Abstract
Patient-derived induced pluripotent stem cells (iPSCs) have become a promising resource for exploring genetics of complex diseases, discovering new drugs, and advancing regenerative medicine. Increasingly, laboratories are creating their own banks of iPSCs derived from diverse donors. However, there are not yet standardized guidelines for qualifying these cell lines, i.e., distinguishing between bona fide human iPSCs, somatic cells, and imperfectly reprogrammed cells. Here, we report the establishment of a panel of 30 iPSCs from CD34+ peripheral blood mononuclear cells, of which 10 were further differentiated in vitro into all three germ layers. We characterized these different cell types with commonly used pluripotent and lineage specific markers, and showed that NES, TUBB3, and OTX2 cannot be reliably used as ectoderm differentiation markers. Our work highlights the importance of marker selection in iPSC authentication, and the need for the field to establish definitive standard assays.
Keywords: Human induced pluripotent stem cell, Trilineage differentiation, Ectoderm marker, NESTIN, PAX6
1. Introduction
Patient-derived induced pluripotent stem cells (iPSCs) retain the genetic characteristics of the donor, exhibit self-renewal, and can be cryopreserved and differentiated into disease-relevant tissue types. These capabilities render iPSCs a highly versatile cellular model enabling a wide range of research studies and experimental designs. For examples, these cells may be used for identifying transcriptomic and metabolomic markers of drug efficacy, screening in drug discovery, and advancing regenerative medicine (Avior et al., 2016; Sterneckert et al., 2014).
iPSCs are reprogrammed from somatic cells through the ectopic expression of defined transcription factors such as POU5F1 (aka OCT4), SOX2, KLF4, and c-MYC (also known as the Yamanaka factors) (Takahashi and Yamanaka, 2006). Since the process of reprogramming may be incomplete, thus generating heterogeneous cell populations, it is essential to validate the quality of iPSC lines generated, and to distinguish bona fide iPSCs from partially reprogrammed cells (Chan et al., 2009). This interrogation includes confirming expression of pluripotency factors, for example POU5F1 or TRA-1–60, and also displaying the ability to differentiate into the three germ layers: endoderm, mesoderm, and ectoderm (Chan et al., 2009). Demonstration of germ layer differentiation has traditionally been performed through a teratoma assay, in which cells considered to be pluripotent would be injected into an immunocompromised mouse, and tested for their ability to create a teratoma with cells from all three germ layers. However, as this assay is labor intensive, costly and time-consuming, in vitro trilineage differentiation, in which specialized media is used to induce lineage-directed differentiation, has become a highly popular alternative (Buta et al., 2013; Vallier et al., 2009).
A wide range of markers has been reported in the literature to be indicative of the three germ layers. Commonly used endoderm markers include SOX17 (Kanai-Azuma et al., 2002), FOXA2 (Burtscher and Lickert, 2009), CXCR4 (D’Amour et al., 2005), and GATA4 (Fujikura et al., 2002). NCAM1 (Evseenko et al., 2010) and TBXT (aka brachyury) (Wilkinson et al., 1990) are often used as mesoderm markers. NES (nestin) (Lendahl et al., 1990) and PAX6 (Zhang et al., 2010) are among the most commonly used markers for ectoderm differentiation. Here we report that several of the ectoderm markers commonly used during validation studies are not specific to ectoderm as there is a high degree of staining in undifferentiated iPSCs derived from CD34+ peripheral blood mononuclear cells (PBMCs). These studies highlight the need for further development of iPSC validation protocols.
2. Materials and methods
2.1. Donors and samples
The iPSCs used to obtain data reported here were generated as part of the Pharmacogenomics of Statin Therapy (POST) Center within the Pharmacogenomics Research Network. Blood samples were drawn from donors recruited at Kaiser Permanente Northern California (Table 1). Written informed consent was obtained from all study subjects, and studies were performed in accordance with all relevant regulatory guidelines. This study was approved by the IRBs at Kaiser Permanente Northern California and the UCSF Benioff Children’s Hospitals.
Table 1.
Demographics of blood donors.
| n | % | |
|---|---|---|
| Total | 30 | 100 |
| Gender | ||
| Female | 18 | 60 |
| Male | 12 | 40 |
| Race/Ethnicity | ||
| White | 23 | 76.7 |
| Black or African American | 3 | 10 |
| Asian | 4 | 13.3 |
2.2. PBMC isolation
After 1:1 dilution in Dulbecco’s Phosphate Buffered Saline containing 2% Fetal Bovine Serum (FBS, Omega Scientific, Cat. # FB-02, Lot # 8462) whole blood from each donor was layered over a Ficoll-Pacque gradient (Fisher Scientific, Cat. # 45001750) in SepMate-50 tubes (StemCell Technologies, Cat. # 85460) and centrifuged at 1200 ×g for 20 min. PBMCs were decanted into a separate tube, washed in PBS supplemented with 2% FBS, and centrifuged for an additional 10 min at 500 × g. PBMC were resuspended in RPMI supplemented with 20% FBS and 10% DMSO, and cryopreserved in liquid nitrogen.
2.3. iPSC generation
PBMC aliquots were thawed and resuspended in StemSpan SFEM II expansion media (StemCell Technologies, Cat. # 09605) with CD34+ expansion supplement (StemCell Technologies, Cat. # 02691). Cells were seeded into an Ultra-Low Attachment 6-well plate (Corning, Cat. # 3471). After 6 days, the CD34+ cells were counted and up to 1 million cells were reprogrammed using a combination of plasmids purchased from Addgene encoding for POU5F1, and shRNA for TP53 (ID: 27077) SOX2, and KLF4 (ID: 27078), L-MYC, and LIN28(ID: 27080), and EBNA1(ID: 37624) (Okita et al., 2013). Cells were transfected using program U-008 on the Lonza Nucleofector 2B device (Lonza, Cat. # AAB-1001), seeded onto mitomycin C treated SNL feeder cells (Applied StemCel,l Cat. # ASF-1327) in SFEM II media plus CD34+ Expansion supplement, and fed every other day with mTeSR1 (StemCell Technologies, Cat. # 85850) until colony establishment.
2.4. iPSC culture
iPSCs were harvested from SNL feeder plates using Magnetic Activated Cell Sorting (MACS) via positive selection for TRA-1–60 surface pluripotency marker (Miltenyi Biotec, Cat. # 130-100-832). iPSCs were seeded at roughly 200,000 cells/well onto 6-well plates coated with Cultrex reduced growth factor basement membrane (Trevigen, Cat. # 3533-001-02). Cells were fed daily with 1.5 mL of mTeSR1 and passaged when approximately 80% confluent using ReLeSR (StemCell Technologies, Cat. # 05872), which leaves behind differentiated cells.
2.5. Live cell staining
Alkaline Phosphatase detection was executed using the Alkaline Phosphatase Live Stain kit (Life Technologies, Cat. # A14353) according to manufacturer’s instructions. Cells were visualized using a Keyence BZ-X700 series microscope. SSEA-4 detection was carried out using the StainAlive SSEA-4 mouse anti-human antibody (Reprocell, Cat. # 09–0097). Antibody was diluted 1:200 in mTeSR1 and cells were incubated in antibody containing medium for 30 min in standard culture conditions. The staining medium was removed and the cells washed gently 2 times with mTeSR1. Fresh media was added and the cells were visualized using a Keyence BZ-X700 series microscope.
2.6. iPSC characterization
Karyotype analysis was performed by the Cedars Sinai RMI iPSC Core. PluriTest and KaryoStat assays were performed by Thermo Fisher Scientific.
2.7. Cellular differentiation assay
Ten iPSC lines underwent in vitro directed differentiation in four independent experiments with 1 to 4 lines in each batch. Cells were seeded onto 12-well plates coated with Cultrex at 400,000 cells/well for both the ectoderm and endoderm conditions and 100,000 cells/well for the mesoderm condition and treated the appropriate differentiation media using the STEMdiff Trilineage Differentiation Kit (StemCell Technologies, Cat. # 05230) according to manufacturer instructions. Differentiated cells were then collected for RNA isolation or fixed with 4% paraformaldehyde for immunostaining.
2.8. Immunostaining and flow cytometry analyses
80% confluent cells were detached from the plates into single cell suspension using Accutase (StemCell Technologies, Cat. # 07920). Cells were washed in DPBS, then fixed with 4% paraformaldehyde in PBS and incubated for 15 min. Fixed cells were washed with DPBS, resuspended with DPBS+1%BSA, and stored at 4 °C for up to 7 days prior to analysis. For intracellular markers, fixed cells were treated with Permeabilization Buffer (eBioscience, Cat. # 00-8333-56). Cells were transferred to Falcon 96-well V bottom plates (Fisher Scientific, Cat. # 08772212) for immunostaining. Antibody incubations were performed on ice in the dark for 45 min in 100 ul/well of CAS-Block (Thermo Fisher Scientific, Cat. # 008120) supplemented with 1% BSA and 5% goat serum (plus 0.2% Triton for intracellular markers). Cells were then washed twice with 200 ul/well DPBS (cell surface markers) or Permeabilization Buffer (intracellular markers) before resuspended in 100 ul/well DPBS +1% BSA. Flow cytometry analysis was performed on the BD LSRFortessa with a High Throughput Sampler, with markers quantified in 9000 gated events on average. The antibodies used for detecting iPSC, endoderm, mesoderm, and ectoderm markers, as well as corresponding isotype controls, are listed in Table 2.
Table 2.
Antibodies used in the study.
| Antigen recognized | Clone | Fluorochrome | Isotype | Supplier | Catalog # |
|---|---|---|---|---|---|
| POU5F1 | 3A2A20 | PE | Mouse IgG2b | BioLegend | 653703 |
| POU5F1 | 3A2A20 | AF488 | Mouse IgG2b | BioLegend | 653705 |
| TRA-1–60 | TRA-1–60-R | PE | Mouse IgM | BioLegend | 330609 |
| SSEA-1 | HI98 | PE | Mouse IgM | BioLegend | 301905 |
| SOX17 | P7–969 | AF647 | Mouse IgGl | BD Biosciences | 562594 |
| FOXA2 | N17–280 | PE | Mouse IgGl | BD Biosciences | 561589 |
| NCAM1 | HCD56 | AF647 | Mouse IgGl | BioLegend | 318313 |
| NES | 25/NESTIN | PE | Mouse IgGl | BD Biosciences | 561230 |
| NES | 10C2 | PE | Mouse IgGl | BioLegend | 656805 |
| TUBB3 | TUJ1 | AF647 | Mouse IgG2a | BD Biosciences | 560394 |
| OTX2 | 246826 | AF488 | Mouse IgG2b | R&D Systems | IC1979G |
| PAX6 | 018–1330 | AF647 | Mouse IgG2a | BD Biosciences | 562249 |
| Isotype control | MM-30 | PE | Mouse IgM | BioLegend | 401611 |
| Isotype control | MPC-11 | PE | Mouse IgG2b | BioLegend | 400313 |
| Isotype control | MOPC-173 | AF647 | Mouse IgG2a | BD Biosciences | 558053 |
| Isotype control | M0PC-21 | AF647 | Mouse IgGl | BD Biosciences | 557714 |
| Isotype control | M0PC-21 | PE | Mouse IgGl | BD Biosciences | 554680 |
| Isotype control | 133303 | AF488 | Mouse IgG2b | R&D Systems | IC0041G |
2.9. qPCR
RNA was isolated via QIAcube (serial number: 50946) and RNeasy Mini Kit protocol “Purification of total RNA from 5 × 106 − 1 × 107 animal cells (with QIAshreddder homogenization)” (Firmware Version FIW-50–001-J_FW_MB.hex and program version FIW-50–002-G_PLC_MB.prs or higher) following manufacturer’s instructions. cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosysystems, Cat. # 4368814) according to manufacturer’s instructions. Quantitative PCR with the Taqman™ primers POU5F1 (Thermofisher, Cat. # 4448892), PAX6 (Thermofisher, Cat. # 4448892), NES (Thermofisher, Cat. # 4448892), TUBB3 (Thermofisher, Cat. # 4448892), and OTX2 (Thermofisher, Cat. # 4448892) was carried out by a liquid handling robotic system and run via Quantstudio 12K Flex (serial number: 285880333).
2.10. Statistical analysis
Two-tailed paired t-tests were used to determine statistically significant differences between cell types. Analyses were performed using GraphPad Prism 7.
3. Results
3.1. Generation and validation of blood-derived human iPSCs
The CD34+ PBMC isolated from 30 individuals were nucleofected by episomal vectors expressing POU5F1, SOX2, KLF4, L-MYC, LIN28, EBNA1, and shRNA for TP53 (Okita et al., 2013). This integration-free approach minimizes genetic manipulation of the iPSCs. After emergence of colonies with distinct iPSC morphology (i.e. embryonic stem cell-like compact colonies comprised of cells with large nuclei and well-defined borders (Chan et al., 2009)), TRA-1–60 expressing cells were selected using Magnetic Activated Cell Sorting (MACS) to obtain one iPSC line per donor in a pooled culture (Yang et al., 2015). Compared to clonally derived iPSCs, this method has been shown to be more efficient, potentially better standardized, and less variable than clonal selection.
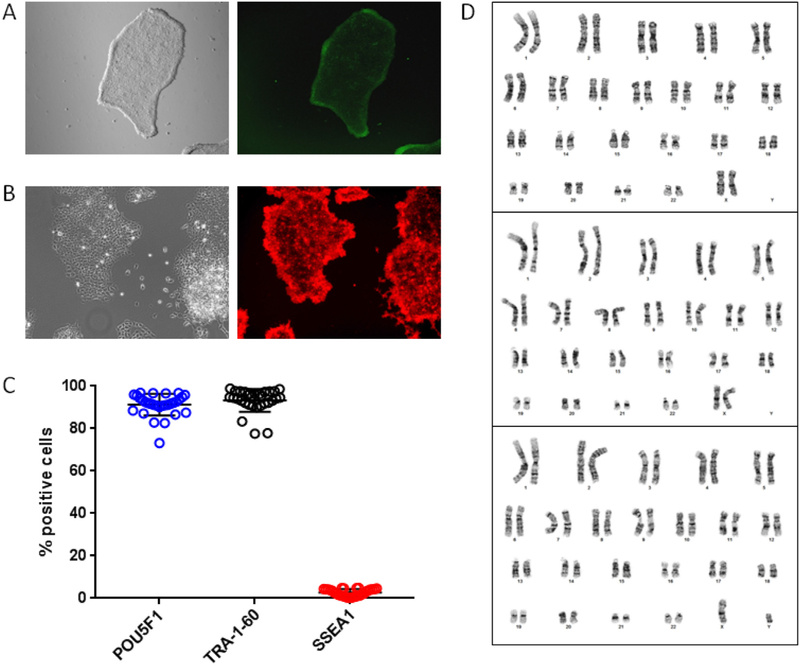
To confirm the identity of the established iPSC lines, colonies were subject to live staining of alkaline phosphatase (AP, Fig. 1A) or SSEA-4 expression (Fig. 1B). In addition, expression of pluripotency markers POU5F1 and TRA-1–60, and the differentiation marker SSEA-1, were quantified by immunostaining and flow cytometry in all 30 iPSC lines. Of the cells within an individual line, 91.3% (range 73.2–96.9%) were positive for POU5F1 expression, 93.3% (range 77.6–98.9%) were positive for TRA-1–60, and 2.7% (range 0.2–4.9%) were positive for SSEA-1 (Fig. 1C and S1). All lines were > 90% positive for POU5F1 and/or TRA-1–60, and < 5% were positive for SSEA-1. Karyotype analyses of three representative lines showed no genetic abnormalities (Fig. 1D). Five iPSC lines (including 3 of the 10 lines underwent in vitro trilineage differentiation) were also tested by PluriTest and KaryoStat, and all of them were found to be pluripotent with no chromosomal aberrations (Fig. S2).
Fig. 1.
Validation of blood-derived human iPSCs. Live stain of representative iPSC colonies showing A) alkaline phosphatase activity, and B) SSEA-4 expression. C) Flow cytometry analysis of pluripotency markers POU5F1 and TRA-1–60, and differentiation marker SSEA-1 in iPSCs (mean ± SD, n = 30). See also Fig. S1 for flow cytometry data on isotype controls. D) Karyotype analysis of three iPSC lines showed normal chromosomal content. See also Fig. S2 for additional characterization of pluripotency marker expression and genomic aberrations.
3.2. Validation of iPSCs via in vitro trilineage differentiation
Next, to confirm the pluripotency of the iPSC lines, we performed a trilineage differentiation assay on 10 lines using the STEMdiff Trilineage Differentiation kit. To authenticate generation of each lineage, endoderm differentiated cells were stained for SOX17 and FOXA2, mesoderm cells were stained for NCAM1, and ectoderm cells were stained for NES, with the degree of immunostaining quantified by flow cytometry. Matched iPSCs, from whom the differentiated cells were derived, were also stained for lineage specific markers as negative controls. After differentiation, cellular morphology was dramatically altered from the typical iPSC phenotype, Fig. S3. Importantly, after directed endoderm differentiation, we found evidence of both SOX17 and FOXA2 positive cells in each of the 10 lines tested, with on average46.3% (range 10–75.7%) of cells/line expressing SOX17, and 45.3% (range 14.8–68.9%) of cells/line expressing FOXA2 (Fig. 2A and Fig. S4). On average 84% (range 26.6–99.5%) of cells/line were NCAM1 positive after directed mesoderm differentiation (Fig. 2A and Fig. S4). For both the endoderm and mesoderm markers, we observed little to no staining of the undifferentiated iPSC lines (2.0% positive cells/marker/line on average).
Fig. 2.
Expression of trilineage differentiation markers. A) Matched iPSCs and differentiated endoderm, mesoderm, ectoderm cells were analyzed by immunostaining followed by flow cytometry for expression of markers specific for each germ layer (mean ± SEM, n = 7–10 cell lines). See also Fig. S1 for isotype control data, Fig. S3 for morphological changes after differentiation, Fig. S4 for paired data, and Fig. S5 for independent validation of iPSC NES staining. B) Representative flow cytometry plots showing co-immunostaining of pluripotency marker POU5F1 with each of the four candidate ectoderm markers in undifferentiated iPSCs. See also Fig. S6 for complete dataset. C, D) Expression of pluripotent marker POU5F1 (C) and four candidate ectoderm markers (D) quantified by qPCR in matched iPSCs and ectoderm differentiated cells (mean ± SEM, n = 8). Gene expression is shown as fold change of ectoderm differentiated cells compared to iPSCs, with dotted line indicating fold change of 1. * P < .05, *** P < .001, **** P < .0001, two-tailed paired t-tests.
After directed differentiation into ectoderm, we observed a high degree of NES positive cells (90%, range 62.2–99.2% positive cells/line). However, surprisingly a similar number of NES positive cells were also detected in the undifferentiated iPSCs (96.5%, range 89.6–98.7% positive cells/line). To test the consistency of this result, we repeated the analysis using a second antibody against NES that was derived from a different clone, and found similarly very high levels of NES positive cells/line in both the ectoderm and undifferentiated iPSC lines (Fig. 2A and Fig. S4). Since NES is a commonly used ectoderm marker, we performed a verification study in which an independent lab tested NES expression in an independent iPSC line before and after ectoderm differentiation using several NES antibodies. Again, the undifferentiated iPSC line had > 80% NES positive cells/line, with the degree of staining indistinguishable between the differentiated and undifferentiated cells (Fig. S5).
Based on these results, we next sought to test whether other commonly reported ectoderm markers may have similar non-specific staining patterns. Since the ectoderm differentiation medium tested creates neuroectoderm like cells, we assessed TUBB3 (aka TUJ1), OTX2, and PAX6. Similar to our observations with NES, we found high levels of TUBB3 and OTX2 positive cells in iPSC lines before and after differentiation, with no difference in their levels between the iPSC and ectoderm lines (Fig. 2A and Fig. S4). In contrast, while the number of PAX6 positive cells/line was relatively low (12.2%, range 0.7–40.9%) after ectoderm directed differentiation, it was statistically significantly higher than in the undifferentiated iPSC lines (0.1%, range 0–0.9%, p = .03). We confirmed our findings by performing co-immunostaining of the pluripotency marker POU5F1 with either NES, TUBB3, OTX2, or PAX6 in undifferentiated iPSCs. While pluripotent iPSCs (i.e. POU5F1 positive cells) express various levels of NES, TUBB3, and OTX2 protein, we found no detectable PAX6 expression (Fig. 2B and Fig. S6).
In the absence of a robust ectoderm-specific marker, to validate that the ectoderm directed differentiation was successful at yielding the expected cell type, we performed qPCR of POU5F1 and the four commonly used ectoderm markers we tested. Transcript levels of pluripotent marker POU5F1 are known to be reduced after ectoderm differentiation (Daily et al., 2017), and as expected, these were reduced by 92 ± 2% (P < .001) (Fig. 2C). Of the four candidate ectoderm markers (NES, TUBB3, OTX2, and PAX6), we found significantly elevated expression levels of OTX2 (11.6 ± 2.6 fold increase, P < .0001) and PAX6 (1143.5 ± 592.5 fold increase, P < .0001) after ectoderm differentiation (Fig. 2D). To ensure that the expression profile was not impacted by the cell type the iPSCs were originated from, we examined our RNAseq data from six fibroblasts-derived and two CD34+ cell-derived iPSC lines and found comparable expression levels of markers selected for self-renewal and ectoderm (Fig. S7).
4. Discussion
The use of iPSC lines in biomedical research has increased exponentially in recent years. However, as this technology remains under development, the lack of standardized guidelines for validating whether newly established human iPSCs are bona fide lines remains a challenge in the field. For example, while teratoma assays were once considered the gold standard for authenticating new lines, recent studies have called into question whether these time-consuming and expensive as-says are still necessary (Buta et al., 2013). In addition, while traditional iPSC lines were established from fibroblasts derived from skin biopsies, more recently generation of iPSC lines from blood derived cells has grown in popularity due to the ease of primary cell isolation. Here, we report that during authentication of a newly established iPSC repository generated from blood derived cells, we discovered that many of the commonly used markers for ectoderm show high levels of expression in undifferentiated iPSC lines.
Using cell lines from 10 donors, we found comparable levels of NES, TUBB3, and OTX2 in iPSCs with and without differentiation. For NES specifically (one of the most commonly reported ectoderm markers), we tested multiple antibodies in multiple cell lines across two independent laboratories, and found similar high degrees of staining in the undifferentiated state. Based on these findings, we conclude that NES, TUBB3 and OTX2 are not ectoderm specific markers. Notably, NES and OTX2 are currently recommended in the STEMDiff Trilineage Differentiation Kit and Human Pluripotent Stem Cell Functional Identification Kit (R&D Systems) as ectoderm markers, respectively.
Although we failed to identify a difference in NES transcript levels between undifferentiated iPSCs and differentiated ectoderm cells, Hoffman et al. (2017) previously reported that NES transcript levels are elevated in neural progenitor cells (NPCs) compared to undifferentiated iPSCs, suggesting that NES may be a suitable neuroectoderm marker. Notably, NPCs are at a more differentiated state compared to the ectoderm that we characterized here. Through NES immunocytochemistry of iPSCs and NPCs we found that iPSCs do express detectable level of NES protein, but to a much lower extent than NPCs (Fig. S8). Thus, when using relatively insensitive methods such as immunohistochemistry, this difference in expression levels may be leveraged to enable use of NES expression as a NPC marker. However, flow cytometry is substantially more sensitive and quantitative compared to immunocytochemistry, and in particular, is able to distinguish even low levels of NES signal compared to an IgG incubated control. Based on our findings, investigators should avoid NES as an ectoderm marker when using immunostaining and flow cytometry to access trilineage differentiation efficiency. Furthermore, care is advised if using NES as a neuroectoderm marker.
In the case of TUBB3, RNAseq data made available by Daily et al. also shows no evidence of increased transcript levels in ectoderm differentiated cells compared to stem cell lines (Daily et al., 2017). These findings are consistent with our results indicating that while TUBB3 is widely used as a marker for neuronal differentiation, it is not suitable as an ectoderm marker in iPSC trilineage validation studies.
Interestingly, we found that while OTX2 transcript levels were significantly upregulated after ectoderm differentiation, the degree of cell surface protein expression in the undifferentiated iPSCs rendered this protein non-useful as an ectoderm marker. Thus, validation of new markers at the protein level must be tested irrespective of whether differences are observed at the mRNA level.
Of the markers tested, PAX6 was the only ectoderm marker that showed increased expression in the differentiated versus undifferentiated cells. While the overall percent positive cells/line was relatively low, consistent with the wide range of differentiation capacity reported among iPSCs (Carcamo-Orive et al., 2017; Kajiwara et al., 2012; Kilpinen et al., 2017; Kyttala et al., 2016), it is possible that with further modification to the protocol, such as changing cell seeding density, greater staining could be obtained. In addition, it has been reported that further differentiating the cells from ectoderm to NPCs using SMAD signaling pathway inhibitors increases NES and PAX6 transcript and protein levels (Chambers et al., 2009; Hoffman et al., 2017).
A caveat of our study is that we only rigorously tested trilineage differentiation using the STEMdiff Trilineage Differentiation Kit. Thus, it would be of interest to assess whether other commercially available kits and reported protocols yield similar results.
In summary, we have shown that NES, TUBB3, and OTX2 cannot be reliably used as ectoderm differentiation markers when flow cytometry is utilized for measurement. These findings also highlight the importance of including relevant negative controls during validation studies. Investigators should be cautious in selecting markers until definitive standard assays for iPSC pluripotency are established.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health Pharmacogenomics Research Network (PGRN) [P50 GM115318]. The Center for Cellular Reprogramming at the University of Florida is partially supported by the National Institutes of Health NIGMS grant to promote personalized medicine [R24 GM119977]. The funders had no role in the study design, data analysis, or manuscript preparation. The authors would like to thank Brendan Neilan for his technical assistance. This study would not have been possible without the contributions of the POST study participants.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2019.101434.
References
- Avior Y, Sagi I, Benvenisty N, 2016. Pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Mol. Cell Biol 17, 170–182. 10.1038/nrm.2015.27. [DOI] [PubMed] [Google Scholar]
- Burtscher I, Lickert H, 2009. Foxa2 regulates polarity and epithelialization in the endoderm germ layer of the mouse embryo. Development 136, 1029–1038. 10.1242/dev.028415. [DOI] [PubMed] [Google Scholar]
- Buta C, David R, Dressel R, Emgard M, Fuchs C, Gross U, Healy L, Hescheler J, Kolar R, Martin U, Mikkers H, Muller FJ, Schneider RK, Seiler AE, Spielmann H, Weitzer G, 2013. Reconsidering pluripotency tests: do we still need teratoma assays? Stem Cell Res. 11, 552–562. 10.1016/j.scr.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcamo-Orive I, Hoffman GE, Cundiff P, Beckmann ND, D’Souza SL, Knowles JW, Patel A, Papatsenko D, Abbasi F, Reaven GM, Whalen S, Lee P, Shahbazi M, Henrion MYR, Zhu K, Wang S, Roussos P, Schadt EE, Pandey G, Chang R, Quertermous T, Lemischka I, 2017. Analysis of transcriptional variability in a large human iPSC library reveals genetic and non-genetic determinants of heterogeneity. Cell Stem Cell 20, 518–532.e9. 10.1016/j.stem.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L, 2009. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol 27, 275–280. 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EM, Ratanasirintrawoot S, Park IH, Manos PD, Loh YH, Huo H, Miller JD, Hartung O, Rho J, Ince TA, Daley GQ, Schlaeger TM, 2009. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat. Biotechnol 27, 1033–1037. 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- D’Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE, 2005. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol 23, 1534–1541. 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- Daily K, Ho Sui SJ, Schriml LM, Dexheimer PJ, Salomonis N, Schroll R, Bush S, Keddache M, Mayhew C, Lotia S, Perumal TM, Dang K, Pantano L, Pico AR, Grassman E, Nordling D, Hide W, Hatzopoulos AK, Malik P, Cancelas JA, Lutzko C, Aronow BJ, Omberg L, 2017. Molecular, phenotypic, and sample-associated data to describe pluripotent stem cell lines and derivatives. Sci. Data 4, 170030. 10.1038/sdata.2017.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evseenko D, Zhu Y, Schenke-Layland K, Kuo J, Latour B, Ge S, Scholes J, Dravid G, Li X, MacLellan WR, Crooks GM, 2010. Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A 107, 13742–13747. 10.1073/pnas.1002077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura J, Yamato E, Yonemura S, Hosoda K, Masui S, Nakao K, Miyazaki Ji J, Niwa H, 2002. Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev. 16, 784–789. 10.1101/gad.968802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman GE, Hartley BJ, Flaherty E, Ladran I, Gochman P, Ruderfer DM, Stahl EA, Rapoport J, Sklar P, Brennand KJ, 2017. Transcriptional signatures of schizophrenia in hiPSC-derived NPCs and neurons are concordant with post-mortem adult brains. Nat. Commun 8, 2225. 10.1038/s41467-017-02330-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara M, Aoi T, Okita K, Takahashi R, Inoue H, Takayama N, Endo H, Eto K, Toguchida J, Uemoto S, Yamanaka S, 2012. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc. Natl. Acad. Sci. U. S. A 109, 12538–12543. 10.1073/pnas.1209979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai-Azuma M, Kanai Y, Gad JM, Tajima Y, Taya C, Kurohmaru M, Sanai Y, Yonekawa H, Yazaki K, Tam PP, Hayashi Y, 2002. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development 129, 2367–2379. [DOI] [PubMed] [Google Scholar]
- Kilpinen H, Goncalves A, Leha A, Afzal V, Alasoo K, Ashford S, Bala S, Bensaddek D, Casale FP, Culley OJ, Danecek P, Faulconbridge A, Harrison PW, Kathuria A, McCarthy D, McCarthy SA, Meleckyte R, Memari Y, Moens N, Soares F, Mann A, Streeter I, Agu CA, Alderton A, Nelson R, Harper S, Patel M, White A, Patel SR, Clarke L, Halai R, Kirton CM, Kolb-Kokocinski A, Beales P, Birney E, Danovi D, Lamond AI, Ouwehand WH, Vallier L, Watt FM, Durbin R, Stegle O, Gaffney DJ, 2017. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature 546, 370–375. 10.1038/nature22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyttala A, Moraghebi R, Valensisi C, Kettunen J, Andrus C, Pasumarthy KK, Nakanishi M, Nishimura K, Ohtaka M, Weltner J, Van Handel B, Parkkonen O, Sinisalo J, Jalanko A, Hawkins RD, Woods NB, Otonkoski T, Trokovic R, 2016. Genetic variability overrides the impact of parental cell type and determines iPSC differentiation potential. Stem Cell Rep. 6, 200–212. 10.1016/j.stemcr.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendahl U, Zimmerman LB, McKay RD, 1990. CNS stem cells express a new class of intermediate filament protein. Cell 60, 585–595. [DOI] [PubMed] [Google Scholar]
- Okita K, Yamakawa T, Matsumura Y, Sato Y, Amano N, Watanabe A, Goshima N, Yamanaka S, 2013. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells 31, 458–466. 10.1002/stem.1293. [DOI] [PubMed] [Google Scholar]
- Sterneckert JL, Reinhardt P, Scholer HR, 2014. Investigating human disease using stem cell models. Nat. Rev. Genet 15, 625–639. 10.1038/nrg3764. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S, 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Vallier L, Touboul T, Brown S, Cho C, Bilican B, Alexander M, Cedervall J, Chandran S, Ahrlund-Richter L, Weber A, Pedersen RA, 2009. Signaling pathways controlling pluripotency and early cell fate decisions of human induced pluripotent stem cells. Stem Cells 27, 2655–2666. 10.1002/stem.199. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, Herrmann BG, 1990. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature 343, 657–659. 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- Yang W, Liu Y, Slovik KJ, Wu JC, Duncan SA, Rader DJ, Morrisey EE, 2015. Generation of iPSCs as a pooled culture using magnetic activated cell sorting of newly reprogrammed cells. PLoS One 10, e0134995. 10.1371/journal.pone.0134995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Huang CT, Chen J, Pankratz MT, Xi J, Li J, Yang Y, Lavaute TM, Li XJ, Ayala M, Bondarenko GI, Du ZW, Jin Y, Golos TG, Zhang SC, 2010. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell 7, 90–100. 10.1016/j.stem.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.