Abstract
Sindbis virus (SINV) is an enveloped, single-stranded RNA virus, which is transmitted via mosquitos to a wide range of vertebrate hosts. SINV produced by vertebrate, baby hamster kidney (BHK) cells is more than an order of magnitude less infectious than SINV produced from mosquito (C6/36) cells. The cause of this difference is poorly understood. In this study, charge detection mass spectrometry was used to determine the masses of intact SINV particles isolated from BHK and C6/36 cells. The measured masses are substantially different: 52.88 MDa for BHK derived SINV and 50.69 MDa for C6/36 derived. Further analysis using several mass spectrometry-based methods and biophysical approaches indicates that BHK derived SINV has a substantially higher mass than C6/36 derived because in the lipid bilayer, there is a higher portion of lipids containing long chain fatty acids. The difference in lipid composition could influence the organization of the lipid bilayer. As a result, multiple stages of the viral lifecycle may be affected including assembly and budding, particle stability during transmission, and fusion events, all of which could contribute to the differences in infectivity.
Keywords: alphavirus, mass spectrometry, lipid, glycan
Graphical Abstract
During viral infection, a mammalian host cell releases between 80 and 10,000 infectious virus particles per cell.1 However, the total number of virus particles released is higher because not all virus particles are infectious. Specific infectivity is the ratio of the number of infectious particles to the total number of particles; a higher number indicates that a larger fraction of the particles can complete an infectious cycle.2,3 Particle-to-PFU (plaque forming unit) has also been used in the literature, and this is the ratio of total particle to infectious particles. Reported specific infectivity values range from approximately 1 for a bacteriophage to 10−3 to 10−4 for picornaviruses and Varicella-Zoster virus.4,5 Some non-infectious particles may fail to complete the infection cycle because of defects (e.g., assembly errors in genome packaging and protein association, maturation defects, and genome replication errors) or because of experimental constraints (e.g., purification methods, cell culture conditions, and techniques used to determine infectious and total particles).6 It has also been proposed that non-infectious particles are intentionally synthesized to promote viral infections by being decoys for immune cells,6 to promote particle aggregation,7,8 and to aid in genetic cooperativity during viral infection.9
Alphaviruses are arboviruses which are transmitted by an arthropod vector, primarily a mosquito, to a vertebrate host. Virus particles produced in vertebrate cell cultures (avian or mammalian) typically have a specific infectivity between 0.001 and 0.0125, in other words, <2% of the particles produced are infectious.10 In contrast, the specific infectivity values for virus particles produced in a mosquito cell culture are between 0.2 and 1, indicating that 20–100% of the particles produced are infectious.10,11 It has been proposed that this wide range of virion-specific infectivity may be important for maintaining viral fitness as the virus circulates in the different vector-host cycle.12,13 Currently there are no therapeutics for alphavirus infections. Drug development currently focuses on both the genome replication cycle and inhibiting steps of entry and exit.14 One strategy is to determine the factors that influence infectivity and identify compounds that will decrease specific infectivity, thereby possibly attenuating the severity of the disease and symptoms.
The alphavirus particle contains a positive-sense, single-stranded RNA genome that is capped and has a poly(A) tail.15–17 There are six structural proteins: Capsid, E3, E2, 6K, TF, and E1. Cryo-electron microscopy (Cryo-EM) has been used to determine the structure of alphavirus particles and the organization of the structural proteins.18–20 The viral genome is encapsulated in a shell of capsid protein (CP) forming the nucleocapsid core. The core is surrounded by a host-derived lipid bilayer, in which the viral glycoprotein spikes are embedded. These surface spikes are made up predominantly of E2 and E1, and in some viruses, E3 remains noncovalently associated with the spikes.21 It has been determined that the TransFrame (TF) protein is present in the virion, but its location has not yet been identified in cryo-EM structures.22 The 6K protein, in contrast, is found predominantly inside the host cell and not in the particle, and 6K has been proposed to have a role in budding and assembly.23 No major difference in the amount of viral proteins from the virions derived from the two hosts has been identified.16,24
When comparing vertebrate and arthropod derived particles, there are known differences between the two cell lines that impact virion assembly and that could contribute to the infectivity difference. For example, proteomic analysis revealed that a host-derived ribosomal 40S small subunit is present in arthropod-derived particles; in contrast, only a small fraction of vertebrate-derived particles contain the 40S ribosomal subunit.25 It was further demonstrated that the particles containing the ribosomal subunit have a higher specific infectivity, enhanced translation and viral synthesis, and produced less type I interferon than virions that did not contain the host-derived ribosomal subunit.
There are also differences in the glycans and lipids that may contribute to the infectivity difference between the two hosts. It has been shown that the N-linked glycans attached to E2 and E1 glycoproteins are host dependent.26–28 The infectivity could be influenced by the exposed glycans.29 Several cellular processes may result in differences in the lipid composition of viruses derived from vertebrate and insect cells. Vertebrate cells and particles produced from these cells contain cholesterol.30 Mosquito cells, in contrast, are cholesterol auxotrophs; they do not produce cholesterol but will utilize it if supplied in the tissue culture media.31 Arthropods do produce different sterols which can function in place of cholesterol. These differences could also affect the infectivity. Vertebrate cells contain elongases and desaturates, enzymes which are missing in invertebrate cells. These enzymes result in fatty acid chains which are, on average, longer and contain more double bonds than lipids typically found in invertebrate cells.32–34 Furthermore, the lipid composition of both the internal organelle membranes and the plasma membrane of vertebrate and invertebrates is different; these differences in lipid membrane compositions could also affect the infectivity,30 because alphaviruses bud predominantly from the plasma membrane in vertebrate cells, but in mosquito cells, they bud from internal vesicles and the plasma membrane.35–37
In this work, we used a combination of mass spectrometry and bioanalytical techniques to characterize the components of the Sindbis virus (SINV) produced from vertebrate (BHK) and arthropod (C6/36) cells. Charge detection mass spectrometry (CDMS) was used to measure the total masses. SINV particles derived from BHK were found to be around 2 MDa more massive than C6/36 derived particles. We used a variety of techniques to explore the source(s) of the mass difference. A glycomics study indicated that the BHK derived SINV has larger carbohydrate moieties compared to C6/36 derived particles, but this only partially accounts for the difference in the mass. An investigation of the lipids revealed that the BHK derived SINV has higher quantities of longer-chain lipids compared to C6/36 derived SINV. These differences do account for the differences in the masses of the BHK and C6/36 derived SINV. The incorporation of longer-chain, unsaturated fatty acids in the BHK derived SINV could affect the organization and stability of the lipid bilayer, which in turn may affect several stages in the viral life cycle, and thus contribute to the observed difference in infectivity.
RESULTS AND DISCUSSION
The Total Masses of SINV Particles Derived from BHK and C6/36.
To give us insight into possible compositional differences between BHK derived and C6/36 derived particles, we measured their total masses using CDMS. CDMS is a single particle method where the mass is determined from simultaneous measurement of the mass to charge ratio (m/z) and charge of each ion. Multiplying m/z and the charge yields the mass. CDMS can provide masses of much larger species than conventional mass spectrometry which is usually limited to molecular weights well below 1 MDa.38–46
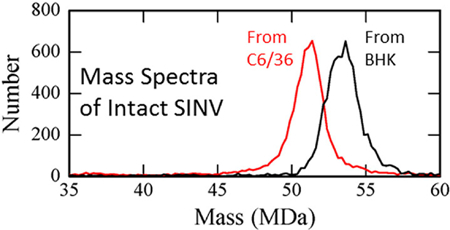
CDMS spectra of intact SINV particles from BHK and C6/36 cells are shown in Figure 1. The BHK-derived particles (spectrum shown in black) are around 2 MDa heavier than the C6/36-derived (red spectrum). CDMS measurements were performed for four BHK preparations and six C6/36 preparations. The results are presented in Supplementary Figure S1 which shows the center masses determined for the different preparations plotted against preparation number. The center masses were determined by fitting the measured peaks with a Gaussian. The small differences in the center masses measured for different preparations (standard deviations of the mean <0.15 MDa) are dwarfed by the large, systematic difference in the masses of the C6/36 and BHK derived particles (2.2 MDa). These results indicate that the composition of the particles is relatively invariant with preparation compared to the substantial composition differences between the particles from different cell lines. For some preparations, CDMS measurements were performed for multiple time points over more than 20 days. The measured masses showed no systematic variation with time.
Figure 1.
Representative CDMS spectra of wild-type SINV derived from C6/36 (mosquito) cells in red and from BHK (vertebrate) cells in black. The C6/36 spectrum (red) contains 9226 individual ion measurements and the BHK spectrum (black) contains 6267.
The average mass of SINV particles derived from the measurements for the BHK cells is 53.46 ± 0.13 MDa, while the average mass for SINV particles derived from C6/36 cells is 51.25 ± 0.14 MDa. In both cases, the uncertainty is the standard deviation of the mean. Native mass spectrometry is known to overestimate the masses of large protein complexes by up to around a percent because of adduct formation.47 In previous CDMS measurements for bacteriophage P22, the measured mass (52.18 MDa) was around 1.1% larger than the expected mass (51.61 MDa).41 If we assume the same deviation here, the masses corrected for adduct formation are 52.88 MDa and 50.69 MDa for BHK and C6/36 derived particles, respectively.
Peaks in the CDMS spectra for both BHK and C6/36 derived SINV are relatively broad compared to peaks in spectra acquired for other viruses.42 The peak width has contributions from the mass heterogeneity (i.e., different virus ions have different masses) and from the resolution of the measurement. For CDMS, the mass resolution is determined by the uncertainties in the m/z and charge measurements, both of which have been well characterized.48,49 The full widths at half-maximum (fwhm) of the measured peaks were determined from Gaussian fits. The average fwhm’s were 2.37 MDa and 2.22 MDa for BHK and C6/36 derived SINV particles, respectively. The mass resolution in the relevant mass range should be around 1 MDa. The excess peak width is mainly due to heterogeneity in the particle masses. The contributions to the peak width from the mass heterogeneity and mass resolution are uncorrelated, so the fwhm’s due to mass heterogeneity are around 2.1 MDa and 2.0 MDa and for BHK and C6/36 derived virions, respectively. Bacteriophage P22, measured previously by CDMS, had a mass of 52.18 MDa and a fwhm of 1.33 MDa. In this case, the mass heterogeneity after accounting for the mass resolution is much smaller and attributed entirely to the distribution of packaged DNA which varies slightly from particle to particle.50 In the case of SINV, the genome length is well-defined and not a source of heterogeneity. Therefore, the mass heterogeneity for SINV must arise from other sources.
Comparison with Expected Total Masses.
The expected mass of SINV was calculated using sequence data and the results of previous compositional studies.15,20,25,26,51 Table 1 gives the masses and stoichiometry of the components for SINV produced from BHK and C6/36 cells used to calculate the expected masses. SINV consists of three regions: the nucleocapsid core, lipid bilayer, and viral glycoprotein spikes. The nucleocapsid consists of the genomic material and CP. The genome for SINV is single-stranded RNA, 11.7 kilobases long with a sequence mass of 3.816 MDa. The CP has a sequence mass of 29.356 kDa. The nucleocapsid has T = 4 symmetry, therefore it contains 240 copies of the CP yielding a capsid mass of 10.87 MDa.51 The glycoprotein spikes consist of 240 copies of E2 and E1 (46.917 kDa and 47.371 kDa, respectively, for just the protein and not the associated carbohydrate moieties). E3 is not associated with purified SINV, in contrast to what is observed with Semliki Forest.22 The amount of TF incorporated into the virion is estimated to be between 12 and 24 copies.23
Table 1.
Calculation of the Expected Masses of SINV Protein and Nucleic Acid Components
| component | mass of one copy (kDa) | stoichiometry | BHK mass (MDa) | C6/36 mass (MDa) |
|---|---|---|---|---|
| ssRNA genome | 3816 | 1 | 3.82 | 3.82 |
| CP | 29.356 | 240 | 7.05 | 7.05 |
| deglycosylated E1 | 47.371 | 240 | 11.37 | 11.37 |
| deglycosylated E2 | 46.917 | 240 | 11.26 | 11.26 |
| TF | 8.001 | 24 | 0.19 | 0.19 |
| 40S ribosomal subunit | 1200 | 0.1 and 1 | 0.12 | 1.2 |
| total protein and genome mass | 33.81 | 34.89 | ||
| CDMS measured | 52.88 | 50.69 | ||
| difference | 19.07 | 15.80 |
Some of the nucleocapsids contain a host derived 40S ribosomal small subunit. The ribosomal subunit has been shown to be present in >90% of C6/36 derived virions, but only ∼10% of the BHK derived virions.25 Since the 40S subunit has a mass of 1.2 MDa, two peaks separated by 1.2 MDa might be expected in the mass distributions. However, the measured peaks due to the SINV particles are more than 2 MDa wide mainly because of sample heterogeneity. Simulations of the expected peak shape taking into account the heterogeneity and experimental resolution show that addition of a components separated by 1.2 MDa in an intensity ratio of 90:10 does not even lead to a discernible shoulder. There is only a slight increase in the width of the composite peak due to the presence of the minor component.
To confirm that our purification method was not preferentially pelleting BHK derived particles that contained the ribosomal subunit, we determined the abundance of the ribosomal subunit in our sample by determining the amount of rRNA and vRNA as described previously.25 Our pelleted material was consistent with 10% of the BHK derived particles and over 95% of the C6/36 derived particles having a ribosomal subunit. Thus, the calculated masses, not accounting for glycosylation or the lipid bilayer, are 33.81 MDa for BHK derived particles and 34.98 MDa for C6/36 derived (see Table 1). Subtracting these values from the corrected measured masses (52.88 MDa and 50.69 MDa for BHK and C6/36 derived particles, respectively) leaves 19.07 MDa that is unattributed for the BHK particles and 15.80 MDa that is unattributed for the C6/36 particles. The difference between these values, 3.27 MDa, is larger than the difference in the corrected measured masses (2.19 MDa) because the ribosomal subunit is more abundant for the C6/36-derived particles. The unattributed masses (19.07 MDa for BHK particles and 15.80 MDa for C6/36) are at least partly accounted for by the glycans attached to E1 and E2. While the sites of glycosylation are conserved in particles produced from all hosts, the glycans (oligomannose, hybrid, or complex) are dictated by the host.26,52
To investigate whether the observed mass difference between BHK and C6/36 derived particles was partly due to glycosylation of E2 and E1, and not to truncation or proteolytic cleavage during assembly, we ran purified particles on SDS-PAGE followed by Coomassie staining to separate and detect the E2 and E1 proteins. As seen in Figure 2, the migration of E2 and E1 from BHK cells is slower than the corresponding glycoproteins from C6/36 cells (compare lanes to glycosylation, PNGase F was used to hydrolyze the glycans. After the glycans were cleaved, the migration of E1 and E2 from both hosts was identical (compare lanes 1 and 3). Thus, as anticipated, the glycoproteins from different hosts have different glycan compositions.
Figure 2.
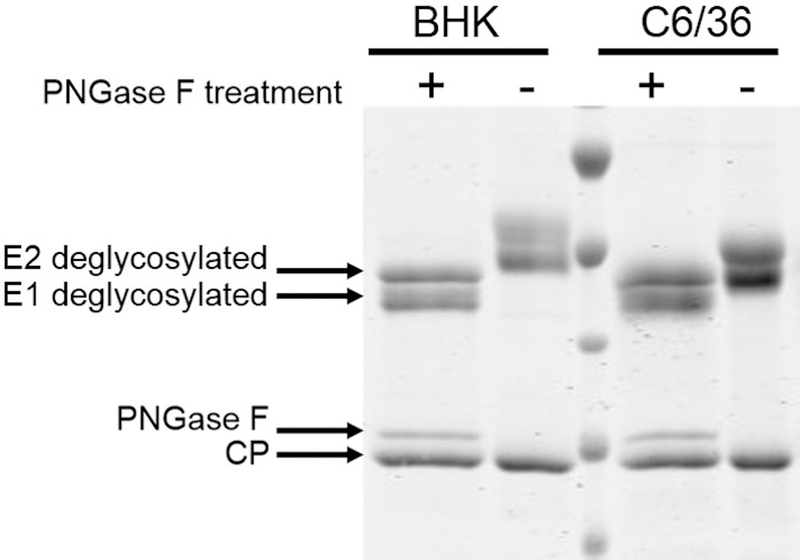
Coomassie stained SDS-PAGE gel of SINV from BHK cells and C6/36 cells. Lanes 2 and 5 (from the left) contain a wild-type virus from BHK and C6/36 cells, respectively. The E1 and E2 bands migrate differently, and the CP is the same for both samples. Lanes 1 and 4 contain SINV derived from BHK and C6/36 cells that has been treated with PNGase F, an enzyme that removes the N-linked glycans. After PNGase F treatment, E1 and E2 migrate the same. The small band above the capsid protein band is PNGase F.
Glycan Masses for BHK and C6/36 Derived SINV Particles.
To determine if the glycans present on E2 and E1 were responsible for the mass differences between BHK and C6/36 derived SINV particles, we determined their mass and identity. There are three types of N-linked glycans, oligomannose, hybrid, and complex. Oligomannoses, or simple sugars, are composed of N-acetylglucosamine and mannose sugar moieties. Hybrid glycans have a similar base branching to oligomannose glycans and only have mannose on the mannose 1–6 core and one or two additional moieties (fucose, galactose, or sialic acid) on the 1–3 arm. Complex glycans are larger than hybrid or oligomannose and consist of antenna stemming from the core containing mannose, fucose, galactose, N-acetylglucosamine, or sialic acid.53 Glycan structure was determined by denaturing and digesting particles from BHK and C6/36. The resulting glycopeptides were analyzed by ultra performance liquid chromatography (UPLC), mass spectrometry, and tandem mass spectrometry. The resulting mass and fragmentation data were used to assign and confirm glycopeptides and glycan structures (see Experimental Methods section).
Table 2 gives a list of the glycans found for each site of SINV derived from C6/36 and BHK cells. More detailed information is provided in Supplementary Tables 1 and 2, which show the confirmed glycopeptides grouped according to the parent peptide. For some of the sites (e.g., E1–139 and E2–196 on SINV from C6/36 cells), only a single glycopeptide was found with only two different glycans. On the other hand, some sites (e.g., E2–196 on BHK derived SINV) were represented by multiple glycopeptides with a wide range of different glycans. In these cases, the relative abundances for the same glycans across different glycopeptides were not necessarily strongly correlated; this may be a consequence of the glycopeptides having different ionization efficiencies.
Table 2.
Summary of Glycans Found for SINV Derived from C6/36 and BHK Cellsa
| C6/36 |
BHK |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| site | GlcNAc | Man | Fuc | SA | site | GlcNAc | Man | Fuc | SA |
| E1-139 | 2 | 4 | E1-139 | 2 | 3 | ||||
| 2 | 6 | 2 | 4 | ||||||
| 2 | 8 | ||||||||
| 2 | 9 | ||||||||
| E1-245 | 2 | 3 | E1-245 | 2 | 5 | ||||
| 2 | 4 | 2 | 6 | ||||||
| 2 | 5 | 2 | 7 | ||||||
| 2 | 6 | 2 | 9 | ||||||
| 2 | 7 | 5 | 5 | 1 | |||||
| 2 | 8 | 5 | 6 | ||||||
| 2 | 9 | 5 | 6 | 1 | |||||
| 5 | 7 | ||||||||
| 5 | 7 | 1 | |||||||
| 6 | 7 | ||||||||
| E2–196 | 2 | 7 | E2–196 | 2 | 5 | ||||
| 2 | 8 | 2 | 6 | ||||||
| 2 | 7 | ||||||||
| 2 | 8 | ||||||||
| 2 | 3 | 1 | |||||||
| 2 | 4 | 1 | |||||||
| 2 | 5 | 1 | |||||||
| 3 | 2 | 1 | 1 | ||||||
| 3 | 4 | 1 | |||||||
| 4 | 3 | 1 | |||||||
| 3 | 3 | 1 | 1 | ||||||
| 4 | 4 | 1 | |||||||
| 3 | 4 | 1 | 1 | ||||||
| 4 | 5 | 1 | |||||||
| 4 | 4 | 1 | 1 | ||||||
| 3 | 6 | 1 | 1 | ||||||
| 4 | 5 | 1 | 1 | ||||||
| 3 | 7 | 1 | 1 | ||||||
| 4 | 6 | 1 | 1 | ||||||
| 4 | 7 | 1 | 1 | ||||||
| E2–318 | 2 | 3 | E2–318 | 2 | 11 | ||||
| 2 | 4 | 2 | 13 | ||||||
| 2 | 5 | ||||||||
| 2 | 6 | ||||||||
| 2 | 7 | ||||||||
| 2 | 8 | ||||||||
| 2 | 9 | ||||||||
The number of copies of each sugar subunit, N-acetylglucosamine (GlcNAc), mannose (Man), fucose (Fuc), and sialic acid (SA), are listed for the glycans found for each site. More detailed information is provided in Supplementary Tables 1 and 2.
SINV derived from C6/36 has oligomannose at all four glycosylation sites (E1–139, E1–245, E2–196, and E2–318,) (see Table 2 and Supplementary Table 1). For SINV derived from BHK cells, a mixture of oligomannose and complex glycans was found (see Table 2 and Supplementary Table 2). Two sites, E1–139 and E2–318, are predominantly modified with oligomannose, and the other two sites, E1–245 and E2–196, are heterogeneous, with a mixture of oligomannose and complex glycans. The addition of N-linked glycosylation initially occurs in the endoplasmic reticulum with the addition of an oligomannose-type glycan, which may be further processed in the Golgi. Insect cells generally lack the enzymes required for further processing to create complex and hybrid glycan structures, consistent with our observation that complex glycans were only identified from SINV produced in mammalian cells.26–28,54,55 This further emphasizes the fact that the virions encounter significantly different processing environments in different host cell types.
To estimate the average mass of the glycans at each glycosylation site, we took the relative intensities of each glycopeptide ion detected for that site and multiplied it by the mass of the associated glycan and then summed the resulting values. There are 240 copies of each glycan. Therefore, to calculate the total glycan mass per particle for each site, we multiplied the average glycan mass for each site by 240. The results are summarized in Table 3. The total glycan mass per particle is the sum of the glycan masses for the four sites. The total glycan masses are 1.884 MDa for BHK derived SINV particles and 1.357 MDa for BHK derived (see Table 3). The difference between the total glycan masses for the different hosts (0.527 MDa) does not account for the difference between their unattributed masses (3.27 MDa), suggesting there are additional factors that contribute to this mass difference.
Table 3.
Total Glycan Masses Determined by UPLC-MS/ MS
| host | E1-139 glycan mass (MDa) | E1-245 glycan mass (MDa) | E2-196 glycan mass (MDa) | E2-318 glycan mass (MDa) | total glycan mass (MDa) | difference (MDa) |
|---|---|---|---|---|---|---|
| BHK | 0.378 | 0.51 | 0.407 | 0.589 | 1.884 | 0.527 |
| C6/36 | 0.287 | 0.323 | 0.394 | 0.353 | 1.357 |
Are There Any Additional Proteins or Nucleic Acid Components in BHK Derived SINV?
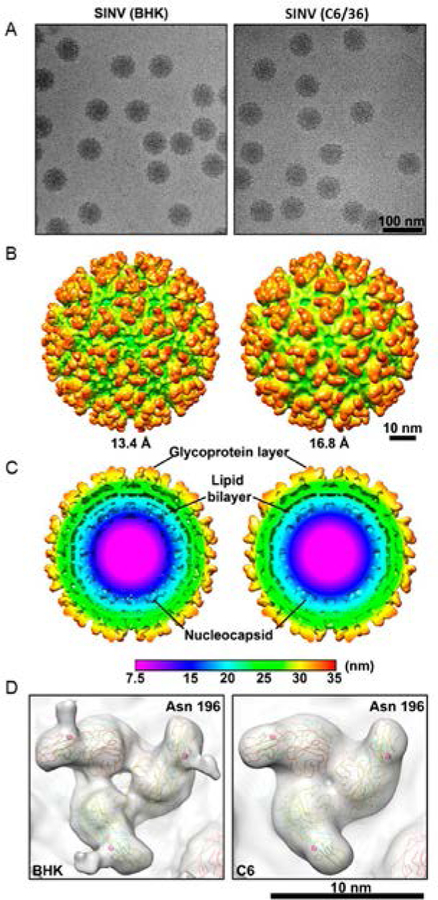
To confirm that there are no structural differences due to additional proteins or RNA components that could account for the extra mass in BHK derived SINV, we evaluated the structures of SINV from BHK and C6/36 cells by cryo-EM. Representative cryo-EM images of the two virion samples are shown in Figure 3A. Both samples show uniform, ∼70 nm spherical particles. The 3D reconstructions (Figure 3B) were computed to 13.4 Å for SINV from BHK cells (left in Figure 3B) and to 16.8 Å for SINV from C6/36 cells. Similar to previously published cryo-EM structures,19 there are 80 glycoprotein spikes on the particle surface, and they are arranged with T = 4 icosahedral symmetry. Looking at the inside of the particles, both samples have an inner core with icosahedral T = 4 symmetry, which is also consistent with previously published results. Thus, there are no major morphological differences between the samples derived from the two cell lines. Cross sections through the three-dimensional (3D) reconstructions are shown in Figure 3C. The nucleocapsid extends from the center to around 20 nm, and it is the same size for both BHK and C6/36 derived virions. The lipid bilayer extends from around 20 nm to around 27 nm in both cases. The glycoprotein spikes extend from around 27 nm to around 35 nm. The cross sections reveal no significant differences between the BHK and C6/36 derived virions. A direct comparison of molecules in the lipid bilayer is not possible at this resolution, and at higher resolution, the analysis would still be speculative because of the amount of disorder and the low density in this region.
Figure 3.
(A) Cryo-EM images of SINV isolated from BHK cells (left) and C6/36 cells (right). (B) 3D reconstruction of SINV from BHK (left) and C6/36 (right). The resolution is 13.4 Å for SINV from BHK and 16.8 Å SINV from C6/36. (C) Cross section through the 3D reconstructions showing the distinct layers of the nucleocapsid core, lipid bilayer, and glycoproteins. The color scale shows the distance from the center. (D) A portion of the reconstruction showing the glycosylation site E2–196. BHK derived virus shows additional density on the spike consistent with the glycan analysis in Table 3.
Cryo-EM analyses have demonstrated that focused reconstructions,56,57 or imposing icosahedral symmetry on portions of the virion particle such as the core or the glycoproteins,58,59 can identify asymmetric features in the particles. This strategy would be beneficial if we knew where the additional density is located in the particle. However, our current structures do not give a location for any extra density.
If the difference in mass between BHK and C6/36 derived particles was due to E3 or TF proteins, there would need to be ∼260–300 copies of these proteins per particle. If the difference was due to additional copies of capsid protein or glycoproteins, there would need to be ∼90 additional capsid proteins or ∼60 additional glycoprotein spikes. Such large numbers of additional copies would likely lead to distorted particles, which would be evident in the cryo images. Even if the extra proteins were not arranged symmetrically, they would be evident in the cryo-EM images and the density would be evident during image analysis. Finally, if there were significant increases in number of capsid proteins, glycoproteins, or TF, this would be evident when the samples were checked for purity with SDS-PAGE. Previous work has shown that released alphaviruses may have assembly defects leading to incomplete cores.59 The decreased number of capsid proteins, and potentially glycoproteins, would lead to heterogeneity in the particles that could account, at least partially, for the widths of the CDMS peaks in Figure 1.
An additional 2–3 copies of the 40S ribosomal small subunit per BHK derived particle could account for the mass difference between the BHK and C6/36 derived particles. However, our cryo-EM structures did not show increased density inside the BHK particles. In addition, qRT-PCR analysis was performed to determine the ratio of viral genome and rRNA. The BHK samples showed 1 − log less rRNA compared to viral RNA, consistent with previous results showing only 10% of the BHK samples contain the ribosomal subunit.25
The glycan analysis described above showed a difference in carbohydrate moieties at residue E2–196, which is located at the distal tip of the glycoprotein spike. A close-up view of the glycoprotein spikes for BHK and C6/36 derived particles shows a difference in density at this residue (Figure 3D). The lack of distinct density in the C6/36 derived particles may be due to the lower resolution or an increase in the flexibility of the glycan at this position.
Composition of the Lipid Membrane.
From our analysis so far, the difference between the measured and calculated masses (the unattributed mass) is 17.18 MDa for BHK derived SINV and 14.44 MDa for C6/36 derived SINV (see Table 4). The unattributed mass corresponds to 32.5% of the total measured mass for BHK derived SINV and 28.5% for C6/36 derived. These values are consistent with reports that the lipid bilayer contributes 26–32% of the total mass, depending on the host cell.15,60 The unattributed mass must be at least partly due to the lipid bilayer because it is an essential component of SINV particles (see Figure 3C) that is not yet included in the mass calculations. If the difference between the masses of the C6/36 and BHK derived SINV particles is entirely due to the lipid bilayer, the lipid bilayer for the BHK particles must contribute an additional 2.74 MDa. Chemical and compositional differences between the lipids bilayers may contribute to the observed mass difference. Chemical diversity arises from the presence of different types of lipids in the bilayers, while compositional differences result from different ratios of the component lipids. Mammalian cells contain elongases and desaturases, which increase the lipid diversity and broaden the composition compared to invertebrate cells.61
Table 4.
Calculation of Unattributed Massa
| host | calculated mass (protein + nucleic acid + glycans) (MDa) | measured by CDMS (MDa) | Unattributed mass (MDa) | unattributed mass/measured mass (%) |
|---|---|---|---|---|
| BHK | 35.69 | 52.88 | 17.18 | 32.50 |
| C6/36 | 36.25 | 50.69 | 14.44 | 28.50 |
The difference between the measured mass and the mass calculated from the protein, nucleic acid, and glycan components.
To investigate whether the lipids from BHK derived and C6/36 derived SINV particles are significantly different, we selectively extracted the lipid membranes from purified particles and analyzed them with high-performance liquid chromatography coupled with tandem mass spectrometry. We screened for 89 targeted lipids, with acyl chains between 16 and 22 carbons long (the lipid-screening library is given in Supplementary Table 3). Of the 89 lipids screened, 42 were detected in both SINV generated by BHK and C6/36 cells, meaning no major chemical differences between the lipid bilayers were detected in this targeted screen. However, significant differences in the lipid concentrations for BHK and C6/36 derived SINV samples were measured in 25 of the 42 detected lipids. Figure 4 shows concentrations for four fatty acids that were present in significantly higher quantities in the BHK derived samples. The mean concentrations and standard deviations for the 25 lipids where significant concentration differences were observed are shown in Supplementary Table 4. For 24 of the 25 lipids, the concentration in BHK was higher than in C6/36. The concentration ratios (BHK/C6) range from 603 to 0.63.
Figure 4.
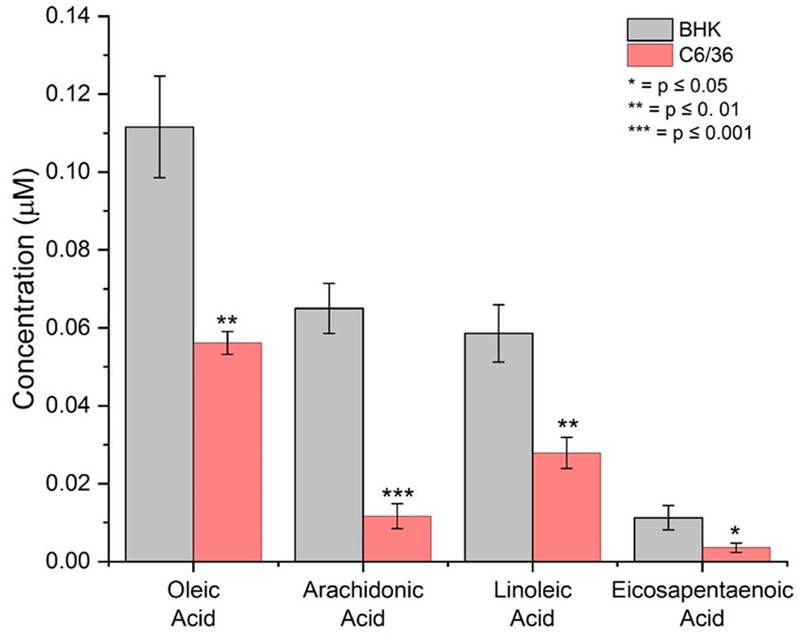
Differences in the concentrations of four fatty acids detected in SINV from BHK and C6/36. Error bars are ± the standard error of the mean for three independent measurements. The concentration units are μM/g of virus.
It is important to note that the C6/36 cells were grown in fetal bovine serum (FBS) prior to viral infection and FBS contains high quantities of long-chain unsaturated fatty acids. Mosquito cells do not contain the enzymes needed to produce these lipids, but they have been shown to act as lipid auxotrophs, so it is likely that the cells simply incorporated them rather than making them de novo. It is also possible that, in addition to having more lipids with long chain fatty acids, there are different amounts of sterol and short chain fatty acid containing lipids from SINV produced in the two hosts. Quantifying the composition of the lipid molecules is not trivial and is beyond the scope of this work. We have determined multiple differences in the levels of long chain fatty acids for BHK and C6/36 derived viral particles, demonstrating a compositional difference in the lipid bilayers in the SINV from different hosts. This compositional difference may directly or indirectly affect the organization and packing of the lipid bilayers.62
To determine how much longer each lipid molecule would need to be to account for the 2.74 MDa mass difference between BHK and C6/36 derived SINV, the number of lipid molecules in the SINV bilayer was estimated. Assuming a lipid molecule surface area of 0.5 nm2 and using the surface area of the lipid bilayer from our cryo-EM reconstructions, a value of 34,000 lipid molecules was obtained. The observed mass difference would correspond to each lipid molecule in BHK derived particles containing, on average, five extra methylene groups (−CH2−). This difference is not large enough to be detectable in our cryo-EM reconstructions (Figure 3).
We focused our lipid analysis on the presence of long chain fatty acids because elongases and desaturases are present in BHK cells but not C6/36. It is important to note that along with the length of hydrocarbon groups, the derivatization of lipid head groups and the presence and absence of other phospholipids and sterol containing lipids could also contribute to the difference in the mass of the virus particles. In addition, there may be other lipids present in low abundance that could contribute to differences in infectivity.
CONCLUSIONS
CDMS measurements indicate that SINV particles derived from C6/36 and from BHK cells have significantly different masses. Based on the measurements described above, differences in the composition of the lipid bilayers are a major contributor to the mass difference. There is a higher portion of lipids containing longer chain fatty acids in BHK derived SINV particles.
An infectious particle is one that enters the host, replicates, and assembles new particles that are able to go on to infect a new cell; multiple stages of the lifecycle contribute to infectivity. Particles derived from vertebrate cells typically have a lower specific infectivity compared to particles derived from mosquito cells even though the same viral proteins and genome at the same stoichiometry are incorporated into the viral particles. Is there anything about the particles that can account for the difference in infectivity?
Most particles from mosquito cells contain a 40S ribosomal small subunit host factor, whereas most mammalian derived particles do not contain this factor. In addition, this work and other studies have identified different glycans on the E2 and E1 proteins depending on the cell line used to generate the particles. Our results here may provide additional insight into the difference in infectivity between viruses derived from different host cells. We have determined that the lipid bilayer of mammalian derived SINV particles has lipid molecules that are slightly longer, on average, than the lipids found in bilayers of C6/36 derived SINV particles. The lipid bilayer composition could affect the specific infectivity of the virus in several ways, all which require further investigation. First, longer chain lipids will have a different packing density and also change the rigidity of a bilayer. Consequently, particle stability during transmission may be affected. Second, a more rigid bilayer will also influence the interactions between the viral glycoproteins and the membrane. These effects might manifest during particle budding when the glycoprotein-embedded bilayer wraps itself around the core. In addition, the interactions between the bilayer and spikes could be altered during fusion between the particle and host membranes. Cholesterol and sphingomyelin affect viral entry and exit, and delipidation of cholesterol from cells lines produces viral particles with altered specific infectivity, presumably because particle stability was altered.63,64 Previous work by Sokoloski et al.65 and Brown and Hernandez (summarized in ref 66) demonstrate that alphaviruses derived from both Aedes aegypti and Aedes albopictus have a higher specific infectivity compared to viruses derived from a panel of vertebrate cells,65 supporting a correlation between lipid composition and virus infectivity.
Now that we know the components that are different between virions produced from different cell types, we can begin to dissect the mechanisms that contribute to their differences in infectivity. Long term, identifying specific mechanisms that contribute to infectivity will be important as we search for panviral compounds to treat arbovirus infections.
EXPERIMENTAL METHODS
Virus Preparation, Titer Determination, and RNA Quantification.
Baby hamster kidney (BHK) cells and C6/36 mosquito cells were grown in minimum essential medium supplemented with non-essential amino acids, penicillin-streptomycin, amphotericin B, L-glutamine, and 10% fetal bovine serum (Corning Cellgro, Manassas, VA). Cells were grown in a 37 °C (BHK) or 28 °C (C6/36) incubator in a controlled humidity environment with 5% CO2.
Cells were exposed to wild-type SINV (strain TE12) with an multiplicity of infection of between 1 and 5 for 45 min at room temperature while rocking.67 Initial inoculum was removed, and cells were washed with PBS before adding serum-free media (SFM) (Gibco 11681–020) supplemented with non-essential amino acids, penicillin-streptomycin-amphotericin B, and L-glutamine. Infections proceeded for 18–23 h (BHK) or 4 days (C6/36). For C6/36 cells, 5 mL of supplemented SFM was added after 2 days. The virus was harvested by collecting the media and clarified by centrifuging at 3000×g for 5 min at room temperature. The supernatant was removed and stored at 4 °C for no more than 24 h. The virus was pelleted by centrifuging at 5300× g for 16 h at 15 °C. The pellet, often not visible, was resuspended with a small volume (30–50 μL of 200 mM ammonium acetate). Samples were stored at 4 °C for up to 4 weeks. Sample purity was determined via Coomassie stained SDS-PAGE, sample morphology was examined by transmission electron microscopy, viral titers were determined via standard plaque assay using BHK-21 cells, the abundance of the 40S ribosomal small subunit was determined by qRT-PCR,65,68 and virus concentration was measured by absorbance at 260 nm using an extinction coefficient of 7.0 mg/mL.35
Charge Detection Mass Spectrometry.
CDMS is a single particle technique where the m/z (mass to charge ratio) and charge are simultaneously measured for individual ions. The m/z and charge are then multiplied to yield the mass of each ion. This process is repeated for thousands of ions, and the masses are binned to obtain a mass spectrum. The CDMS instrument39,69,70 and data analysis methods48,49 have been described in detail elsewhere. Briefly, ions are produced by a nanoelectrospray source (Advion Biosciences, Ithaca, NY, USA) and introduced into the instrument through a heated capillary. Ions pass through three differentially pumped regions to remove the background gas: The first region contains an ion funnel, the second an RF hexapole, and the third an RF quadrupole. The nominal ion energy, 100 eV/charge, is set by the DC potential applied to the hexapole. Ions that exit the quadrupole are focused into a dual hemispherical deflection energy analyzer (HDA) which transmits ions with a narrow band of kinetic energies centered on 100 eV/charge. The transmitted ions are focused into a modified cone trap with a detector tube between the end-caps.
When an ion enters the detector tube, it induces a charge which is detected by a charge-sensitive preamplifier with a cryogenically cooled JFET at the input. For these experiments, an ion triggered trapping scheme was used. Initially both end-caps are at ground, the potential on the rear end-cap is raised, and then when the signal from an ion entering the detector tube exceeds a threshold, the potential on the front end-cap is raised to trap the ion. The threshold used here corresponds to around 75 elementary charges (e). The trapped ion oscillates back and forth through the charge detector tube, inducing a periodic signal, which is amplified, digitized, and transferred to a computer for analysis. After a predetermined time, the end-caps are grounded, and the trap is emptied. The trapping time for these experiments was 100 ms. The periodic signal is amplified, digitized, and transferred to a computer for analysis by a Fortran program using fast Fourier transforms. The m/z is determined from the frequency of the fundamental, and the charge is determined from the magnitudes of the fundamental and second harmonic.
PNGase F Treatment.
SINV C6/36 and BHK were treated with PNGase F according to the New England Biolabs’ protocol.71 The virus was mixed with glycoprotein denaturing buffer, 0.5% sodium dodecyl sulfate, 40 mM dithiothreitol, and water and heated to 95 °C for 10 min. The denatured virus was then mixed with water, GlycoBuffer 2, NP-40, and PNGase F (New England Biolabs, Ipswich, MA) and allowed to react at 37 °C overnight.
Trypsin Digestion.
SINV C6/36 and BHK were denatured using 8 M urea in 100 mM ammonium bicarbonate (Sigma-Aldrich, St. Louis, MO). Denatured proteins were reduced with 4 mM tris (2-carboxyethyl) phosphine (Sigma-Aldrich, St. Louis, MO) for 1 h at 56°C and then alkylated using 8 mM iodoacetamide (Sigma-Aldrich, St. Louis, MO) for 1 h at room temperature. Alkylated proteins were digested at 37°C using trypsin (Promega, Madison, WI). Trypsin digestion was performed overnight at a urea concentration of 1 M and 100 mM ammonium bicarbonate. The digested peptides were desalted using Omix C18 micro pipet tips (Agilent, Santa Clara, CA) and lyophilized to dryness. The desalted peptides were either analyzed directly or treated with PNGase F (New England Biolabs, Ipswich, MA) to release the glycans. Glycan release was performed in 25 mM ammonium bicarbonate at 37°C overnight.
Glycan Analysis by Mass Spectrometry.
Digested peptides were dissolved in aqueous 0.1% formic acid (Fischer Scientific, Hampton, NH) and loaded at 5 μL/min onto an Ultra Performance Liquid Chromatography (UPLC) C18 loading column (Symmetry C18, 180 μm × 20 mm, Waters, Milford, MA). The peptides were separated using a UPLC C18 column (1.8 μm HSS T3, 75 μm × 250 mm, Waters, Milford, MA) with a linear gradient of 3% buffer B to 34% buffer B over 60 min at a flow rate of 300 nL/min. Buffer B was acetonitrile with 0.1% formic acid (Fischer Scientific, Hampton, NH). Peptides eluting from the separation column were ionized by electrospray and analyzed by a Synapt G2S (Waters, Milford, MA) mass spectrometer. The Synapt was operated in the ion mobility supplemented MSE mode. In this mode, mass spectra are acquired while alternating between low and high collision energy as peptides elute from the column. The low collision energy spectra are used to identify the precursor ion masses, and the high collision energy spectra provide the fragment ion masses. Collisional activation occurs after the ions have passed through the ion mobility cell, so if the peptides are not sufficiently separated by UPLC, precursor ions and fragment ions can be correlated by their drift times.
The results for the PNGase F treated peptides were analyzed first to determine the masses and sequences of the unglycosylated peptides. The results were exported as tables containing mass, charge, m/z, intensity, drift time, and retention time, along with other information not used for this analysis. To identify glycopeptides, the fragment spectra (MS2) were searched for glycan specific fragment ions (reporter ions). The reporter ions for mannose and N-acetylglucosamine are singly charge ions with m/z values of ∼163.06 Da and ∼204.09 Da, respectively.72 The retention times for the reporter ions were then used to identify the parent glycopeptides. A Fortran program identified masses within a ±0.5 min retention time window of the reporter ion and compared them to masses of known glycopeptides. Cases where the parent mass agreed with a known glycopeptide mass to within a specified deviation were added to a glycopeptide list. For each member on the list, we then went back to the MS2 spectra and manually confirmed that the fragmentation pattern was consistent with the proposed glycan. Glycopeptides that could not be confirmed in this way were removed from the list. The confirmed glycopeptides are grouped according to the bare peptide mass in Supplementary Tables 1 and 2.
Cryo-EM and Image Reconstruction.
Cryo-EM specimens were prepared by applying 4 μL of virus solution to glow-discharged continuous carbon film-coated copper grids and vitrified using FEI Vitrobot (Mark III). The frozen-hydrated specimen was transferred into a JEOL JEM-3200FS TEM using a Gatan 626 cryo-holder. Low-dose cryo-micrographs (≤20 e−/Å2) were collected at a nominal magnification of 30,000× using a DE-12 direct electron detection camera (Direct Electron LP, San Diego, CA) with energy filter slit set to 20 eV. The magnification yields a pixel size of 1.904 Å at the image space. Particle images were extracted from micrographs using e2boxer.py.73 Image processing, CTF correction, and 3D reconstruction were performed using the gold standard protocol in auto3dem.74 In total 10,873 particles were used for SINV from BHK to yield a 13.4 Å 3D reconstruction, and 5676 particles were used for SINV from C6/36 to yield a 16.8 Å 3D reconstruction. The electron density maps were rendered using UCSF Chimera.75
HPLC Lipid Extraction.
Samples were prepared for lipid extraction by adding 1 mL of high-pressure liquid chromatography (HPLC)-grade methanol (Fisher, Fair Lawn, NJ, USA) to each sample in 1.5 mL microcentrifuge tubes. Samples were spiked with 500 pmol of deuterium-labeled N-arachidonoyl ethanolamine (d8AEA; Cayman Chemical, Ann-Arbor, MI, USA). Tubes were vortexed and left on ice for 2 h. Samples were centrifuged at 18,000g at 4 °C for 10 min. The supernatant was decanted into 3 mL of HPLC-grade water (Fisher). Lipids were then partially purified on C18 solid-phase extraction columns (Agilent, Palo Alto, CA, USA) arranged in a vacuum manifold, as previously described.76–78 5 mL of HPLC methanol was added to each column. After the methanol had run through the column, 2.5 mL of HPLC water was added. The supernatant/water solution was loaded onto the column. Afterward, 5 mL of HPLC H2O was added to the column to wash off impurities. A series of five elutions with 1.5 mL of 40%, 60%, 75%, 85%, and 100% methanol were collected and stored at −80 °C until analysis.
HPLC-Mass Spectrometry Lipid Analysis.
Lipids were analyzed using an Applied Biosystems API 3000 triple quadrupole mass spectrometer with electrospray ionization (Foster City, CA, USA). 20 μL from each elution was analyzed using an XDB-C18 reversed-phase HPLC analytical column (Agilent) and optimized mobile phase gradients. Mobile phase A: 20% methanol, 80% water (v/v) and 1 mM ammonium acetate (Sigma, St. Louis, MO, USA). Mobile phase B: 100% methanol, 1 mM ammonium acetate. Two Shimadzu 10ADvp pumps (Columbia, MD, USA) provided the pressure for gradient elution. Levels of each compound were determined by running each sample using a multiple reactions monitoring (MRM) method tailored for each group of structurally similar compounds. Analysis of the HPLC/MS/MS data was performed using Analyst software (Applied Biosystems), as previously described.76–78 Chromatograms were generated by determining the retention time of analytes with a [M + 1] or [M − 1] parent peak and a fragmentation peak corresponding to programmed values for each lipid. The retention time was then compared to the retention time of a standard for the suspected compound. If the retention times matched, then the concentration of the compound was determined by calculating the area under the curve for the unknown and comparing it to the calibration curve obtained from the standards. Therefore, unknown lipids are matched to known standards according to the retention time from the analytical column and their mass fingerprint. Extraction efficiency was calculated with a d8AEA spiked recovery vial as a standard. For each individual lipid, mean concentrations in mol/L adjusted for percent recovery were compared between groups using a one-way ANOVA. All of the statistical tests were carried out using SPSS Statistics (IBM, Armonk, NY, USA). 89 lipids were screened in each sample, including lipoamines, 2-acyl glycerols, prostaglandins, and free fatty acids (see Supplementary Table 3), and 42 of these lipids were detected in all 6 samples.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the support of the National Science Foundation through grant numbers CHE-1531823 (M.F.J.) and DA00668 (H.B.B.) and the National Institutes of Health through grant number R01 GM117207-01 (D.E.C.). We also gratefully acknowledge the support of the Indiana University Office of the Vice President for Research (S.M.).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfec-dis.8b00356.
One figure showing mass measured by CDMS as a function of preparation number. Two tables showing glycan assignments. A table showing the lipid screening library, and a table giving results for the 25 lipids with significantly concentration differences between the two hosts (PDF)
The authors declare the following competing financial interest(s): The authors except D.E.C. and M.F.J. declare no competing interests. D.E.C. and M.F.J. are associated with a company that is developing charge detection mass spectrometry.
REFERENCES
- (1).Milo R, Jorgensen P, Moran U, Weber G, and Springer M (2010) BioNumbers -the database of key numbers in molecular and cell biology. Nucleic Acids Res 38 (suppl. 1), D750–D753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Bhattacharya T, Newton ILG, and Hardy RW (2017) Wolbachia elevates host methyltransferase expression to block an RNA virus early during infection. PLoS Pathog 13, e1006427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Byrd EA, and Kielian M (2017) An Alphavirus E2Membrane-Proximal Domain Promotes Envelope Protein Lateral Interactions and Virus Budding. mBio 8, e01564–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Carpenter JE, Henderson EP, and Grose C (2009) Enumeration of an extremely high particle-to-PFU ratio for Varicella-zoster virus. J. Virol 83, 6917–6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Flint SJ, and Enquist LW (2015) Principles of Virology, ASM Press, Washington, DC. [Google Scholar]
- (6).Klasse PJ (2015) Molecular determinants of the ratio of inert to infectious virus particles. Prog. Mol. Biol. Transl. Sci 129, 285–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hirst GK, and Pons MW (1973) Mechanism of influenza recombination. II. Virus aggregation and its effect on plaque formation by so-called non-infective virus. Virology 56, 620–631. [DOI] [PubMed] [Google Scholar]
- (8).Brooke CB (2014) Biological activities of ‘non-infectious’ influenza A virus particles. Future Virol 9, 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Altan-Bonnet N (2016) Extracellular vesicles are the Trojan horses of viral infection. Curr. Opin. Microbiol 32, 77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Sokoloski KJ, Hayes CA, Dunn MP, Balke JL, Hardy RW, and Mukhopadhyay S (2012) Sindbis virus infectivity improves during the course of infection in both mammalian and mosquito cells. Virus Res 167, 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Hernandez R, Sinodis C, and Brown DT (2005) Sindbis virus: propagation, quantification, and storage. Curr. Protoc. Microbiol 00, 15B.1.1–15B.1.34. [DOI] [PubMed] [Google Scholar]
- (12).Weaver SC, Brault AC, Kang W, and Holland JJ (1999) Genetic and fitness changes accompanying adaptation of an arbovirus to vertebrate and invertebrate cells. J. Virol 73, 4316–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Coffey LL, Vasilakis N, Brault AC, Powers AM, Tripet F, and Weaver SC (2008) Arbovirus evolution in vivo is constrained by host alternation. Proc. Natl. Acad. Sci. U. S. A 105, 6970–6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Abdelnabi R, Neyts J, and Delang L (2015) Towards antivirals against chikungunya virus. Antiviral Res 121, 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Strauss JH, and Strauss EG (1994) The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev 58, 491–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Jose J, Snyder JE, and Kuhn RJ (2009) A structural and functional perspective of alphavirus replication and assembly. Future Microbiol 4, 837–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Sokoloski KJ, Haist KC, Morrison TE, Mukhopadhyay S, and Hardy RW (2015) Noncapped alphavirus genomic RNAs and their role during infection. J. Virol 89, 6080–6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, and Strauss JH (2002) Structure of Dengue virus: implication for flavivirus organization, maturation and fusion. Cell 108, 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zhang Y, Corver J, Chipman PR, Zhang W, Pletnev SV, Sedlak D, Baker TS, Strauss JH, Kuhn RJ, and Rossmann MG (2003) Structures of immature flavivirus particles. EMBO J 22, 2604–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Zhang W, Mukhopadhyay S, Pletnev SV, Baker TS, Kuhn RJ, and Rossmann MG (2002) Placement of the structural proteins in Sindbis virus. J. Virol 76, 11645–11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Ziemiecki A, and Garoff H (1978) Subunit composition of the membrane glycoprotein complex of semliki forest virus. J. Mol. Biol 122, 259–269. [DOI] [PubMed] [Google Scholar]
- (22).Snyder JE, Kulcsar KA, Schultz KLW, Riley CP, Neary JT, Marr S, Jose J, Griffin DE, and Kuhn RJ (2013) Functional Characterization of the Alphavirus TF protein. J. Virol 87, 8511–8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Firth AE, Chung BYW, Fleeton MN, and Atkins JF (2008) Discovery of frameshifting in alphavirus 6K resolves a 20-year enigma. Virol. J 5, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Mackenzie-Liu D, Sokoloski KJ, Purdy S, and Hardy RW (2018) Encapsidated host factors in alphavirus particles influence midgut infection of Aedes aegypti. Viruses 10, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Sokoloski KJ, Snyder AJ, Liu NH, Hayes CA, Mukhopadhyay S, and Hardy RW (2013) Encapsidation of host-derived factors correlates with enhanced infectivity of Sindbis virus. J. Virol 87, 12216–12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Pletnev SV, Zhang W, Mukhopadhyay S, Fisher BR, Hernandez R, Brown DT, Baker TS, Rossmann MG, and Kuhn RJ (2001) Locations of carbohydrate sites on alphavirus glycoproteins show that E1 forms an icosahedral scaffold. Cell 105, 127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Sefton BM (1977) Immediate glycosylation of Sindbis virus membrane proteins. Cell 10, 659–668. [DOI] [PubMed] [Google Scholar]
- (28).Keegstra K, Sefton B, and Burke D (1975) Sindbis virus glycoproteins: effect of the host cell on the oligosaccharides. J. Virol 16, 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Lancaster C, Pristatsky P, Hoang VM, Casimiro DR, Schwartz RM, Rustandi R, and Ha S (2016) Characterization of N-glycosylation profiles from mammalian and insect cell derived chikungunya VLP. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci 1032, 218–223. [DOI] [PubMed] [Google Scholar]
- (30).Kalvodova L, Sampaio JL, Cordo S, Ejsing CS, Shevchenko A, and Simons K (2009) The lipidomes of vesicular stomatitis virus, semliki forest virus, and the host plasma membrane analyzed by quantitative shotgun mass spectrometry. J. Virol 83, 7996–8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Olsen BN, Bielska AA, Lee T, Daily MD, Covey DF, Schlesinger PH, Baker NA, and Ory DS (2013) The structural basis of cholesterol accessibility in membranes. Biophys. J 105, 1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Chen S, He H, Liu X, and Ariga H (2017) Tissue expression profiles and transcriptional regulation of elongase of very long chain fatty acid 6 in bovine mammary epithelial cells. PLoS One 12, e0175777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Zheng X, Tocher DR, Dickson CA, Bell JG, and Teale AJ (2005) Highly unsaturated fatty acid synthesis in vertebrates: New insights with the cloning and characterization of a Δ6 desaturase of Atlantic salmon. Lipids 40, 13–24. [DOI] [PubMed] [Google Scholar]
- (34).Hashimoto K, Yoshizawa AC, Okuda S, Kuma K, Goto S, and Kanehisa M (2008) The repertoire of desaturases and elongases reveals fatty acid variation in 56 eukaryotic genomes. J. Lipid Res 49, 183–191. [DOI] [PubMed] [Google Scholar]
- (35).Miller ML, and Brown DT (1993) The distribution of Sindbis virus proteins in mosquito cells as determined by immunofluorescence and immunoelectron microscopy. J. Gen. Virol 74, 293–298. [DOI] [PubMed] [Google Scholar]
- (36).Karpf AR, and Brown DT (1998) Comparison of Sindbis virus-induced pathology in mosquito and vertebrate cell cultures. Virology 240, 193–201. [DOI] [PubMed] [Google Scholar]
- (37).Jose J, Taylor AB, and Kuhn RJ (2017) Spatial and temporal analysis of alphavirus replication and assembly in mammalian and mosquito cells. mBio 8, e02294–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Fuerstenau SD, and Benner WH (1995) Molecular weight determination of megadalton-DNA electrospray ions using charge detection mass spectrometry. Rapid Commun. Mass Spectrom 9, 1528–1538. [DOI] [PubMed] [Google Scholar]
- (39).Contino NC, and Jarrold MF (2013) Charge detection mass spectrometry for single ions with a limit of detection of 30 charges. Int. J. Mass Spectrom 345–347, 153–159. [Google Scholar]
- (40).Keifer DZ, Pierson EE, Hogan JA, Bedwell GJ, Prevelige PE, and Jarrold MF (2014) Charge detection mass spectrometry of bacteriophage P22 procapsid distributions above 20 MDa. Rapid Commun. Mass Spectrom 28, 483–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Keifer DZ, Motwani T, Teschke CM, and Jarrold MF (2016) Measurement of the accurate mass of a 50 MDa infectious virus. Rapid Commun. Mass Spectrom 30, 1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Keifer DZ, Pierson EE, and Jarrold MF (2017) Charge detection mass spectrometry: weighing heavier things. Analyst 142, 1654–1671. [DOI] [PubMed] [Google Scholar]
- (43).Doussineau T, Désert A, Lambert O, Taveau J-C, Lansalot M, Dugourd P, Bourgeat-Lami E, Ravaine S, Duguet E, and Antoine R (2015) Charge detection mass spectrometry for the characterization of mass and surface area of composite nanoparticles. J. Phys. Chem. C 119, 10844–10849. [Google Scholar]
- (44).Doussineau T, Mathevon C, Altamura L, Vendrely C, Dugourd P, Forge V, and Antoine R (2016) Mass determination of entire amyloid fibrils by using mass spectrometry. Angew. Chem., Int. Ed 55, 2340–2344. [DOI] [PubMed] [Google Scholar]
- (45).Elliott AG, Harper CC, Lin H-W, and Williams ER (2017) Mass, mobility and MSn measurements of single ions using charge detection mass spectrometry. Analyst 142, 2760–2769. [DOI] [PubMed] [Google Scholar]
- (46).Elliott AG, Harper CC, Lin H-W, Susa AC, Xia Z, and Williams ER (2017) Simultaneous measurements of mass and collisional cross-section of single ions with charge detection mass spectrometry. Anal. Chem 89, 7701–7708. [DOI] [PubMed] [Google Scholar]
- (47).Lu J, Trnka MJ, Roh SH, Robinson PJ, Shiau C, Fujimori DG, Chiu W, Burlingame AL, and Guan S (2015) Improved peak detection and deconvolution of native electrospray mass spectra from large protein complexes. J. Am. Soc. Mass Spectrom 26, 2141–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Pierson EE, Contino NC, Keifer DZ, and Jarrold MF (2015) Charge detection mass spectrometry for single ions with an uncertainty in the charge measurement of 0.65 e. J. Am. Soc. Mass Spectrom 26, 1213–1220. [DOI] [PubMed] [Google Scholar]
- (49).Keifer DZ, Shinholt DL, and Jarrold MF (2015) Charge detection mass spectrometry with almost perfect charge accuracy. Anal. Chem 87, 10330–10337. [DOI] [PubMed] [Google Scholar]
- (50).Casjens S, and Hayden M (1988) Analysis in vivo of the bacteriophage P22 headful nuclease. J. Mol. Biol 199, 467–474. [DOI] [PubMed] [Google Scholar]
- (51).Paredes AM, Simon MN, and Brown DT (1992) The mass of the Sindbis virus nucleocapsid suggests it has T = 4 icosahedral symmetry. Virology 187, 329–332. [DOI] [PubMed] [Google Scholar]
- (52).Hsieh P, Rosner MR, and Robbins PW (1983) Host dependent variation of asparagine-linked oligosaccharides at individual glycosylation sites of Sindbis virus glycoproteins. J. Biol. Chem 258, 2548–2554. [PubMed] [Google Scholar]
- (53).Stanley P, Schachter H, and Taniguchi N (2009) N-Glycans, In Essentials of Glycobiology (Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, and Etzler ME, Eds.) 2nd ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, Chapter 8. [PubMed] [Google Scholar]
- (54).Lancaster C, Pristatsky P, Hoang VM, Casimiro DR, Schwartz RM, Rustandi R, and Ha S (2016) Characterization of N-glycosylation profiles from mammalian and insect cell derives chikungunya VLP. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci 1032, 218–223. [DOI] [PubMed] [Google Scholar]
- (55).Mayne JT, Bell JR, Strauss EG, and Strauss JH (1985) Pattern of glycosylation of Sindbis virus envelope proteins synthesized in hamster and chicken cells. Virology 142, 121–133. [DOI] [PubMed] [Google Scholar]
- (56).Prasad BV, Hardy ME, Dokland T, Bella J, Rossmann MG, and Estes MK (1999) X-ray crystallographic structure of the Norwalk virus capsid. Science 286, 287–290. [DOI] [PubMed] [Google Scholar]
- (57).von Loeffelholz O, Natchiar SK, Djabeur N, Myasnikov AG, Kratzat H, Ménétret JF, Hazemann I, and Klaholz BP (2017) Focused classification and refinement in high-resolution cryo-EM structural analysis of ribosome complexes. Curr. Opin. Struct. Biol 46, 140–148. [DOI] [PubMed] [Google Scholar]
- (58).Therkelsen MD, Klose T, Vago F, Jiang W, Rossmann MG, and Kuhn RJ (2018) Flaviviruses have imperfect icosahedral symmetry. Proc. Natl. Acad. Sci. U. S. A 115, 11608–11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Wang JC, Mukhopadhyay S, and Zlotnick A (2018) Geometric defects and icosahedral viruses. Viruses 10, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Strauss E, and Strauss J (1973) Togaviruses, In The Molecular Biology of Animal Viruses, pp 112–123, Marcel Dekker, New York [Google Scholar]
- (61).Guillou H, Zadravec D, Martin PG, and Jacobsson A (2010) The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res 49, 186–199. [DOI] [PubMed] [Google Scholar]
- (62).Sousa IP, Carvalho CAM, Ferreira DF, Weissmüller G, Rocha GM, Silva JL, and Gomes AMO (2011) Envelope lipid-packing as a critical factor for the biological activity and stability of alphavirus particles isolated from mammalian and mosquito cells. J. Biol. Chem 286, 1730–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Kielian M, Chatterjee PK, Gibbons DL, and Lu YE (2000) Specific roles for lipids in virus fusion and exit examples from the alphaviruses, In Subcellular Biochemistry, Vol. 34, pp 409–455, Springer, Boston, MA: [DOI] [PubMed] [Google Scholar]
- (64).Hafer A, Whittlesey R, Brown DT, and Hernandez R (2009) Differential incorporation of cholesterol by Sindbis virus grown in mammalian or insect cells. J. Virol 83, 9113–9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Sokoloski KJ, Hayes CA, Dunn MP, Balke JL, Hardy RW, and Mukhopadhyay S (2012) Sindbis virus infectivity improves during the course of infection in both mammalian and mosquito cells. Virus Res 167, 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Hernandez R, and Brown DT (2010) Growth and maintenance of mosquito cell lines. Curr. Protoc. Microbiol 17, A.4J.1–A.4J.8. [DOI] [PubMed] [Google Scholar]
- (67).Lustig S, Jackson AC, Hahn CS, Griffin DE, Strauss EG, and Strauss JH (1988) Molecular basis of Sindbis virus neurovirulence in mice. J. Virol 62, 2329–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Sokoloski KJ, Snyder AJ, Liu NH, Hayes CA, Mukhopadhyay S, and Hardy RW (2013) Encapsidation of host-derived factors correlates with enhanced infectivity of Sindbis virus. J. Virol 87, 12216–12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Contino NC, Pierson EE, Keifer DZ, and Jarrold MF (2013) Charge detection mass spectrometry with resolved charge states. J. Am. Soc. Mass Spectrom 24, 101–108. [DOI] [PubMed] [Google Scholar]
- (70).Pierson EE, Keifer DZ, Contino NC, and Jarrold MF (2013) Probing higher order multimers of pyruvate kinase with charge detection mass spectrometry. Int. J. Mass Spectrom 337, 50–56. [Google Scholar]
- (71).Maley F, Trimble RB, Tarentino AL, and Plummer TH (1989) Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem 180, 195–204. [DOI] [PubMed] [Google Scholar]
- (72).Mechref Y (2012) Use of CID/ETD mass spectrometry to analyze glycopeptides. Curr. Protoc. Protein Sci 68, 12.11.1–12.11.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Galaz-Montoya JG, Flanagan J, Schmid MF, and Ludtke SJ (2015) Single particle tomography in EMAN2. J. Struct. Biol 190, 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Yan X, Sinkovits RS, and Baker TS (2007) AUTO3DEM–an automated and high throughput program for image reconstruction of icoashedral particles. J. Struct. Biol 157, 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- (76).Leishman E, Murphy M, Mackie K, and Bradshaw HB (2018) Delta(9)-Tetrahydrocannabinol changes the brain lipidome and transcriptome differentially in the adolescent and the adult. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 1863, 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Stuart JM, Paris JJ, Frye C, and Bradshaw HB (2013) Brain levels of prostaglandins, endocannabinoids, and related lipids are affected by mating strategies. Int. J. Endocrinol 2013, 436252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Tortoriello G, Rhodes BP, Takacs SM, Stuart JM, Basnet A, Raboune S, Widlanski TS, Harkany T, and Bradshaw HB (2013) Targeted lipidomics in Drosophila melanogaster identifies novel 2-monoacylglycerols and N-acyl amides. PLoS One 8, e67865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.