Abstract
Topoisomerase II (topo II) inhibitors are commonly used as chemotherapy to treat multiple types of cancer, though their use is also associated with the development of therapy related acute leukemias. While the chromosome-damaging effects of etoposide, a topo II poison, have been proposed to act through a threshold mechanism, little is known about the chromosome damaging effects and dose responses for the catalytic inhibitors of the enzyme. The current study was designed to further investigate the potencies and concentration-response relationships of several topoisomerase II inhibitors, including the topoisomerase II poison etoposide, as well as catalytic inhibitors aclarubicin, merbarone, ICRF−154 and ICRF−187 using both a traditional in vitro micronucleus assay as well as a flow-cytometry based version of the assay. Benchmark dose (BMD) analysis was used to identify models that best fit the data and estimate a BMD, in this case the concentration at which a one standard deviation increase above the control frequency would be expected. All of the agents tested were potent in inducing micronuclei in human lymphoblastoid TK6 cells, with significant increases seen at low micromolar, and in the cases of aclarubicin and etoposide, at low nanomolar concentrations. Use of the anti-kinetochore CREST antibody with the microscopy-based assay demonstrated that the vast majority of the micronuclei originated from chromosome breakage. In comparing the two versions of the micronucleus assay, significant increases in micronucleated cells were observed at similar or lower concentrations using the traditional microscopy-based assay. BMD modeling of the data exhibited several advantages and proved to be a valuable alternative for concentration-response analysis producing points of departure comparable to those derived using traditional no-observed or lowest-observed genotoxic effect level (NOGEL or LOGEL) approaches.
Keywords: topoisomerase II inhibitors, dose-response, human cells, micronucleus, flow cytometry, benchmark dose
Introduction
Type II DNA topoisomerases are important nuclear enzymes that relieve topological stress during DNA replication, transcription, repair, and mitosis (1–4). The enzyme’s catalytic cycle involves covalent binding of the enzyme to DNA, forming a double stranded break and a cleavage complex through which another DNA duplex can pass. Following strand passage, the double stranded break is religated and the enzyme is released from the DNA (Fig 1) (4–5). Due to the formation of the protected double-stranded break, disruptions in the enzyme’s catalytic cycle have the ability to lead to multiple types of chromosomal alterations including cancer-related translocations (1–4).
Figure 1.
Topo II catalytic cycle. The sites of action of the topo II inhibitors used in current study are shown. Adapted from Mondrala and Eastmond (5).
A number of compounds are known to disrupt or inhibit topoisomerase II (topo II) including some important classes of chemotherapeutic agents. These can act at various stages of the catalytic cycle (Fig 1) (1,6). Topo II poisons, such as etoposide, act to stabilize the cleavage complex and inhibit the religation step, an important step leading to the formation of unprotected double stranded breaks. Catalytic inhibitors, on the other hand, affect other parts of the topo II catalytic cycle and do not directly stabilize the cleavage complex, though have been shown to have clastogenic effects in vitro and in vivo (6–8).
While several drugs targeting topo II are front line therapies for the treatment of various types of cancer, one limitation of their use is increased risk for development of treatment-related acute leukemia (1–4, 6). These leukemias are secondary to the original cancers for which the topo II inhibitors were originally prescribed and have characteristically short median latency periods of approximately 2–3 years (9–12). Topo II poisons etoposide and doxorubicin have been associated with treatment-related acute myelogenous leukemia (t-AML), typically of monocytic or myelomonocytic origin, caused by balanced translocations involving the MLL (mixed lineage leukemia; also known as KMT2A) gene on chromosome band 11q23, which encodes a transcriptional coactivator that regulates gene expression during hematopoiesis (9,12). Similarly, mitoxantrone, has been associated with development of a different subtype of t-AML, acute promyelocytic leukemia, as a result of a reciprocal translocation fusing the retinoic acid receptor alpha gene (RARA) from chromosome 17 to the promyelocytic leukemia gene (PML) on chromosome 15 resulting in the stable expression of a PML-RARA fusion protein (10,11). In addition, there is some concern that exposure to naturally occurring topo II poisons such as genistein and other bioflavonoids in utero may play a role in development of infant AML (13,14). While most topo II inhibitors associated with leukemia fall under the category of topo II poisons, there is also evidence of similar leukemogenic effects in patients treated with the catalytic inhibitors ICRF-154 and bimolane (12,15)
The goal of the current study is to more thoroughly investigate concentration-response relationships of a variety of topo II inhibitors to better understand the concentrations at which damage occurs and how different mechanisms of inhibition of topo II may affect the dose-response curves. To do so, we examined the concentration-responses of the topo II poison, etoposide, as well as two catalytic inhibitors that act prior to the formation of the cleavable complex (alcarubicin and merbarone) and two that act after the religation step (ICRF−154 and ICRF−187). In addition, these studies compared the results of a traditional in vitro micronucleus assay technique with those from a more recently developed flow cytometry-based micronucleus assay, and used benchmark dose modeling to evaluate the results.
Methods
Cell culture and treatments
The human lymphoblastoid cell line TK6 was maintained in RPMI 1640 medium (GIBCO; Carlsbad, CA) containing 10% iron-supplemented calf serum (Hyclone; Logan, UT) with 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (Fisher Scientific; Pittsburg, PA) at 37 °C in an atmosphere of 5% CO2/95% air. Exponentially growing cells with a doubling time of ~14 hrs were treated with various concentrations of each of the following topo II inhibitors: alcarubicin (Sigma; St. Louis, MO), merbarone (NCI; Bethesda, MD), ICRF−154 (NCI; Bethesda, MD), ICRF−187 (NCI; Bethesda, MD), and etoposide (Sigma; St. Louis, MO). All compounds were dissolved in dimethylsulfoxide (DMSO) with a final DMSO concentration of 0.1% in the culture flasks. Cells were harvested at 24 hours after treatment.
In vitro micronucleus assay with CREST staining
The procedure for the in vitro micronucleus assay was performed as previously described (16) with minor modifications. Cells were treated with varying concentrations of each topo II inhibitor as well as 4.5 μg/mL cytochalasin B for 24 hours before the cells were harvested for slide preparation. Aliquots of the cell suspension were centrifuged directly onto slides and then briefly air-dried and fixed in 100% methanol. Prepared slides were then stained with CREST primary antibody, followed by a FITC-conjugated secondary antibody (both obtained from Antibodies Inc.; Davis, CA), with DAPI used as a DNA counterstain. Slides were then coded and 1000 binucleated cells per test concentration were scored in a blind fashion for the presence of kinetochore-positive (K+) and kinetochore-negative (K-) micronuclei representing micronuclei formed from chromosome loss and chromosome breakage, respectively. Means and standard deviations were calculated with data from 2–3 replicate experiments.
Micronucleus assay by flow-cytometry
Staining, instrumentation, and gating for the MN assay by flow-cytometry was performed as previously described by Avelsevich et al (17). Briefly, at time of harvest, cells previously treated in the absence of cytochalasin B were stained with ethidium monoazide (EMA). A photoactivation step resulted in covalent binding of EMA with DNA from necrotic and late-stage apoptotic cells. Following this, the cells were lysed and stained with SYTOX-Green, which binds to all DNA, resulting in a suspension of nuclei and micronuclei with differentially stained DNA to distinguish between dead or dying cells (EMA+) and live cells (EMA-/SYTOX+). Data from 20,000 EMA-/SYTOX+ cells per sample were acquired and analyzed using a Becton Dickinson FACSort flow cytometer and Cell Quest software. Micronuclei were enumerated based on size (Forward Scatter) and DNA content.
Statistical analysis and benchmark dose modeling
For MN data using the in vitro micronucleus assay with CREST staining, concentration-related increases in micronucleated cells were determined using the Cochran–Armitage test for trend in binomial proportions. Following a positive response in the trend test, a one-tailed Fisher’s exact test was used to compare individual treatments against the respective DMSO-treated controls. For data obtained from the flow-cytometry based assay, an ANOVA test was performed and Dunnett’s T-test was used to compare individual treatments to the control.
BMD modeling of the micronucleus frequency was conducted using U.S. EPA BMD software (version 2.4.0, 2013). The benchmark response was defined as one control standard deviation (BMD1SD) and its lower 95th percentile confidence limit (BMDL1SD) according to U.S. EPA (2012) guidance for continuous data (18). Data were fit to Exponential (3, 4, and 5), Hill, Linear, Polynomial, and Power models assuming constant variance. The factors collectively taken into consideration for selecting the best-fit model included the global goodness-of-fit p value (must be≥ .1); lowest AIC value (a statistical estimate of model quality), Chi-square residual values of less than 2 at each dose level, visual fit, and the margin between the BMD1SD and BMDL1SD (18).
Results
Micronucleus induction by topo II inhibitors
Strong concentration-dependent increases in the induction of micronuclei, formed primarily from chromosome breakage, were seen with all of the topo II inhibitors tested. The results for each of the chemicals are briefly described below.
A strong monotonic increase in MN was seen with the pre-cleavage complex catalytic inhibitor aclarubicin. Statistically significant increases were observed beginning at the 12.5 nM test concentration, where an approximate 2-fold increase in MN was observed compared to controls when measured using the flow-based assay. The maximum amount of MN observed at the highest test concentration was 5%, representing a 7-fold increase (Figure 2A). These values and the fold increase were mirrored quite closely when MN were scored manually with microscopy (Figure 2B). The CREST data showed that most (approximately 83–90%) of the MN induced were kinetochore-negative and formed due to chromosome breakage (Figure 7A). The effects seen in TK6 cells treated with aclarubicin occurred at nanomolar concentrations with approximately 55% cytotoxicity as measured by relative population doubling or relative increases in cell count occurring at 12.5 nM (Table 1).
Figure 2.
A. Percent micronuclei in TK6 cells treated with aclarubicin measured using an in vitro flow cytometry-based micronucleus assay. B. Frequency of total micronucleated cells using a microscopy-based assay. *Statistically significant vs. the DMSO controls (P ≤ 0.05; Dunnett’s T-test for the flow results; and Fisher’s exact test for the manual scoring).
Figure 7.
A-E. Frequency of micronucleated cells in TK6 cells treated with the tested topo II inhibitors measured using a in vitro microscopy-based micronucleus assay are represented in the bar graph as percent micronucleated cells (# per hundred). The percentages of micronucleated cells originating from chromosome loss (kinetochore-positive (K+)) and chromosome breakage (kinetochore-negative (K-)) are also shown. The means and standard deviations are shown. *Statistically significant vs. the DMSO controls (Fisher’s exact test; P ≤ 0.05).
Table 1.
Measures of Cytotoxicity for TK6 cells treated with Topo II inhibitors
| Concentration | Relative Population Doubling (RPD) | Relative Increase in Cell Counts (RICC) | Relative Cytokinesis-Blocked Proliferation Index (RCBPI) |
|---|---|---|---|
| Aclarubicin (nM) | |||
| 0 | 100.00 | 100.00 | 100 |
| 3.125 | 98.74 | 98.07 | 92.47 |
| 6.25 | 82.84 | 73.56 | 80.54 |
| 12.5 | 50.11 | 36.09 | 67.67 |
| 25 | 26.25 | 16.92 | 51.37 |
| 50 | 19.89 | 11.77 | 31.97 |
| Merbarone (μM) | |||
| 0 | 100.00 | 100.00 | 100 |
| 2.5 | 97.52 | 96.07 | 92.67 |
| 5 | 87.97 | 82.01 | 73.69 |
| 10 | 40.24 | 27.07 | 63.61 |
| 20 | 7.93 | 4.42 | 52.96 |
| Etoposide (nM) | |||
| 0 | 100.00 | 100.00 | 100 |
| 12.5 | 86.76 | 79.12 | 89.42 |
| 25 | 76.18 | 64.82 | 76.00 |
| 50 | 59.56 | 45.45 | 66.03 |
| 100 | 9.23 | 5.23 | 57.06 |
| 200 | 0* | 0* | 33.76 |
| ICRF-154 (μM) | |||
| 0 | 100.00 | 100.00 | 100 |
| 3.125 | 103.13 | 104.78 | 95.41 |
| 6.25 | 101.60 | 102.38 | 94.40 |
| 12.5 | 98.29 | 97.95 | 80.88 |
| 25 | 49.32 | 37.00 | 73.42 |
| 50 | 0* | 0* | 57.36 |
| ICRF-187 (μM) | |||
| 0 | 100.00 | 100.00 | 100 |
| 0.6125 | 84.48 | 76.23 | 97.64 |
| 1.25 | 88.09 | 81.96 | 94.75 |
| 2.5 | 57.50 | 43.69 | 81.39 |
| 5 | 23.59 | 14.64 | 63.06 |
| 10 | 0* | 0* | 50.21 |
A minimum value of 0 used in cases where the cell counts after treatments became lower than initial pre-treatment counts.
Compared to aclarubicin, effects seen with merbarone, the other precleavage complex catalytic inhibitor, occurred at much higher concentrations and the compound induced considerably more MN (Figure 3A). A statistically significant 3.5-fold increase (8.5% total MN) was seen at 5 μM where <20% cytotoxicity was observed. Doubling of the test concentration to 10 μM led to an average 11-fold increase in MN. While continued increases in MN were observed at concentrations above 10 μM, cytotoxicity greatly exceeded 60% (Table 1). Using the microscopy-based method (Figure 3B), a statistically significant increase was seen at the lowest test concentration of 2.5 μM, but overall increases measured on a fold basis were roughly comparable to those seen in the flow-based assay across the range.
Figure 3.
A. Percentages of micronuclei in TK6 cells treated with merbarone measured using a flow cytometry-based micronucleus assay. B. Frequency of total micronucleated cells using a microscopy-based assay. *Statistically significant vs. the DMSO controls (P ≤ 0.05; Dunnett’s T-test for the flow results; and Fisher’s exact test for the manual scoring).
The topo II poison etoposide induced significant increases in micronuclei across the entire concentration range tested using both the flow cytometry and microscopy based micronucleus assays (Figures 4A and 4B). While test concentrations of 100 nM or greater were associated with cytotoxicity exceeding 60%, increases in MN were seen at lower concentrations including an approximate 8-fold increase seen at 50 nM.
Figure 4.
A. Percentages of micronuclei in TK6 cells treated with etoposide measured using a flow cytometry-based micronucleus assay. B. Frequency of total micronucleated cells using a microscopy-based assay. *Statistically significant vs. the DMSO controls (P ≤ 0.05; Dunnett’s T-test for the flow results; and Fisher’s exact test for the manual scoring).
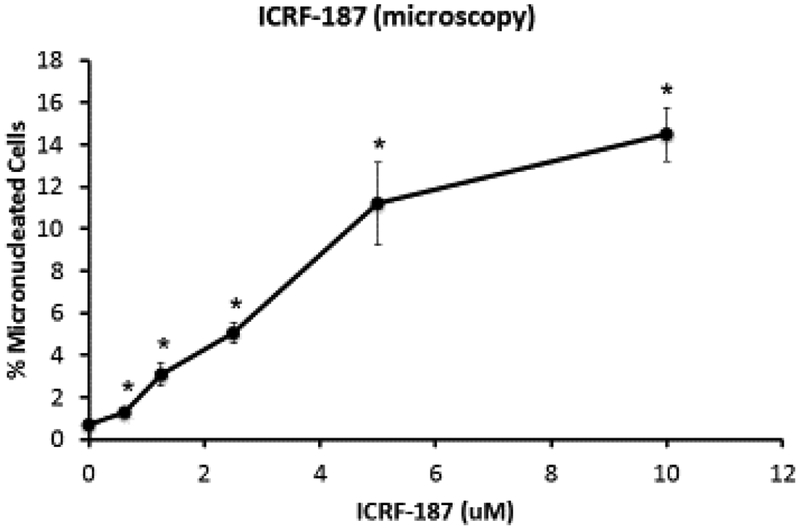
Lastly, increases in MN were also seen in TK6 cells treated with both ICRF−154 and ICRF−187, which act as post-cleavage complex catalytic inhibitors at test concentrations as low as 6.25 μM for ICRF-154 and 1.25 μM for ICRF-187 when measured by flow-cytometry (Figures 5A and 6A). With the exception of the highest concentrations tested for both compounds, the statistically significant increases in MN were observed at concentrations that were associated with approximately 60% cytotoxicity or lower (Table 1). Again, the majority of micronuclei induced were due to chromosome breakage as shown with large increases in K-MN using CREST staining (Figures 7D and 7E).
Figure 5.
A. Percentages of micronuclei in TK6 cells treated with ICRF 154 measured using a flow cytometry-based micronucleus assay. B. Frequency of total micronucleated cells using a microscopy-based assay. *Statistically significant vs. the DMSO controls (P ≤ 0.05; Dunnett’s T-test for the flow results; and Fisher’s exact test for the manual scoring).
Figure 6.
A. Percentages of micronuclei in TK6 cells treated with ICRF 154 measured using a flow cytometry-based micronucleus assay. B. Frequency of total micronucleated cells using a microscopy-based assay. *Statistically significant vs. the DMSO controls (P ≤ 0.05; Dunnett’s T-test for the flow results; and Fisher’s exact test for the manual scoring).
Comparison of flow-cytometry and manual scoring IVMN assays
Since one of the goals of this study was to assess if a flow-cytometry based micronucleus assay would help better understand dose-response relationships in vitro, we compared both assays using the same test compounds and concentrations (Figures 2–6). While the overall concentration responses for each of the test compounds were similar when comparing the two assays, there were some notable differences. First, the actual MN frequency in both the controls and treated cells tended to be higher when measured by flow-cytometry than compared to microscopy, presumably due to the detection of early apoptotic cells in the flow cytometry assay. When compared on a fold-change basis, however, roughly similar increases in MN were observed when using the two different types of assay. Another significant difference between the methods was that for 3 of the 5 compounds tested, significant increases in MN frequency were observed at lower concentrations using the microscopy-based assay compared to the flow-cytometry assay. These results suggested that the microscopy-based assay might be more sensitive than the flow-based assay.
To rule out that the differences in sensitivity observed were not simply due to differences in statistical approaches used to analyze the flow-and manual-scoring datasets, we used BMD modeling to estimate points of departure (PoD) within the concentration ranges tested for each compound. Using this approach, the differences between the flow-cytometry and microscopy-based assays were much less pronounced than when simply comparing no observed-or lowest observed genotoxic effect levels (NOGEL and LOGEL, respectively) (Table 2). For instance, merbarone, ICRF-187, and aclarubicin all had comparably similar (2-fold or less) BMD and BMDL estimates when comparing results of the two different types of assay; however, for ICRF−154 and etoposide, manual scoring was still more sensitive than the flow-based assay with BMD/BMDL estimates one-third to one-fifth of those seen with the flow assay.
Table 2.
Comparison of NOGEL/LOGEL values with BMD1SD and BMDL1SD for topo II inhibitors using microscopy and flow-cytometry based micronucleus assays
| Microscopy | Flow-cytometry | |||||||
|---|---|---|---|---|---|---|---|---|
| NOGEL | LOGEL | BMD1SD | BMDL1SD | NOGEL | LOGEL | BMD1SD | BMDL1SD | |
| Aclarubicin | 6.25 nM | 12.5 nM | 4.9 nM | 2.6 nM | 6.25 nM | 12.5 nM | 4.1 nM | 3.1 nM |
| Merbarone | N/A | 2.5 μM | 1.8 μM | 1.2 μM | 2.5 μM | 5 μM | 3.2 μM | 2.5 μM |
| Etoposide | N/A | 12.5 nM | 6.2 nM | 3 nM | N/A | 12.5 nM | 29 nM | 13 nM |
| ICRF-154 | N/A | 3.13 μM | 2.4 μM | 1.6 μM | 3.13 μM | 6.25 μM | 8 μM | 6 μM |
| ICRF-187 | N/A | 0.61 μM | 0.7 μM | 0.4 μM | 0.61 μM | 1.25 μM | 0.7 μM | 0.3 μM |
Discussion
Overall, all of the topo II inhibitors induced concentration-dependent increases in micronuclei in the TK6 cells. In examining the results from the two methods of MN scoring, the increases appear to be linear or curvilinear at the test concentrations included in our studies. It should be noted that in most cases, cytotoxicity at the highest test concentrations for the five compounds often exceeded values generally considered in an acceptable range (<55% cytotoxicity; OECD, 2016) (19), but with the exception of aclarubicin, statistically-significant increases in MN were seen at concentrations where relative population doubling (RPD) or relative increases in cell counts (RICC) were 75% or higher (Table 1).
CREST staining in manually scored cells revealed that a large majority of MN were kinetochore-negative in cells treated with all five inhibitors, indicating that most micronuclei were formed from chromosome breakage (Figure 7). It should be noted that increases in chromosome loss were also seen with all of the compounds tested, but the increases occurred at the higher test concentrations and at much lower magnitude than the increases in chromosome breakage.
In terms of potency, bisdioxopiperazines ICRF−154 and ICRF−187 as well as merbarone induced significant increases in MN at low micromolar concentrations (3.125 μM, 0.6125 μM, and 2.5 μM respectively). These concentrations were roughly 500-fold greater than those at which the other catalytic inhibitor tested, aclarubicin, induced significant increases in micronuclei. In fact, the 6.25 nM aclarubicin concentration was similar to the 12.5 nM concentration of the topo II poison etoposide in terms of cytotoxicity and magnitude of micronucleus induction.
The mechanism underlying the clastogenic effects induced by topo II poisons such as etoposide is fairly well understood, as stabilization of the cleavage complex and interference with religation would also lead to persistence of the otherwise transient double stranded break that occurs during the enzyme’s catalytic cycle (20). Subsequent removal of the covalently bound topo II found in the cleavage complex can occur by either endonucleolytic cleavage (21), enzyme-mediated hydrolysis (22–23), or proteasomal degradation (24) exposing an unprotected DNA double strand break that can result in chromosomal breaks and translocations.
While the clastogenic effects induced by the catalytic inhibitors seen here are in agreement with previously reported findings, the mechanism by which chromosome breaks occur for the catalytic inhibitors is not as well understood (7–8, 25–29). Compounds such as ICRF−154 and ICRF−187 are believed to trap topo II in a “closed-clamp” formation where the double strand break has been properly ligated and the enzyme is no longer covalently bound to the DNA. The inhibited enzyme is unable to be converted back to a catalytically active form, and the enzyme continues to encircle the DNA duplex (26–27). The resulting depletion of active topo II caused by these compounds could then lead to the observed clastogenic responses as the enzyme is no longer available to relieve the torsional strain associated with DNA replication and transcription, or allow for the decatenation of sister chromatids during mitosis.
In our study, aclarubicin was the one catalytic inhibitor tested that behaved considerably differently than the others. It is believed that aclarubicn acts through intercalation, thereby preventing topo II from binding to DNA (28). The low nanomolar concentrations at which effects are seen, however, seem inconsistent with the results seen with other compounds believed to act through similar mechanisms (29). It seems likely, as has been reported by others (28, 31–32), that aclarubicin has additional targets in the cell and topo II is not be the only enzyme or process affected.
As shown in Table 1, substantial differences were seen using the different measures of cytotoxicity/cell proliferation. For manual scoring by microscopy using cytochalasin B, the OECD TG 487 recommends using the CBPI as the measurement of cytotoxicity (19). Compared to the two other measures, RPD and RICC, the CBPI showed substantially less cytotoxicity, particularly at higher concentrations. The reason for the observed differences is not known. However, previous studies have shown that treatment with ICRF 154 and bimolane, both topo II catalytic inhibitors, can produce binucleated cells in the absence of cytochalasin B (7, 33). These earlier results indicate that the CBPI may not be the most accurate measure of cytotoxicity for this class of compounds.
For concentration-response data from the flow-cytometry based assay, NOGEL values were identified for 4 of the 5 inhibitors studied (Table 2). In contrast using the microscopy-based assay, the MN frequency at the lowest concentrations tested for all of the inhibitors, except for aclarubicin, were significantly increased compared to the control, and as a result, NOGEL values could not be identified. While assay sensitivity may contribute to some of the differences seen, it should be noted that different statistical approaches were used to analyze data from the two different types of assay. The data from the microscopy-based assay was analyzed by conducting a binomial trend test followed by a Fisher’s exact test to compare individual treatments to DMSO-treated controls. This statistical analysis approach has been largely derived from earlier work evaluating chromosomal aberration frequencies where the numbers of cells scored were quite low. Using the microscopy-based assay with 1000 cells scored and two to three combined replicates at each concentration, this approach for analysis of data begins to become quite sensitive though statistically significant increases still occur at what intuitively seem to be biologically relevant ranges of 70–80% above the control frequencies. When using this approach with the flow-cytometry data, however, increases in micronuclei as low as 8–10% above the control frequency would be concluded to be significantly elevated with low associated p-values due to the large numbers of cells scored (e.g. 20,000 cells per concentration). These small increases above the control frequency also lie well within the inter-replicate range of controls. For this reason, a different type of analysis was used; in this case, following a positive ANOVA result, a Dunnett’s T-test was used to analyze the flow data, a common statistical approach and consistent with the one recommended by Johnson et al (34). With ANOVA and the Dunnett’s test, the sample size is based on the number of experiments rather than the number of cells analyzed per experiment. The use of this approach resulted in statistically significant increases at responses deemed to be more biologically relevant. The use of different approaches likely contributed to the different NOGEL values that were obtained using the microscopy and flow-cytometry based assays in our parallel experiments, making it more difficult to make direct comparisons between the two assays. Because of this, an alternative approach of BMD modeling was explored to describe the concentration-response data for the five topo II inhibitors tested.
As mentioned previously, BMD modeling has been used in other fields of toxicology and risk assessment to overcome pitfalls when estimating PoDs, and more recently in dose-response modeling of genotoxicity data (34–35). BMD modeling is advantageous since it considers the entire range of the experimental data and is less influenced by sample size and dose-spacing (18,36). The BMR specified here was one standard deviation from controls as specified by the US EPA for modeling of continuous data (18). Selection of specific response rates is currently an area of active discussion. Recent reports from working groups utilizing the BMD approach with genotoxicity data have used response rates of 5% or 10%. Based on our experience, a 5–10% increase above the control frequency falls well within normal assay variability and would yield a highly conservative PoD estimate (35, 37). Table 2 compares NOGEL or LOGEL values for each of the compounds tested to BMD1SD estimates and their associated lower confidence bounds (BMDL1SD). BMD1SD estimates were in a range where minimal but significant increases are expected to be observed. In most cases, the BMD1SD estimates fell between the NOGEL and the LOGEL values or, when a NOGEL was not available, between the control and the LOGEL, thus within the range of observation and consistent with the goal of having BMDLs agree, on average, with NOAELs (or in this case NOGELs) (38). Also, while using the NOGEL/LOGEL approach was problematic when comparing PoD estimates between manual and flow-cytometric studies, BMD1SD and BMDL1SD values were often quite comparable between the two assays, as seen with merbarone, ICRF−187, and aclarubicin.
Our results indicate that BMD modeling is an appropriate and useful method for quantitatively describing dose-response data for micronucleus induction, which combined with appropriate in vitro and in vivo mechanistic information, could help identify biologically relevant PoD estimates. This is in agreement with several recently published reports highlighting the use of BMD modeling for genotoxicity data with model compounds utilizing large curated datasets (36–37,39). In addition, BMD modeling can provide reliable PoD estimates for dose response studies even when relatively few doses are available. Further studies will be useful to refine study design to identify an optimal number of doses, dose spacing, and confirming one standard deviation or another value as the appropriate benchmark response.
Highlights.
Topoisomerase II inhibitors were effective inducers of micronuclei.
Micronuclei originated primarily from chromosome breakage.
Most catalytic inhibitors of topoisomerase II were less potent than etoposide.
Manual scoring was similar or more sensitive than flow cytometry for detecting micronuclei.
Benchmark dose modeling proved to be valuable for dose-response analysis.
Acknowledgements
We thank the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment, National Cancer Institute (Bethesda, MD) for generously providing several of the topo II inhibitors used in this study. Support for P.G. was provided in part by a NRSA Institutional Training grant (T32 ES018827) from the NIEHS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest: None
References
- 1.Pendleton M, Lindsey RH Jr, Felix CA, Grimwade D, Osheroff N. Topoisomerase II and leukemia. Ann N Y Acad Sci 2014;1310: 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McClendon AK, Osheroff N. DNA topoisomerase II, genotoxicity, and cancer. Mutat Res. 2007;623: 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deweese JE, Osheroff N. The DNA cleavage reaction of topoisomerase II: wolf in sheep’s clothing. Nucleic Acids Res. 2009;37: 738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vos SM, Tretter EM, Schmidt BH, Berger JM. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat Rev Mol Cell Biol. 2011;12: 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mondrala S, Eastmond DA. Topoisomerase II inhibition by the bioactivated benzene metabolite hydroquinone involves multiple mechanisms. Chem Biol Interact. 2010;184: 259–268. [DOI] [PubMed] [Google Scholar]
- 6.Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nat Rev Cancer. 2009;9: 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vuong MC, Hasegawa LS, Eastmond DA. A comparative study of the cytotoxic and genotoxic effects of ICRF−154 and bimolane, two catalytic inhibitors of topoisomerase II. Mutat Res. 2013;750: 63–71. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Eastmond DA. Catalytic inhibitors of topoisomerase II are DNA-damaging agents: induction of chromosomal damage by merbarone and ICRF−187. Environ Mol Mutagen. 2002;39: 348–356. [DOI] [PubMed] [Google Scholar]
- 9.Mauritzson N, Albin M, Rylander L, Billström R, Ahlgren T, Mikoczy Z, Björk J, Strömberg U, Nilsson PG, Mitelman F, Hagmar L, Johansson B. Pooled analysis of clinical and cytogenetic features in treatment-related and de novo adult acute myeloid leukemia and myelodysplastic syndromes based on a consecutive series of 761 patients analyzed 1976–1993 and on 5098 unselected cases reported in the literature 1974–2001. Leukemia. 2002;16: 2366–2378. [DOI] [PubMed] [Google Scholar]
- 10.Joannides M, Mays AN, Mistry AR, Hasan SK, Reiter A, Wiemels JL, Felix CA, Lo Coco F, Osheroff N, Solomon E, Grimwade D. Molecular pathogenesis of secondary acute promyelocytic leukemia. Mediterr J Hematol Infect Dis. 2011;3: e2011045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rashidi A, Fisher SI. Therapy-related acute promyelocytic leukemia: a systematic review. Med Oncol. 2013;30: 625. [DOI] [PubMed] [Google Scholar]
- 12.Li Y-S, Zhao Y-L, Jiang Q-P, Yang C-L. Specific chromosome changes and non-occupational exposure to potentially carcinogenic agents in acute leukemia in china. Leuk Res. 1989;13: 367–376. [DOI] [PubMed] [Google Scholar]
- 13.Strick R, Strissel PL, Borgers S, Smith SL, Rowley JD. Dietary bioflavonoids induce cleavage in the MLL gene and may contribute to infant leukemia. Proc Natl Acad Sci U S A. 2000;97: 4790–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spector LG, Xie Y, Robison LL, Heerema NA, Hilden JM, Lange B, Felix CA, Davies SM, Slavin J, Potter JD, Blair CK, Reamna GH, Ross JA. Maternal diet and infant leukemia: the DNA topoisomerase II inhibitor hypothesis: a report from the children’s oncology group. Cancer Epidemiol Biomarkers Prev. 2005;14: 651–655. [DOI] [PubMed] [Google Scholar]
- 15.Xue Y, Lu D, Guo Y, Lin B. Specific chromosomal translocations and therapy-related leukemia induced by bimolane therapy for psoriasis. Leuk Res. 1992;16: 1113–1123. [DOI] [PubMed] [Google Scholar]
- 16.Eastmond DA, Tucker JD. Identification of aneuploidy-inducing agents using cytokinesis-blocked human lymphocytes and an antikinetochore antibody. Environ Mol Mutagen. 1989;13: 34–43. [DOI] [PubMed] [Google Scholar]
- 17.Avlasevich SL, Bryce SM, Cairns SE, Dertinger SD. In vitro micronucleus scoring by flow cytometry: differential staining of micronuclei versus apoptotic and necrotic chromatin enhances assay reliability. Environ Mol Mutagen. 2006;47: 56–66. [DOI] [PubMed] [Google Scholar]
- 18.Epa US, ORD, NCEA. Benchmark Dose Technical Guidance. 2014; Available: https://www.epa.gov/risk/benchmark-dose-technical-guidance [Google Scholar]
- 19.OECD (2010), Test No. 487: In Vitro Mammalian Cell Micronucleus Test, OECD Publishing, Paris, 10.1787/9789264091016-en. [DOI] [Google Scholar]
- 20.Lynch A, Harvey J, Aylott M, Nicholas E, Burman M, Siddiqui A, Walker S, Rees R. Investigations into the concept of a threshold for topoisomerase inhibitor-induced clastogenicity. Mutagenesis. 2003;18: 345–353. [DOI] [PubMed] [Google Scholar]
- 21.Hartsuiker E, Neale MJ, Carr AM. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol Cell. 2009;33: 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. A human 5’-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461: 674–678. [DOI] [PubMed] [Google Scholar]
- 23.Borda MA, Palmitelli M, Verón G, González-Cid M, de Campos Nebel M. Tyrosyl-DNA-phosphodiesterase I (TDP1) participates in the removal and repair of stabilized-Top2α cleavage complexes in human cells. Mutat Res. 2015;781: 37–48. [DOI] [PubMed] [Google Scholar]
- 24.Mao Y, Desai SD, Ting CY, Hwang J, Liu LF. 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes. J Biol Chem. 2001;276: 40652–40658. [DOI] [PubMed] [Google Scholar]
- 25.Larsen AK, Escargueil AE, Skladanowski A. Catalytic topoisomerase II inhibitors in cancer therapy. Pharmacol Ther. 2003;99: 167–181. [DOI] [PubMed] [Google Scholar]
- 26.Tanabe K, Ikegami Y, Ishida R, Andoh T. Inhibition of topoisomerase II by antitumor agents bis(2,6-dioxopiperazine) derivatives. Cancer Res. 1991;51: 4903–4908. [PubMed] [Google Scholar]
- 27.Andoh T, Ishida R. Catalytic inhibitors of DNA topoisomerase II. Biochim Biophys Acta. 1998;1400: 155–171. [DOI] [PubMed] [Google Scholar]
- 28.Sørensen BS, Sinding J, Andersen AH, Alsner J, Jensen PB, Westergaard O. Mode of action of topoisomerase II-targeting agents at a specific DNA sequence. Uncoupling the DNA binding, cleavage and religation events. J Mol Biol. 1992;228: 778–786. [DOI] [PubMed] [Google Scholar]
- 29.Monnot M, Mauffret O, Simon V, Lescot E, Psaume B, Saucier JM, et al. DNA-drug recognition and effects on topoisomerase II-mediated cytotoxicity. A three-mode binding model for ellipticine derivatives. J Biol Chem. 1991;266: 1820–1829. [PubMed] [Google Scholar]
- 30.Snyder RD, Arnone MR. Putative identification of functional interactions between DNA intercalating agents and topoisomerase II using the V79 in vitro micronucleus assay. Mutat Res. 2002;503: 21–35. [DOI] [PubMed] [Google Scholar]
- 31.Pang B, Qiao X, Janssen L, Velds A, Groothuis T, Kerkhoven R, Niewland M, Ovaa H, Rottenberg S, van Tellingen O, Janssen J, Huijgens P, Zwart W, Neefjes J. Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin. Nat Commun. 2013;4: 1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pang B, de Jong J, Qiao X, Wessels LFA, Neefjes J. Chemical profiling of the genome with anti-cancer drugs defines target specificities. Nat Chem Biol. 2015;11: 472–480. [DOI] [PubMed] [Google Scholar]
- 33.Roy SK, Eastmond DA. Bimolane induces multiple types of chromosomal aberrations in human lymphocytes in vitro. Mutat Res. 2011;726: 181–187. [DOI] [PubMed] [Google Scholar]
- 34.Johnson GE, Soeteman-Hernández LG, Gollapudi BB, Bodger OG, Dearfield KL, Heflich RH, Hixon JG, Lovell DP, MacGregor JT, Pottenger LH, Thompson CM, Abraham L, Thybaud V, Tanir JY, Zeiger E, van Benthem J, White PA. Derivation of point of departure (PoD) estimates in genetic toxicology studies and their potential applications in risk assessment. Environ Mol Mutagen. 2014;55: 609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gollapudi BB, Johnson GE, Hernandez LG, Pottenger LH, Dearfield KL, Jeffrey AM, Julien E, Kim JH, Lovell DP, Macgregor JT, Moor MM, van Benthem J, White PA, Zeiger E, Thybaud V. Quantitative approaches for assessing dose-response relationships in genetic toxicology studies. Environ Mol Mutagen. 2013;54: 8–18. [DOI] [PubMed] [Google Scholar]
- 36.Davis JA, Gift JS, Zhao QJ. Introduction to benchmark dose methods and U.S. EPA’s benchmark dose software (BMDS) version 2.1.1. Toxicol Appl Pharmacol. 2011;254: 181–191. [DOI] [PubMed] [Google Scholar]
- 37.Bemis JC, Wills JW, Bryce SM, Torous DK, Dertinger SD, Slob W. Comparison of in vitro and in vivo clastogenic potency based on benchmark dose analysis of flow cytometric micronucleus data. Mutagenesis. 2016;31: 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haber LT, Dourson ML, Allen BC, Hertzberg RC, Parker A, Vincent MJ, Maier A, Boobis AR. Benchmark dose (BMD) modeling: current practice, issues, and challenges. Crit Rev Toxicol. 2018;48: 387–415. [DOI] [PubMed] [Google Scholar]
- 39.MacGregor JT, Frötschl R, White PA, Crump KS, Eastmond DA, Fukushima S, Guérard M, Hayashi M, Soeteman-Hernández LG, Johnson GE, Kasamatsu T, Levy DD, Morita T, Müller L, Schoeny R, Schuler MJ, Thybaud V. IWGT report on quantitative approaches to genotoxicity risk assessment II. Use of point-of-departure (PoD) metrics in defining acceptable exposure limits and assessing human risk. Mutat Res Genet Toxicol Environ Mutagen. 2015;783: 66–78 [DOI] [PubMed] [Google Scholar]