Abstract
Osteoclasts employ highly specialized intracellular trafficking controls for bone resorption and organelle homeostasis. The sorting nexin Snx10 is a PI3P-binding protein which localizes to osteoclast early endosomes. Osteoclasts from humans and mice lacking functional Snx10 are severely dysfunctional. They show marked impairments in endocytosis, extracellular acidification, ruffled border formation and bone resorption, suggesting that Snx10 regulates membrane trafficking. To better understand how SNX10 regulates vesicular formation and trafficking in osteoclasts, we set out on a search for Snx10 partners. We performed a yeast two-hybrid (Y2H) screening and identified FKBP12. FKBP12 is expressed in RANKL-stimulated RAW264.7 monocytes, co-immunoprecipitates with Snx10 and co-localizes with Snx10 in osteoclasts. We also found that FKBP12, Snx10 and EEA1 are present in the same sub-cellular fractions obtained by centrifugation in sucrose gradients, which confirms localization FKBP12 to early endosomes.
Taken together, these results indicate that Snx10 and FKBP12 are partners and suggest that Snx10 and FKBP12 are involved in the regulation of endosome/lysosome homeostasis via the synthesis. These findings may suggest novel therapeutic approaches to control bone loss by targeting essential steps in osteoclast membrane trafficking
Keywords: Osteoclast, Resorption, vesicular trafficking
INTRODUCTION
Bone homeostasis is dependent on bidirectional regulations of, and communications between, bone forming osteoblasts and bone resorbing osteoclasts (See Chen et al., 2018 [Chen et al., 2018]) for a recent review). Osteoclasts are multinucleated and formed by the differentiation and fusion of mononuclear hematopoietic precursor cells from the monocyte lineage. Osteoclast differentiation is mediated by macrophage colony-stimulating factor (M-CSF) and receptor activator of nuclear factor kB ligand (RANKL), the former necessary for proliferation and survival of the mononucleated cells, whereas the latter triggers differentiation along the osteoclastic lineage [Chen et al., 2018; Stattin et al., 2017]. Inability to form osteoclasts or loss/altered osteoclast function leads to osteopetrosis, a heterogeneous group of rare disorders characterized by increased bone mass [Chen et al., 2018; Sobacchi et al., 2013; Stattin et al., 2017]. We have shown previously that the gene encoding sorting nexin 10 (Snx10) is required for osteoclast formation and resorption activity [Zhu et al., 2012] and that Snx10 is a newly identified locus associated with human osteopetrosis [Ye et al., 2013]. Furthermore, using mice harboring an Snx10-inactivating insertion, we showed that global Snx10-deficiency caused osteopetrorickets as a result of not only failed osteoclast activity but also loss of gastric acid production and calcium absorption [Ye et al., 2015].
Several different mutations in Snx10 have been shown to cause autosomal recessive osteopetrosis [Stattin et al., 2017]. As a member of the sorting nexin family of proteins, SNX10 plays crucial roles in cargo sorting in the endosomal pathway [Teasdale and Collins, 2012]. We have reported that Snx10 mRNA levels got elevated during RANKL-stimulated osteoclastogenesis and that Snx10 silencing does not prevent osteoclast differentiation but inhibits osteoclastic resorption activity and tartrate-resistant acid phosphatase (TRAP) secretion. Snx10-deficient osteoclasts fail to form endosomes, to form a ruffled border and to secrete acid, indicating that SNX10 has an essential role in osteoclast vesicle trafficking and osteoclastic resorption [Ye et al., 2015; Zhu et al., 2012]. Put together, these results suggest that SNX10 is involved in osteoclast function rather than its formation [Chen et al., 2018; Stattin et al., 2017; Ye et al., 2015]. This is consistent with the known high dependency of osteoclasts function on vesicular trafficking pathways [Coxon and Taylor, 2008].
The Ca (2+)/calmodulin-sensitive phosphatase calcineurin is an essential downstream mediator for osteoclast differentiation. Regulation of osteoclast differentiation and function is predominantly controlled by calcineurin/NFATc1 signaling pathway in which NFATc1 acts as a critical molecular switch [Hirotani et al., 2004; Sun et al., 2007]. Recently, FKBP12 was shown to regulate calcineurin activity by facilitating dephosphorylation of proteins involved in actin reorganization, ion channel regulation, endocytosis, and vesicle trafficking and recycling [Caraveo et al., 2017].
Identification of osteoclastic genes and the underlying molecular mechanisms that are required for normal osteoclast formation and function will not only provide improved diagnosis and clinical outcomes but it will also lead to finding novel potential therapeutic targets in treating bone diseases. To better understand how SNX10 regulates vesicular formation and trafficking in osteoclasts, we set out on a search for partners of Snx10. For that, we resorted to yeast two-hybrid (Y2H) screening. Of a total of 83.9 million interactions, three genes came back as strong partner candidates with very high confidence in the interaction. FKBP12 is one of them. FKBP12 is expressed in RANKL-stimulated RAW264.7 monocytes, co-immunoprecipitates with Snx10 and co-localizes with Snx10 in osteoclasts. We also found that FKBP12, Snx10 and EEA1 are present in the same sub-cellular fractions obtained by centrifugation in sucrose gradients, which confirms localization FKBP12 to early endosomes.
MATERIALS AND METHODS
Yeast 2 Hybrid Screening
In collaboration with Hybrigenics (Paris, France), we performed an ULTImate Y2HTM screen to identify potential partners of Snx10 and to characterize specific functions for Snx10. We provided a 2417 bp mouse Snx10 cDNA cloned into Kan/Neo resistant plasmid pCMV6 using RsrII-NotI sites to Hybrigenics for generating reference bait SNX10 protein in yeast. This clone generated a 201 amino acid long SNX10 protein (Met1-Ser201). Hybrigenics cloned Snx10 cDNA in their vector as N-LexA-bait-C fusion. The construct was in frame with the Gal4 activation domain. This bait was screened using prey cDNA library of mouse adult brain in Y2H system. The Prey Library used was a Mouse Adult Brain library provided by Hybrigenics. We chose to use that library because there was no osteoclast library available at this time. Also, we found that Snx10 is highly expressed in several mouse tissues, including adult brain.
Hybrigenics computed a predicted biological score (PBS) for each interaction to assess the reliability of interaction in Y2H. This score represents the probability of an interaction to be non-specific and was derived based on the comparison between the number of independent prey fragments found for an interaction versus the background noise (i.e. the chance of finding them at random). This value varies between 0 and 1 and several thresholds are arbitrarily defined for ranking the results in four categories A-D (the highest confidence rank being A). PBS was adjusted by integrating the PBS of other interactions from Hybrigenics database in which interaction domains of the involved proteins are known to be present. As an example, reciprocal interactions found in independent screens are technically very reliable and tagged as A, B or C (Hybrigenics Services, Paris, France). PBS of D is assigned to interactions identified through one unique prey fragment or multiple identical ones and may reflect a false-positive or an unsuitable interaction for Y2H.
Animals
All animal studies were approved by the Institutional Animal Care and Use Committee at our institution and were in compliance with all federal and local guidelines. All mice were of the 129/C57 mixed background.
Cells
RAW 264.7 (TIB-71, mouse macrophage/monocytes) was purchased from ATCC. Cells were cultured in DMEM/1.5 g/L sodium bicarbonate (JRH Biosciences, Lenexa, KS) supplemented with 10% non-heat inactivated FBS (BioWhittaker, Cambrex, Walkersville, MD). To induce osteoclast differentiation, cells were cultured for 5 days in medium supplemented with 50 ng/ml of RANKL, (PeproTech Inc., Rocky Hill, NJ), with changes of medium and RANKL every other day. Bone marrow mononuclear cells (BMM) were collected from 2-week-old mice and cultured in a-MEM medium (Invitrogen, Carlsbad, CA) with 10% non-heat inactivated FBS (Bio Whittaker). To stimulate osteoclast differentiation, cells were cultured for 5 days in the presence of 50 ng/ml soluble RANKL (PeproTech Inc, Rocky Hill, NJ) and 25 ng/ml soluble M-CSF (PeproTech Inc.) with changes of medium, RANKL, and M-CSF every other day.
Immunofluorescence
Immunofluorescence analysis was done following standard protocols. Briefly, cells were seeded on cover slips, treated with RANKL or RANKL and M-CSF to stimulate osteoclast differentiation, washed with PBS, fixed with 4% of Paraformaldehyde and permeabilized with 0.1% of Saponin. For Snx10 detection we used a goat anti-mouse Snx10 primary antibody (1:500), for FKBP12 staining we incubated the cells with a rabbit anti-FKBP12 antibody (Mouse anti-FKBP12: sc-133067; dilution 1: 200), followed by incubation with a TRITC- or FITC-conjugated secondary antibody (dilution 1:1,000). Cells were then counter stained with DAPI (40,6-diamidino-2-phenylindole) prior to mounting on glass cover slips. Images were obtained with a Fluorescent Microscope (Olympus BX43).
Sucrose flotation gradient
RAW 264.7 mouse macrophages/monocytes were cultured in DMEM with or without RANKL (in 10 cm dishes). After osteoclast differentiation, cells were washed thrice with DMEM and resuspended in 0.5 ml of homogenization buffer (250 mM sucrose, 1mM EDTA and 1 mM phenylmethylsulfonyl fluoride (PMSF). Cells were then gently detached using a cell scraper, lysed, and further processed for analysis by a sucrose flotation assay as previously described [Yu and Lai, 2005]. Briefly, cells were collected and lysed in homogenization buffer and the cell suspensions were subjected to centrifugation (1000×g for 10 min), to pellet the nuclei and obtain the post-nuclear supernatant (PNS), which contains all intracellular organelles in suspension. After centrifugation the supernatant was collected and adjusted to a concentration of 25% sucrose and 1mM EDTA in 1 ml total volume. In 1 ml increments, 2.4 ml of a 45% sucrose solution was transferred to the bottom of a SW41Ti tube and sequentially overlaid with 5.2 ml of 35% sucrose solution, 3.9 ml of 25% sucrose solution and 1ml of PNS in 25% sucrose. Following centrifugation (100,000×g), 1 ml fractions were collected from top (light) to bottom (heavy) and densities were measured by refractometry. These fractions were further analyzed for protein analysis expression by western blot analysis.
Western blotting and Immunoprecipitation
Western blotting was performed to monitor expression of Snx10, FKBP12 and EEA1. Briefly, cells were lysed on ice in lysis buffer (10 mM Tris-HCl pH7.4, 150 mM NaCl, 5 mM EDTA, 0.2% Nonidet P-40) supplemented with protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN) and centrifuged at 15,000 g for 10 minutes at 4°C. Protein concentration was determined by Bio-Rad protein assay kit (Bio-Rad, Hercules, CA). Lysates were mixed with SDS sample buffer and heated to 90°C for 5 minutes. Proteins were separated by electrophoresis on SDS-PAGE gels and transferred onto PVDF membranes (0.4 uM) in 25 mM Tris, 192 mM glycine and 20% methanol at 100 V for 1.5 hour. Blots were incubated with primary antibodies at 4°C overnight followed by the incubation with secondary antibodies. Blots were then developed with horseradish peroxidase substrate (West Femto Solution, Pierce, Rockford, IL) for 5 minutes at room temperature, and analyzed using GelDoc 200 (Bio-Rad, Hercules, CA). For immunoprecipitation, the cells were suspended in lysis buffer, the lysates were incubated with anti-Snx10 or anti FKBP12 antibodies for 2h at 4°C and mixed with protein G-Sepharose beads (20 ul of 50% slurry) overnight at 4 °C on a rotator. The beads were then washed three times with lysis buffer, mixed with SDS sample buffer, and heated to 90 °C for 5 min followed by SDS-PAGE. The samples were blotted onto polyvinylidene difluoride membranes and then probed with FKBP12 or Snx10 antibodies and developed with horseradish peroxidase substrate (West Femto Solution, Pierce, Rockford, IL) for 5 minutes at room temperature, and analyzed using GelDoc 200 (Bio-Rad, Hercules, CA).
FKbp12 silencing
RAW 264.7 cells were cultured on a 6-well plate (2×105 cells/well) and transfected with 10nM FKbp12 siRNA (Origene, cat # SR425778) for 48 hours. At that time, cells were scraped and plated again on a 6-well plate (8 ×104 cells/well) containing Dulbeccos’s Modified Eagle Medium (DMEM) supplemented with 10% Fetal Bovine Serum, 100 U/ml penicillin and 100 U/ml streptomycin. Cells were then incubated at 37˚C and allowed to adhere overnight (24h). The following day, the culture medium was replaced with fresh medium containing 100ng/ml of receptor activator of nuclear factor-kappa B ligand (RANKL, 100ng/ml) (Peprotech, cat # 315-11).We considered this as day 0 of differentiation. Medium and RANKL were replaced on day 3. At day 5 (complete osteoclast differentiation) cells were washed with PBS and used for expression analysis.
RNA isolation and qPCR analysis
Total RNA was extracted using TRIzol Reagent (Invitrogen, CA). 0.05-0.2mg of total RNA in a 20 µl reaction was used for RT-PCR analysis using the TaqMan® RNA-to-CT™ 1-Step Kit (Applied Biosystems). Quantitative real-time PCR was performed using QuantStudio™ 3 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The PCR amplification was performed in the following cycling conditions, RT reaction was carried out at 48 ˚C for 15 min, with initial denaturation at 95 ˚C for 15 min, followed by denaturation at 95 ˚C for 15 sec, followed by 40 cycles of annealing and elongation at 60 ˚C for 1 min. The expression fold change was calculated using, 2-ΔΔCt comparative cycle threshold method. All reactions were carried out in triplicate. RT-PCR was analysed for the following genes such as, FKbp12 (Mm01243847_g1), TRAP (Mm00475698_m1), SNX-10 (Mm00511052_g1), and MMP-9 (Mm00442991_m1). β-actin (Mm 02619580_g1) serves as an internal control.
RESULTS
Y2H screening identifies FKBP12 as a partner of Snx10
Of a total of 83.9 million interactions, 46 passed stringent statistical and control criteria as positive interactions and were ranked according to their assigned probability scores (Predicted Biological Score (PBS), e-values from 0-1 reflecting the probability of the interaction being non-specific). Out of the 46 processed clones three came back with the highest “A” PBS indicating very high confidence in the interaction. FKBP12 is one of these three clones with 9 hits representing 3 independent clones (Fig 1; FKBP12, Supplemental Figures 1 and 2). The interaction of Snx10 with these 3 clones was so powerful that use of 3-aminotriazole (3-AT, a chemical used to reduce Y2H background) was not needed. This result indicated with high confidence that Snx10 interacts with FKBP12.
Figure 1: Y2H screening identifies FKBP12 as a partner of Snx10.
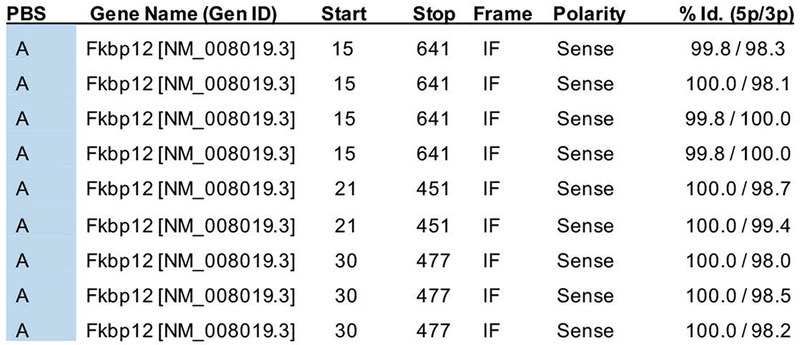
The Predicted Biological Score (PBS) indicates interaction reliability and is primarily based on the comparison between the number of independent prey fragments found for an interaction and the chance of finding them at random (background noise). A PBS of A indicates the highest confidence. FKBP12 had 9 hits representing 3 independent clones: 1. Clone 15-641 (4 hits), 2. Clone 21-451 (2 hits) and 3. Clone 20-477 (3 hits). These numbers indicate the position of the 5p and 3p prey fragment ends, relative to the position of the ATG start codon (A=0). The degree of identity of all these fragments relative to the reference gene (FKBP12) is 100%.
FKBP12 and Snx10 are expressed in early endosomes
The Y2H screen was performed using a mouse brain library. In order to confirm those results in mouse osteoclasts, we performed sucrose gradient separation analysis. If FKBP12 interacts with Snx10, after centrifugation both proteins should be present in the same endosomal fraction/s. In sucrose gradient separation, endosomes migrate differentially based on the size, shape, and density as well as the density and viscosity of the gradient. After centrifugation, the top fractions contain lighter vesicles (early endosomes), whereas the bottom fractions contain heavier vesicles (late endosomes/lysosomes). We performed sucrose flotation assay, collected 12 × 1 ml fractions and then subjected each fraction to Western Blot analysis. Early Endosome Antigen 1 (EEA1) was used as a control for early endosomes.
Based on sucrose gradient studies, FKBP12 and Snx10 co-localize in fractions 1, 2 and 3 (lighter vesicles, early endosomes) in osteoclasts (Fig 2).
Figure 2: FKBP12 and Snx10 are expressed in the same (lighter) sucrose gradient fractions.
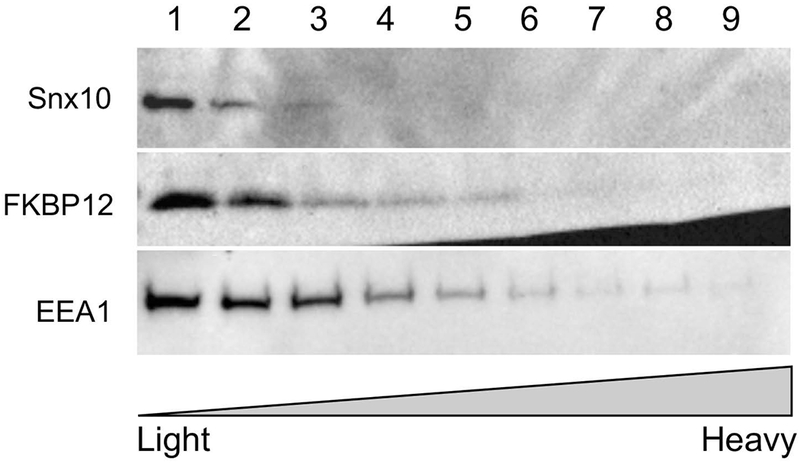
Collected gradient fractions 1–12 of differentiated RAW264.7 cells were analyzed by SDS-PAGE and Western blotting for the distribution of organelle markers: early endosome antigen 1 (EEA1; EE marker), FKBP12 and Snx10. Fractions 1-3 contained the majority of the FKBP12 signal.
FKBP12 and Snx10 co-immunoprecipitate and colocalize in osteoclasts
To confirm these findings, we performed co-immunoprecipitation analysis on protein extracts from differentiated RAW264.7 cells using Snx10 and FKBP12 antibodies. The results (Fig 3) indicate that, 1) FKBP12 is expressed in undifferentiated and RANKL-stimulated RAW264.7 cells and 2) that Snx10 and FKBP12 co-immunopreciopitate in osteoclasts. Co-localization of Snx10 and FKBP12 was confirmed by immunohistochemistry in both multinucleated osteoclasts and gastric zymogenic cells (Fig 4).
Figure 3. FKBP12 and Snx10 coprecipitate in osteoclast protein extracts.
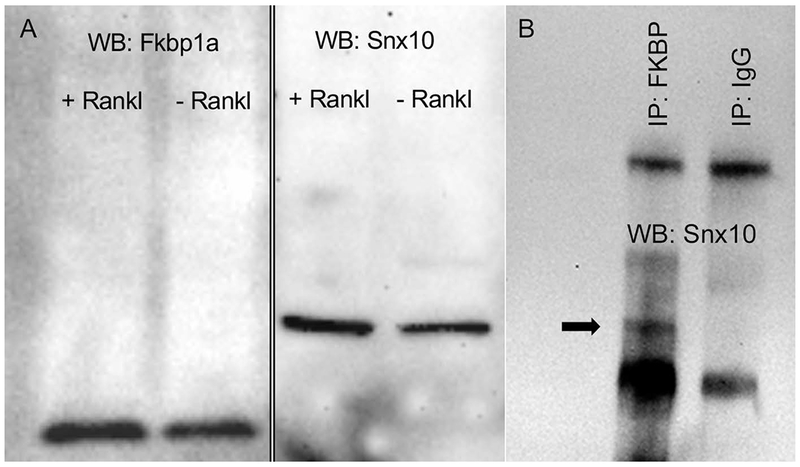
FKBP12 is expressed in unstimulated and RANKL-stimulated RAW264.7 cells (2a). Snx10 interacts with FKBP12 in RANKL-stimulated RAW264.7 cells (2b). Immunoprecipitation was done using an α-FKBP12 antibody. Immunoprecipitates were subjected to western blot analysis using an α-Snx10 antibody. The black arrow (2b) indicates a 25KD band corresponding to Snx10.
Figure 4. Immunofluorescent analysis confirms of Snx10 and FKBP12 co-localize to osteoclasts.
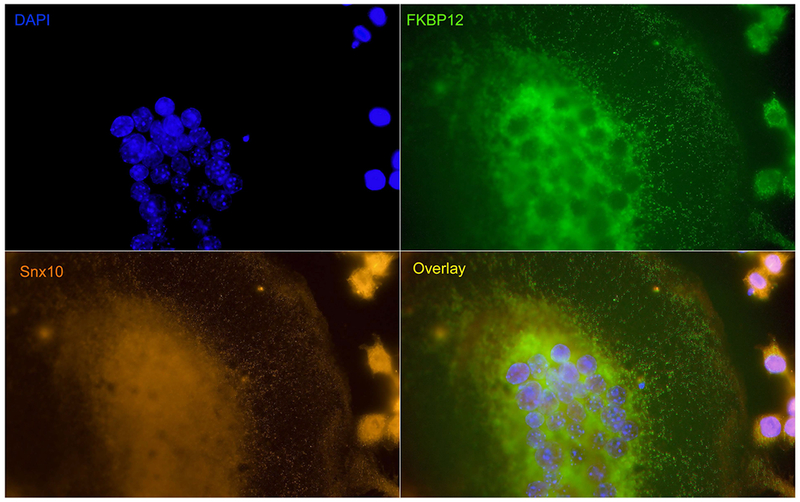
RANKL and M-CSF stimulated mouse bone marrow mononuclear cells were fixed and stained with Snx10 (orange) and FKBP12 (green) antibodies. DAPI staining indicate nucleus. The superimposed image (overlay) indicates that both proteins colocalize to endosomes.
FKBP12 silencing inhibits osteoclast differentiation
Snx10 is required for osteoclast differentiation and activity in vitro and in vivo. Since FKBP12 colocalizes and interacts with Snx10, we next wanted to know if FKBP12 expression was also required for osteoclast differentiation, in vitro. To answer that question, we silenced FKBP12 in RAW264.7 cells and stimulated them with RANKL to undergo osteoclast differentiation. The results (Table 1) indicate that, in FKBP12-silenced cells (53% reduction), expression of specific osteoclast genes is reduced by 16% (TRAP), 60% (Snx10) and 25% (MMP9). These results show that FKBP12 expression is required for osteoclast differentiation
Table 1. FKBP12 silencing inhibits expression of osteoclastic genes.
Relative expression of osteoclast genes TRAP, Snx10 and MMP9 is reduced in FKBP12 silenced cells
| Relative Expression | ||||
|---|---|---|---|---|
| FKBP12 | TRAP | Snx10 | MMP9 | |
| No RANKL | 1.111 +/− 0.65 | 1.054 +/− 0.41 | 1.064 +/− 0.44 | 1.002 +/− 0.08 |
| RANKL | 1.032 +/− 0.32 | 15.576 +/− 6.41 | 1.066 +/− 0.22 | 632.682 +/− 223.42 |
| RANKL/FKBP12 si | 0.492 +/− 0.23 | 13.104 +/− 3.97 | 0.434 +/− 0.12 | 476.657 +/− 49.00 |
DISCUSSION
SNX10 and FKBP12 showed highest confidence of interaction in Y2H which strongly suggests that this interaction is physiologically relevant. As compared to validation of their interaction in in vitro assays, which required larger amounts of purified proteins, Y2H has provided a strong physiological methodological approach. Since its inception about 30 years ago, the Y2H has not only bridged the methodological gap in discovering protein interactions, it has evolved as a method to identify and validate therapeutic targets and as a tool in drug discovery [Fields and Song, 1989; Hamdi and Colas, 2012]. The fundamental discoveries that a) transcription factors contain at least two functional domains, one for DNA binding and the other for transcription activation and b) these domains can interact to activate transcription, led to Y2H. The interaction causes transcription factor activation and the resulting interaction can be quantified by a transcribed reporter gene [Fields and Song, 1989]. To remove the false interaction signals in Y2H, positive and negative reference interaction sets are used. These positive and negative sets are known to interact and not to interact, respectively, [Hamdi and Colas, 2012]. ULTImate screens (Hybrigenics Services, Paris, France), which we utilized as Y2H, has already incorporated positive and negative reference interaction sets to eliminate false interactions. Y2H has had previous success of using FKBP12 in this method. The first so-called yeast three hybrid system had utilized Y2H signal by expressing FKBP12 as prey and then obtaining FPBK12 clones by screening a cDNA library using this as bait [Hamdi and Colas, 2012; Licitra and Liu, 1996].
The eukaryotic FK506 binding proteins belong to the immunophilin family and primarily acts as folding chaperones for proteins containing proline residues. Of the fifteen identified human FKBPs, FKBP12, which catalyzes the peptidyl prolyl cis–trans isomerization, is the smallest and most extensively studied [Liu et al., 2017]. FKBP12 (a 12 kDa enzyme consisting of 108 amino acids) is abundantly expressed in the cytoplasm and has been shown to possess several functions in signal transduction by interacting with different cellular targets that impact processes such as T-cell activation and the regulation of cancer progression and cell growth [Liu et al., 2017]. FKBP12 binds to different cellular receptors or targets under selective conditions to influence processes such as major calcium-release channels in the sarcoplasmic and endoplasmic reticula and channel gating, whereas its interaction with TGF-β type I receptor inhibits receptor-mediated signal transduction. In addition, FKBP12 inhibits cellular activity of epidermal growth factor receptor by modulating the receptor’s phosphorylation [Liu et al., 2017]. Recently, FKBP12-mediated degradation of generic oncogene MDM2 and the expression levels of FKBP12 in cancer cells have been correlated with the potential treatment outcomes in radiotherapy and chemotherapy [Liu et al., 2017].
While Y2H protein interactions results are of great value, alternative methods should be used to support the interaction. Therefore, we have used sucrose gradient separation of SNX10 and FKBP12 and their co-immunoprecipitation and immunofluorescence studies to substantiate and quantitate their interaction by Y2H. A high level of interaction of SNX10 with FKBP12 using mouse brain library in Y2H and in vitro assays in this study is not unusual and can be explained as follows.
It is well established that Snx family of proteins mediates endosomal sorting, endocytosis, recycling of membrane proteins, and endosomes to Golgi apparatus trafficking [Worby and Dixon, 2002]. Although the Snx family members are diverse cytoplasmic and membrane-associated proteins, they are unified by a common PX domain, a phospholipid-binding motif. Protein sorting and membrane trafficking by these members involve protein-protein complexes and protein-lipid interactions [Worby and Dixon, 2002].
With regards to osteoclasts, formation and maintenance of the ruffled border are achieved by endocytic vesicles originating in the basolateral membrane of the osteoclast [Coxon and Taylor, 2008]. While studying the effect of Snx10 deficiency on endosomal trafficking, vesicle formation and resorption in Snx10 deficient mice, we have reported that Snx10 deficiency prevented endocytosis, caused severely impaired ruffled border formation, and blocked bone resorption activity [Ye et al., 2015]. Out data also suggested that Snx10 appears to mediate both bone resorption and stomach acidification by regulating vesicular trafficking [Ye et al., 2015].
While FKBP12 was widely known to influence processes such as major calcium-release channels in the sarcoplasmic and endoplasmic reticula and channel gating [Liu et al., 2017], the recent findings that FKBP12 regulates calcineurin activity by facilitating dephosphorylation of proteins involved in actin reorganization, ion channel regulation, and in particular endocytosis and vesicle trafficking and recycling [Caraveo et al., 2017] are relevant to this study for SNX10 interaction. These findings about FKBP12 has therapeutic implications as well because Calcineurin is an essential Ca2+-dependent phosphatase and its increased activity is associated with α-synuclein (α-syn) toxicity, a protein associated with several neurodegenerative diseases including Parkinson’s. Calcineurin activity can be reduced with genetic tools or with drug Tacrolimus (FK506), producing a protective outcome [Caraveo et al., 2017]. It is interesting to note that both Calcineurin and FKBP12 are present at very high levels in the brain relative to any peripheral tissue [Dawson et al., 1994].
Snx10 and FKBP12 both eluted in early endosomal sucrose fraction. Both proteins seem to be involved with endosome homeostasis and vesicular trafficking because when they impair vesicular trafficking, giant vacuoles are formed. For example, overexpression of Snx10 induces giant vacuoles in mammalian cells [Qin et al., 2006]. FKBP12 was shown to interact with calcineurin in the absence of Tacrolimus (FK506) and contribute to α-syn toxicity in a calcineurin-dependent pathway [Caraveo et al., 2017]. Toxic levels of α-syn impair vesicle trafficking, resulting in the aggregation of α-syn into functionally diverse large vesicles [Cooper et al., 2006]. As FKBP12 enables calcineurin-dependent dephosphorylation of substrates, which impairs vesicle trafficking, low doses of Tacrolimus restored the block in vesicle trafficking as evidenced by the reduced vesicles [Caraveo et al., 2017]. The calcineurin/NFAT signaling pathway is of key importance in the regulation of osteoclast differentiation and calcineurin is an essential downstream effector of the RANKL-induced signal transduction pathway leading to osteoclast differentiation. Calcineurin is inhibited by either of the immunosuppressant drugs cyclosporin A and FK506 inhibiting the RANKL-induced differentiation into mature osteoclasts [Hirotani et al., 2004; Igarashi et al., 2004]. NFAT family members get up-regulated in response to RANKL stimulation but NFATc1 is the master regulatory transcription factor for osteoclast differentiation because its activation in osteoclast precursor cells by itself, in the absence of RANKL in vitro and in vivo, is sufficient to initiate a genetic program that results in the mature functional osteoclasts [Caraveo et al., 2017; Hirotani et al., 2004; Igarashi et al., 2004].
FKBP12 regulates calcineurin activity by dephosphorylation of proteins involved in vesicle recycling [Caraveo et al., 2017]. FKBP12 binds to the immunosuppressants FK506 and rapamycin. While binding of FKBP12 to FK506 and calcineurin forms a ternary complex to inhibit the serine/threonine phosphatase activity of calcineurin, impacting several cellular processes, the complex of FKBP12 and rapamycin interacts with mechanistic target of rapamycin (mTOR) and inhibits its roles in regulating cell growth [Jiang et al., 2017]. mTOR is a serine/threonine protein kinase belonging to the phosphoinositide 3-kinase (PI3K)-related kinase family and dysregulation of its catalytic subunit protein complex mTORC1 is probably involved in several skeletal diseases including osteoarthritis and osteoporosis [Chen and Long, 2018]. Role of mTORC1 in regulating the osteoclast lineage is not fully understood. It is likely that mTORC1 inhibits osteoclast differentiation through suppression of NF-kB and NFATc1, both of which are critical transcription factors of osteoclastogenesis [Chen and Long, 2018]. Using mouse genetic models, mTORC1 has been implicated as a key proliferation-to-differentiation switch during osteoclastogenesis mTORC1 is required but must be dynamically regulated to activate during proliferation and inactivate during differentiation [Wan, 2018]. Calcineurin inhibits mTORC1 via dephosphorylation whereas mTORC1 inhibits NFATc1 via phosphorylation and an important signaling cascade has been identified in which calcineurin inhibits mTORC1, thus blocking its inhibition of NFATc1, leading to osteoclast differentiation [Wan, 2018].
While activation of the NFATc1 transcription factor is both necessary and sufficient for osteoclastogenesis, it is worth mentioning that many other transcription factors affect various aspects of osteoclast development and function and they are likely to cooperate under the regulation of osteoclast-specific genes [Heather A. Carey, 2018; Hirotani et al., 2004]. In fact, ETs family transcription factor PU.1, and a known co-partner of PU.1 in osteoclasts MITF (microphthalmia-associated transcription factor), bind to 18 transcription factors with known functions in osteoclast differentiation, suggesting that PU.1 regulation of osteoclast differentiation is by promoting the expression of downstream pro-osteoclastogenic transcription factors and suppressing the expression of those negatively regulating osteoclast differentiation [Heather A. Carey, 2018]. Our understanding of the complex signaling pathways that administer osteoclast differentiation is far from complete [Chen and Long, 2018] and interaction of SNX10 and FKBP12, as reported in our study, is certainly indicative of that assumption.
Supplementary Material
Acknowledgments
DISCLOSURES
Ricardo Battaglino, Prakash Jha, Farhath Sultana, Weimin Liu, and Leslie Morse declare that they have no conflict of interest. This study received support from: The National Institute of Arthritis and Musculoskeletal and Skin Diseases R01AR064793 and National Institute on Disability, Independent Living, and Rehabilitation Research (NIDILRR 90SI5007-01-02).
REFERENCES
- Caraveo G, Soste M, Cappelleti V, Fanning S, van Rossum DB, Whitesell L, Huang Y, Chung CY, Baru V, Zaichick S, Picotti P, Lindquist S. 2017. FKBP12 contributes to alpha-synuclein toxicity by regulating the calcineurin-dependent phosphoproteome. Proc Natl Acad Sci U S A 114:E11313–E11322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Long F. 2018. mTOR signaling in skeletal development and disease. Bone Res 6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wang Z, Duan N, Zhu G, Schwarz EM, Xie C. 2018. Osteoblast-osteoclast interactions. Connect Tissue Res 59:99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, Cao S, Caldwell KA, Caldwell GA, Marsischky G, Kolodner RD, Labaer J, Rochet JC, Bonini NM, Lindquist S. 2006. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science 313:324–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coxon FP, Taylor A. 2008. Vesicular trafficking in osteoclasts. Semin Cell Dev Biol 19:424–33. [DOI] [PubMed] [Google Scholar]
- Dawson TM, Steiner JP, Lyons WE, Fotuhi M, Blue M, Snyder SH. 1994. The immunophilins, FK506 binding protein and cyclophilin, are discretely localized in the brain: relationship to calcineurin. Neuroscience 62:569–80. [DOI] [PubMed] [Google Scholar]
- Fields S, Song O. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245–6. [DOI] [PubMed] [Google Scholar]
- Hamdi A, Colas P. 2012. Yeast two-hybrid methods and their applications in drug discovery. Trends Pharmacol Sci 33:109–18. [DOI] [PubMed] [Google Scholar]
- Carey Heather A. BEHI, Geisler Jennifer A., Nickel Mara C., Cabrera Jennifer, Ghosh Sankha, Jiang Yue, Yan Jing, Lee James, Makam Sandeep, Young Nicholas A., Valiente Giancarlo R., Jarjour Wael N., Huang Kun, Rosol Thomas J., Toribio Ramiro E., Charles Julia F., Ostrowski Michael C. & Sharma Sudarshana M. 2018. Enhancer variants reveal a conserved transcription factor network governed by PU.1 during osteoclast differentiation. Bone Researchvolume 6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotani H, Tuohy NA, Woo JT, Stern PH, Clipstone NA. 2004. The calcineurin/nuclear factor of activated T cells signaling pathway regulates osteoclastogenesis in RAW264.7 cells. J Biol Chem 279:13984–92. [DOI] [PubMed] [Google Scholar]
- Igarashi K, Hirotani H, Woo JT, Stern PH. 2004. Cyclosporine A and FK506 induce osteoclast apoptosis in mouse bone marrow cell cultures. Bone 35:47–56. [DOI] [PubMed] [Google Scholar]
- Jiang D, Cho W, Li Z, Xu X, Qu Y, Jiang Z, Guo L, Xu G. 2017. MiR-758-3p suppresses proliferation, migration and invasion of hepatocellular carcinoma cells via targeting MDM2 and mTOR. Biomed Pharmacother 96:535–544. [DOI] [PubMed] [Google Scholar]
- Licitra EJ, Liu JO. 1996. A three-hybrid system for detecting small ligand-protein receptor interactions. Proc Natl Acad Sci U S A 93:12817–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Xiong J, Yi S, Zhang H, Zhou S, Gu L, Zhou M. 2017. FKBP12 enhances sensitivity to chemotherapy-induced cancer cell apoptosis by inhibiting MDM2. Oncogene 36:1678–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin B, He M, Chen X, Pei D. 2006. Sorting nexin 10 induces giant vacuoles in mammalian cells. J Biol Chem 281:36891–6. [DOI] [PubMed] [Google Scholar]
- Sobacchi C, Schulz A, Coxon FP, Villa A, Helfrich MH. 2013. Osteopetrosis: genetics, treatment and new insights into osteoclast function. Nat Rev Endocrinol 9:522–36. [DOI] [PubMed] [Google Scholar]
- Stattin EL, Henning P, Klar J, McDermott E, Stecksen-Blicks C, Sandstrom PE, Kellgren TG, Ryden P, Hallmans G, Lonnerholm T, Ameur A, Helfrich MH, Coxon FP, Dahl N, Wikstrom J, Lerner UH. 2017. SNX10 gene mutation leading to osteopetrosis with dysfunctional osteoclasts. Sci Rep 7:3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Peng Y, Zaidi N, Zhu LL, Iqbal J, Yamoah K, Wang X, Liu P, Abe E, Moonga BS, Epstein S, Zaidi M. 2007. Evidence that calcineurin is required for the genesis of bone-resorbing osteoclasts. Am J Physiol Renal Physiol 292:F285–91. [DOI] [PubMed] [Google Scholar]
- Teasdale RD, Collins BM. 2012. Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: structures, functions and roles in disease. Biochem J 441:39–59. [DOI] [PubMed] [Google Scholar]
- Wan HDHY. 2018. mTORC1 impedes osteoclast differentiation via calcineurin and NFATc1. Communications Biology 1:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby CA, Dixon JE. 2002. Sorting out the cellular functions of sorting nexins. Nat Rev Mol Cell Biol 3:919–31. [DOI] [PubMed] [Google Scholar]
- Ye L, Morse LR, Battaglino RA. 2013. Snx10: a newly identified locus associated with human osteopetrosis. IBMS Bonekey 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Morse LR, Zhang L, Sasaki H, Mills JC, Odgren PR, Sibbel G, Stanley JR, Wong G, Zamarioli A, Battaglino RA. 2015. Osteopetrorickets due to Snx10 deficiency in mice results from both failed osteoclast activity and loss of gastric acid-dependent calcium absorption. PLoS Genet 11:e1005057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu GY, Lai MM. 2005. The ubiquitin-proteasome system facilitates the transfer of murine coronavirus from endosome to cytoplasm during virus entry. J Virol 79:644–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu CH, Morse LR, Battaglino RA. 2012. SNX10 is required for osteoclast formation and resorption activity. J Cell Biochem 113:1608–15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


