Abstract
Processing and maturation of precursor RNA species is coupled to RNA polymerase II transcription. Co‐transcriptional RNA processing helps to ensure efficient and proper capping, splicing, and 3′ end processing of different RNA species to help ensure quality control of the transcriptome. Many improperly processed transcripts are not exported from the nucleus, are restricted to the site of transcription, and are in some cases degraded, which helps to limit any possibility of aberrant RNA causing harm to cellular health. These critical quality control pathways are regulated by the highly dynamic protein–protein interaction network at the site of transcription. Recent work has further revealed the extent to which the processes of transcription and RNA processing and quality control are integrated, and how critically their coupling relies upon the dynamic protein interactions that take place co‐transcriptionally. This review focuses specifically on the intricate balance between 3′ end processing and RNA decay during transcription termination.
This article is categorized under:
RNA Turnover and Surveillance > Turnover/Surveillance Mechanisms
RNA Processing > 3' End Processing
RNA Processing > Splicing Mechanisms
RNA Processing > Capping and 5' End Modifications
Keywords: 3′ end processing, capping, co‐transcriptional, quality control, RNA processing, RNAPII, splicing, termination, transcription
1. INTRODUCTION
RNA polymerase II (RNAPII) is one of three major eukaryotic DNA‐dependent RNA polymerases and is responsible for transcribing several RNA species, including messenger RNA (mRNA), noncoding RNA (ncRNA), and small nuclear/nucleolar RNAs (sn/snoRNAs) (Tan‐Wong et al., 2012; Wyers et al., 2005). Plants have two additional RNA polymerases which were derived from RNAPII that have been reviewed in depth previously (Haag & Pikaard, 2011; Zhou & Law, 2015). Transcription by RNAPII consists of three basic phases: initiation, RNAPII recruitment to the promoter, and synthesis of the first few RNA nucleotides; elongation, RNAPII moving further into the gene in the context of chromatin and extending the nascent RNA transcript; and termination, release of RNAPII and the fully processed nascent RNA transcript from the template DNA. Each of these steps requires a distinct set of proteins that aid and regulate RNAPII to ensure proper transcription and gene expression.
The majority of RNAPII transcripts need to be processed before serving their cellular purpose, and their class‐specific processing occurs in tandem with the transcription of nascent RNA. RNA processing machinery is recruited to RNAPII at the site of transcription and the success of processing is inherently linked to the progression of transcription and regulatory proteins the two processes share. The nature and extent of processing depends on the species of RNA, but the major processes are: 5′ end capping (Cho, Takagi, Moore, & Buratowski, 1997; McCracken et al., 1997), splicing (Carrillo Oesterreich, Preibisch, & Neugebauer, 2010; Misteli & Spector, 1999), and 3′ end processing (Ahn, Kim, & Buratowski, 2004; Kim, Ahn, Krogan, Greenblatt, & Buratowski, 2004; Licatalosi et al., 2002). Under normal biological conditions, if RNA is not properly co‐transcriptionally processed, it will be degraded often at or near the site of transcription. Although not often thought of in the context of quality control, proper processing ensures that the necessary transcripts are able to be exported from the nucleus and translated and is therefore essential for the maintenance of the transcriptome, and ultimately the proteome. Because it is so critical in determining the fate of nascent RNA, co‐transcriptional processing mechanisms and their regulation will be highlighted in this review as primary steps in quality control. RNA transcripts that contain transcription errors or are improperly processed pose a threat to cell health. If these erroneous and/or unprocessed transcripts are not degraded, they can cause disease through multiple mechanisms such as nonfunctional protein expression, DNA damage through R‐loop accumulation, and downregulation of functional protein expression through nuclear and/or cytoplasmic mRNA decay pathways (Bresson & Tollervey, 2018). Degradation of erroneous transcripts in the nucleus is an important quality control strategy, and its intricate relationship with RNA processing will be prominently featured in this review.
The proteins that regulate co‐transcriptional processing and degradation need to be recruited to the site of transcription at the appropriate time and perhaps withdraw immediately after their purpose is served to clear the way for the next wave of RNAPII interacting proteins. Proteins move on and off RNAPII during the transcription cycle, creating a highly dynamic interaction network at the site of transcription that governs the quality of the transcribed RNA (Kim, Ahn, et al., 2004; Mayer et al., 2010, 2012). This flexible precision is critical to ensure quality control of nascent transcripts in order to maintain proper gene expression. Altogether, this review will focus on the major protein groups that regulate quality control of RNA through balancing co‐transcriptional RNA processing with degradation during eukaryotic RNAPII transcription termination. For more information on posttranscriptional RNA quality control refer to: Halbeisen, Galgano, Scherrer, and Gerber (2008), Inada (2013), and/or Schaefke, Sun, Li, Fang, and Chen (2018).
2. RECRUITMENT 101: UTILIZING THE C‐TERMINAL DOMAIN AND PAUSING
2.1. Phosphorylation of the RNAPII CTD
RNAPII is distinguished from RNA polymerases I and III by the conserved C‐terminal domain (CTD) of its largest subunit, Rpb1, which plays an integral role in the recruitment of proteins to RNAPII. The CTD consists of repeats of the peptide Y1S2P3T4S5P6S7, and is conserved from fungi to humans with variations in repeat number (Ahearn, Bartolomei, West, Cisek, & Corden, 1987; Eick & Geyer, 2013; Stiller & Hall, 2002; Yang, Hager, & Stiller, 2014). Five out of seven of the residues in this repeat can be phosphorylated, and the dynamic phosphorylation status of these residues throughout the transcription cycle is responsible for the specific recruitment of numerous regulatory proteins. Proteins with CTD‐binding domains have binding affinity preferences for different phosphorylation patterns; as the pattern is modified throughout the transcription cycle, the CTD acts as a “landing pad” (Buratowski, 2003, 2009; Greenleaf, 1993) for the dynamic interactors at the site of transcription. Indeed, deletion of the RNAPII CTD has been shown to inhibit co‐transcriptional processing of nascent RNA (Fong & Bentley, 2001; McCracken et al., 1997).
RNAPII exists in a hypo‐phosphorylated state when it is recruited to the promoter by the preinitiation complex. However, very early in transcription CTD serine 5 (Ser5) levels rise and peak in early elongation. Ser5 phosphorylation is linked to recruiting both elongation factors and RNA processing proteins, such as the 5′ end capping complex (Ghosh, Shuman, & Lima, 2011; Komarnitsky, Cho, & Buratowski, 2000) and the spliceosome (Nojima et al., 2018). Ser5 levels have also been shown to peak at actively spliced exons (Nojima et al., 2015) helping to recruit the spliceosome and regulate splicing (Harlen et al., 2016). As elongation proceeds, Ser5 phosphorylation levels fall, while serine 2 (Ser2) levels rise. This shift in the phosphorylation dynamic is integral in continued regulation of splicing, and recruitment of transcription termination factors and proteins involved in 3′ end processing and polyadenylation of the nascent RNA (Ahn et al., 2004; Davidson, Muniz, & West, 2014). Phosphorylation of both Tyr‐1 and Thr‐4 in the CTD repeats has also been shown to play a regulatory role in RNAPII transcription termination suggesting that this step, which is intimately coupled to mRNA 3′ end processing, is tightly regulated by the CTD phosphorylation state (Hsin, Sheth, & Manley, 2011; Mayer et al., 2012; Nemec et al., 2017; Schreieck et al., 2014). Finally, Ser‐7 phosphorylation has been implicated in snRNA processing and recruitment of the Integrator complex (Egloff et al., 2007). Thus, the CTD has been shown to activate all three major RNA processing pathways, fitting in with the “recruitment model” (Bentley, 2014) of coupling RNA processing to transcription. Zaborowska, Egloff, & Murphy (2016) review the “CTD code” in depth.
2.2. Pausing of RNAPII
The “kinetic competition model” poses that as transcription occurs and the nascent RNA is extended, RNA‐binding proteins are able to bind the transcript in a sequence‐specific (or nonspecific) manner (Dujardin et al., 2013; Nilsen & Graveley, 2010). The rate at which these binding sequences within the nascent RNA are synthesized during transcription elongation may allow competition between RNA‐binding regulatory proteins that play important roles in RNA processing, RNAPII termination, and quality control checkpoints (Roberts, Gooding, Mak, Proudfoot, & Smith, 1998). Since the rate of transcription has an impact on the kinetic recruitment of proteins to the site of transcription, an important consideration is RNAPII pausing. Pausing, or accumulation of RNAPII on DNA, can occur at any stage of transcription and provide a window of opportunity for recruitment of factors involved in the modulation of gene expression and co‐transcriptional RNA processing (Henriques et al., 2013; Svejstrup, 2007). RNAPII pausing is associated with the three major co‐transcriptional processing pathways, RNA capping (Rasmussen & Lis, 1993), splicing (Alexander, Innocente, Barrass, & Beggs, 2010), and 3′ end processing, as well as RNA proofreading and degradation (Glover‐Cutter, Kim, Espinosa, & Bentley, 2008; Kireeva et al., 2008; Nudler, 2012). This coupling of RNA processing to RNAPII activity may ensure that the nascent RNA is protected from degradation and efficiently matures into a functional mRNA (Figure 1).
Figure 1.
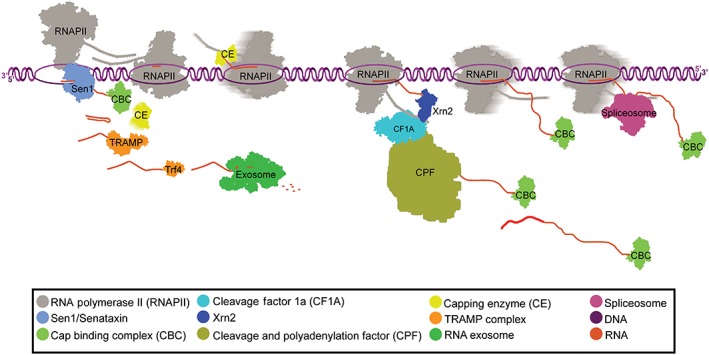
RNA processing and degradation: Processing and degradation factors are recruited to the site of transcription. Shown are two polymerases moving in opposite directions demonstrating each of the major RNA processing pathways (as described in the key)
In metazoans, RNAPII pausing proximal to the promoter is a regulatory step in transcription for the majority of protein coding genes (Muse et al., 2007; Nechaev et al., 2010) and is thought to be used as a rate‐limiting elongation checkpoint that can hold back RNAPII to give a “window of opportunity” for the recruitment of factors needed for transcription elongation and/or co‐transcriptional RNA processing (Adelman & Lis, 2012; Henriques et al., 2013; Valen et al., 2011). Pausing may provide a timing opportunity and an interaction surface to facilitate capping, as interactions have been reported between the capping machinery and the pause‐regulatory factor/positive transcription elongation factor, DSIF (Spt4/5) (Adelman & Lis, 2012; Mandal et al., 2004; Moore, Schwartzfarb, Silver, & Yu, 2006). Positive elongation factor b (P‐TEFb) has been shown to phosphorylate DSIF and NELF to trigger pause release. The regulation of promoter proximal pause release may ensure that RNAPII does not proceed into productive elongation before it is appropriately modified for binding by the RNA processing factors and may provide a binding platform later in transcription for complexes carrying out 3′ end processing (Buratowski, 2009). Of note, recent findings have suggested that RNAPII release from pausing at highly abundant promoter proximal pause sites may occur through premature transcription termination up to 99% of the time (Erickson, Sheridan, Cortazar, & Bentley, 2018; Steurer et al., 2018). These findings suggest that termination is the predominant form of RNAPII removal from promoter proximal pause sites rather than release into productive elongation.
It is possible that the promoter proximal pause site could serve as a key location for mRNA 5′ capping quality control. This possibility is supported in elegant work showing that inhibition of the Cdk9 kinase in Schizosaccharomyces pombe leads to significant decreases in the phosphorylation of the elongation factor Spt5, which has been implicated in both promoter proximal pausing and the recruitment of capping enzyme (CE) components (Booth, Parua, Sansó, Fisher, & Lis, 2018; Pei, Schwer, & Shuman, 2003; Viladevall et al., 2009). The biological mechanisms that could underlie a large degree of RNAPII turnover at promoter proximal pause sites have not been explored, but this topic remains controversial since other groups have reported measurement of stable RNAPII pausing as a potential mechanism to poise the transcription machinery for rapid induction. Regardless, rapid RNAPII removal from promoter proximal pause sites could involve unique termination mechanisms such as potential termination coupled RNA degradation by the exosome, which to date has not been explored in metazoan cells (Fox, Gao, Smith‐Kinnaman, Liu, & Mosley, 2015; Lemay et al., 2014). RNAPII has also been shown to pause at a variety of splice sites throughout the genome and also once it reaches the 3′ end of genes and various polyadenylation sites (Kwak, Fuda, Core, & Lis, 2013; Mayer et al., 2015; Nojima et al., 2015). Pausing is an important intermediate step leading to termination in many mammalian genes, providing the opportunity for the termination machinery to be recruited to chromatin (Andrulis et al., 2002; Gusarov & Nudler, 1999). Additionally, it has been shown that RNAPII pausing sites change positions when alternative polyadenylation sites are used in cells (Fusby et al., 2016). Details regarding the role of pausing during each processing step are provided in their respective sections. Pausing and its effects on transcription have recently been reviewed in Adelman and Lis (2012), Chen, Smith, and Shilatifard (2018), and Mayer, Landry, and Churchman (2017).
3. THE RNA EXOSOME: THE CLEAN‐UP CREW
Proper maturation and processing of the mRNA lends protective features to the transcripts that keep them from being degraded. The major quality control mechanism for aberrant RNA is degradation. However, degradation is not solely restricted to incorrectly made or processed RNAs, as it also occurs during routine processing in the case of many ncRNAs. Arguably, the most significant role for degradation in RNA quality control is its role in the removal of RNAs produced from pervasive transcription. There are both nuclear and cytoplasmic RNA degradation systems, but this review will only focus on the nuclear mechanisms due to their connection to transcription and co‐transcriptional RNA processing.
Processing transcripts while they are still attached to chromatin provides additional checkpoints for the removal of unprocessed and potentially deleterious transcripts and avoids wasteful transcription. The RNA exosome is a multi‐subunit 3′–5′ exonuclease complex that has been shown to have a large number of regulatory roles in RNA biology including, 3′ end processing and the degradation of ncRNAs and unstable transcripts (Allmang et al., 1999; Chlebowski, Lubas, Jensen, & Dziembowski, 2013; Mitchell, Petfalski, Shevchenko, Mann, & Tollervey, 1997). The exosome is responsible for degrading mRNA transcripts that have been improperly processed (Bitton et al., 2015; Bousquet‐Antonelli, Presutti, & Tollervey, 2000; Gudipati et al., 2012; Schneider, Kudla, Wlotzka, Tuck, & Tollervey, 2012; Szczepinska et al., 2015), including: lncRNAs (Pefanis et al., 2015; Wlotzka, Kudla, Granneman, & Tollervey, 2011), cryptic unstable transcripts (CUTs) (Davis & Ares, 2006; Szczepinska et al., 2015; Wyers et al., 2005) and their human counterparts promoter upstream transcripts (PROMPTs) (Preker et al., 2008, 2011), and heterochromatin‐forming repetitive elements such as rRNA and centromeres (Buhler, Haas, Gygi, & Moazed, 2007; Houseley, Kotovic, El Hage, & Tollervey, 2007; Vasiljeva, Kim, Terzi, Soares, & Buratowski, 2008). Defective transcripts that are destined for exosomal degradation often accumulate and result in the retention of RNAPII at the site of transcription (de Almeida, Garcia‐Sacristan, Custodio, & Carmo‐Fonseca, 2010; Eberle et al., 2010; Hilleren & Parker, 2001). Exosome‐dependent degradation at the site of transcription could have an array of consequences such as increases in the local concentrations of nucleotides, which could facilitate RNAPII transcription at nearby genes.
The exosome has two catalytic subunits responsible for degradation of RNA, Dis3 and Rrp6, with Rrp6 able to function independently of the exosome core particle (Chlebowski et al., 2013). However, the exosome requires all subunits plus cofactors for optimal activity and appropriate substrate selection. Four such cofactors are the Trf4/5‐Air1/2‐Mtr4 polyadenylation (TRAMP) complex, the yeast Nrd1‐Nab3‐Sen1 (NNS) complex, and the human nuclear exosome targeting (NEXT) and poly‐A tail exosome targeting (PAXT) complexes. The exosome and its cofactors were recently reviewed in Zinder and Lima (2017). The TRAMP complex interacts with the exosome to increase both the hydrolytic activity of Rrp6 (Callahan & Butler, 2010) and to add short polyA tails to RNA substrates in order to make them more accessible for degradation (Schmidt & Butler, 2013). TRAMP function is generally coupled to that of the NNS complex (Arigo, Eyler, Carroll, & Corden, 2006; Schulz et al., 2013; Thiebaut, Kisseleva‐Romanova, Rougemaille, Boulay, & Libri, 2006), which is further discussed later in this review. The exosome has an established role in posttranscriptional quality control (Lemieux et al., 2011; Schneider et al., 2012; Wang, Stevenson, Kearsey, Watt, & Bahler, 2008), but there is also evidence showing that Rrp6 and the exosome are recruited to transcribed genes (Andrulis et al., 2002; Hessle, von Euler, Gonzalez de Valdivia, & Visa, 2012; Hieronymus, Yu, & Silver, 2004; Lim, Boyle, Chinen, Dale, & Lei, 2013), and that the exosome interacts with elongating RNAPII in metazoans (Andrulis et al., 2002).
Transcriptome analysis of quality control mutants revealed an important role for nucleases in removing aberrant mRNA species (Davis & Ares, 2006; Gudipati et al., 2012; Schneider et al., 2012; Wyers et al., 2005). The accumulation of RNA as a result of exosome mutations can have a variety of negative effects on the transcriptome. Aberrant RNA accumulation can negatively impact the cell in multiple ways that include: competing with properly processed RNA for RNA binding proteins (Coy, Volanakis, Shah, & Vasiljeva, 2013), activating antiviral defense mechanisms that trigger autoimmunity (Eckard et al., 2014), or producing DNA–RNA hybrids (R‐loops) that induce double‐stranded DNA breaks and chromatin instability (Wahba, Gore, & Koshland, 2013). Mutations in the exosome have been linked to a variety of human pathologies, particularly to spinal motor neuron disorders such as pontocerebellar hypoplasia type 1B (Boczonadi et al., 2014; J. Wan et al., 2012). It has also been seen that Rrp6 mediates the transcriptional silencing of HIV‐1 promoter and that the loss of Rrp6 function leads to a loss in inhibition of the HIV‐1 promoter (Wagschal et al., 2012). Additionally, a mouse model with a mutation in the exosome presents with B lymphocyte dysfunction (Pefanis et al., 2014). It has been postulated that the phenotype is due to the fact that the exosome is required for class switch recombination and somatic hypermutation, both of which are necessary for antibody diversity. The multitude of disease phenotypes produced from defects in the exosome highlights the importance of proper quality control of nascent RNA transcripts. The following sections will highlight the different RNA processing reactions and their interconnectedness with degradation for proper RNA quality control. There are many recent reviews that cover RNA degradation and decay in more detail, and a selection is listed here: Bresson and Tollervey (2018), Karousis, Nasif, and Muhlemann (2016), Palumbo, Farina, and Paci (2015), Schmid and Jensen (2018), and Schmidt and Butler (2013).
4. IN BRIEF: 5′ CAPPING AND SPLICING
4.1. Co‐transcriptional capping and the initiation–elongation transition
Shortly after the 5′ end of the nascent transcript emerges from the RNA exit channel, a 7‐methyl guanosine cap is added (Perales & Bentley, 2009; Rasmussen & Lis, 1993). There are three steps to the capping process: conversion of the tri‐phosphate group at the 5′ end of the nascent transcript to a diphosphate group; attachment of a GMP molecule; and methylation of the N7 atom of the guanosine base to produce a mature cap (Furuichi & Shatkin, 2000; Shuman, 1995). In yeast, two proteins (Cet1 and Ceg1) form a heterodimer capping complex, while mammals have one bifunctional enzyme for the first two steps (Itoh, Yamada, Kaziro, & Mizumoto, 1987; Yue et al., 1997). In the nucleus, the 5′ cap protects against 5′–3′ exonucleolysis (C. J. Wilusz, Wormington, & Peltz, 2001) and plays roles in processes such as pre‐mRNA splicing (Izaurralde et al., 1994), 3′ end formation (Flaherty, Fortes, Izaurralde, Mattaj, & Gilmartin, 1997), and RNA export (Izaurralde et al., 1995). A combination of cryo‐electron microscopy (Cryo‐EM) and crosslinking mass spectrometry (XL‐MS) determined the open and closed states of the CE in yeast and illuminated the position CE takes at the end of the RNAPII exit tunnel with its active sites facing the nascent RNA (Martinez‐Rucobo et al., 2015). Their model provides a structural basis for understanding how capping continuously protects the 5′ end of the RNA from exonucleases. For more information on the function of the cap and cap‐binding proteins refer to Cougot, van Dijk, Babajko, and Seraphin (2004) and Topisirovic, Svitkin, Sonenberg, and Shatkin (2011).
Nuclear decay systems take advantage of co‐transcriptional RNA processing to assess the quality of nascent RNA and degrade any nonoptimal transcripts (Schmid & Jensen, 2018). Mutations that lead to improper capping, whether directly or indirectly, result in mRNA decay and can cause premature termination of transcription (Jimeno‐Gonzalez, Haaning, Malagon, & Jensen, 2010). These degradative quality control mechanisms are known to involve the 5′–3′ exonuclease Xrn2 (also known as Rat1), the decapping endonuclease Rai1 (also known as Dxo), and the Rai1 homolog Dxo1 that has both decapping and 5′–3′ exoRNAse activity (Brannan et al., 2012; Chang et al., 2012; Jiao et al., 2010; S. Xiang et al., 2009). Rai1 is already known to bind and stimulate the activity of Xrn2 for RNAPII transcription termination (Kim et al., 2004; Xue et al., 2000). Rai1 has been shown to convert the 5′‐triphosphate into monophosphates that can target these transcripts Xrn2 for degradation (Jiao et al., 2010; S. Xiang et al., 2009). The mammalian Rai1 homolog, DOXO, has pyrophosphatase, decapping, and exoribonuclease activity, and has been shown to have the ability to prepare both uncapped and unmethylated‐capped RNAs for degradation (Jiao, Chang, Kilic, Tong, & Kiledjian, 2013; S. Xiang et al., 2009). Co‐deletion of yeast Rai1 and Dxo1 leads to accumulation of incompletely capped RNAs; depletion of the human homolog DXO leads to accumulation of aberrantly capped, unspliced, and inefficiently 3′ end cleaved RNAs (Chang et al., 2012; Jiao et al., 2013). So, it is thought that Rai1 and Dxo1 play critical roles in terminating RNAPII and degrading improperly capped nascent transcripts (Chang et al., 2012; S. Xiang et al., 2009). However, it is unknown how this process precisely intersects with the control of promoter proximal pause release and/or premature RNAPII termination. The process of decapping is also a major regulatory step for mRNA degradation, but occurs in the cytoplasm, and so it is not extensively reviewed here, but is addressed in the following publications: Coller and Parker (2004), Franks and Lykke‐Andersen (2008), Grudzien‐Nogalska and Kiledjian (2017), Song, Li, and Kiledjian (2010), and Zhai and Xiang (2014).
4.2. Co‐transcriptional splicing
The splicing of introns out of mRNA occurs at many genes in eukaryotes coordinately with RNAPII transcription elongation (Carrillo Oesterreich et al., 2016). Alternative splice variants provide the opportunity for expanded diversity within the proteome without extensive genomic expansion. In order to ensure proper splicing, this processing event is often directly coupled to transcription. Electron microscopy studies in both Drosophila melanogaster and Chironomus tentans demonstrated that splicing does occur co‐transcriptionally (Bauren & Wieslander, 1994; Beyer & Osheim, 1988). Sequencing experiments in human (Ameur et al., 2011; Tilgner et al., 2012; Windhager et al., 2012), mouse (Khodor, Menet, Tolan, & Rosbash, 2012), yeast (Carrillo Oesterreich et al., 2010), and fly (Khodor et al., 2011) model systems have also shown that an extensive amount of splicing occurs co‐transcriptionally. In their 2018 paper, Burke et al. (2018) developed a sequencing method to globally profile spliceosome‐bound pre‐mRNA, intermediates, and spliced mRNA at single nucleotide resolution through combining biochemical purification of endogenous spliceosomes sequencing. This new method provides a tool for quantifiable studies of previously hard to identify RNA species and allowed measurement of differential splicing between three yeast species. Additionally, this method provides the ability to investigate splicing regulation through intron retention, which was previously not possible due to the transient nature of these RNA species and mutations that had to be made for stabilization. Further advances in our understanding of splicing have occurred through recently solved structures characterizing the spliceosome in different forms (Agafonov et al., 2016; Bertram, Agafonov, Dybkov, et al., 2017; Bertram, Agafonov, Liu, et al., 2017; Finci et al., 2018; Galej et al., 2016; Nguyen et al., 2015, 2016; Ohi, Ren, Wall, Gould, & Walz, 2007; Pomeranz Krummel, Oubridge, Leung, Li, & Nagai, 2009; R. Wan et al., 2016; Yan et al., 2015). More in‐depth reviews of structural studies of the spliceosome can be found in Fica and Nagai (2017) and Nguyen et al. (2016). Although not all splicing occurs co‐transcriptionally, it is likely that most spliceosomes assemble on the nascent transcript (Pandya‐Jones & Black, 2009) with support from co‐immunoprecipitation studies using antibodies specific for active spliceosomes that showed 80% of active spliceosomes to be bound to chromatin in HeLa cells (Girard et al., 2012). Herzel, Ottoz, Alpert, and Neugebauer (2017) recently reviewed co‐transcriptional spliceosome assembly and function.
Intron‐containing mRNA is particularly prone to nuclear degradation (Kilchert et al., 2015). Mutations that delay splicing exacerbate this effect, and so unspliced or improperly spliced pre‐mRNAs have been shown to be rapidly degraded (Danin‐Kreiselman, Lee, & Chanfreau, 2003; Gudipati et al., 2012; Lemieux et al., 2011). However, the precise mechanisms that target intron containing mRNA for degradation and the factors involved in this nuclear quality control pathway are still unknown (Bresson & Tollervey, 2018). Failure to splice pre‐mRNA may trigger any of the following responses in addition to others: transcription downregulation (Damgaard et al., 2008); failure of mRNA export (M. J. Luo & Reed, 1999; Reed & Hurt, 2002); pre‐mRNA degradation from the 5′ and 3′ ends by either Xrn2 and the nuclear exosome (Bousquet‐Antonelli et al., 2000; Kufel, Bousquet‐Antonelli, Beggs, & Tollervey, 2004) or via endonucleolysis by Swt1 (Skruzny et al., 2009); and posttranscriptional anchoring of the unspliced pre‐mRNA at the NPC via the Mlp proteins (Galy et al., 2004). It has also been shown that defects in snRNP assembly lead to degradation by two distinct quality control mechanisms, either through the actions of the TRAMP complex and the RNA exosome or via decapping and 5′ to 3′ decay by Xrn1 (Shukla & Parker, 2014). Additionally, depletion of either core splicing or transcription termination components in Drosophila has been shown to increase the production of circular RNAs which occur through backsplicing reactions (Liang et al., 2017). Considering that many splicing reactions have been shown to occur efficiently during transcription elongation, backsplicing should be a rare event that occurs only when splicing rates are slowed to an extent allowing production of a downstream splice site donor before removal of an upstream intron with an intact splice acceptor (J. E. Wilusz, 2018). While backsplicing could be a normal scenario for some poorly spliced transcripts, the creation of circular RNAs may also serve as a fail‐safe and/or quality control mechanism for a larger number of mRNAs when splicing and/or termination pathways are disrupted. This review does not delve in depth into the mechanisms of splicing, however, the following reviews on this topic are recommended: Kaida (2016), Sperling (2017), J. E. Wilusz (2018), and Woodward, Mabin, Gangras, and Singh (2017).
5. THE END OF THE LINE: 3′ END PROCESSING AND DEGRADATION
The mechanisms of termination of RNAPII transcription are still not fully understood in eukaryotes although there is a wide array of knowledge on the factors that are required for termination to occur (Fong et al., 2015; Zhang, Rigo, & Martinson, 2015). Termination of transcription occurs when RNAPII stops nucleotide addition to the nascent RNA and both the RNA transcript and RNAPII are released from the DNA template. Termination is highly dynamic and can occur at multiple sites within a single gene and is also coupled to mRNP export (Gilbert & Guthrie, 2004; Johnson, Cubberley, & Bentley, 2009; W. Luo, Johnson, & Bentley, 2006). In order to be dynamic and flexible, termination is an extremely complex process with pathways that depend on numerous regulatory proteins (>100 proteins involved at protein coding genes in humans) (Shi et al., 2009). These multiple pathways for termination involve many proteins required for both RNAPII termination and RNA processing (Arndt & Reines, 2015; Porrua & Libri, 2015; Shi et al., 2009), as termination is inherently tied to co‐transcriptional mRNA 3′ end processing. Many termination factors interact with RNA processing and degradation enzymes and will be discussed below. The decision as to which termination pathway is utilized has a large influence on the future of an RNA: stabilization and protection versus degradation.
5.1. Cleavage and polyadenylation of mRNAs
The major 3′ end processing pathway used in the context of mRNAs, but also utilized for some ncRNAs, is cleavage followed by polyadenylation of the resulting 3′‐OH by polyA polymerase (Figure 2; Kuehner, Pearson, & Moore, 2011; Mischo & Proudfoot, 2013; K. Xiang, Tong, & Manley, 2014). Addition of the polyA tail is an important mechanism to regulate RNA processing, both for transcripts that will be exported and those fated for degradation (Box1). In metazoans, the polyA polymerase interacts with a protein complex known as the cleavage and polyadenylation specificity factor (CPSF) which contains subunits responsible for recognition of the AAUAAA hexanucleotide. The metazoan cleavage complex is also known to contain: cleavage stimulation factor, cleavage factor I, and cleavage factor II. Similarly, in yeast the cleavage and polyadenylation factor (CPF) is made up of various sub‐assemblies of protein complexes and interacts with a cleavage factor known as cleavage factor Ia. Recent Cryo‐EM, XL‐MS, and non‐covalent nanoelectrospray ionization mass spectrometry (nanoESI‐MS) studies of yeast CPF revealed a high degree of modularity within CPF, which has also been suggested for mammalian CPSF, by extensive biochemical analysis (Casanal et al., 2017; Hernández & Robinson, 2007). Three modules were identified for S. cerevisiae CPF: nuclease, phosphatase, and polyA polymerase. The polyA polymerase module is very similar to the minimal mammalian components of CPSF needed for polyadenylation, suggesting high conservation between the yeast and metazoan machinery (Schonemann et al., 2014). Several components of the full cleavage and polyadenylation factor‐cleavage factor (CPF‐CF) complex both bind to RNAPII and recognize termination and processing signals in the 3′ UTR of the nascent RNA (Baejen et al., 2014; Pearson & Moore, 2014; K. Xiang et al., 2014). RNAs polyadenylated by CPF‐CF are rapidly exported to the cytoplasm if they pass quality control steps (Mouaikel et al., 2013; Zenklusen, Larson, & Singer, 2008). Mutants in the CPF‐CF pathway in yeast can lead to accumulation of polyA RNA in the nucleus as a consequence of defective coupling of transcription termination and mRNA export (Amberg, Goldstein, & Cole, 1992; Hammell et al., 2002). Furthermore, it was recently shown in human cells that cleavage via the CPSF processing endonuclease CPSF73 is required for efficient termination of protein‐coding genes, as loss of CPSF73 activity lead to termination defects and increased read‐through transcription (Eaton et al., 2018). The increased read‐through transcription in mutants in the CPF complex can also lead to increased production of exonuclease‐resistant circular RNAs, discussed in more detail below (Liang et al., 2017).
Figure 2.
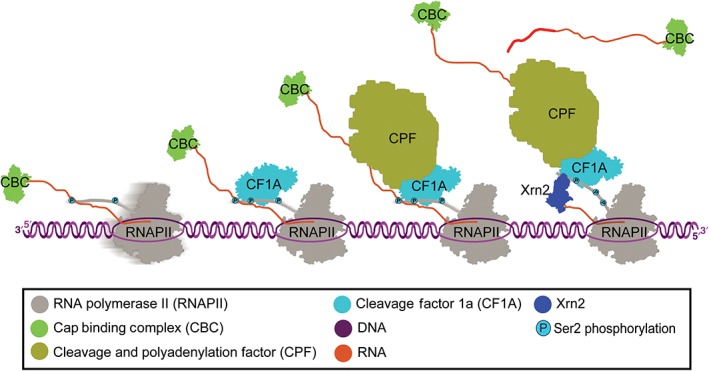
Cleavage and polyadenylation: Cap binding complex (CBC) binds 5′ guanosine cap. Cleavage factor 1A (CF1A) is recruited to Ser2 phosphorylated CTD of RNAPII. Cleavage and polyadenylation factor (CPF) is recruited and cleaves RNA after polyadenylation signal. Poly(A) polymerase polyadenylates 3′ end of RNA following cleavage. Xrn2 degrades 5′ end of uncapped RNA and removes RNAPII from the DNA template
BOX 1. POLY A BINDING PROTEIN NUCLEAR 1: A COMMON RESOURCE FOR RNA PROCESSING AND DECAY.
A good example of the interconnectedness of processing and degradation for quality control is polyA binding protein nuclear 1 (PABPN1). PABPN1 interacts with polyA polymerase to aid in the addition of the polyA tail to transcripts (Kerwitz et al., 2003). One way in which PAPBN1 functions in quality control of RNAs is through regulation of alternative cleavage and polyadenylation (APA) (Jenal et al., 2012). Similar to alternative splicing, APA can provide a way in which multiple transcripts can be produced from a single gene, with as many as 50% of human genes derived in this manner (Tian, Hu, Zhang, & Lutz, 2005). Moreover, usage of alternative polyadenylation sites can lead to differing lengths in 3′ untranslated regions (UTRs), which can have effects on mRNA stability, localization, and translation efficiency by changing targets for RNA binding proteins and miRNAs (Andreassi & Riccio, 2009; Fabian, Sonenberg, & Filipowicz, 2010). APA has been described in more detail in the following reviews Di Giammartino, Nishida, and Manley (2011), Elkon, Ugalde, and Agami (2013), Lutz and Moreira (2011), and Tian and Manley (2013). PABPN1 and polyA polymerases have also been shown to have a role in RNA decay via the exosome (Beaulieu, Kleinman, Landry‐Voyer, Majewski, & Bachand, 2012; Bresson, 2015; Bresson & Conrad, 2013; Lee & Glaunsinger, 2009). This functional connection between PABPN1 and the exosome has been further supported by the discovery of a protein that physically links PABN1 to the human NEXT complex (Meola et al., 2016). The fact that this PABN1 plays important roles in both 3′ end processing and targeting transcripts to the exosome highlights how quality control is managed through the balance between processing and degradation.
Mutations in 3′ end processing factors often cause decreases in transcript levels (Baejen et al., 2017; Hilleren & Parker, 2001; Libri et al., 2002; Milligan, Torchet, Allmang, Shipman, & Tollervey, 2005; Torchet et al., 2002). One reason for this decrease could be interference with polyA tail addition, exposing the mRNA to 3′‐exonucleocytic attack. This concept is supported by the fact that co‐deletion of exosome components with 3′ end processing mutants leads to restoration of stable, yet likely incorrectly processed, mRNAs (Burkard & Butler, 2000; Libri et al., 2002; Milligan et al., 2005; Pefanis et al., 2015; Tan‐Wong et al., 2012). The outcome of pre‐mRNA 3′ end formation is determined by relative efficiencies of polyadenylation versus RNA decay. It has been shown that polyA polymerase interacts with Rrp6 (Burkard & Butler, 2000) and the exosome might directly influence the activity of the polyA machinery (Milligan et al., 2005; Saguez et al., 2008). RNAPII pauses downstream of the polyA site to allow time for co‐transcriptional cleavage and polyadenylation (Glover‐Cutter et al., 2008; Gromak, West, & Proudfoot, 2006). It is possible that this pause favors backtracking, leading to a potential “reverse torpedo” mechanism to favor coupled termination and RNA degradation via the exosome (Fox et al., 2015; Lemay et al., 2014; Proudfoot, 2016). This model could be coupled with a posttranscriptional RNA cleavage and polyadenylation event to restrict exosome degradation (Bresson & Tollervey, 2018).
5.2. Two models for RNAPII release from the DNA template
Following cleavage and polyadenylation of the mRNA, RNAPII transcribes downstream ~150 nucleotides before being released from the DNA template (Creamer et al., 2011). The exact mechanisms responsible for RNAPII release during termination are still under investigation. It is proposed that pausing at the end of a transcription unit couples 3′ end processing to termination (Kuehner et al., 2011; Mayer et al., 2015; Nojima et al., 2015; Richard & Manley, 2009). Cleavage of the mRNA is important for RNAPII release from the template, as cleavage defective mutants tend to be impaired for termination, resulting in transcription read‐through phenotypes (Sadowski, Dichtl, Hubner, & Keller, 2003). There are two models proposed for release of RNAPII from the DNA: an allosteric model and a torpedo model. In the allosteric model, binding of the termination complex results in a conformational change in the elongation complex as elongation factors are lost, leading to a decrease in processivity (reviewed in Richard & Manley, 2009). Support for this is shown by the fact that RNAPII loses elongation factors before being released (Ahn et al., 2004; Baejen et al., 2017; Kim, Ahn, et al., 2004; Mayer et al., 2010). Also, in vitro studies have indicated that transcript cleavage, a key requirement for the torpedo model, is not required for RNAPII transcription termination (Zhang et al., 2015). In the torpedo model, cleavage of the 3′ end of the mRNA by CPF‐CF provides an entry point for the Rat1/Xrn2 5′–3′ exonuclease to degrade the nascent RNA up to RNAPII and displace the elongation complex (Brannan et al., 2012; Kim, Krogan, et al., 2004; Pearson & Moore, 2013; West, Gromak, & Proudfoot, 2004).
Support for the torpedo model has been shown in yeast and humans through the occurrence of termination read‐through transcription when Rat1 activity is defective, and through the use of in vitro termination assays with yeast Rat1 (Park, Kang, & Kim, 2015). However, Rat1 and its interacting partner Rai1 may be facilitated in vivo through the CTD binding activity of Rtt103, a CTD‐interaction domain protein that has been shown to recruit Rat1/Rai1 to 3′ end of genes to facilitate the dismantling of the elongation complex (Dengl & Cramer, 2009; Kim, Krogan, et al., 2004; Lunde et al., 2010; W. Luo et al., 2006). The human homologs of Rtt103, RPRD1a and 1b (also known as Kub5/Hera), have been implicated in a wide array of functions including interaction with Xrn2 and the serine 5 CTD phosphatase RPAP2 that has also been implicated in loss of RNAPII occupancy on protein coding genes in yeast (Hunter et al., 2016; Morales et al., 2014; Ni et al., 2014). A unified version of both the torpedo and allosteric models has also been proposed where a complex containing both Xrn2/Rat1 and CPF‐CF assembles at polyA sites mediating cleavage, nascent RNA degradation, and termination through an allosteric change in the elongation complex (Baejen et al., 2017; Lunde et al., 2010; W. Luo et al., 2006). Depletion of the Rat1/Xrn2 exonuclease or the CPF subunits Cpsf73 or Symplekin in Drosophila cells can cause circular RNA production through the backsplicing of read‐through RNA transcripts with retained introns from upstream genes (Liang et al., 2017). It is unclear, however, if the backsplicing reactions are coupled with fail‐safe mechanisms that might facilitate RNAPII termination under conditions in which termination factors are limited. Perhaps RNAPII pausing associated with RNA backsplicing events (as reported for canonical splicing events) may provide an opportunity for alternative termination mechanisms to occur such as those discussed in the next section involving the helicase Senataxin/Sen1.
5.3. Other termination mechanisms
A number of other termination mechanisms have been described that employ additional factors for RNAPII termination coordinated with RNA processing and/or RNA decay, often in cases of premature or non‐polyA‐dependent transcription termination. These pathways may also recruit a number of proteins and/or protein complexes that are involved in the polyA‐dependent termination pathway described above; however, we will focus on the unique players involved in these pathways in the following section.
A well‐characterized alternate termination pathway exists in budding yeast, the NNS‐dependent pathway (Figure 3). This pathway is responsible for termination at genes encoding snRNAs and snoRNAs (Steinmetz, Conrad, Brow, & Corden, 2001) and various types of pervasive transcripts such as yeast CUTs (Arigo, Eyler, et al., 2006; Schulz et al., 2013; Thiebaut et al., 2006; Wyers et al., 2005). Nrd1‐Nab3 binding sites have been shown to be enriched in regions upstream of promoters of CUTs and antisense transcription units (Cakiroglu, Zaugg, & Luscombe, 2016; Carroll, Pradhan, Granek, Clarke, & Corden, 2004; Schulz et al., 2013; Wlotzka et al., 2011). NNS is generally considered to terminate shorter transcripts (typically less than 1,000 nucleotides) compared to the traditional polyA‐dependent pathway (Creamer et al., 2011; Gudipati, Villa, Boulay, & Libri, 2008; Schulz et al., 2013; Vasiljeva, Kim, Terzi, et al., 2008). However, Nrd1‐Nab3‐dependent termination has also been shown to provide a fail‐safe for transcripts that read past a polyA site, restricting mRNAs where 3′ end formation has failed (Rondon, Mischo, Kawauchi, & Proudfoot, 2009; Vasiljeva & Buratowski, 2006). Degradation intermediates originating from unspliced RNA species have also been UV‐crosslinked to Nrd1, Nab3, or Trf4 (Wlotzka et al., 2011). It is not understood how Nrd1 is specifically recruited to aberrant RNAs in order to mediate their degradation by the exosome, but there is some evidence that Nrd1 is generally recruited to all RNAPII transcripts perhaps through its protein–protein interaction with the RNAPII CTD (Mayer et al., 2012; Schulz et al., 2013). Recently, Bresson, Tuck, Staneva, & Tollervey (2017) demonstrated that NNS and the TRAMP complex were targeted to transcripts that were being downregulated in response to glucose starvation. This suggests that NNS could be a mechanism for selective degradation of RNAs in order to change transcriptional programming under different cellular and environmental signaling pathways.
Figure 3.
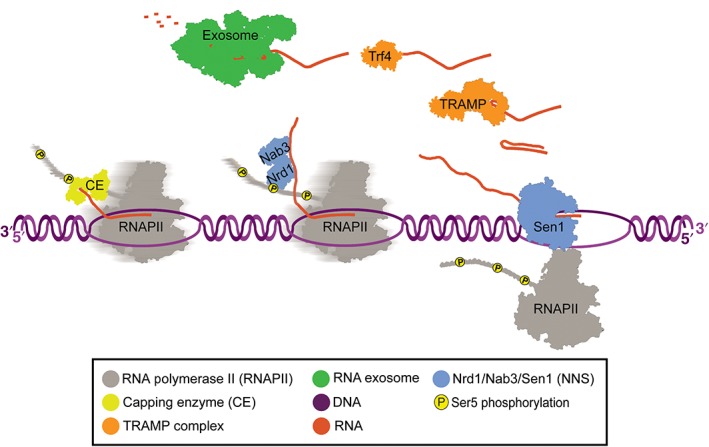
Nrd1‐Nab3‐Sen1 termination: Nrd1 is recruited to the site of transcription by Ser5 phosphorylated CTD. Nab3 and Nrd1 form a heterodimer and bind to RNA via their RNA recognition motif domains. Nab3 and Nrd1 are thought to be able to recruit Sen1, which then catches RNAPII and unwinds the DNA/RNA hybrid (also known as R‐loop). TRAMP unwinds RNA and its subunit Trf4 polyadenylates the 3′ end of RNA for processing and/or degradation. The exosome complex then degrades the RNA 3′–5′
With the major exception of sn/snoRNA transcripts, many of the RNAs terminated by the NNS pathway are rapidly degraded after transcription by the RNA exosome and can only be fully detected when the exosome is perturbed. Increased recruitment of Nrd1 during transcription, even if it does not induce termination, can destabilize a transcript (Honorine, Mosrin‐Huaman, Hervouet‐Coste, Libri, & Rahmouni, 2011; Vasiljeva, Kim, Terzi, et al., 2008). Timely termination of CUTs is important for preventing transcription interference with the coding transcriptome. Nrd1 and Nab3 recognize specific sequence motifs on the RNA that are crucial to their specificity (Carroll, Ghirlando, Ames, & Corden, 2007; Creamer et al., 2011; Porrua et al., 2012; Wlotzka et al., 2011) and are often clustered with AU‐rich sequences, contributing to termination efficiency (Porrua et al., 2012). Nrd1 also interacts with phosphorylated Ser5 on the RNAPII CTD (Heo et al., 2013; Kubicek et al., 2012; Tudek et al., 2014; Vasiljeva, Kim, Mutschler, Buratowski, & Meinhart, 2008), which is the predominant phosphoform during early elongation (Kim et al., 2011; Komarnitsky et al., 2000; Mayer et al., 2010; Tietjen et al., 2010). Nrd1 and Nab3 may act to ensure efficient and specific Sen1 recruitment since it is present at relatively low levels and appears to recognize RNA indiscriminately (Creamer et al., 2011; Ghaemmaghami et al., 2003; Porrua & Libri, 2013). Cleavage of the primary transcript has not been demonstrated for this pathway, although the CF protein Pcf11 has been implicated in NNS (Grzechnik, Gdula, & Proudfoot, 2015). Human Pcf11 is also involved in snRNA gene termination (O'Reilly et al., 2014). The CTD phosphatases RPAP2/Rtr1 and Ssu72 both have also been implicated in sn/snoRNA termination control in metazoans while Ssu72 plays a major, although mechanistically uncharacterized, role in NNS termination in yeast (Dichtl et al., 2002; Egloff, Zaborowska, Laitem, Kiss, & Murphy, 2012; Loya, O'Rourke, & Reines, 2012; O'Reilly et al., 2014). Upon release of the RNA transcript from RNAPII as a result of termination aided by Sen1, the exosome can facilitate either the processing or degradation of the transcript (Vasiljeva & Buratowski, 2006). In budding yeast, the exosome is particularly connected to the NNS‐termination pathway, due to the type of unstable transcripts typically produced. Rrp6 and Dis3 trim the 3′ end of snRNA and snoRNA precursors to convert them into mature species (Allmang et al., 1999; Gudipati et al., 2012; van Hoof, Lennertz, & Parker, 2000) and completely degrade CUTs (Arigo, Eyler, et al., 2006; Gudipati et al., 2012; Thiebaut et al., 2006; Wyers et al., 2005). Nrd1 also recruits TRAMP through direct recognition of a CTD mimic in Trf4 of the TRAMP complex (Tudek et al., 2014). Efficient degradation and processing require the TRAMP complex which both catalyzes polyadenylation and facilitates degradation by the exosome.
Although a pathway directly homologous to the yeast NNS pathway has not been identified in higher eukaryotes, PROMPTs are produced through early RNAPII termination and degraded by the exosome (Preker et al., 2008, 2011). Furthermore, the human NEXT complex has been shown to play a role in RNA surveillance, cryptic transcript degradation, and the termination and 3′ end processing of snRNAs (Hrossova et al., 2015; Lubas et al., 2011). NEXT contains hMTR4, the zinc‐finger protein ZCCHC8, the RNA‐binding factor RBM7. Unlike NNS, which is highly sequence‐specific, RBM7 seems only to prefer U‐rich regions and tends to be promiscuous (Hrossova et al., 2015; Lubas et al., 2015). NEXT mainly targets unprocessed transcripts and recent publications have provided evidence for a PAXT connection (Meola et al., 2016). Both NEXT and PAXT have been shown to have physical linkages to the cap‐binding complex and its associated factors (Andersen et al., 2013; Lubas et al., 2015; Meola et al., 2016), suggesting mechanisms for recruiting the exosome to capped transcripts. TRAMP, NEXT, and PAXT all contain hMTR4, suggesting a possible mechanism in which the exosome can target different transcripts through substitution of adaptors containing hMTR4 (Meola et al., 2016). The conservation of TRAMP and the role of the NEXT complex in recruiting the exosome to short transcripts suggest comparable mechanisms for early RNAPII termination and coupled RNA decay exist throughout eukaryotes.
Speed of transcription affects the genomic position at which RNAPII termination occurs and accordingly, pausing of RNAPII may provide the opportunity for Sen1 to locate RNAPII and aid in the termination of transcription via the NNS pathway (Hazelbaker, Marquardt, Wlotzka, & Buratowski, 2013; Jamonnak et al., 2011; Schaughency, Merran, & Corden, 2014). Sen1 has also been shown to trigger forward translocation of stalled RNAPII complexes (Han, Libri, & Porrua, 2017). Sen1 also functions independently of NNS in yeast because: (a) Sen1 has been shown to bind to a number of polyA‐dependent termination sites (Creamer et al., 2011; Jamonnak et al., 2011); (b) Sen1 is sufficient to displace the elongation complex DNA in vitro through interaction with the nascent RNA which requires its ATP‐dependent helicase activity (Han et al., 2017; Leonaite et al., 2017; Porrua & Libri, 2013); and (c) Sen1 inactivation (using an anchor away approach) leads to RNAPII accumulation at the 3′ end of both coding and noncoding genes (Schaughency et al., 2014). Human and yeast Senataxin/Sen1 have been shown to trigger RNAPII termination to resolve R‐loops (Mischo et al., 2011; Skourti‐Stathaki, Kamieniarz‐Gdula, & Proudfoot, 2014; Skourti‐Stathaki, Proudfoot, & Gromak, 2011), which occur in numerous cellular contexts (see Santos‐Pereira & Aguilera, 2015 for review). R‐loops can cause various types of genome instability and have recently been found to be required for efficient double strand break repair (Ohle et al., 2016). R‐loops can also be formed by circular RNAs that are retained in the nucleus (Conn et al., 2017). However, it remains to be seen if the circular RNAs that are retained in the nucleus can facilitate RNAPII termination through existing R‐loop resolving pathways. Interestingly, an R‐loop resolution pathway has been characterized in human cells line in which the RNA‐binding protein SMN binds to arginine 1810 symmetric dimethylation on the RNAPII CTD for stable Senataxin recruitment. Recruitment of Senataxin through SMN leads to both R‐loop resolution and recruitment of Xrn2 to trigger premature RNAPII termination (Zhao et al., 2016). It is possible that circular RNAs could work in a similar way to form R‐loops at particular genomic regions thereby recruiting R‐loop binding proteins and Senataxin to specific sites to trigger RNAPII termination. R‐loop resolution pathways may have both basal and fail‐safe roles in the regulation and protection of the genome (Chédin, 2016; Santos‐Pereira & Aguilera, 2015). Recent work has also shown that yeast Sen1 levels are tightly controlled by the anaphase‐promoting complex likely for overall genome maintenance and control of RNAPII termination (Mischo et al., 2018). In addition, it was shown that overexpression of Sen1 leads to cellular fitness defects. It remains to be determined if Xrn2 is required for Sen1/Senataxin regulated RNAPII termination and vice versa, although previous studies have indicated that both Senataxin and Xrn2 may be required for torpedo function at a model polyA‐dependent terminator (Kawauchi, Mischo, Braglia, Rondon, & Proudfoot, 2008; Mischo et al., 2011; Rondon et al., 2009; Skourti‐Stathaki et al., 2011), as is suggested for other R‐loop resolution pathways (Zhao et al., 2016).
In metazoans, the multi‐subunit Integrator complex is responsible for 3′ end formation and processing of snRNAs (Baillat et al., 2005). Integrator has been shown to be recruited to a Ser2‐P/Ser7‐P CTD phosphoform of RNAPII with Ser5‐P being inhibitory for recruitment (Egloff et al., 2010). As such, the recruitment of Integrator may have a dependence on the activity of the two conserved Ser5‐P CTD phosphatases RPAP2 (known as Rtr1 in yeast) and Ssu72 which have both been shown to regulate Ser5‐P levels in both yeast and mammalian cells (Egloff et al., 2012; Hunter et al., 2016; Mosley et al., 2009; Ni et al., 2014). In fact, knockdown studies for either phosphatase resulted in decreased snRNA processing efficiency, indicating that native levels of both RPAP2 and Ssu72 are required for proper recruitment/regulation of Integrator (Egloff et al., 2012; O'Reilly et al., 2014; Wani et al., 2014). Incorrect processing by Integrator could lead to exosome‐dependent degradation of the nascent snRNA similar to defects caused by improper snRNP formation in SMN‐deficient cells (Shukla & Parker, 2014). The Integrator complex has also been implicated in the termination of numerous other ncRNAs including promoter proximal transcripts in a mechanism that terminates DSIF (Spt4/Spt5)‐associated RNAPII elongation complexes and enhancer RNAs (Lai, Gardini, Zhang, & Shiekhattar, 2015; Skaar et al., 2015). Considering that polyA‐dependent termination factors are temporally recruited after dissociation of the core elongation machinery, which includes DSIF, it is possible that Integrator could also carry out premature RNAPII termination of pervasive transcripts in metazoans similar to the NNS termination pathway in yeast (Mayer et al., 2010, 2012). It has been shown that Senataxin and Xrn2 are not required for RNAPII termination at U1 and U2 snRNA genes (O'Reilly et al., 2014). However, it has not been determined if RNAPII termination of pervasive transcripts in mammalian cells requires Senataxin and/or Xrn2; but both proteins have been implicated in kinetic competition models of termination that could be facilitated by RNAPII pausing as a consequence of engagement of RNA processing machinery such as Integrator (Fong et al., 2015; Hazelbaker et al., 2013). The kinetic competition models provide a strong mechanistic foundation for the intimate coupling of RNA processing and RNAPII termination (Box 2).
BOX 2. THE RACE BETWEEN RNAPII AND TERMINATION FACTORS.
The DNA:RNA helicase Sen1 and the 5′–3′ exonuclease Xrn2 are required for various termination pathways. In models of kinetic competition between RNAPII and Sen1/Xrn2, it has been proposed that slow‐moving RNAPII will be able to be caught earlier in transcription, while fast‐moving RNAPII will not be caught until later in the gene. Pausing of RNAPII could provide an increased probability for termination via either Xrn2 or Sen1 (see the figure below) (Mischo & Proudfoot, 2013).
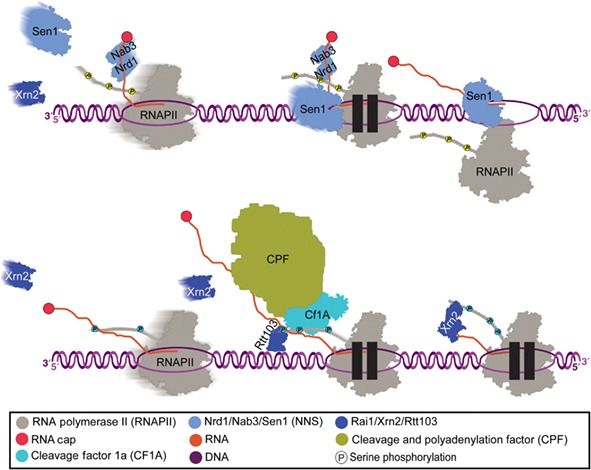
Kinetic competition between RNAPII and Sen1/Xrn2. The rate of transcription varies, due to factors that influence RNAPII kinetics and passage through chromatin, and these variations in rate can affect the termination window. In general, it is thought that slower elongation rates promote earlier termination, while faster elongation rates lead to termination spreading further downstream. The faster RNAPII moves along a gene, the harder it will be for the termination machinery to catch up, and how long it takes for RNAPII to be caught will help to determine what termination pathway is used. Two termination‐associated proteins thought to be tasked with catching RNAPII are Xrn2 (also known as Rat1) and Sen1 (yeast homolog of Senataxin). Pausing of RNAPII would promote termination via either Xrn2 or Sen1 by providing the ability for them to catch up to the elongation complex (Mischo & Proudfoot, 2013)
Use of RNAPII trigger loop mutants (fast mutant: rpb1‐E1103G, slow mutant: rpb1‐N488D) has demonstrated that slow elongation leads to earlier termination, while fast elongation lead to later termination (Malagon et al., 2006). Growth and termination defects in some Xrn2 mutant cells can be overcome by RNAPII slow mutants (Fong et al., 2015; Jimeno‐Gonzalez et al., 2010). Similarly, Sen1 mutants display read‐through termination defects which can be suppressed by the introduction of slow RNAPII (Hazelbaker et al., 2013). In both cases, the slow RNAPII mutants provide a larger window of opportunity for proper termination by the mutant termination factors. In complementary work, it has recently been shown that Sen1 protein levels are modulated by the cell cycle (Mischo et al., 2018). Overexpression of Sen1 leads to a decrease in ncRNA production and an increase in efficiency of mRNA termination, with changes observed in both the occupancy of RNAPII and the position of termination (Mischo et al., 2018). The toxicity of Sen1 overexpression is likely caused by excess termination. This model is supported by the fact that mutations in other termination factors suppress this phenotype (Mischo et al., 2018).
The mechanisms of transcription termination including some additional examples of unique pathways are reviewed in more detail in the following publications: Kuehner et al. (2011), Porrua, Boudvillain, and Libri (2016), and Proudfoot (2016).
6. CONCLUSIONS
Recent work has begun to reveal how intimately the processes of transcription and RNA processing are coupled, particularly in the regulation of RNA quality control pathways. The clearest example of this important connection is the requirement for both 5′ and 3′ exonucleases for proper control of RNAPII termination and RNA transport. The rate of RNAPII transcription and phosphorylation of the RNAPII CTD are both critical for the recruitment of the RNA processing machinery and this coupling of processes in return likely ensures proper quality control of the transcribed RNAs to maintain proper gene expression and cellular health. As discussed in this review, capping, splicing, and 3′ end processing are inherently tied to the site of transcription, while the basal transcription machinery coordinately plays a significant role in managing checkpoints of RNA quality control and degradation in the nucleus. Future work is needed to define the proteins and mechanisms involved in the coupled transcription—RNA quality checkpoints which remain poorly understood throughout eukaryotes.
Cutting‐edge structural biology and sequencing method development have greatly contributed to a gain in the understanding of the molecular mechanisms that underpin how these processes work both individually and cooperatively. Cryo‐EM in particular has helped to make a number of recent advances in mechanistic knowledge of the transient, and subsequently hard to study, RNA processing pathways. However, there are still many open questions when it comes to understanding the mechanisms of these processes, and full structures of the mRNA 3′ end processing machinery have not been reported. The precise interplay between RNA processing machine assembly/disassembly and RNAPII transcription elongation is still poorly understood. As discussed, a number of recent observations of RNAPII pausing at splice sites and termination sites suggest an intimate crosstalk between RNAPII progression and RNA processing that likely provides coordinated regulation of alternative splicing and polyadenylation.
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
RELATED WIREs ARTICLE
Rrp6: Integrated roles in nuclear RNA metabolism and transcription termination
ACKNOWLEDGMENTS
We sincerely thank the members of the Mosley lab for critical discussions and comments. A.L.M. acknowledges support from NIH grant R01 GM099714 and NSF grant MCB 1515748. S.A.P. is supported by NIH T32 HL007910.
Peck SA, Hughes KD, Victorino JF, Mosley AL. Writing a wrong: Coupled RNA polymerase II transcription and RNA quality control. WIREs RNA. 2019;10:e1529. 10.1002/wrna.1529
[The copyright line for this article was changed on 18 March 2019 after original online publication.]
Funding information National Heart and Lung Institute, Grant/Award Number: T32 HL007910; National Institute of General Medical Sciences, Grant/Award Number: GM099714; National Science Foundation, Grant/Award Number: MCB 1515748
References
FURTHER READING
- Adelman, K. , Marr, M. T. , Werner, J. , Saunders, A. , Ni, Z. , Andrulis, E. D. , & Lis, J. T. (2005). Efficient release from promoter‐proximal stall sites requires transcript cleavage factor TFIIS. Molecular Cell, 17(1), 103–112. 10.1016/j.molcel.2004.11.028 [DOI] [PubMed] [Google Scholar]
- Arigo, J. T. , Carroll, K. L. , Ames, J. M. , & Corden, J. L. (2006). Regulation of yeast NRD1 expression by premature transcription termination. Molecular Cell, 21(5), 641–651. 10.1016/j.molcel.2006.02.005 [DOI] [PubMed] [Google Scholar]
- Berman, H. M. , Westbrook, J. , Feng, Z. , Gilliland, G. , Bhat, T. N. , Weissig, H. , … Bourne, P. E. (2000). The Protein Data Bank. Nucleic Acids Research, 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candelli, T. , Challal, D. , Briand, J. B. , Boulay, J. , Porrua, O. , Colin, J. , & Libri, D. (2018). High‐resolution transcription maps reveal the widespread impact of roadblock termination in yeast. EMBO Journal, 37(4), e97490 10.15252/embj.201797490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, A. C. , & Cramer, P. (2011). Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature, 471(7337), 249–253. 10.1038/nature09785 [DOI] [PubMed] [Google Scholar]
- Colin, J. , Candelli, T. , Porrua, O. , Boulay, J. , Zhu, C. , Lacroute, F. , … Libri, D. (2014). Roadblock termination by reb1p restricts cryptic and readthrough transcription. Molecular Cell, 56(5), 667–680. 10.1016/j.molcel.2014.10.026 [DOI] [PubMed] [Google Scholar]
- Danko, C. G. , Hah, N. , Luo, X. , Martins, A. L. , Core, L. , Lis, J. T. , … Kraus, W. L. (2013). Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Molecular Cell, 50(2), 212–222. 10.1016/j.molcel.2013.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, L. , Kerr, A. , & West, S. (2012). Co‐transcriptional degradation of aberrant pre‐mRNA by Xrn2. EMBO Journal, 31(11), 2566–2578. 10.1038/emboj.2012.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk, S. , Weir, J. R. , Hentschel, J. , Reichelt, P. , Bonneau, F. , & Conti, E. (2014). The molecular architecture of the TRAMP complex reveals the organization and interplay of its two catalytic activities. Molecular Cell, 55(6), 856–867. 10.1016/j.molcel.2014.07.020 [DOI] [PubMed] [Google Scholar]
- Fuchs, G. , Voichek, Y. , Benjamin, S. , Gilad, S. , Amit, I. , & Oren, M. (2014). 4sUDRB‐seq: Measuring genomewide transcriptional elongation rates and initiation frequencies within cells. Genome Biology, 15(5), R69 10.1186/gb-2014-15-5-r69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, M. , Rajashankar, K. R. , & Lima, C. D. (2010). Structure of the Saccharomyces cerevisiae Cet1‐Ceg1 mRNA capping apparatus. Structure, 18(2), 216–227. 10.1016/j.str.2009.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani, S. , Yogesha, S. D. , Mayfield, J. , Zhang, M. , Zhang, Y. , Matthews, W. L. , … Zhang, Y. J. (2016). Structure of Saccharomyces cerevisiae Rtr1 reveals an active site for an atypical phosphatase. Science Signaling, 9(417), ra24 10.1126/scisignal.aad4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. D. , Choe, J. , & Seo, Y. S. (1999). The sen1(+) gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry, 38(44), 14697–14710. [DOI] [PubMed] [Google Scholar]
- Larson, D. R. , Zenklusen, D. , Wu, B. , Chao, J. A. , & Singer, R. H. (2011). Real‐time observation of transcription initiation and elongation on an endogenous yeast gene. Science, 332(6028), 475–478. 10.1126/science.1202142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon, J. C. , Wind, M. , Saunders, L. , Hock, M. B. , & Reines, D. (1998). Mutations in RNA polymerase II and elongation factor SII severely reduce mRNA levels in Saccharomyces cerevisiae . Molecular and Cellular Biology, 18(10), 5771–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna, R. , Jimeno, S. , Marin, M. , Huertas, P. , Garcia‐Rubio, M. , & Aguilera, A. (2005). Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Molecular Cell, 18(6), 711–722. 10.1016/j.molcel.2005.05.001 [DOI] [PubMed] [Google Scholar]
- Mazza, C. , Segref, A. , Mattaj, I. W. , & Cusack, S. (2002). Large‐scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap‐binding complex. EMBO Journal, 21(20), 5548–5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag, A. , & Steitz, J. A. (2012). Tri‐snRNP‐associated proteins interact with subunits of the TRAMP and nuclear exosome complexes, linking RNA decay and pre‐mRNA splicing. RNA Biology, 9(3), 334–342. 10.4161/rna.19431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauhut, R. , Fabrizio, P. , Dybkov, O. , Hartmuth, K. , Pena, V. , Chari, A. , … Luhrmann, R. (2016). Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science, 353(6306), 1399–1405. 10.1126/science.aag1906 [DOI] [PubMed] [Google Scholar]
- Reines, D. , Conaway, R. C. , & Conaway, J. W. (1999). Mechanism and regulation of transcriptional elongation by RNA polymerase II. Current Opinion in Cell Biology, 11(3), 342–346. 10.1016/S0955-0674(99)80047-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reines, D. , Ghanouni, P. , Gu, W. , Mote, J., Jr. , & Powell, W. (1993). Transcription elongation by RNA polymerase II: Mechanism of SII activation. Cellular & Molecular Biology Research, 39(4), 331–338. [PubMed] [Google Scholar]
- Rodríguez‐Molina, J. B. , Tseng, S. C. , Simonett, S. P. , Taunton, J. , & Ansari, A. Z. (2016). Engineered covalent inactivation of TFIIH‐kinase reveals an elongation checkpoint and results in widespread mRNA stabilization. Molecular Cell, 63(3), 433–444. 10.1016/j.molcel.2016.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, K. , Gabunilas, J. , Gillespie, A. , Ngo, D. , & Chanfreau, G. F. (2016). Common genomic elements promote transcriptional and DNA replication roadblocks. Genome Research, 26(10), 1363–1375. 10.1101/gr.204776.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller, J. M. , Falk, S. , Fromm, L. , Hurt, E. , & Conti, E. (2018). Structure of the nuclear exosome captured on a maturing preribosome. Science, 360(6385), 219–222. 10.1126/science.aar5428 [DOI] [PubMed] [Google Scholar]
- Sigurdsson, S. , Dirac‐Svejstrup, A. B. , & Svejstrup, J. Q. (2010). Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Molecular Cell, 38(2), 202–210. 10.1016/j.molcel.2010.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somesh, B. P. , Reid, J. , Liu, W. F. , Sogaard, T. M. , Erdjument‐Bromage, H. , Tempst, P. , & Svejstrup, J. Q. (2005). Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell, 121(6), 913–923. 10.1016/j.cell.2005.04.010 [DOI] [PubMed] [Google Scholar]
- Steinmetz, E. J. , Ng, S. B. , Cloute, J. P. , & Brow, D. A. (2006). Cis‐ and trans‐acting determinants of transcription termination by yeast RNA polymerase II. Molecular and Cellular Biology, 26(7), 2688–2696. 10.1128/MCB.26.7.2688-2696.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydow, J. F. , Brueckner, F. , Cheung, A. C. , Damsma, G. E. , Dengl, S. , Lehmann, E. , … Cramer, P. (2009). Structural basis of transcription: Mismatch‐specific fidelity mechanisms and paused RNA polymerase II with frayed RNA. Molecular Cell, 34(6), 710–721. 10.1016/j.molcel.2009.06.002 [DOI] [PubMed] [Google Scholar]
- Tous, C. , Rondon, A. G. , Garcia‐Rubio, M. , Gonzalez‐Aguilera, C. , Luna, R. , & Aguilera, A. (2011). A novel assay identifies transcript elongation roles for the Nup84 complex and RNA processing factors. EMBO Journal, 30(10), 1953–1964. 10.1038/emboj.2011.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turowski, T. W. , & Tollervey, D. (2015). Cotranscriptional events in eukaryotic ribosome synthesis. WIREs RNA, 6(1), 129–139. 10.1002/wrna.1263 [DOI] [PubMed] [Google Scholar]
- Uptain, S. M. , Kane, C. M. , & Chamberlin, M. J. (1997). Basic mechanisms of transcript elongation and its regulation. Annual Review of Biochemistry, 66, 117–172. 10.1146/annurev.biochem.66.1.117 [DOI] [PubMed] [Google Scholar]
- Veloso, A. , Kirkconnell, K. S. , Magnuson, B. , Biewen, B. , Paulsen, M. T. , Wilson, T. E. , & Ljungman, M. (2014). Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Research, 24(6), 896–905. 10.1101/gr.171405.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wind, M. , & Reines, D. (2000). Transcription elongation factor SII. BioEssays, 22(4), 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wind‐Rotolo, M. , & Reines, D. (2001). Analysis of gene induction and arrest site transcription in yeast with mutations in the transcription elongation machinery. Journal of Biological Chemistry, 276(15), 11531–11538. 10.1074/jbc.M011322200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, K. , Nagaike, T. , Xiang, S. , Kilic, T. , Beh, M. M. , Manley, J. L. , & Tong, L. (2010). Crystal structure of the human symplekin‐Ssu72‐CTD phosphopeptide complex. Nature, 467(7316), 729–733. 10.1038/nature09391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K. , Fischer, T. , Porter, R. L. , Dhakshnamoorthy, J. , Zofall, M. , Zhou, M. , … Grewal, S. I. (2011). Clr4/Suv39 and RNA quality control factors cooperate to trigger RNAi and suppress antisense RNA. Science, 331(6024), 1624–1627. 10.1126/science.1198712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Fu, J. , & Gilmour, D. S. (2005). CTD‐dependent dismantling of the RNA polymerase II elongation complex by the pre‐mRNA 3′‐end processing factor, Pcf11. Genes & Development, 19(13), 1572–1580. 10.1101/gad.1296305 [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES
- Adelman, K. , & Lis, J. T. (2012). Promoter‐proximal pausing of RNA polymerase II: Emerging roles in metazoans. Nature Reviews Genetics, 13(10), 720–731. 10.1038/nrg3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agafonov, D. E. , Kastner, B. , Dybkov, O. , Hofele, R. V. , Liu, W. T. , Urlaub, H. , … Stark, H. (2016). Molecular architecture of the human U4/U6.U5 tri‐snRNP. Science, 351(6280), 1416–1420. 10.1126/science.aad2085 [DOI] [PubMed] [Google Scholar]
- Ahearn, J. M., Jr. , Bartolomei, M. S. , West, M. L. , Cisek, L. J. , & Corden, J. L. (1987). Cloning and sequence analysis of the mouse genomic locus encoding the largest subunit of RNA polymerase II. Journal of Biological Chemistry, 262(22), 10695–10705. [PubMed] [Google Scholar]
- Ahn, S. H. , Kim, M. , & Buratowski, S. (2004). Phosphorylation of serine 2 within the RNA polymerase II C‐terminal domain couples transcription and 3′ end processing. Molecular Cell, 13(1), 67–76. [DOI] [PubMed] [Google Scholar]
- Alexander, R. D. , Innocente, S. A. , Barrass, J. D. , & Beggs, J. D. (2010). Splicing‐dependent RNA polymerase pausing in yeast. Molecular Cell, 40(4), 582–593. 10.1016/j.molcel.2010.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang, C. , Kufel, J. , Chanfreau, G. , Mitchell, P. , Petfalski, E. , & Tollervey, D. (1999). Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO Journal, 18(19), 5399–5410. 10.1093/emboj/18.19.5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg, D. C. , Goldstein, A. L. , & Cole, C. N. (1992). Isolation and characterization of RAT1: An essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes & Development, 6(7), 1173–1189. [DOI] [PubMed] [Google Scholar]
- Ameur, A. , Zaghlool, A. , Halvardson, J. , Wetterbom, A. , Gyllensten, U. , Cavelier, L. , & Feuk, L. (2011). Total RNA sequencing reveals nascent transcription and widespread co‐transcriptional splicing in the human brain. Nature Structural & Molecular Biology, 18(12), 1435–1440. 10.1038/nsmb.2143 [DOI] [PubMed] [Google Scholar]
- Andersen, P. R. , Domanski, M. , Kristiansen, M. S. , Storvall, H. , Ntini, E. , Verheggen, C. , … Jensen, T. H. (2013). The human cap‐binding complex is functionally connected to the nuclear RNA exosome. Nature Structural & Molecular Biology, 20(12), 1367–1376. 10.1038/nsmb.2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassi, C. , & Riccio, A. (2009). To localize or not to localize: mRNA fate is in 3′UTR ends. Trends in Cell Biology, 19(9), 465–474. 10.1016/j.tcb.2009.06.001 [DOI] [PubMed] [Google Scholar]
- Andrulis, E. D. , Werner, J. , Nazarian, A. , Erdjument‐Bromage, H. , Tempst, P. , & Lis, J. T. (2002). The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature, 420(6917), 837–841. 10.1038/nature01181 [DOI] [PubMed] [Google Scholar]
- Arigo, J. T. , Eyler, D. E. , Carroll, K. L. , & Corden, J. L. (2006). Termination of cryptic unstable transcripts is directed by yeast RNA‐binding proteins Nrd1 and Nab3. Molecular Cell, 23(6), 841–851. 10.1016/j.molcel.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Arndt, K. M. , & Reines, D. (2015). Termination of transcription of short noncoding RNAs by RNA polymerase II. Annual Review of Biochemistry, 84, 381–404. 10.1146/annurev-biochem-060614-034457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baejen, C. , Andreani, J. , Torkler, P. , Battaglia, S. , Schwalb, B. , Lidschreiber, M. , … Cramer, P. (2017). Genome‐wide analysis of RNA polymerase II termination at protein‐coding genes. Molecular Cell, 66(1), 38–49 .e36. 10.1016/j.molcel.2017.02.009 [DOI] [PubMed] [Google Scholar]
- Baejen, C. , Torkler, P. , Gressel, S. , Essig, K. , Soding, J. , & Cramer, P. (2014). Transcriptome maps of mRNP biogenesis factors define pre‐mRNA recognition. Molecular Cell, 55(5), 745–757. 10.1016/j.molcel.2014.08.005 [DOI] [PubMed] [Google Scholar]
- Baillat, D. , Hakimi, M. A. , Naar, A. M. , Shilatifard, A. , Cooch, N. , & Shiekhattar, R. (2005). Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C‐terminal repeat of RNA polymerase II. Cell, 123(2), 265–276. 10.1016/j.cell.2005.08.019 [DOI] [PubMed] [Google Scholar]
- Bauren, G. , & Wieslander, L. (1994). Splicing of Balbiani ring 1 gene pre‐mRNA occurs simultaneously with transcription. Cell, 76(1), 183–192. [DOI] [PubMed] [Google Scholar]
- Beaulieu, Y. B. , Kleinman, C. L. , Landry‐Voyer, A. M. , Majewski, J. , & Bachand, F. (2012). Polyadenylation‐dependent control of long noncoding RNA expression by the poly(A)‐binding protein nuclear 1. PLoS Genetics, 8(11), e1003078 10.1371/journal.pgen.1003078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley, D. L. (2014). Coupling mRNA processing with transcription in time and space. Nature Reviews Genetics, 15(3), 163–175. 10.1038/nrg3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram, K. , Agafonov, D. E. , Dybkov, O. , Haselbach, D. , Leelaram, M. N. , Will, C. L. , … Stark, H. (2017). Cryo‐EM structure of a pre‐catalytic human spliceosome primed for activation. Cell, 170(4), 701–713. 10.1016/j.cell.2017.07.011 [DOI] [PubMed] [Google Scholar]
- Bertram, K. , Agafonov, D. E. , Liu, W. T. , Dybkov, O. , Will, C. L. , Hartmuth, K. , … Luhrmann, R. (2017). Cryo‐EM structure of a human spliceosome activated for step 2 of splicing. Nature, 542(7641), 318–323. 10.1038/nature21079 [DOI] [PubMed] [Google Scholar]
- Beyer, A. L. , & Osheim, Y. N. (1988). Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes & Development, 2(6), 754–765. [DOI] [PubMed] [Google Scholar]
- Bitton, D. A. , Atkinson, S. R. , Rallis, C. , Smith, G. C. , Ellis, D. A. , Chen, Y. Y. , … Bahler, J. (2015). Widespread exon skipping triggers degradation by nuclear RNA surveillance in fission yeast. Genome Research, 25(6), 884–896. 10.1101/gr.185371.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boczonadi, V. , Muller, J. S. , Pyle, A. , Munkley, J. , Dor, T. , Quartararo, J. , … Horvath, R. (2014). EXOSC8 mutations alter mRNA metabolism and cause hypomyelination with spinal muscular atrophy and cerebellar hypoplasia. Nature Communications, 5, 4287 10.1038/ncomms5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth, G. T. , Parua, P. K. , Sansó, M. , Fisher, R. P. , & Lis, J. T. (2018). Cdk9 regulates a promoter‐proximal checkpoint to modulate RNA polymerase II elongation rate in fission yeast. Nature Communications, 9, 543 10.1038/s41467-018-03006-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet‐Antonelli, C. , Presutti, C. , & Tollervey, D. (2000). Identification of a regulated pathway for nuclear pre‐mRNA turnover. Cell, 102(6), 765–775. 10.1016/S0092-8674(00)00065-9 [DOI] [PubMed] [Google Scholar]
- Brannan, K. , Kim, H. , Erickson, B. , Glover‐Cutter, K. , Kim, S. , Fong, N. , … Bentley, D. L. (2012). mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Molecular Cell, 46(3), 311–324. 10.1016/j.molcel.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresson, S. M. , & Tollervey, D. (2018). Surveillance‐ready transcription: Nuclear RNA decay as a default fate. Open Biology, 8(3), e170270 10.1098/rsob.170270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresson, S. M. , Tuck, A. , Staneva, D. , & Tollervey, D. (2017). Nuclear RNA decay pathways aid rapid remodeling of gene expression in yeast. Molecular Cell, 65(5), 787–800 .e785. 10.1016/j.molcel.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresson, S. M. , & Conrad, N. K. (2013). The human nuclear poly(A)‐binding protein promotes RNA hyperadenylation and decay. PLoS Genetics, 9(10), e1003893 10.1371/journal.pgen.1003893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresson, S. M. , Hunter, O. V. , Hunter, A. C. , & Conrad, N. K. (2015). Canonical poly(A) polymerase activity promotes the decay of a wide variety of mammalian nuclear RNAs. PLoS Genetics, 11(10), e1005610 10.1271/journal.pgen.1005610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler, M. , Haas, W. , Gygi, S. P. , & Moazed, D. (2007). RNAi‐dependent and ‐independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell, 129(4), 707–721. 10.1016/j.cell.2007.03.038 [DOI] [PubMed] [Google Scholar]
- Buratowski, S. (2003). The CTD code. Nature Structural Biology, 10(9), 679–680. 10.1038/nsb0903-679 [DOI] [PubMed] [Google Scholar]
- Buratowski, S. (2009). Progression through the RNA polymerase II CTD cycle. Molecular Cell, 36(4), 541–546. 10.1016/j.molcel.2009.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard, K. T. D. , & Butler, J. S. (2000). A nuclear 3′–5′ exonuclease involved in mRNA degradation interacts with poly(A) polymerase and the hnRNA protein Npl3p. Molecular and Cellular Biology, 20(2), 604–616. 10.1128/Mcb.20.2.604-616.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, J. E. , Longhurst, A. D. , Merkurjev, D. , Sales‐Lee, J. , Rao, B. , Moresco, J. J. , … Madhani, H. D. (2018). Spliceosome profiling visualizes operations of a dynamic RNP at nucleotide resolution. Cell, 173(4), 1014–1030. 10.1016/j.cell.2018.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakiroglu, S. A. , Zaugg, J. B. , & Luscombe, N. M. (2016). Backmasking in the yeast genome: Encoding overlapping information for protein‐coding and RNA degradation. Nucleic Acids Research, 44(17), 8065–8072. 10.1093/nar/gkw683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan, K. P. , & Butler, J. S. (2010). TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. Journal of Biological Chemistry, 285(6), 3540–3547. 10.1074/jbc.M109.058396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo Oesterreich, F. , Herzel, L. , Straube, K. , Hujer, K. , Howard, J. , & Neugebauer, K. M. (2016). Splicing of nascent RNA coincides with intron exit from RNA polymerase II. Cell, 165, 372–381. 10.1016/j.cell.2016.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo Oesterreich, F. , Preibisch, S. , & Neugebauer, K. M. (2010). Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Molecular Cell, 40(4), 571–581. 10.1016/j.molcel.2010.11.004 [DOI] [PubMed] [Google Scholar]
- Carroll, K. L. , Ghirlando, R. , Ames, J. M. , & Corden, J. L. (2007). Interaction of yeast RNA‐binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA, 13(3), 361–373. 10.1261/rna.338407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, K. L. , Pradhan, D. A. , Granek, J. A. , Clarke, N. D. , & Corden, J. L. (2004). Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Molecular and Cellular Biology, 24(14), 6241–6252. 10.1128/MCB.24.14.6241-6252.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanal, A. , Kumar, A. , Hill, C. H. , Easter, A. D. , Emsley, P. , Degliesposti, G. , … Passmore, L. A. (2017). Architecture of eukaryotic mRNA 3′‐end processing machinery. Science, 358(6366), 1056–1059. 10.1126/science.aao6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, J. H. , Jiao, X. F. , Chiba, K. , Oh, C. , Martin, C. E. , Kiledjian, M. , & Tong, L. (2012). Dxo1 is a new type of eukaryotic enzyme with both decapping and 5′–3′ exoribonuclease activity. Nature Structural and Molecular Biology, 19(10), 1011–1017. 10.1038/nsmb.2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chédin, F. (2016). Nascent connections: R‐loops and chromatin patterning. Trends in Genetics, 32(12), 828–838. 10.1016/j.tig.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F. X. , Smith, E. R. , & Shilatifard, A. (2018). Born to run: Control of transcription elongation by RNA polymerase II. Nature Reviews Molecular Cell Biology, 19(7), 464–478. 10.1038/s41580-018-0010-5 [DOI] [PubMed] [Google Scholar]
- Chlebowski, A. , Lubas, M. , Jensen, T. H. , & Dziembowski, A. (2013). RNA decay machines: The exosome. Biochimica et Biophysica Acta, 1829(6–7), 552–560. 10.1016/j.bbagrm.2013.01.006 [DOI] [PubMed] [Google Scholar]
- Cho, E. J. , Takagi, T. , Moore, C. R. , & Buratowski, S. (1997). mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy‐terminal domain. Genes & Development, 11(24), 3319–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, J. , & Parker, R. (2004). Eukaryotic mRNA decapping. Annual Review of Biochemistry, 73(1), 861–890. 10.1146/annurev.biochem.73.011303.074032 [DOI] [PubMed] [Google Scholar]
- Conn, V. M. , Hugouvieux, V. , Nayak, A. , Conos, S. A. , Capovilla, G. , Cildir, G. , … Conn, S. J. (2017). A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R‐loop formation. Nature Plants, 3, 17053 10.1038/nplants.2017.53 [DOI] [PubMed] [Google Scholar]
- Cougot, N. , van Dijk, E. , Babajko, S. , & Seraphin, B. (2004). Cap‐tabolism. Trends in Biochemical Sciences, 29(8), 436–444. 10.1016/j.tibs.2004.06.008 [DOI] [PubMed] [Google Scholar]
- Coy, S. , Volanakis, A. , Shah, S. , & Vasiljeva, L. (2013). The Sm complex is required for the processing of non‐coding RNAs by the exosome. PLoS One, 8(6), e65606 10.1371/journal.pone.0065606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creamer, T. J. , Darby, M. M. , Jamonnak, N. , Schaughency, P. , Hao, H. , Wheelan, S. J. , & Corden, J. L. (2011). Transcriptome‐wide binding sites for components of the Saccharomyces cerevisiae non‐poly(A) termination pathway: Nrd1, Nab3, and Sen1. PLoS Genetics, 7(10), e1002329 10.1371/journal.pgen.1002329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard, C. K. , Kahns, S. , Lykke‐Andersen, S. , Nielsen, A. L. , Jensen, T. H. , & Kjems, J. (2008). A 5′ splice site enhances the recruitment of basal transcription initiation factors in vivo. Molecular Cell, 29(2), 271–278. 10.1016/j.molcel.2007.11.035 [DOI] [PubMed] [Google Scholar]
- Danin‐Kreiselman, M. , Lee, C. Y. , & Chanfreau, G. (2003). RNAse III‐mediated degradation of unspliced pre‐mRNAs and lariat introns. Molecular Cell, 11(5), 1279–1289. [DOI] [PubMed] [Google Scholar]
- Davidson, L. , Muniz, L. , & West, S. (2014). 3′ end formation of pre‐mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes & Development, 28(4), 342–356. 10.1101/gad.231274.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, C. A. , & Ares, M., Jr. (2006). Accumulation of unstable promoter‐associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae . Proceedings of the National Academy of Sciences of the United States of America, 103(9), 3262–3267. 10.1073/pnas.0507783103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida, S. F. , Garcia‐Sacristan, A. , Custodio, N. , & Carmo‐Fonseca, M. (2010). A link between nuclear RNA surveillance, the human exosome and RNA polymerase II transcriptional termination. Nucleic Acids Research, 38(22), 8015–8026. 10.1093/nar/gkq703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengl, S. , & Cramer, P. (2009). Torpedo nuclease Rat1 is insufficient to terminate RNA polymerase II in vitro. Journal of Biological Chemistry, 284(32), 21270–21279. 10.1074/jbc.M109.013847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino, D. C. , Nishida, K. , & Manley, J. L. (2011). Mechanisms and consequences of alternative polyadenylation. Molecular Cell, 43(6), 853–866. 10.1016/j.molcel.2011.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl, B. , Blank, D. , Ohnacker, M. , Friedlein, A. , Roeder, D. , Langen, H. , & Keller, W. (2002). A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Molecular Cell, 10(5), 1139–1150. [DOI] [PubMed] [Google Scholar]
- Dujardin, G. , Lafaille, C. , Petrillo, E. , Buggiano, V. , Gomez Acuña, L. I. , Fiszbein, A. , … Kornblihtt, A. R. (2013). Transcriptional elongation and alternative splicing. Biochimica et Biophysica Acta, 1829(1), 134–140. 10.1016/j.bbagrm.2012.08.005 [DOI] [PubMed] [Google Scholar]
- Eaton, J. D. , Davidson, L. , Bauer, D. L. V. , Natsume, T. , Kanemaki, M. T. , & West, S. (2018). Xrn2 accelerates termination by RNA polymerase II, which is underpinned by CPSF73 activity. Genes & Development, 32(2), 127–139. 10.1101/gad.308528.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle, A. B. , Hessle, V. , Helbig, R. , Dantoft, W. , Gimber, N. , & Visa, N. (2010). Splice‐site mutations cause Rrp6‐mediated nuclear retention of the unspliced RNAs and transcriptional down‐regulation of the splicing‐defective genes. PLoS One, 5(7), e11540 10.1371/journal.pone.0011540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckard, S. C. , Rice, G. I. , Fabre, A. , Badens, C. , Gray, E. E. , Hartley, J. L. , … Stetson, D. B. (2014). The SKIV2L RNA exosome limits activation of the RIG‐I‐like receptors. Nature Immunology, 15(9), 839–845. 10.1038/ni.2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff, S. , O'Reilly, D. , Chapman, R. D. , Taylor, A. , Tanzhaus, K. , Pitts, L. , … Murphy, S. (2007). Serine‐7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science, 318(5857), 1777–1779. 10.1126/science.1145989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff, S. , Szczepaniak, S. A. , Dienstbier, M. , Taylor, A. , Knight, S. , & Murphy, S. (2010). The integrator complex recognizes a new double mark on the RNA polymerase II carboxyl‐terminal domain. Journal of Biological Chemistry, 285(27), 20564–20569. 10.1074/jbc.M110.132530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff, S. , Zaborowska, J. , Laitem, C. , Kiss, T. , & Murphy, S. (2012). Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Molecular Cell, 45(1), 111–122. 10.1016/j.molcel.2011.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick, D. , & Geyer, M. (2013). The RNA polymerase II carboxy‐terminal domain (CTD) code. Chemical Reviews, 113(11), 8456–8490. 10.1021/cr400071f [DOI] [PubMed] [Google Scholar]
- Elkon, R. , Ugalde, A. P. , & Agami, R. (2013). Alternative cleavage and polyadenylation: Extent, regulation and function. Nature Reviews Genetics, 14(7), 496–506. 10.1038/nrg3482 [DOI] [PubMed] [Google Scholar]
- Erickson, B. , Sheridan, R. M. , Cortazar, M. , & Bentley, D. L. (2018). Dynamic turnover of paused Pol II complexes at human promoters. Genes & Development, 32(17–18), 1215–1225. 10.1101/gad.316810.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian, M. R. , Sonenberg, N. , & Filipowicz, W. (2010). Regulation of mRNA translation and stability by microRNAs. Annual Review of Biochemistry, 79, 351–379. 10.1146/annurev-biochem-060308-103103 [DOI] [PubMed] [Google Scholar]
- Fica, S. M. , & Nagai, K. (2017). Cryo‐electron microscopy snapshots of the spliceosome: Structural insights into a dynamic ribonucleoprotein machine. Nature Structural & Molecular Biology, 24(10), 791–799. 10.1038/nsmb.3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finci, L. I. , Zhang, X. , Huang, X. , Zhou, Q. , Tsai, J. , Teng, T. , … Larsen, N. A. (2018). The cryo‐EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre‐mRNA substrate competitive mechanism of action. Genes & Development, 32(3–4), 309–320. 10.1101/gad.311043.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty, S. M. , Fortes, P. , Izaurralde, E. , Mattaj, I. W. , & Gilmartin, G. M. (1997). Participation of the nuclear cap binding complex in pre‐mRNA 3′ processing. Proceedings of the National Academy of Sciences of the United States of America, 94(22), 11893–11898. 10.1073/pnas.94.22.11893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, N. , & Bentley, D. L. (2001). Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: Different functions for different segments of the CTD. Genes & Development, 15(14), 1783–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, N. , Brannan, K. , Erickson, B. , Kim, H. , Cortazar, M. A. , Sheridan, R. M. , … Bentley, D. L. (2015). Effects of transcription elongation rate and Xrn2 exonuclease activity on RNA polymerase II termination suggest widespread kinetic competition. Molecular Cell, 60(2), 256–267. 10.1016/j.molcel.2015.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, M. J. , Gao, H. , Smith‐Kinnaman, W. R. , Liu, Y. , & Mosley, A. L. (2015). The exosome component Rrp6 is required for RNA polymerase II termination at specific targets of the Nrd1‐Nab3 pathway. PLoS Genetics, 11(2), e1004999 10.1371/journal.pgen.1004999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks, T. M. , & Lykke‐Andersen, J. (2008). The control of mRNA decapping and P‐body formation. Molecular Cell, 32(5), 605–615. 10.1016/j.molcel.2008.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi, Y. , & Shatkin, A. J. (2000). Viral and cellular mRNA capping: Past and prospects. Advances in Virus Research, 55, 135–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusby, B. , Kim, S. , Erickson, B. , Kim, H. , Peterson, M. L. , & Bentley, D. L. (2016). Coordination of RNA polymerase II pausing and 3′ end processing factor recruitment with alternative polyadenylation. Molecular and Cellular Biology, 36(2), 295–303. 10.1128/MCB.00898-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galej, W. P. , Wilkinson, M. E. , Fica, S. M. , Oubridge, C. , Newman, A. J. , & Nagai, K. (2016). Cryo‐EM structure of the spliceosome immediately after branching. Nature, 537(7619), 197–201. 10.1038/nature19316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy, V. , Gadal, O. , Fromont‐Racine, M. , Romano, A. , Jacquier, A. , & Nehrbass, U. (2004). Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell, 116(1), 63–73. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami, S. , Huh, W. , Bower, K. , Howson, R. W. , Belle, A. , Dephoure, N. , … Weissman, J. S. (2003). Global analysis of protein expression in yeast. Nature, 425(6959), 737–741. 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- Ghosh, A. , Shuman, S. , & Lima, C. D. (2011). Structural insights to how mammalian capping enzyme reads the CTD code. Molecular Cell, 43(2), 299–310. 10.1016/j.molcel.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, W. , & Guthrie, C. (2004). The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Molecular Cell, 13(2), 201–212. [DOI] [PubMed] [Google Scholar]
- Girard, C. , Will, C. L. , Peng, J. , Makarov, E. M. , Kastner, B. , Lemm, I. , … Luhrmann, R. (2012). Post‐transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nature Communications, 3, 994 10.1038/ncomms1998 [DOI] [PubMed] [Google Scholar]
- Glover‐Cutter, K. , Kim, S. , Espinosa, J. , & Bentley, D. L. (2008). RNA polymerase II pauses and associates with pre‐mRNA processing factors at both ends of genes. Nature Structural & Molecular Biology, 15(1), 71–78. 10.1038/nsmb1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf, A. L. (1993). Positive patches and negative noodles: Linking RNA processing to transcription? Trends in Biochemical Sciences, 18(4), 117–119. [DOI] [PubMed] [Google Scholar]
- Gromak, N. , West, S. , & Proudfoot, N. J. (2006). Pause sites promote transcriptional termination of mammalian RNA polymerase II. Molecular and Cellular Biology, 26(10), 3986–3996. 10.1128/MCB.26.10.3986-3996.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grudzien‐Nogalska, E. , & Kiledjian, M. (2017). New insights into decapping enzymes and selective mRNA decay. WIREs RNA, 8(1), e1379 10.1002/wrna.1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzechnik, P. , Gdula, M. R. , & Proudfoot, N. J. (2015). Pcf11 orchestrates transcription termination pathways in yeast. Genes & Development, 29(8), 849–861. 10.1101/gad.251470.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipati, R. K. , Villa, T. , Boulay, J. , & Libri, D. (2008). Phosphorylation of the RNA polymerase II C‐terminal domain dictates transcription termination choice. Nature Structural & Molecular Biology, 15(8), 786–794. 10.1038/nsmb.1460 [DOI] [PubMed] [Google Scholar]
- Gudipati, R. K. , Xu, Z. , Lebreton, A. , Seraphin, B. , Steinmetz, L. M. , Jacquier, A. , & Libri, D. (2012). Extensive degradation of RNA precursors by the exosome in wild‐type cells. Molecular Cell, 48(3), 409–421. 10.1016/j.molcel.2012.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarov, I. , & Nudler, E. (1999). The mechanism of intrinsic transcription termination. Molecular Cell, 3(4), 495–504. [DOI] [PubMed] [Google Scholar]
- Haag, J. R. , & Pikaard, C. S. (2011). Multisubunit RNA polymerases IV and V: Purveyors of non‐coding RNA for plant gene silencing. Nature Reviews Molecular Cell Biology, 12(8), 483–492. 10.1038/nrm3152 [DOI] [PubMed] [Google Scholar]
- Halbeisen, R. E. , Galgano, A. , Scherrer, T. , & Gerber, A. P. (2008). Post‐transcriptional gene regulation: From genome‐wide studies to principles. Cellular and Molecular Life Sciences, 65(5), 798–813. 10.1007/s00018-007-7447-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell, C. M. , Gross, S. , Zenklusen, D. , Heath, C. V. , Stutz, F. , Moore, C. , & Cole, C. N. (2002). Coupling of termination, 3′ processing, and mRNA export. Molecular and Cellular Biology, 22(18), 6441–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Z. , Libri, D. , & Porrua, O. (2017). Biochemical characterization of the helicase Sen1 provides new insights into the mechanisms of non‐coding transcription termination. Nucleic Acids Research, 45(3), 1355–1370. 10.1093/nar/gkw1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlen, K. M. , Trotta, K. L. , Smith, E. E. , Mosaheb, M. M. , Fuchs, S. M. , & Churchman, L. S. (2016). Comprehensive RNA polymerase II interactomes reveal distinct and varied roles for each phospho‐CTD residue. Cell Reports, 15(10), 2147–2158. 10.1016/j.celrep.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbaker, D. Z. , Marquardt, S. , Wlotzka, W. , & Buratowski, S. (2013). Kinetic competition between RNA polymerase II and Sen1‐dependent transcription termination. Molecular Cell, 49(1), 55–66. 10.1016/j.molcel.2012.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques, T. , Gilchrist, D. A. , Nechaev, S. , Bern, M. , Muse, G. W. , Burkholder, A. , … Adelman, K. (2013). Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Molecular Cell, 52(4), 517–528. 10.1016/j.molcel.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo, D. H. , Yoo, I. , Kong, J. , Lidschreiber, M. , Mayer, A. , Choi, B. Y. , … Kim, M. (2013). The RNA polymerase II C‐terminal domain‐interacting domain of yeast Nrd1 contributes to the choice of termination pathway and couples to RNA processing by the nuclear exosome. Journal of Biological Chemistry, 288(51), 36676–36690. 10.1074/jbc.M113.508267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández, H. , & Robinson, C. V. (2007). Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nature Protocols, 2(3), 715–726. 10.1038/nprot.2007.73 [DOI] [PubMed] [Google Scholar]
- Herzel, L. , Ottoz, D. S. M. , Alpert, T. , & Neugebauer, K. M. (2017). Splicing and transcription touch base: Co‐transcriptional spliceosome assembly and function. Nature Reviews Molecular Cell Biology, 18(10), 637–650. 10.1038/nrm.2017.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessle, V. , von Euler, A. , Gonzalez de Valdivia, E. , & Visa, N. (2012). Rrp6 is recruited to transcribed genes and accompanies the spliced mRNA to the nuclear pore. RNA, 18(8), 1466–1474. 10.1261/rna.032045.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus, H. , Yu, M. C. , & Silver, P. A. (2004). Genome‐wide mRNA surveillance is coupled to mRNA export. Genes & Development, 18(21), 2652–2662. 10.1101/gad.1241204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren, P. , & Parker, R. (2001). Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3′‐end formation of nascent transcripts. RNA, 7(5), 753–764. 10.1017/S1355838201010147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honorine, R. , Mosrin‐Huaman, C. , Hervouet‐Coste, N. , Libri, D. , & Rahmouni, A. R. (2011). Nuclear mRNA quality control in yeast is mediated by Nrd1 co‐transcriptional recruitment, as revealed by the targeting of rho‐induced aberrant transcripts. Nucleic Acids Research, 39(7), 2809–2820. 10.1093/nar/gkq1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley, J. , Kotovic, K. , El Hage, A. , & Tollervey, D. (2007). Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO Journal, 26(24), 4996–5006. 10.1038/sj.emboj.7601921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrossova, D. , Sikorsky, T. , Potesil, D. , Bartosovic, M. , Pasulka, J. , Zdrahal, Z. , … Vanacova, S. (2015). RBM7 subunit of the NEXT complex binds U‐rich sequences and targets 3′‐end extended forms of snRNAs. Nucleic Acids Research, 43(8), 4236–4248. 10.1093/nar/gkv240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin, J. P. , Sheth, A. , & Manley, J. L. (2011). RNAP II CTD phosphorylated on threonine‐4 is required for histone mRNA 3′ end processing. Science, 334(6056), 683–686. 10.1126/science.1206034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, G. O. , Fox, M. J. , Smith‐Kinnaman, W. R. , Gogol, M. , Fleharty, B. , & Mosley, A. L. (2016). Phosphatase Rtr1 regulates global levels of serine 5 RNA polymerase II C‐terminal domain phosphorylation and cotranscriptional histone methylation. Molecular and Cellular Biology, 36(17), 2236–2245. 10.1128/MCB.00870-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada, T. (2013). Quality control systems for aberrant mRNAs induced by aberrant translation elongation and termination. Biochimica et Biophysica Acta, 1829(6–7), 634–642. 10.1016/j.bbagrm.2013.02.004 [DOI] [PubMed] [Google Scholar]
- Itoh, N. , Yamada, H. , Kaziro, Y. , & Mizumoto, K. (1987). Messenger‐RNA guanylyltransferase from Saccharomyces cerevisiae—Large‐scale purification, subunit functions, and subcellular‐localization. Journal of Biological Chemistry, 262(5), 1989–1995. [PubMed] [Google Scholar]
- Izaurralde, E. , Lewis, J. , Gamberi, C. , Jarmolowski, A. , McGuigan, C. , & Mattaj, I. W. (1995). A cap‐binding protein complex mediating U snRNA export. Nature, 376(6542), 709–712. 10.1038/376709a0 [DOI] [PubMed] [Google Scholar]
- Izaurralde, E. , Lewis, J. , McGuigan, C. , Jankowska, M. , Darzynkiewicz, E. , & Mattaj, I. W. (1994). A nuclear cap binding protein complex involved in pre‐mRNA splicing. Cell, 78(4), 657–668. [DOI] [PubMed] [Google Scholar]
- Jamonnak, N. , Creamer, T. J. , Darby, M. M. , Schaughency, P. , Wheelan, S. J. , & Corden, J. L. (2011). Yeast Nrd1, Nab3, and Sen1 transcriptome‐wide binding maps suggest multiple roles in post‐transcriptional RNA processing. RNA, 17(11), 2011–2025. 10.1261/rna.2840711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal, M. , Elkon, R. , Loayza‐Puch, F. , van Haaften, G. , Kühn, U. , Menzies, F. M. , … Agami, R. (2012). The poly(A)‐binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell, 149(3), 538–553. 10.1016/j.cell.2012.03.022 [DOI] [PubMed] [Google Scholar]
- Jiao, X. , Chang, J. H. , Kilic, T. , Tong, L. , & Kiledjian, M. (2013). A mammalian pre‐mRNA 5′ end capping quality control mechanism and an unexpected link of capping to pre‐mRNA processing. Molecular Cell, 50(1), 104–115. 10.1016/j.molcel.2013.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, X. , Xiang, S. , Oh, C. , Martin, C. E. , Tong, L. , & Kiledjian, M. (2010). Identification of a quality‐control mechanism for mRNA 5′‐end capping. Nature, 467(7315), 608–611. 10.1038/nature09338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno‐Gonzalez, S. , Haaning, L. L. , Malagon, F. , & Jensen, T. H. (2010). The yeast 5′–3′ exonuclease Rat1p functions during transcription elongation by RNA polymerase II. Molecular Cell, 37(4), 580–587. 10.1016/j.molcel.2010.01.019 [DOI] [PubMed] [Google Scholar]
- Johnson, S. A. , Cubberley, G. , & Bentley, D. L. (2009). Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Molecular Cell, 33(2), 215–226. 10.1016/j.molcel.2008.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida, D. (2016). The reciprocal regulation between splicing and 3′‐end processing. WIREs RNA, 7(4), 499–511. 10.1002/wrna.1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karousis, E. D. , Nasif, S. , & Muhlemann, O. (2016). Nonsense‐mediated mRNA decay: Novel mechanistic insights and biological impact. WIREs RNA, 7(5), 661–682. 10.1002/wrna.1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi, J. , Mischo, H. , Braglia, P. , Rondon, A. , & Proudfoot, N. J. (2008). Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes & Development, 22(8), 1082–1092. 10.1101/gad.463408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerwitz, Y. , Kuhn, U. , Lilie, H. , Knoth, A. , Scheuermann, T. , Friedrich, H. , … Wahle, E. (2003). Stimulation of poly(A) polymerase through a direct interaction with the nuclear poly(A) binding protein allosterically regulated by RNA. EMBO Journal, 22(14), 3705–3714. 10.1093/emboj/cdg347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor, Y. L. , Menet, J. S. , Tolan, M. , & Rosbash, M. (2012). Cotranscriptional splicing efficiency differs dramatically between Drosophila and mouse. RNA, 18(12), 2174–2186. 10.1261/rna.034090.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor, Y. L. , Rodriguez, J. , Abruzzi, K. C. , Tang, C. H. , Marr, M. T., 2nd , & Rosbash, M. (2011). Nascent‐seq indicates widespread cotranscriptional pre‐mRNA splicing in Drosophila . Genes & Development, 25(23), 2502–2512. 10.1101/gad.178962.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilchert, C. , Wittmann, S. , Passoni, M. , Shah, S. , Granneman, S. , & Vasiljeva, L. (2015). Regulation of mRNA levels by decay‐promoting introns that recruit the exosome specificity factor Mmi1. Cell Reports, 13(11), 2504–2515. 10.1016/j.celrep.2015.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M. , Ahn, S. H. , Krogan, N. J. , Greenblatt, J. F. , & Buratowski, S. (2004). Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO Journal, 23(2), 354–364. 10.1038/sj.emboj.7600053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M. , Krogan, N. J. , Vasiljeva, L. , Rando, O. J. , Nedea, E. , Greenblatt, J. F. , & Buratowski, S. (2004). The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature, 432(7016), 517–522. 10.1038/nature03041 [DOI] [PubMed] [Google Scholar]
- Kim, W. , Bennett, E. J. , Huttlin, E. L. , Guo, A. , Li, J. , Possemato, A. , … Gygi, S. P. (2011). Systematic and quantitative assessment of the ubiquitin‐modified proteome. Molecular Cell, 44(2), 325–340. 10.1016/j.molcel.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva, M. L. , Nedialkov, Y. A. , Cremona, G. H. , Purtov, Y. A. , Lubkowska, L. , Malagon, F. , … Kashlev, M. (2008). Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Molecular Cell, 30(5), 557–566. 10.1016/j.molcel.2008.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky, P. , Cho, E. J. , & Buratowski, S. (2000). Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & Development, 14(19), 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek, K. , Cerna, H. , Holub, P. , Pasulka, J. , Hrossova, D. , Loehr, F. , … Stefl, R. (2012). Serine phosphorylation and proline isomerization in RNAP II CTD control recruitment of Nrd1. Genes & Development, 26(17), 1891–1896. 10.1101/gad.192781.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehner, J. N. , Pearson, E. , & Moore, C. (2011). Unravelling the means to an end: RNA polymerase II transcription termination. Nature Reviews Molecular Cell Biology, 12(5), 283–294. 10.1038/nrm3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel, J. , Bousquet‐Antonelli, C. , Beggs, J. D. , & Tollervey, D. (2004). Nuclear pre‐mRNA decapping and 5′ degradation in yeast require the Lsm2‐8p complex. Molecular and Cellular Biology, 24(21), 9646–9657. 10.1128/MCB.24.21.9646-9657.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak, H. , Fuda, N. J. , Core, L. J. , & Lis, J. T. (2013). Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science, 339, 950–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, F. , Gardini, A. , Zhang, A. , & Shiekhattar, R. (2015). Integrator mediates the biogenesis of enhancer RNAs. Nature, 525(7569), 399–403. 10.1038/nature14906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. J. , & Glaunsinger, B. A. (2009). Aberrant herpesvirus‐induced polyadenylation correlates with cellular messenger RNA destruction. PLoS Biology, 7(5), e1000107 10.1371/journal.pbio.1000107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay, J. F. , Larochelle, M. , Marguerat, S. , Atkinson, S. , Bahler, J. , & Bachand, F. (2014). The RNA exosome promotes transcription termination of backtracked RNA polymerase II. Nature Structural & Molecular Biology, 21(10), 919–926. 10.1038/nsmb.2893 [DOI] [PubMed] [Google Scholar]
- Lemieux, C. , Marguerat, S. , Lafontaine, J. , Barbezier, N. , Bahler, J. , & Bachand, F. (2011). A pre‐mRNA degradation pathway that selectively targets intron‐containing genes requires the nuclear poly(A)‐binding protein. Molecular Cell, 44(1), 108–119. 10.1016/j.molcel.2011.06.035 [DOI] [PubMed] [Google Scholar]
- Leonaite, B. , Han, Z. , Basquin, J. , Bonneau, F. , Libri, D. , Porrua, O. , & Conti, E. (2017). Sen1 has unique structural features grafted on the architecture of the Upf1‐like helicase family. EMBO Journal, 36(11), 1590–1604. 10.15252/embj.201696174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, D. , Tatomer, D. C. , Luo, Z. , Wu, H. , Yang, L. , Chen, L. L. , … Wilusz, J. E. (2017). The output of protein‐coding genes shifts to circular RNAs when the pre‐mRNA processing machinery is limiting. Molecular Cell, 68(5), 940–954 .e943. 10.1016/j.molcel.2017.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri, D. , Dower, K. , Boulay, J. , Thomsen, R. , Rosbash, M. , & Jensen, T. H. (2002). Interactions between mRNA export commitment, 3′‐end quality control, and nuclear degradation. Molecular and Cellular Biology, 22(23), 8254–8266. 10.1128/mcb.22.23.8254-8266.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi, D. D. , Geiger, G. , Minet, M. , Schroeder, S. , Cilli, K. , McNeil, J. B. , & Bentley, D. L. (2002). Functional interaction of yeast pre‐mRNA 3′ end processing factors with RNA polymerase II. Molecular Cell, 9(5), 1101–1111. [DOI] [PubMed] [Google Scholar]
- Lim, S. J. , Boyle, P. J. , Chinen, M. , Dale, R. K. , & Lei, E. P. (2013). Genome‐wide localization of exosome components to active promoters and chromatin insulators in Drosophila . Nucleic Acids Research, 41(5), 2963–2980. 10.1093/nar/gkt037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loya, T. J. , O'Rourke, T. W. , & Reines, D. (2012). A genetic screen for terminator function in yeast identifies a role for a new functional domain in termination factor Nab3. Nucleic Acids Research, 40(15), 7476–7491. 10.1093/nar/gks377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas, M. , Andersen, P. R. , Schein, A. , Dziembowski, A. , Kudla, G. , & Jensen, T. H. (2015). The human nuclear exosome targeting complex is loaded onto newly synthesized RNA to direct early ribonucleolysis. Cell Reports, 10(2), 178–192. 10.1016/j.celrep.2014.12.026 [DOI] [PubMed] [Google Scholar]
- Lubas, M. , Christensen, M. S. , Kristiansen, M. S. , Domanski, M. , Falkenby, L. G. , Lykke‐Andersen, S. , … Jensen, T. H. (2011). Interaction profiling identifies the human nuclear exosome targeting complex. Molecular Cell, 43(4), 624–637. 10.1016/j.molcel.2011.06.028 [DOI] [PubMed] [Google Scholar]
- Lunde, B. M. , Reichow, S. L. , Kim, M. , Suh, H. , Leeper, T. C. , Yang, F. , … Varani, G. (2010). Cooperative interaction of transcription termination factors with the RNA polymerase II C‐terminal domain. Nature Structural & Molecular Biology, 17(10), 1195–1201. 10.1038/nsmb.1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M. J. , & Reed, R. (1999). Splicing is required for rapid and efficient mRNA export in metazoans. Proceedings of the National Academy of Sciences of the United States of America, 96(26), 14937–14942. 10.1073/pnas.96.26.14937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, W. , Johnson, A. W. , & Bentley, D. L. (2006). The role of Rat1 in coupling mRNA 3′‐end processing to transcription termination: Implications for a unified allosteric‐torpedo model. Genes & Development, 20(8), 954–965. 10.1101/gad.1409106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz, C. S. , & Moreira, A. (2011). Alternative mRNA polyadenylation in eukaryotes: An effective regulator of gene expression. WIREs RNA, 2(1), 22–31. 10.1002/wrna.47 [DOI] [PubMed] [Google Scholar]
- Malagon, F. , Kireeva, M. L. , Shafer, B. K. , Lubkowska, L. , Kashlev, M. , & Strathern, J. N. (2006). Mutations in the Saccharomyces cerevisiae RPB1 gene conferring hypersensitivity to 6‐azauracil. Genetics, 172(4), 2201–2209. 10.1534/genetics.105.052415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal, S. S. , Chu, C. , Wada, T. , Handa, H. , Shatkin, A. J. , & Reinberg, D. (2004). Functional interactions of RNA‐capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proceedings of the National Academy of Sciences of the United States of America, 101(20), 7572–7577. 10.1073/pnas.0401493101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez‐Rucobo, F. W. , Kohler, R. , van de Waterbeemd, M. , Heck, A. J. R. , Hemann, M. , Herzog, F. , … Cramer, P. (2015). Molecular basis of transcription‐coupled pre‐mRNA capping. Molecular Cell, 58(6), 1079–1089. 10.1016/j.molcel.2015.04.004 [DOI] [PubMed] [Google Scholar]
- Mayer, A. , di Iulio, J. , Maleri, S. , Eser, U. , Vierstra, J. , Reynolds, A. , … Churchman, L. S. (2015). Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell, 161(3), 541–554. 10.1016/j.cell.2015.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, A. , Heidemann, M. , Lidschreiber, M. , Schreieck, A. , Sun, M. , Hintermair, C. , … Cramer, P. (2012). CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science, 336(6089), 1723–1725. 10.1126/science.1219651 [DOI] [PubMed] [Google Scholar]
- Mayer, A. , Landry, H. M. , & Churchman, L. S. (2017). Pause & go: From the discovery of RNA polymerase pausing to its functional implications. Current Opinion in Cell Biology, 46, 72–80. 10.1016/j.ceb.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, A. , Lidschreiber, M. , Siebert, M. , Leike, K. , Soding, J. , & Cramer, P. (2010). Uniform transitions of the general RNA polymerase II transcription complex. Nature Structural & Molecular Biology, 17(10), 1272–1278. 10.1038/nsmb.1903 [DOI] [PubMed] [Google Scholar]
- McCracken, S. , Fong, N. , Rosonina, E. , Yankulov, K. , Brothers, G. , Siderovski, D. , … Bentley, D. L. (1997). 5′‐Capping enzymes are targeted to pre‐mRNA by binding to the phosphorylated carboxy‐terminal domain of RNA polymerase II. Genes & Development, 11(24), 3306–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken, S. , Fong, N. , Yankulov, K. , Ballantyne, S. , Pan, G. , Greenblatt, J. , … Bentley, D. L. (1997). The C‐terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature, 385(6614), 357–361. 10.1038/385357a0 [DOI] [PubMed] [Google Scholar]
- Meola, N. , Domanski, M. , Karadoulama, E. , Chen, Y. , Gentil, C. , Pultz, D. , … Jensen, T. H. (2016). Identification of a nuclear exosome decay pathway for processed transcripts. Molecular Cell, 64(3), 520–533. 10.1016/j.molcel.2016.09.025 [DOI] [PubMed] [Google Scholar]
- Milligan, L. , Torchet, C. , Allmang, C. , Shipman, T. , & Tollervey, D. (2005). A nuclear surveillance pathway for mRNAs with defective polyadenylation. Molecular and Cellular Biology, 25(22), 9996–10004. 10.1128/MCB.25.22.9996-10004.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo, H. E. , Chun, Y. , Harlen, K. M. , Smalec, B. M. , Dhir, S. , Churchman, L. S. , & Buratowski, S. (2018). Cell‐cycle modulation of transcription termination factor Sen1. Molecular Cell, 70(2), 312–326. 10.1016/j.molcel.2018.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo, H. E. , Gomez‐Gonzalez, B. , Grzechnik, P. , Rondon, A. G. , Wei, W. , Steinmetz, L. , … Proudfoot, N. J. (2011). Yeast Sen1 helicase protects the genome from transcription‐associated instability. Molecular Cell, 41(1), 21–32. 10.1016/j.molcel.2010.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo, H. E. , & Proudfoot, N. J. (2013). Disengaging polymerase: Terminating RNA polymerase II transcription in budding yeast. Biochimica et Biophysica Acta, 1829(1), 174–185. 10.1016/j.bbagrm.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T. , & Spector, D. L. (1999). RNA polymerase II targets pre‐mRNA splicing factors to transcription sites in vivo. Molecular Cell, 3(6), 697–705. [DOI] [PubMed] [Google Scholar]
- Mitchell, P. , Petfalski, E. , Shevchenko, A. , Mann, M. , & Tollervey, D. (1997). The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell, 91(4), 457–466. [DOI] [PubMed] [Google Scholar]
- Moore, M. J. , Schwartzfarb, E. M. , Silver, P. A. , & Yu, M. C. (2006). Differential recruitment of the splicing machinery during transcription predicts genome‐wide patterns of mRNA splicing. Molecular Cell, 24(6), 903–915. 10.1016/j.molcel.2006.12.006 [DOI] [PubMed] [Google Scholar]
- Morales, J. C. , Richard, P. , Rommel, A. , Fattah, F. J. , Motea, E. A. , Patidar, P. L. , … Boothman, D. A. (2014). Kub5‐Hera, the human Rtt103 homolog, plays dual functional roles in transcription termination and DNA repair. Nucleic Acids Research, 42(8), 4996–5006. 10.1093/nar/gku160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley, A. L. , Pattenden, S. G. , Carey, M. , Venkatesh, S. , Gilmore, J. M. , Florens, L. , … Washburn, M. P. (2009). Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Molecular Cell, 34(2), 168–178. 10.1016/j.molcel.2009.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouaikel, J. , Causse, S. Z. , Rougemaille, M. , Daubenton‐Carafa, Y. , Blugeon, C. , Lemoine, S. , … Libri, D. (2013). High‐frequency promoter firing links THO complex function to heavy chromatin formation. Cell Reports, 5(4), 1082–1094. 10.1016/j.celrep.2013.10.013 [DOI] [PubMed] [Google Scholar]
- Muse, G. W. , Gilchrist, D. A. , Nechaev, S. , Shah, R. , Parker, J. S. , Grissom, S. F. , … Adelman, K. (2007). RNA polymerase is poised for activation across the genome. Nature Genetics, 39(12), 1507–1511. 10.1038/ng.2007.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev, S. , Fargo, D. C. , dos Santos, G. , Liu, L. , Gao, Y. , & Adelman, K. (2010). Global analysis of short RNAs reveals widespread promoter‐proximal stalling and arrest of Pol II in Drosophila . Science, 327(5963), 335–338. 10.1126/science.1181421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemec, C. M. , Yang, F. , Gilmore, J. M. , Hintermair, C. , Ho, Y. H. , Tseng, S. C. , … Ansari, A. Z. (2017). Different phosphoisoforms of RNA polymerase II engage the Rtt103 termination factor in a structurally analogous manner. Proceedings of the National Academy of Sciences of the United States of America, 114(20), E3944–E3953. 10.1073/pnas.1700128114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, T. H. , Galej, W. P. , Bai, X. C. , Savva, C. G. , Newman, A. J. , Scheres, S. H. , & Nagai, K. (2015). The architecture of the spliceosomal U4/U6.U5 tri‐snRNP. Nature, 523(7558), 47–52. 10.1038/nature14548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, T. H. , Galej, W. P. , Fica, S. M. , Lin, P. C. , Newman, A. J. , & Nagai, K. (2016). CryoEM structures of two spliceosomal complexes: Starter and dessert at the spliceosome feast. Current Opinion in Structural Biology, 36, 48–57. 10.1016/j.sbi.2015.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, T. H. , Galej, W. P. , Bai, X. C. , Oubridge, C. , Newman, A. J. , Scheres, S. H. W. , & Nagai, K. (2016). Cryo‐EM structure of the yeast U4/U6.U5 tri‐snRNP at 3.7 A resolution. Nature, 530(7590), 298–302. 10.1038/nature16940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, Z. , Xu, C. , Guo, X. , Hunter, G. O. , Kuznetsova, O. V. , Tempel, W. , … Greenblatt, J. F. (2014). RPRD1A and RPRD1B are human RNA polymerase II C‐terminal domain scaffolds for Ser5 dephosphorylation. Nature Structural & Molecular Biology, 21(8), 686–695. 10.1038/nsmb.2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen, T. W. , & Graveley, B. R. (2010). Expansion of the eukaryotic proteome by alternative splicing. Nature, 463(7280), 457–463. 10.1038/nature08909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima, T. , Gomes, T. , Grosso, A. R. F. , Kimura, H. , Dye, M. J. , Dhir, S. , … Proudfoot, N. J. (2015). Mammalian NET‐Seq reveals genome‐wide nascent transcription coupled to RNA processing. Cell, 161(3), 526–540. 10.1016/j.cell.2015.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima, T. , Rebelo, K. , Gomes, T. , Grosso, A. R. , Proudfoot, N. J. , & Carmo‐Fonseca, M. (2018). RNA polymerase II phosphorylated on CTD serine 5 interacts with the spliceosome during co‐transcriptional splicing. Molecular Cell, 72(2), 369–379. 10.1016/j.molcel.2018.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler, E. (2012). RNA polymerase backtracking in gene regulation and genome instability. Cell, 149(7), 1438–1445. 10.1016/j.cell.2012.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi, M. D. , Ren, L. , Wall, J. S. , Gould, K. L. , & Walz, T. (2007). Structural characterization of the fission yeast U5.U2/U6 spliceosome complex. Proceedings of the National Academy of Sciences of the United States of America, 104(9), 3195–3200. 10.1073/pnas.0611591104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohle, C. , Tesorero, R. , Schermann, G. , Dobrev, N. , Sinning, I. , & Fischer, T. (2016). Transient RNA–DNA hybrids are required for efficient double‐strand break repair. Cell, 167(4), 1001–1013. 10.1016/j.cell.2016.10.001 [DOI] [PubMed] [Google Scholar]
- O'Reilly, D. , Kuznetsova, O. V. , Laitem, C. , Zaborowska, J. , Dienstbier, M. , & Murphy, S. (2014). Human snRNA genes use polyadenylation factors to promote efficient transcription termination. Nucleic Acids Research, 42(1), 264–275. 10.1093/nar/gkt892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo, M. C. , Farina, L. , & Paci, P. (2015). Kinetics effects and modeling of mRNA turnover. WIREs RNA, 6(3), 327–336. 10.1002/wrna.1277 [DOI] [PubMed] [Google Scholar]
- Pandya‐Jones, A. , & Black, D. L. (2009). Co‐transcriptional splicing of constitutive and alternative exons. RNA, 15(10), 1896–1908. 10.1261/rna.1714509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. , Kang, M. , & Kim, M. (2015). Unraveling the mechanistic features of RNA polymerase II termination by the 5′–3′ exoribonuclease Rat1. Nucleic Acids Research, 43(5), 2625–2637. 10.1093/nar/gkv133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, E. , & Moore, C. (2014). The evolutionarily conserved Pol II flap loop contributes to proper transcription termination on short yeast genes. Cell Reports, 9(3), 821–828. 10.1016/j.celrep.2014.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, E. , & Moore, C. L. (2013). Dismantling promoter‐driven RNA polymerase II transcription complexes in vitro by the termination factor Rat1. Journal of Biological Chemistry, 288(27), 19750–19759. 10.1074/jbc.M112.434985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pefanis, E. , Wang, J. , Rothschild, G. , Lim, J. , Chao, J. , Rabadan, R. , … Basu, U. (2014). Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature, 514(7522), 389–393. 10.1038/nature13580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pefanis, E. , Wang, J. , Rothschild, G. , Lim, J. , Kazadi, D. , Sun, J. , … Basu, U. (2015). RNA exosome‐regulated long non‐coding RNA transcription controls super‐enhancer activity. Cell, 161(4), 774–789. 10.1016/j.cell.2015.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, Y. , Schwer, B. , & Shuman, S. (2003). Interactions between fission yeast Cdk9, its cyclin partner Pch1, and mRNA capping enzyme Pct1 suggest an elongation checkpoint for mRNA quality control. Journal of Biological Chemistry, 278, 7180–7188. 10.1074/jbc.M211713200 [DOI] [PubMed] [Google Scholar]
- Perales, R. , & Bentley, D. (2009). "Cotranscriptionality": The transcription elongation complex as a nexus for nuclear transactions. Molecular Cell, 36(2), 178–191. 10.1016/j.molcel.2009.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz Krummel, D. A. , Oubridge, C. , Leung, A. K. , Li, J. , & Nagai, K. (2009). Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature, 458(7237), 475–480. 10.1038/nature07851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrua, O. , Boudvillain, M. , & Libri, D. (2016). Transcription termination: Variations on common themes. Trends in Genetics, 32(8), 508–522. 10.1016/j.tig.2016.05.007 [DOI] [PubMed] [Google Scholar]
- Porrua, O. , Hobor, F. , Boulay, J. , Kubicek, K. , D'Aubenton‐Carafa, Y. , Gudipati, R. K. , … Libri, D. (2012). In vivo SELEX reveals novel sequence and structural determinants of Nrd1‐Nab3‐Sen1‐dependent transcription termination. EMBO Journal, 31(19), 3935–3948. 10.1038/emboj.2012.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrua, O. , & Libri, D. (2013). A bacterial‐like mechanism for transcription termination by the Sen1p helicase in budding yeast. Nature Structural & Molecular Biology, 20(7), 884–891. 10.1038/nsmb.2592 [DOI] [PubMed] [Google Scholar]
- Porrua, O. , & Libri, D. (2015). Transcription termination and the control of the transcriptome: Why, where and how to stop. Nature Reviews Molecular Cell Biology, 16(3), 190–202. 10.1038/nrm3943 [DOI] [PubMed] [Google Scholar]
- Preker, P. , Almvig, K. , Christensen, M. S. , Valen, E. , Mapendano, C. K. , Sandelin, A. , & Jensen, T. H. (2011). PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Research, 39(16), 7179–7193. 10.1093/nar/gkr370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker, P. , Nielsen, J. , Kammler, S. , Lykke‐Andersen, S. , Christensen, M. S. , Mapendano, C. K. , … Jensen, T. H. (2008). RNA exosome depletion reveals transcription upstream of active human promoters. Science, 322(5909), 1851–1854. 10.1126/science.1164096 [DOI] [PubMed] [Google Scholar]
- Proudfoot, N. J. (2016). Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science, 352(6291), aad9926 10.1126/science.aad9926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, E. B. , & Lis, J. T. (1993). In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proceedings of the National Academy of Sciences of the United States of America, 90(17), 7923–7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, R. , & Hurt, E. (2002). A conserved mRNA export machinery coupled to pre‐mRNA splicing. Cell, 108(4), 523–531. [DOI] [PubMed] [Google Scholar]
- Richard, P. , & Manley, J. L. (2009). Transcription termination by nuclear RNA polymerases. Genes & Development, 23(11), 1247–1269. 10.1101/gad.1792809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, G. C. , Gooding, C. , Mak, H. Y. , Proudfoot, N. J. , & Smith, C. W. (1998). Co‐transcriptional commitment to alternative splice site selection. Nucleic Acids Research, 26(24), 5568–5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon, A. G. , Mischo, H. E. , Kawauchi, J. , & Proudfoot, N. J. (2009). Fail‐safe transcriptional termination for protein‐coding genes in S. cerevisiae . Molecular Cell, 36(1), 88–98. 10.1016/j.molcel.2009.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski, M. , Dichtl, B. , Hubner, W. , & Keller, W. (2003). Independent functions of yeast Pcf11p in pre‐mRNA 3′ end processing and in transcription termination. EMBO Journal, 22(9), 2167–2177. 10.1093/emboj/cdg200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saguez, C. , Schmid, M. , Olesen, J. R. , Ghazy, M. A. , Qu, X. , Poulsen, M. B. , … Jensen, T. H. (2008). Nuclear mRNA surveillance in THO/sub2 mutants is triggered by inefficient polyadenylation. Molecular Cell, 31(1), 91–103. 10.1016/j.molcel.2008.04.030 [DOI] [PubMed] [Google Scholar]
- Santos‐Pereira, J. M. , & Aguilera, A. (2015). R loops: New modulators of genome dynamics and function. Nature Reviews Genetics, 16(10), 583–597. 10.1038/nrg3961 [DOI] [PubMed] [Google Scholar]
- Schaefke, B. , Sun, W. , Li, Y. S. , Fang, L. , & Chen, W. (2018). The evolution of posttranscriptional regulation. WIREs RNA, 9, e1485 10.1002/wrna.1485 [DOI] [PubMed] [Google Scholar]
- Schaughency, P. , Merran, J. , & Corden, J. L. (2014). Genome‐wide mapping of yeast RNA polymerase II termination. PLoS Genetics, 10(10), e1004632 10.1371/journal.pgen.1004632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M. , & Jensen, T. H. (2018). Controlling nuclear RNA levels. Nature Reviews Genetics, 19, 518–529. 10.1038/s41576-018-0013-2 [DOI] [PubMed] [Google Scholar]
- Schmidt, K. , & Butler, J. S. (2013). Nuclear RNA surveillance: Role of TRAMP in controlling exosome specificity. WIREs RNA, 4(2), 217–231. 10.1002/wrna.1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C. , Kudla, G. , Wlotzka, W. , Tuck, A. , & Tollervey, D. (2012). Transcriptome‐wide analysis of exosome targets. Molecular Cell, 48(3), 422–433. 10.1016/j.molcel.2012.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonemann, L. , Kuhn, U. , Martin, G. , Schafer, P. , Gruber, A. R. , Keller, W. , … Wahle, E. (2014). Reconstitution of CPSF active in polyadenylation: Recognition of the polyadenylation signal by WDR33. Genes & Development, 28(21), 2381–2393. 10.1101/gad.250985.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreieck, A. , Easter, A. D. , Etzold, S. , Wiederhold, K. , Lidschreiber, M. , Cramer, P. , & Passmore, L. A. (2014). RNA polymerase II termination involves C‐terminal‐domain tyrosine dephosphorylation by CPF subunit Glc7. Nature Structural & Molecular Biology, 21(2), 175–179. 10.1038/nsmb.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, D. , Schwalb, B. , Kiesel, A. , Baejen, C. , Torkler, P. , Gagneur, J. , … Cramer, P. (2013). Transcriptome surveillance by selective termination of noncoding RNA synthesis. Cell, 155(5), 1075–1087. 10.1016/j.cell.2013.10.024 [DOI] [PubMed] [Google Scholar]
- Shi, Y. , Di Giammartino, D. C. , Taylor, D. , Sarkeshik, A. , Rice, W. J. , Yates, J. R., 3rd , … Manley, J. L. (2009). Molecular architecture of the human pre‐mRNA 3′ processing complex. Molecular Cell, 33(3), 365–376. 10.1016/j.molcel.2008.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, S. , & Parker, R. (2014). Quality control of assembly‐defective U1 snRNAs by decapping and 5′‐to‐3′ exonucleolytic digestion. Proceedings of the National Academy of Sciences of the United States of America, 111(32), E3277–E3286. 10.1073/pnas.1412614111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman, S. (1995). Capping enzyme in eukaryotic mRNA synthesis. Progress in Nucleic Acid Research and Molecular Biology, 50, 101–129. [DOI] [PubMed] [Google Scholar]
- Skaar, J. R. , Ferris, A. L. , Wu, X. , Saraf, A. , Khanna, K. K. , Florens, L. , … Pagano, M. (2015). The integrator complex controls the termination of transcription at diverse classes of gene targets. Cell Research, 25(3), 288–305. 10.1038/cr.2015.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti‐Stathaki, K. , Kamieniarz‐Gdula, K. , & Proudfoot, N. J. (2014). R‐loops induce repressive chromatin marks over mammalian gene terminators. Nature, 516(7531), 436–439. 10.1038/nature13787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti‐Stathaki, K. , Proudfoot, N. J. , & Gromak, N. (2011). Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2‐dependent termination. Molecular Cell, 42(6), 794–805. 10.1016/j.molcel.2011.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skruzny, M. , Schneider, C. , Racz, A. , Weng, J. , Tollervey, D. , & Hurt, E. (2009). An endoribonuclease functionally linked to perinuclear mRNP quality control associates with the nuclear pore complexes. PLoS Biology, 7(1), e8 10.1371/journal.pbio.1000008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, M. G. , Li, Y. , & Kiledjian, M. (2010). Multiple mRNA decapping enzymes in mammalian cells. Molecular Cell, 40(3), 423–432. 10.1016/j.molcel.2010.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling, R. (2017). The nuts and bolts of the endogenous spliceosome. WIREs RNA, 8(1), e1377 10.1002/wrna.1377 [DOI] [PubMed] [Google Scholar]
- Steinmetz, E. J. , Conrad, N. K. , Brow, D. A. , & Corden, J. L. (2001). RNA‐binding protein Nrd1 directs poly(A)‐independent 3′‐end formation of RNA polymerase II transcripts. Nature, 413(6853), 327–331. 10.1038/35095090 [DOI] [PubMed] [Google Scholar]
- Steurer, B. , Janssens, R. C. , Geverts, B. , Geijer, M. E. , Wienholz, F. , Theil, A. F. , … Marteijn, J. A. (2018). Live‐cell analysis of endogenous GFP‐RPB1 uncovers rapid turnover of initiating and promoter‐paused RNA polymerase II. Proceedings of the National Academy of Sciences of the United States of America, 115(19), E4368–E4376. 10.1073/pnas.1717920115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller, J. W. , & Hall, B. D. (2002). Evolution of the RNA polymerase II C‐terminal domain. Proceedings of the National Academy of Sciences of the United States of America, 99(9), 6091–6096. 10.1073/pnas.082646199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svejstrup, J. Q. (2007). Contending with transcriptional arrest during RNAPII transcript elongation. Trends in Biochemical Sciences, 32(4), 165–171. 10.1016/j.tibs.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Szczepinska, T. , Kalisiak, K. , Tomecki, R. , Labno, A. , Borowski, L. S. , Kulinski, T. M. , … Dziembowski, A. (2015). DIS3 shapes the RNA polymerase II transcriptome in humans by degrading a variety of unwanted transcripts. Genome Research, 25(11), 1622–1633. 10.1101/gr.189597.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan‐Wong, S. M. , Zaugg, J. B. , Camblong, J. , Xu, Z. , Zhang, D. W. , Mischo, H. E. , … Proudfoot, N. J. (2012). Gene loops enhance transcriptional directionality. Science, 338(6107), 671–675. 10.1126/science.1224350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebaut, M. , Kisseleva‐Romanova, E. , Rougemaille, M. , Boulay, J. , & Libri, D. (2006). Transcription termination and nuclear degradation of cryptic unstable transcripts: A role for the nrd1‐nab3 pathway in genome surveillance. Molecular Cell, 23(6), 853–864. 10.1016/j.molcel.2006.07.029 [DOI] [PubMed] [Google Scholar]
- Tian, B. , Hu, J. , Zhang, H. , & Lutz, C. S. (2005). A large‐scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Research, 33(1), 201–212. 10.1093/nar/gki158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, B. , & Manley, J. L. (2013). Alternative cleavage and polyadenylation: The long and short of it. Trends in Biochemical Sciences, 38(6), 312–320. 10.1016/j.tibs.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietjen, J. R. , Zhang, D. W. , Rodriguez‐Molina, J. B. , White, B. E. , Akhtar, M. S. , Heidemann, M. , … Ansari, A. Z. (2010). Chemical‐genomic dissection of the CTD code. Nature Structural & Molecular Biology, 17(9), 1154–1161. 10.1038/nsmb.1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner, H. , Knowles, D. G. , Johnson, R. , Davis, C. A. , Chakrabortty, S. , Djebali, S. , … Guigo, R. (2012). Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co‐transcriptional in the human genome but inefficient for lncRNAs. Genome Research, 22(9), 1616–1625. 10.1101/gr.134445.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topisirovic, I. , Svitkin, Y. V. , Sonenberg, N. , & Shatkin, A. J. (2011). Cap and cap‐binding proteins in the control of gene expression. WIREs RNA, 2(2), 277–298. 10.1002/wrna.52 [DOI] [PubMed] [Google Scholar]
- Torchet, C. , Bousquet‐Antonelli, C. , Milligan, L. , Thompson, E. , Kufel, J. , & Tollervey, D. (2002). Processing of 3′‐extended read‐through transcripts by the exosome can generate functional mRNAs. Molecular Cell, 9(6), 1285–1296. [DOI] [PubMed] [Google Scholar]
- Tudek, A. , Porrua, O. , Kabzinski, T. , Lidschreiber, M. , Kubicek, K. , Fortova, A. , … Libri, D. (2014). Molecular basis for coordinating transcription termination with noncoding RNA degradation. Molecular Cell, 55(3), 467–481. 10.1016/j.molcel.2014.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valen, E. , Preker, P. , Andersen, P. R. , Zhao, X. , Chen, Y. , Ender, C. , … Jensen, T. H. (2011). Biogenic mechanisms and utilization of small RNAs derived from human protein‐coding genes. Nature Structural & Molecular Biology, 18(9), 1075–1082. 10.1038/nsmb.2091 [DOI] [PubMed] [Google Scholar]
- van Hoof, A. , Lennertz, P. , & Parker, R. (2000). Yeast exosome mutants accumulate 3′‐extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Molecular and Cellular Biology, 20(2), 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva, L. , & Buratowski, S. (2006). Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Molecular Cell, 21(2), 239–248. 10.1016/j.molcel.2005.11.028 [DOI] [PubMed] [Google Scholar]
- Vasiljeva, L. , Kim, M. , Mutschler, H. , Buratowski, S. , & Meinhart, A. (2008). The Nrd1‐Nab3‐Sen1 termination complex interacts with the Ser5‐phosphorylated RNA polymerase II C‐terminal domain. Nature Structural & Molecular Biology, 15(8), 795–804. 10.1038/nsmb.1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva, L. , Kim, M. , Terzi, N. , Soares, L. M. , & Buratowski, S. (2008). Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Molecular Cell, 29(3), 313–323. 10.1016/j.molcel.2008.01.011 [DOI] [PubMed] [Google Scholar]
- Viladevall, L. , St Amour, C. V. , Rosebrock, A. , Schneider, S. , Zhang, C. , Allen, J. J. , … Fisher, R. P. (2009). TFIIH and P‐TEFb coordinate transcription with capping enzyme recruitment at specific genes in fission yeast. Molecular Cell, 33(6), 738–751. 10.1016/j.molcel.2009.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagschal, A. , Rousset, E. , Basavarajaiah, P. , Contreras, X. , Harwig, A. , Laurent‐Chabalier, S. , … Kiernan, R. (2012). Microprocessor, Setx, Xrn2, and Rrp6 co‐operate to induce premature termination of transcription by RNAPII. Cell, 150(6), 1147–1157. 10.1016/j.cell.2012.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba, L. , Gore, S. K. , & Koshland, D. (2013). The homologous recombination machinery modulates the formation of RNA–DNA hybrids and associated chromosome instability. eLife, 2, e00505 10.7554/eLife.00505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, J. , Yourshaw, M. , Mamsa, H. , Rudnik‐Schoneborn, S. , Menezes, M. P. , Hong, J. E. , … Jen, J. C. (2012). Mutations in the RNA exosome component gene EXOSC3 cause pontocerebellar hypoplasia and spinal motor neuron degeneration. Nature Genetics, 44(6), 704–708. 10.1038/ng.2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, R. , Yan, C. , Bai, R. , Wang, L. , Huang, M. , Wong, C. C. , & Shi, Y. (2016). The 3.8 A structure of the U4/U6.U5 tri‐snRNP: Insights into spliceosome assembly and catalysis. Science, 351(6272), 466–475. 10.1126/science.aad6466 [DOI] [PubMed] [Google Scholar]
- Wang, S. W. , Stevenson, A. L. , Kearsey, S. E. , Watt, S. , & Bahler, J. (2008). Global role for polyadenylation‐assisted nuclear RNA degradation in posttranscriptional gene silencing. Molecular and Cellular Biology, 28(2), 656–665. 10.1128/MCB.01531-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani, S. , Yuda, M. , Fujiwara, Y. , Yamamoto, M. , Harada, F. , Ohkuma, Y. , & Hirose, Y. (2014). Vertebrate Ssu72 regulates and coordinates 3′‐end formation of RNAs transcribed by RNA polymerase II. PLoS One, 9(8), e106040 10.1371/journal.pone.0106040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, S. , Gromak, N. , & Proudfoot, N. J. (2004). Human 5′→3′ exonuclease Xrn2 promotes transcription termination at co‐transcriptional cleavage sites. Nature, 432(7016), 522–525. 10.1038/nature03035 [DOI] [PubMed] [Google Scholar]
- Wilusz, C. J. , Wormington, M. , & Peltz, S. W. (2001). The cap‐to‐tail guide to mRNA turnover. Nature Reviews Molecular Cell Biology, 2(4), 237–246. 10.1038/35067025 [DOI] [PubMed] [Google Scholar]
- Wilusz, J. E. (2018). A 360° view of circular RNAs: From biogenesis to functions. WIREs RNA, 9, e1478 10.1002/wrna.1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windhager, L. , Bonfert, T. , Burger, K. , Ruzsics, Z. , Krebs, S. , Kaufmann, S. , … Dolken, L. (2012). Ultrashort and progressive 4sU‐tagging reveals key characteristics of RNA processing at nucleotide resolution. Genome Research, 22(10), 2031–2042. 10.1101/gr.131847.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlotzka, W. , Kudla, G. , Granneman, S. , & Tollervey, D. (2011). The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO Journal, 30(9), 1790–1803. 10.1038/emboj.2011.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward, L. A. , Mabin, J. W. , Gangras, P. , & Singh, G. (2017). The exon junction complex: A lifelong guardian of mRNA fate. WIREs RNA, 8(3), e1477 10.1002/wrna.1411 [DOI] [PubMed] [Google Scholar]
- Wyers, F. , Rougemaille, M. , Badis, G. , Rousselle, J. C. , Dufour, M. E. , Boulay, J. , … Jacquier, A. (2005). Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell, 121(5), 725–737. 10.1016/j.cell.2005.04.030 [DOI] [PubMed] [Google Scholar]
- Xiang, K. , Tong, L. , & Manley, J. L. (2014). Delineating the structural blueprint of the pre‐mRNA 3′‐end processing machinery. Molecular and Cellular Biology, 34(11), 1894–1910. 10.1128/MCB.00084-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, S. , Cooper‐Morgan, A. , Jiao, X. F. , Kiledjian, M. , Manley, J. L. , & Tong, L. (2009). Structure and function of the 5′→3′ exoribonuclease Rat1 and its activating partner Rai1. Nature, 458(7239), 784–788. 10.1038/nature07731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, Y. , Bai, X. X. , Lee, I. , Kallstrom, G. , Ho, J. , Brown, J. , … Johnson, A. W. (2000). Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Molecular and Cellular Biology, 20(11), 4006–4015. 10.1128/Mcb.20.11.4006-4015.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, C. Y. , Hang, J. , Wan, R. X. , Huang, M. , Wong, C. C. L. , & Shi, Y. G. (2015). Structure of a yeast spliceosome at 3.6 Å resolution. Science, 349(6253), 1182–1191. 10.1126/science.aac7629 [DOI] [PubMed] [Google Scholar]
- Yang, C. , Hager, P. W. , & Stiller, J. W. (2014). The identification of putative RNA polymerase II C‐terminal domain associated proteins in red and green algae. Transcription, 5(5), e970944 10.4161/21541264.2014.970944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, Z. , Maldonado, E. , Pillutla, R. , Cho, H. , Reinberg, D. , & Shatkin, A. J. (1997). Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proceedings of the National Academy of Sciences of the United States of America, 94(24), 12898–12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborowska, J. , Egloff, S. , & Murphy, S. (2016). The pol II CTD: New twists in the tail. Nature Structural & Molecular Biology, 23(9), 771–777. 10.1038/nsmb.3285 [DOI] [PubMed] [Google Scholar]
- Zenklusen, D. , Larson, D. R. , & Singer, R. H. (2008). Single‐RNA counting reveals alternative modes of gene expression in yeast. Nature Structural & Molecular Biology, 15(12), 1263–1271. 10.1038/nsmb.1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, L. T. , & Xiang, S. (2014). mRNA quality control at the 5′ end. Journal of Zhejiang University. Science. B, 15(5), 438–443. 10.1631/jzus.B1400070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Rigo, F. , & Martinson, H. G. (2015). Poly(A) signal‐dependent transcription termination occurs through a conformational change mechanism that does not require cleavage at the poly(A) site. Molecular Cell, 59(3), 437–448. 10.1016/j.molcel.2015.06.008 [DOI] [PubMed] [Google Scholar]
- Zhao, D. Y. , Gish, G. , Braunschweig, U. , Li, Y. , Ni, Z. , Schmitges, F. W. , … Greenblatt, J. F. (2016). SMN and symmetric arginine dimethylation of RNA polymerase II C‐terminal domain control termination. Nature, 529(7584), 48–53. 10.1038/nature16469 [DOI] [PubMed] [Google Scholar]
- Zhou, M. , & Law, J. A. (2015). RNA Pol IV and V in gene silencing: Rebel polymerases evolving away from Pol II's rules. Current Opinion in Plant Biology, 27, 154–164. 10.1016/j.pbi.2015.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder, J. C. , & Lima, C. D. (2017). Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes & Development, 31(2), 88–100. 10.1101/gad.294769.116 [DOI] [PMC free article] [PubMed] [Google Scholar]


