 100 nM for 24 h after which the cells were collected for luciferase assay.
Figure S4. Retinoid analogues induce RARβ mRNA expression in HepG2 cells. HepG2 cells were grown in 12-well plates, treated with either ethanol as the vehicle control or retinoid compounds including ATRA at 3 different concentrations in triplicate for 4 h, after which total RNA was analyzed by RT-PCR. In the bar graphs the expression of RARβ relative to18S ribosomal RNA, used as the control, was set to 1.0 in vehicle-treated cells.