Abstract
Tristetraprolin (TTP), the prototype member of the protein family of the same name, was originally discovered as the product of a rapidly inducible gene in mouse cells. Development of a knockout (KO) mouse established that absence of the protein led to a severe inflammatory syndrome, due in part to elevated levels of tumor necrosis factor (TNF). TTP was found to bind directly and with high affinity to specific AU-rich sequences in the 3’-untranslated region of the TNF mRNA. This initial binding led to promotion of TNF mRNA decay and inhibition of its translation. Many additional TTP target mRNAs have since been identified, some of which are cytokines and chemokines involved in the inflammatory response.
There are three other proteins in the mouse with similar activities and domain structures, but whose KO phenotypes are remarkably different. Moreover, proteins with similar domain structures and activities have been found throughout eukaryotes, demonstrating that this protein family arose from an ancient ancestor.
The defining characteristic of this protein family is the tandem zinc finger (TZF) domain, a 64 amino acid sequence with many conserved residues that is responsible for the direct RNA binding. We discuss here many aspects of this protein domain that have been elucidated since the original discovery of TTP, including its sequence conservation throughout eukarya; its apparent continued evolution in some lineages; its functional dependence on many key conserved residues; its “interchangeability” among evolutionarily distant species; and the evidence that RNA binding is required for the physiological functions of the proteins.
Graphical Abstract
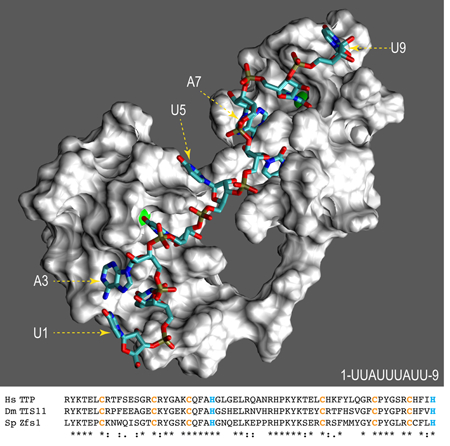
Solution structure model showing a surface representation of the tandem zinc finger domain of human TTP. Zinc atoms are represented by green spheres, and the numbered RNA oligonucleotide by colored sticks. Shown underneath are the aligned domains from human (Hs) TTP, Drosophila (Ds) TIS11, and S. pombe (Sp) Zfs1.
Introduction
Members of the tristetraprolin (TTP) family of mRNA binding proteins can bind directly to AU-rich elements within mRNAs, and then promote decay of those mRNAs and/or inhibit their translation (Brooks and Blackshear 2013, Wells, Perera et al. 2017). The earliest example of this activity was seen with mouse TTP, which was found to bind directly to AU-rich elements within the 3’-untranslated region (UTR) of the mRNA encoding tumor necrosis factor alpha (TNF), promoting decay of the mRNA and inhibiting production and secretion of the TNF protein, a major pro-inflammatory cytokine (Taylor, Carballo et al. 1996, Carballo, Gilkeson et al. 1997, Carballo, Lai et al. 1998). Since then, TTP has been implicated in the control of the stability of many other mRNAs, many of them involved in the regulation of the innate immune response.
TTP is the prototype of a small family of related proteins, totaling four in the mouse and three in humans, that act in essentially the same way in vitro and in cell assays (Lai, Carballo et al. 2000, Lai, Kennington et al. 2003). These proteins are encoded by different genes on different chromosomes, and their complete knock-out (KO) in the mouse leads to dramatically different phenotypes. TTP KO mice exhibit a severe systemic inflammatory syndrome, due in part to the TNF excess (Taylor, Carballo et al. 1996); ZFP36L1-deficient mice die at mid-gestation from failure of chorioallantoic fusion (Stumpo, Byrd et al. 2004); ZFP36L2-deficient mice exhibit defects in hematopoiesis (Stumpo, Broxmeyer et al. 2009); and ZFP36L3 KO mice have defects in placental physiology and overall fertility (Stumpo, Byrd et al. 2004). Since all four proteins behave similarly in transfection assays of RNA binding, and mRNA deadenylation and decay, a major and persistent question concerns how the very specific and different phenotypes of the KO mice are mediated.
We have found members of the TTP family in all four kingdoms of eukarya, using as the defining characteristic the presence of the RNA-binding tandem zinc finger (TZF) domain (Blackshear and Perera 2014, Wells, Perera et al. 2017). We have found that TZF domains from human and very distant organisms, such as lower plants, bind the characteristic RNA binding sites with remarkably similar binding attributes in terms of affinity and target sequence specificity. Thus, it appears that this protein module has maintained essentially the same function for more than a billion years. The purpose of this review is to summarize recent evidence from our laboratory and others on several aspects of this protein domain, including: Its key amino acid residues as applied to its predicted structure and RNA binding; the apparent continued evolution of this domain in some lineages; the concept of interchangeability of this domain among very distant species; the possibilities of regulation of RNA binding by protein modification; and the proposal that all aspects of protein function can be abrogated by point mutations in the TZF domain that prevent RNA binding. For more general recent discussions of the proteins of this family, the reader is referred to the following reviews (Baou, Jewell et al. 2009, Zanocco-Marani 2010, Ross, Brennan-Laun et al. 2012, Sanduja, Blanco et al. 2012, Brooks and Blackshear 2013, Prabhala and Ammit 2015, Guo, Wang et al. 2017, Maeda and Akira 2017, Wells, Perera et al. 2017, Gupta, Bebawy et al. 2018, Park, Lee et al. 2018). For discussions of zinc fingers in general, see the following reviews: (Laity, Lee et al. 2001, Klug 2010, Cassandri, Smirnov et al. 2017, Fu and Blackshear 2017, Abbehausen 2019).
Amino acid frequencies
The TZF domain as discussed here consists of 64 amino acids containing two CCCH zinc fingers, separated by 18 amino acids, with a six-residue conserved sequence leading into each finger. For a current snapshot of residue conservation at each site, we recently searched (on 7–27-18) the GenBank non-redundant protein sequences (nr) database, specifying “eukaryotes”, using a significance cutoff of e-24, with the TZF domain protein sequence from human TTP, comprising amino acids 103–166 of GenBank accession number NP_003398.1. Note that GenBank has updated this sequence by adding six amino acids to the amino terminus, so that the TZF domain would represent residues 109–172 (NP_003398.2). To our knowledge, this amino terminal sequence addition has not been confirmed experimentally, and an initiator methionine at that site is not common in vertebrates other than monkeys. In our search, no gaps were allowed, and we did not investigate low frequency hits for sequencing errors. For the current analysis, we obtained 1237 sequences meeting these criteria. The results of this analysis are presented in Fig. 1A, which shows the relative amino acid frequencies at each site, using numbering systems of 1–64 for the TZF domain, and 103–166 representing the corresponding sequence numbers from the whole protein, NP_003398.1. The actual frequencies and identities of the residues shown schematically in Fig. 1A are shown in Table 2.
Fig.1. Amino acid frequency at each position of the TZF domain in eukaryotes.
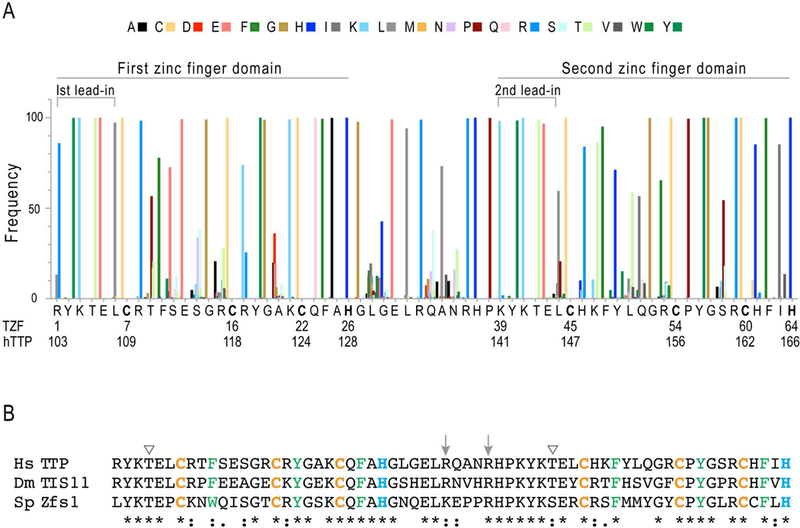
The search parameters for the data summarized in (A) are described in the text. Shown in the histogram are the frequencies of a given amino acid, as percentages of all amino acids at a given site, color coded as shown at the top. At the bottom of the graph is shown the amino acid sequence of the 64-amino acid TZF domain of human TTP, with two numbering systems: One is for the TZF domain itself, with residues listed from 1–64, and the other is from the protein sequence of human TTP from NP_003398.1. Indicated are the lead-in sequences to each finger, and the first and second zinc fingers. The zinc coordinating residues are in bold type. In (B) are the aligned TZF domains from human (Hs) TTP, Drosophila (Ds) Tis11, and S. pombe (Sp) Zfs1. The arrowheads on top indicate possible phosphorylation sites (Thr) within the lead-in regions, and the arrows indicate the two arginines that may contribute to the nuclear localization signal within the linker region. The zinc coordinating cysteines and histidines are in orange and blue, respectively. The aromatic amino acids that interact with the sidechains of H26 or H64, or with RNA bases, are in green. Among the three TZF domains, 68% of the residues are invariant or highly conserved, as indicated by the asterisks and colons, respectively.
Table 2. Residues that form the ARE base binding pockets in the human TTP and ZFP36L2 TZF domains.
The compositions of the binding pockets for each RNA base of the RNA oligomer (1-UUAUUUAUU-9) were obtained from the human TTP simulation solution model, with the equivalent residue in parentheses from the NMR structure of the TZF domain of ZFP36L2 (TIS11d; PDB ID 1RGO) in complex with RNA. Numbers in square parentheses correspond to TZF domain residue positions noted in Fig. 1. Binding pockets are made up largely of residues from equivalent sequence positions within the TZF domains of the two structures. The TZF domain of human TTP is comprised of residues 103–166 from GenBank accession number NP_003398.1; the corresponding residues of human ZFP36L2 (Tis11D) are 153–216 of NP_008818.3. This table has been adapted from (Lai, Perera et al. 2014).
| 2-UAUU-5 Subsite | 6-UAUU-9 Subsite | ||
|---|---|---|---|
| TTP (ZFP36L2) | TTP (ZFP36L2) | ||
| U2 | H139 (H189) [37] K141 (K191) [39] Y142 (Y192) [40] K143 (K193) [41] T144 (T194) [42] E145 (E195) [43] H163 (H213) [61] §F164 (F214) [62] |
U6 | R103 (R153) [1] Y104 (Y154) [2] K105 (K153) [3] T106 (T156) [4] E107 (E157) [5] Q125 (Q175) [23] §F126 (F176) [24] R161 (R211) [59] |
| A3 | E145 (E195) [43] L146 (L196) [44] C147 (C197) [45] ¶H148 (R198) [46] §F164 (F214) [62] |
A7 | E107 (E157) [5] L108 (L158) [6] C109 (C159) [7] R110 (R160) [8] T111 (P161) [9] §F126 (F176) [24] R161 (R211) [59] |
| U4 | C147 (C197) [45] ¶H148 [46] K149 (T199) [47] P157 (P207) [55] §Y158 (Y208) [56] R161 (R211) [59] C162 (C212) [60] H163 (H213) [61] §F164 (F214) [62] |
U8 | C109 (C159) [7] T111 (P161) [9] C118 (C168) [16] R119 (K169) [17] §Y120 (Y170) [18] K123 (K173) [21] C124 (C174) [22] Q125 (Q175) [23] §F126 (F176) [24] |
| U5 | R103 (R153) [1] P157 (P207) [55] §Y158 (Y208) [56] R161 (R211) [59] |
U9 | R119 (K169) [17] §Y120 (Y170) [18] G121 (G171) [19] K123 (K173) [21] |
Side chain involved in stacking with the RNA bases: U2-F164-A3; U4-Y158-U5; U6-F126-A7; U8-Y120-U9 (in ZFP36L2: U2-F214-A3; U4-Y208-U5; U6-F176-A7; U8-Y170-U9).
H148 of TTP is located at the rim of both A3 and U4 pockets. R198 forms part of the wall of the pocket for A3.
One of the most striking findings in this figure is the high percentage of invariant or near-invariant residues, totaling approximately 42 of 64 amino acids (66%). Others that are more variable often have amino acids of one chemical type; for example, in position 63 within the human TTP TZF domain, the native isoleucine was present in 83.7% of sequences, but two other branched chain amino acids, valine (15.1%) and leucine (1.1%), made up the total.
Many of these invariant residues will be discussed further below, in terms of the effects of mutating these residues on function, and their possible effects on structure of the protein-RNA complex. As extreme examples of this, we found that mutating any of the eight cysteines and histidines involved in coordinating the two zinc molecules could completely destroy high affinity RNA binding (Lai, Perera et al. 2014).
Phosphorylation
Three aspects of this analysis will be touched on here. The first is regarding the presence of possible phosphorylation sites within the TZF domain. Even before its function as an mRNA destabilizing protein was known, TTP was shown to be a phosphoprotein whose phosphorylation was stimulated by growth factors and hormones in cultured cells (Taylor, Thompson et al. 1995). Once its identity as an RNA binding protein was understood, phosphorylation of residues within the RNA-binding TZF domain seemed like a potential way to regulate RNA binding affinity. Indeed, Cao et al demonstrated that TTP produced in E. coli bound to a TNF-based probe with higher affinity than TTP purified from mammalian cells (Cao, Dzineku et al. 2003), and TTP expressed in human HEK293 cells and then dephosphorylated by CIAP was able to bind more tightly to a GM-CSF mRNA ARE probe than native, fully phosphorylated TTP from HEK293 cells (Carballo, Cao et al. 2001). Phosphorylation of the intact protein has been implicated in aspects of its activity, sequestration and stability, and discussions of these topics can be found in the following papers and reviews: (Hitti, Iakovleva et al. 2006, Cao, Deterding et al. 2007, Sandler and Stoecklin 2008, Kratochvill, Machacek et al. 2011, Brooks and Blackshear 2013, Ross, Smallie et al. 2015, Clark and Dean 2016).
Focusing on the TZF domain itself, the two serines found in the human TTP TZF domain, at positions 11 and 13, are highly variable in other family member proteins (Figs. 1A, B). There are, however, several other essentially invariant amino acids potentially capable of phosphorylation, including threonines at positions 4 and 42, and tyrosines at positions 2, 18, 40, and 56 (Figs. 1A, B). In the analysis by Cao et al (Cao, Deterding et al. 2007), a major phosphopeptide from human TTP labeled in intact cells was YKTELCRTFSESGR, from the first zinc finger, with the three potential phosphorylated residues underlined. In our survey of 1237 eukaryotic family members, the threonine in the fourth position (T4) in the human TTP TZF domain was 100% conserved (Fig. 1A, B), with two potential outliers (XP_003498309.1 and XP_015265645.1) containing sequencing mistakes, as confirmed by BLASTing against GenBank Sequence Read Archives from those species. Since the two potentially phosphorylatable serines in this phosphopeptide are highly variable among other proteins of this family, it seems possible that phosphorylation of T4 is more likely, and could be an important regulatory event. Unfortunately, in the Cao et al analysis (Cao, Deterding et al. 2007), the specific phosphoamino acid was not identified, and future work will be necessary to determine if T4 is phosphorylated in vivo, and, if so, what the effect is of this modification on TZF domain behavior. Of note, recent analyses of the phosphoproteome of Schizosaccharomyces pombe did not note any phosphorylation sites within the TZF domain of the single TTP family member expressed in that species, Zfs1 (Navarro, Chakravarty et al. 2017, Swaffer, Jones et al. 2018).
A potential conflict was seen in a later analysis by Cao et al (Cao, Deterding et al. 2006) in which an HA-tagged TZF domain peptide was expressed in HEK293 cells, and was not found to be phosphorylated in radioactive labeling experiments. However, we now know that this peptide would be found exclusively in the nucleus (see below), and presumably would not be subject to the same protein kinases as it normally would in the cytoplasm. It is possible to link such a peptide to a nuclear export sequence to cause its cytoplasmic localization, and this puzzle could be solved by this means.
Nuclear localization
Even before a function was determined for TTP, we recognized that it was a nucleo-cytoplasmic shuttling protein whose localization could be controlled by external stimuli (Taylor, Thompson et al. 1996). In those experiments, most immunoreactive TTP resided in the nucleus of serum-deprived, quiescent fibroblasts, but it moved to the cytoplasm within minutes of stimulation of the cells with serum. Later experiments (Phillips, Ramos et al. 2002) identified a nuclear export sequence at the amino terminus of TTP, and at the C-termini of ZFP36L1 and ZFP36L2, all of which interacted with the CRM1 nuclear export protein. In contrast, constructs consisting of the GFP-linked TZF domain alone from TTP and ZFP36L1 were entirely nuclear. We also found that cysteine mutants of either TTP or ZFP36L1 that prevented RNA binding could shuttle normally from the nucleus to the cytoplasm, indicating that the nuclear localization was not dependent on the ability of the proteins to bind RNA.
A potential nuclear localization signal (NLS) within the TZF domain of rat TTP was proposed by Murata et al (Murata, Yoshino et al. 2002), and localized to two arginine residues that are reflected in the essentially invariant residues R32 and R36 in Fig. 1A, B. We confirmed in later studies with the TZF domain of mouse ZFP36L3 that mutation of the two corresponding arginines caused some loss of nuclear localization of the GFP-fusion peptide (Frederick, Ramos et al. 2008). However, we also found in that study that mutation of both arginines to alanines completely prevented RNA binding of the protein, presumably by changing the fundamental structure of the TZF domain in some way. It is therefore not clear whether these residues represent a classical basic NLS. Nonetheless, their almost perfect conservation within TTP family members throughout eukarya, as demonstrated in Fig. 1A and B, suggests that they are of great importance to the function of the proteins in some way, and certainly are required for the RNA binding capability of the TZF domain.
In the present analysis, we identified several proteins from diverse species that appear to contain alternative residues at these sites. Specifically, at site R32, XP_009055670.1 (Lottia gigantea, the owl limpet), XP_016100341.1 (Sinocyclocheilus graham, a fish), and XP_017564357.1 (Pygocentrus nattereri, the red-bellied piranha) contained Q at that site, and 11 others contained K residues. At site R36, EXX57770.1 (Rhizophagus irregularis, a fungus) and GBB99881.1 (Rhizophagus clarus, a related fungus) contained H residues, and XP_022348216.1 (Enhydra lutris kenyoni, a sea otter) contained a Q residue, at the R36 site. No proteins had changes of these types at both sites, and we cannot exclude sequencing errors. It will be interesting to determine whether these naturally occurring variants affect either the RNA binding ability or the nuclear localizing ability of their TZF domains.
Continued evolution
We have argued previously that TTP family members containing TZF domains of the type we have been discussing, with or without linkage to C-terminal CNOT1 binding domains, must have been present in a common ancestor of humans and the most primitive plants and protists more than 1.5 billion years ago (Blackshear and Perera 2014, Wells, Perera et al. 2017). However, the genes encoding these proteins have continued to evolve. Interesting examples of this include the “destruction” of the gene encoding TTP in the bird lineage, presumably during the generation of microchromosomes from bird ancestors (Lai, Stumpo et al. 2013); the generation of a completely new protein in certain species of rodents, derived from the retrotransposition of Zfp36l2 into the X chromosome of a common ancestor of Muridae and Cricetidae rodents approximately 25 million years ago (Gingerich, Stumpo et al. 2016); the generation of a completely new protein in a common ancestor of fish and amphibians, that is highly expressed in the maternal oocyte compartment (De, Lai et al. 1999); and the duplication of several of the genes in teleost fish, leading to the seven family members expressed in modern fish such as Danio rerio.
Evolution has also affected the TZF domain itself. One interesting example that we would like to highlight here has occurred in red algae, a very interesting group to study because of their extreme evolutionary distance from mammals. Many red algae express typical TTP family member proteins of the type we are discussing here, with typical TZF domains linked to C-terminal CNOT1 binding domains. However, a subset of red algae has evolved TZF domains in their single TTP family proteins that contain an “extra” glycine within the C-X8-C region of the first zinc finger (Fig. 2). All of the algae that contain this additional amino acid are from a single subgroup, the Florideophyceae, which separated from the parental line an estimated 943 million years ago (Yang, Boo et al. 2016). We do not yet know whether this mutation has an effect on RNA binding affinity or RNA sequence specificity, although, according to Lai et al (Lai, Perera et al. 2014), “The addition or deletion of a single residue in the C-X8-C region of finger 1 significantly reduced RNA binding…”. This novel change has presumably introduced a survival advantage to the species that have maintained it, and it remains to be seen what that survival advantage is, as well as the effect of this change on RNA binding affinity.
Fig. 2. Selected TZF domains from Florideophyceae and Pezizales.

In (A) are shown the TZF domain sequences from TTP family proteins from a number of red algae. The bar at the right indicates those from Florideophyceae that have an amino acid addition in the C-x8-C interval of the first zinc finger domain. In (B) are shown the TZF domains from TTP family members expressed in Pezizales (fungi), with the sequence from S. pombe shown at the top for comparison. The bar at the right indicates fungi from the Pezizales with an amino acid “missing” from the C-x8-C interval in the second zinc finger domain. The zinc coordinating cysteines and histidines are in yellow and blue, respectively. The aromatic amino acids that interact with the RNA bases are in green.
A second example occurs in fungi. We previously noted the apparent loss of the TZF domain in the most highly evolved subphylum of fungi, the Pezizomycotina (Blackshear and Perera 2014). However, it appears that some members of this group, within the early branching order Pezizales, express otherwise typical TZF domains that are missing an amino acid in the C-X8-C interval of the second zinc finger domain. The most famous member of this group is the Perigord black truffle, Tuber melanosporum. As we previously stated (Lai, Perera et al. 2014), “…addition or deletion of a single residue in finger 1 significantly reduced RNA binding,… whereas similar modifications in finger 2 had little or no effect…. Thus, changes in length (by one residue) of the C-X8-C interval were tolerated in finger 2, but not in finger 1…”. Again, it remains to be seen whether this modification in the Pezizales affects binding affinity and/or RNA sequence specificity in these species. Other than the Pezizales, the other members of the Pezizomycotina seem to lack TZF domain-containing sequences altogether; how this “disappearance” came about in these medically and economically important organisms remains to be determined.
Effect of key conserved residues on activity.
The cysteines and the histidines in the TTP TZF domain are involved in Zn binding (Brewer 1991, Worthington, Amann et al. 1996, Shimberg, Ok et al. 2017), and in the presence of Zn the NMR structure of the ZFP36L2 TZF domain became a folded structure as compared to an unstructured state when zinc-free, even in the absence of RNA (Hudson, Martinez-Yamout et al. 2004). When any of the zinc coordinating cysteines or histidines was mutated, TTP and related family proteins completely lost RNA binding activity (Carballo, Lai et al. 1998, Lai, Kennington et al. 2002, Choi, Lai et al. 2014).
In addition to the requirement for the zinc coordinating residues CCCH in each finger, the highly conserved aromatic residues within the C-X8-C (F10 and F48, Fig.1 and Table 1), C-X5-C (Y18 and Y56), and C-X3-H (F24 and F62) intervals in each finger are critical for ARE binding. Replacement of any one of these with a non-aromatic residue has been shown to abolish the protein’s ability to bind RNA (Lai, Kennington et al. 2002). The NMR structure of the TZF domain from the human family member ZFP36L2 (PDB code 1RGO) revealed that the aromatic side chains of Y18, Y56, F24 and F62 intercalate with two of the U1U2A3U4U5U6A7U8U9 RNA bases to stabilize binding (Hudson, Martinez-Yamout et al. 2004). In addition, the aromatic side chains of F10 and F48 within the C-X8-C intervals have been shown to be involved in stacking interactions with the aromatic sidechains of the zinc coordinating residues H48 and H26, respectively (Hudson, Martinez-Yamout et al. 2004).
Table 1.
Amino acid occurrence frequencies at positions 1–64 of the TZF domain and effects of substitutions on RNA binding. The zinc-coordinating cysteines and histidines are in wheat and blue, respectively.
| Feature | TZF residues | Frequencies (next or most frequent) |
Binding |
Single mutation (% RNA binding) |
|
|---|---|---|---|---|---|
| Position | hTTP | ||||
| First lead-in | 1 | R | 85.93 (L: 13.34) | U6 | E (10); L (58) |
| 2 | Y | 99.84 | U6 | n/a | |
| 3 | K | 100 | U6 | E (0); L (4) | |
| 4 | T | 99.84 | U6 | n/a | |
| 5 | E | 100 | U6, A7 | K (26); I (23) | |
| 6 | L | 97.25 | A7 | P (42) | |
| Zinc coordinating | 7 | C | 100 | A7, U8 | H (0); S (0) |
| Cx8C | 8 | R | 98.30 | A7 | E (36); L (26) |
| 9 | T | 20.78 (P: 56.67; S: 17.06) | A7, U8 | n/a | |
| 10 | F | 77.85 (W and Y: 21.59) | Y (110); Q (10) | ||
| 11 | S | 12.77 (E: 77.59) | A (99); D (104) | ||
| 12 | E | 99.19 | n/a | ||
| 13 | S | 38.97 (N: 34.03) | n/a | ||
| 14 | G | 99.03 | E (80) | ||
| 15 | R | 10.43 (T: 27.88) | E (99); L (99) | ||
| Zinc coordinating | 16 | C | 100 | U8 | H (0); R (0); Y (0) |
| Cx5C | 17 | R | 25.55 (K: 74.05) | U8 | E (72) |
| 18 | Y | 100 | U8 | F (80) | |
| 19 | G | 98.87 | R (103) | ||
| 20 | A | 19.97 (T: 27.88) | n/a | ||
| 21 | K | 99.11 | U8 | D (18) | |
| Zinc coordinating | 22 | C | 100 | D (0); H (0); R (0); S (0) | |
| Cx3H | 23 | Q | 100 | U6 | V (86) |
| 24 | F | 99.43 (Y: 0.57) | U6, A7 | Y (80); N (0) | |
| 25 | A | 99.84 | N (0) | ||
| Zinc coordinating | 26 | H | 100 | C (0), K (0) | |
| Linker | 27 | G | 97.66 | S (38) | |
| 28 | L | 12.37 (I: 19.48) | n/a | ||
| 29 | G | 11.96 (H: 42.76) | C (102) | ||
| 30 | E | 99.11 | R (1); V (0) | ||
| 31 | L | 94.18 | T (0) | ||
| 32 | R | 98.87 (K:0.89) | E (19); I (36) | ||
| 33 | Q | 10.51 (S: 38.08) | n/a | ||
| 34 | A | 9.46 (L: 73.24) | n/a | ||
| 35 | N | 16.09 (S: 27.89) | n/a | ||
| 36 | R | 99.68 (H:0.24) | E (10); I (36) | ||
| 37 | H | 100 | U2 | E (5), L (10) | |
| 38 | P | 99.84 | N (64) | ||
| Second lead-in | 39 | K | 98.14 | U2 | E (1); L (37) |
| 40 | Y | 98.46 | U2 | F (150); Q (13) | |
| 41 | K | 99.92 | U2 | E (0); L (21) | |
| 42 | T | 99.11 | U2 | N (3) | |
| 43 | E | 96.69 | U2, A3 | K (110); I (46) | |
| 44 | L | 59.58 (P: 20.61) | A3 | S (73); P (118) | |
| Zinc coordinating | 45 | C | 100 | A3, U4 | R (0) |
| Cx8C | 46 | H | 10.11 (R: 83.99) | A3, U4 | E (110); L (48) |
| 47 | K | 10.67 (T: 86.26) | U4 | n/a | |
| 48 | F | 95.15 | Y (110) | ||
| 49 | Y | 15.20 (H: 71.22, W: 11.64) | A (14); K (94) | ||
| 50 | L | 11.24 (T: 58.77) | n/a | ||
| 51 | Q | 10.67 (I: 56.67) | n/a | ||
| 52 | G | 99.84 | K (51) | ||
| 53 | R | 9.30 (F: 65.48; L: 1.94) | L (99) | ||
| Zinc coordinating | 54 | C | 100 | S (0) | |
| Cx5C | 55 | P | 99.43 | U4 | G (26) |
| 56 | Y | 99.92 | U4 | E (0); Q (0) | |
| 57 | G | 99.84 | A (76); K (38) | ||
| 58 | S | 18.27 (P: 54.49) | n/a | ||
| 59 | R | 99.92 | U6, U4 | E (3); L (8) | |
| Zinc coordinating | 60 | C | 100 | H (0) | |
| Cx3H | 61 | H | 85.29 (C: 10.35) | U2 | L (108) |
| 62 | F | 99.76 | U2, A3 | Y (91) | |
| 63 | I | 85.29 (V:13.66) | N (12) | ||
| Zinc coordinating | 64 | H | 100 | C (0), L (0) | |
Frequencies: This column shows the frequency of the human TTP residue at this site as a percentage of all amino acids seen at this site. Residues and percentages in parentheses indicate the most frequent (other than in TTP) or the next most frequent.
Binding pocket: This column shows whether a residue at a given position within the human TTP TZF domain was a member of the binding pocket for an RNA base of the RNA oligomer U1U2A3U4U5U6A7U8U9.
Single mutation (%RNA binding): A single amino acid substitution made at the indicated position within the TZF domain from full-length human TTP, and the resulting effect of the mutation on RNA binding, as compared to wild-type TTP, are indicated in the parentheses as percentages of the wild-type control. n/a: Not assayed.
The root-mean-square deviation (RMSD) of the backbone atoms from the well-ordered RNA-bound NMR structure of the TZF domain of ZFP36L2 (TIS11d, PDB ID 1RGO) was reported to be 0.67 Å, indicating that the reported structure was well resolved (Hudson, Martinez-Yamout et al. 2004). Since that original structure appeared to be of high quality, we have used molecular dynamics (MD) simulations (Karplus and McCammon 2002) to model many structures of TZF domains from other TTP family members. MD simulations are widely used to obtain solution structure models that attempt to mimic the motion of proteins, other biological macromolecules such as DNA and RNA, and their assemblies at the atomic level in their physiological environments. Using starting conformations based either on experimental techniques such as X-ray crystallography, NMR, and Cryo EM, or created from a homology model, MD trajectories are calculated in a solvent environment by solving Newton’s equations of motion to generate molecular conformations at a desired temperature (Karplus and McCammon 2002). Such conformations can be used to deduce connections between structure and functions. Limitations of the technique include reliance on the quality of interacting potential functions used in solving Newton’s equations, as well as inadequate conformation sampling.
Solution structures of the wild-type human TTP TZF domain and many selected mutants were created (Lai, Perera et al. 2014) using the RNA-bound NMR structure of the TZF domain of ZFP36L2 as a template, and by mutating residues of ZFP36L2 with Coot (Emsley, Lohkamp et al. 2010), using lengthy all-atom solution MD trajectory calculations with the PMEMD module of the Amber.11 package (http://ambermd.org/). It was determined from RMSD values observed in the latter part of the simulation that the MD trajectories reached equilibration. When compared with the ZFP36L2 NMR structure, the final wild-type RNA-bound solution simulation model showed an RMSD value of 2.17 Å, while the values were 0.82 Å and 1.19 Å for each individual zinc finger. Slightly different RMSD values were observed for the RNA-free TZF domain (2.59 Å, 1.15 Å, and 0.85 Å for the total peptide and the two zinc fingers, respectively). The RMSDs of RNA-bound TZF domains indicate that the ZFP36L2 NMR structure and the simulation model of the TTP TZF domain are rather similar, and that the backbone structures that already formed in the presence of zinc (Hudson, Martinez-Yamout et al. 2004) are not much influenced when bound to RNA.
The electrostatic surface potential (ESP, Fig. 3) of the RNA binding surface was generally positively charged (blue), for both the ZFP36L2 and TTP TZF domains. There are only three negatively charged RNA backbone phosphates (connecting U1-U2, U5-U6, and U8-U9) that are in direct contact with the peptide surface. Both peptides showed rather similar binding surface features. The binding pockets formed by the TZF domain residues for some of the RNA bases are also similar for both TZF domains (Table 2). The interactions between the peptide and the RNA are mainly between the bases of the RNA and the TZF domain residues in various binding pockets (Hudson, Martinez-Yamout et al. 2004).
Fig. 3. Electrostatic surface potential of the human ZFP36L2 and TTP TZF domains.
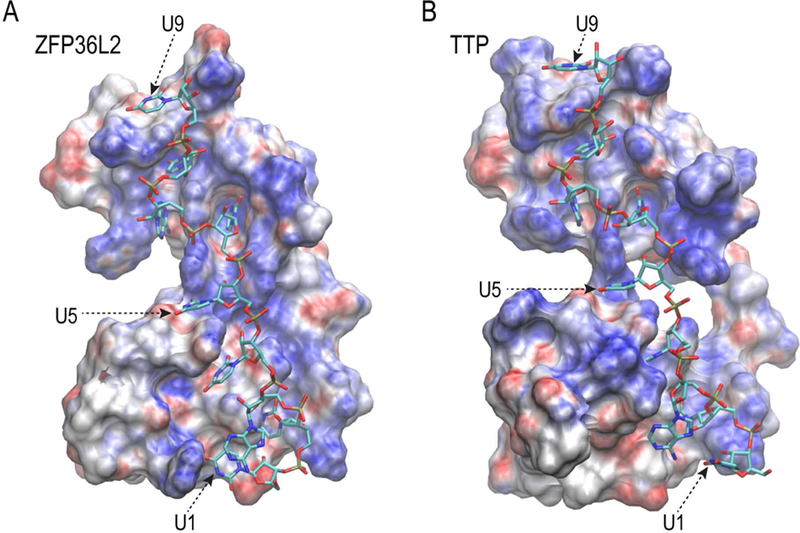
The electrostatic surface potentials (ESP) of the interaction surfaces of the (A) ZFP36L2 and (B) human TTP TZF domains are shown with the RNA oligomer (sticks). The ESPs (shown between −10 to + 10 J/K) were produced using APBS (Jurrus, Engel et al. 2018) with AMBER charges from simulations, and the figures were generated using VMD (Humphrey, Dalke et al. 1996).
With the appreciation of sequence conservation and the help of the NMR structure of the ZFP36L2 TZF domain (PDB code 1RGO), as well as the simulation solution structures of the TTP TZF domain, the TZF domain-RNA recognition mechanism can be better understood by studying the interactions between the RNA nucleotides and the key residues of the TTP TZF domain, by comparing the WT sequence to a panel of single residue mutations of the TTP TZF domain (Lai, Perera et al. 2014). Some of the effects of these single amino acid mutations on the interactions between TTP and RNA are summarized in Tables 1 and 2.
1. The lead-in sequences.
The original NMR structure analysis indicated that zinc finger 1 (ZF1) of ZFP36L2 bound to U6A7U8U9 and zinc finger 2 (ZF2) bound to U2A3U4U5 sub-sites of the AU-rich element (ARE) 9-mer (Hudson, Martinez-Yamout et al. 2004). The NMR structure of ZFP36L2 and the simulation solution structure model of the human TTP TZF domain (Lai, Perera et al. 2014) revealed that all of the six residues in the lead-in sequences of TTP ZF1 and ZF2 participate in forming the binding pockets for the ARE bases (Table 1). Residues R1, T4, E5, L6, K39, T42, E43, and L44 within the human TTP TZF domain also appear to interact directly with the ARE bases or their backbone riboses and phosphates.
The lead-in sequences to each finger, RYKTEL and KYKTEL, have been highly conserved during evolution (Fig. 1A, Table 1). Family member proteins from fungi and certain protists often contain a leucine at position 1 (Figs 1 and 2, Table 1) instead of the arginine generally found in vertebrates. An R to L substitution at position 1 in the human TTP TZF domain led to a moderate decrease in ARE binding (Table 1), presumably due to the loss of interaction between the charged side chain of R with the U5-U6 backbone phosphate. However, the model suggested that the hydrophobic portion of the leucine side chain could still contribute to the stability of the binding pockets of both U5 and U6, so that the substitution of leucine could preserve the biological function of the protein. In ZF2, position 39 is invariably a lysine in vertebrates, and a lysine or arginine in many single cell organisms (Fig. 2). Similar to R1, K39 interacted with the uridine backbone phosphate in the model (U1-U2) and stabilized these bases’ binding pockets. For all family members, a basic residue seems to be more important at this position of ZF2 than in ZF1 (Table 1).
An aromatic residue seems to be required at positions 2 and 40 for effective RNA binding (Table 1).
A substitution with an acidic residue in either position 3 or 41 made the protein unable to bind RNA at all. The lysine side chains of K3 and K41 probably contribute to the stability of the two fingers by interacting with other residues. Specifically, there is a particularly important sidechain-sidechain interaction between K3 and E30 in the linker region to stabilize ZF1, and a K3L substitution prevented RNA binding. In ZF2, the sidechain of K41 appears to interact with H61 at position C+1 of the C-X3-H interval to stabilize the U2 binding pocket.
The side chains of T4 and T42 also interacted with the backbones of the first residues of their respective lead-in sequences, apparently a critical connection for the correct turn of the TZF domain peptide backbone. A T42N substitution eliminated RNA binding. A modification of the threonine side chain, such as by phosphorylation, could potentially lead to the same outcome.
The simulation solution model reveals a salt bridge comprising sidechain interactions between E5 and R59, with the latter being the residue at the C+5 position of the C-X5-C interval of ZF2. This connection between E5 and R59 should play a critical role in keeping the two zinc fingers oriented towards each other properly (Fig. 4) (Lai, Perera et al. 2014). Reversing the charge by way of an E5K substitution greatly decreased RNA binding. However, there was no apparent equivalent interaction between E43 and K21 of the C-X5-C interval of ZF1, and an E43K substitution did not affect RNA binding (Table 1).
Fig. 4. Ribbon diagram of the peptide backbone of the human TTP TZF domain in complex with its 9-base binding site.
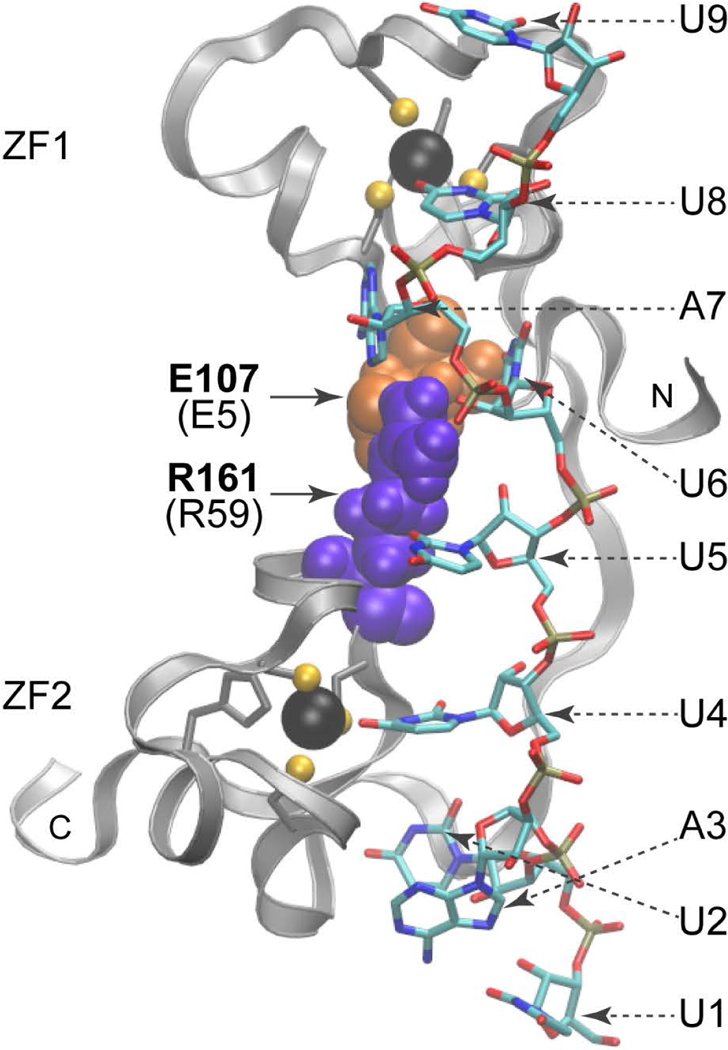
Shown are the side chains of E107 (E5 in Fig. 1A), denoted as orange spheres, in the lead-in sequence to ZF1; and R161 (R59 in Fig. 1A), shown as purple spheres, at the C+5 position of the ZF2 C-x5-C region. Dashed arrows indicate the nucleosides (sticks). Note that U5, U6, and A7 interact with E107 (E5) and R161(R59). Zinc atoms (black spheres) and the zinc coordinating residues (ball and stick) of each finger are also displayed. The N- and C-termini of the TZF domain peptides are indicated. Adapted from (Lai, Perera et al. 2014), with permission.
The last residues of each lead-in sequence are components of the two hydrophobic clusters that play important roles in TZF domain stability (Hudson, Martinez-Yamout et al. 2004). One of these consists of a cluster formed by L6 from the first lead-in sequence, A25 at position C+3 in the C-X3-H interval of ZF1, and three residues from the linker region: G27, L28, and L31. Note that A25, G27, and L31 are highly conserved (Fig. 1A, Table 1). The other cluster includes K41, E43, and L44 from the second lead-in sequence, I63 at position C+3 in the C-X3-H interval of ZF2, and P68, a residue that falls outside of the TZF domain. Note that in the second lead-in sequence, a leucine appears at position 44 with a 60% frequency in all TZF domains surveyed (Fig. 1A, Table 1). A proline appears in that position about 21% of the time, e.g., in the TZF domain of rodent ZFP36L3. In human TTP, an L44P substitution did not change the RNA binding ability. A proline at position 6 is rare in eukaryotes (Fig. 1A, Table 1), but it is found in S. pombe (Fig. 2B) and other members of the Schizosaccharomyces genus, along with a few other fungi. Nonetheless, an L6P substitution in the ZF1 of human TTP decreased the RNA binding substantially.
2. The C-X8-C intervals.
Several resides in the C-X8-C intervals do not participate directly in forming binding pockets with the RNA bases, including amino acids C+3 to C+8 in both fingers (Tables 1 and 2) (Hudson, Martinez-Yamout et al. 2004, Lai, Perera et al. 2014).
The ZFP36L2 NMR structure demonstrated that its two ZF domains adopted virtually identical folds, whether in the RNA-free or RNA-bound state (Hudson, Martinez-Yamout et al. 2004). However, in the case of TTP, in the absence of RNA, only ZF1 adopted a stable fold (Blackshear, Lai et al. 2003, Deveau and Massi 2016). Even when RNA was present, the simulation model indicated that the two fingers from TTP are structurally distinct, largely due to a unique second aromatic residue, Y49 at C+4, which appears to interact with the backbone of I63 in the C-X3-H interval of ZF2 (Lai, Perera et al. 2014). That interaction results in the “over-winding” of the α helix in the C-X8-C interval of ZF2 as compared to ZF1. In their study of the TTP TZF domain using NMR spectroscopy, Deveau and Massi (Deveau and Massi 2016) showed that the α helix in the C-X8-C interval of ZF2 was indeed shorter than the cognate interval in ZF1. Even though Y49 does not directly interact with the RNA bases, a Y49A substitution drastically decreased TTP binding to RNA, as well as its ability to promote deadenylation and decay of a target mRNA (Lai, Perera et al. 2014).
Amid the ten amino acids (11–15 in ZF1 and 49–53 in ZF2) that do not directly interact with RNA, the loss of a residue, as illustrated in Fig. 2B, might result in little or no effect on the protein’s ability to bind RNA. In the case of TTP, the deletion of S11 in ZF1 probably indirectly affected the orientation of R8, leading to the destabilization of the A7 binding pocket and a decrease in RNA binding (70% of normal binding). However, the parallel deletion of Y49 in ZF2 seemed to be well tolerated (97% of normal binding), perhaps due to the maintenance of the hydrophobic environment contributed by L44 from the lead-in sequence, L50 from the C-X8-C interval, and I63 from the C-X3-H interval of ZF2.
Substituting the highly conserved basic residues (positions 8 and 46) with a leucine at C+1 of either finger decreased the binding to RNA (Table 1), whereas changing the highly conserved glycine at C+7 (position 14 or 52) to an acidic or basic residue affected the binding differently (Table 1).
3. The C-X5-C intervals.
In the C-X5-C intervals, there were differences in tolerance when highly conserved residues were replaced. For example, at C+1 in ZF1, a charge reversal mutation was well tolerated (Table 1), whereas in ZF2, a P55G substitution caused a change in the finger’s orientation, decreasing RNA binding substantially. There was no change in RNA binding when G19 at C+3 in ZF1 was replaced by an R, but in ZF2 the G57K substitution greatly decreased binding (Table 1). Substitution of the basic residues at C+5 of either finger reduced RNA binding substantially, especially when R59 was changed to an E or an L, in each case abolishing RNA binding (Table 1). As mentioned above, the sidechain-sidechain interaction between E6 in the lead-in sequence to ZF1, and R59 in ZF2, is a major determinant of correct inter-finger orientation (Fig. 4) (Lai, Perera et al. 2014) and for the stability of the fingers. In addition, the R59 sidechain also interacts with the U5 base and the U6 ribose, as indicated by the NMR structure of ZFP36L2 TZF and the simulation model of TTP (Hudson, Martinez-Yamout et al. 2004, Lai, Perera et al. 2014).
4. The C-X3-H intervals.
Although the residues at the C+1 position within the C-X3-H intervals of both fingers are highly conserved (Fig. 1A, Table 1), the replacement of either with residues of different characteristics (Q23V or H61L) did not affect TTP binding to the RNA (Table 1). At the C+3 positions, there are almost always hydrophobic residues (with the exception of the two proteins from S. cerevisiae and many other yeasts, which contain a K at C+3 of ZF2). Each of the C+3 hydrophobic amino acids can serve as a core residue for one of the two hydrophobic clusters that stabilize the two fingers (Hudson, Martinez-Yamout et al. 2004). RNA binding ability was entirely lost when A25 was replaced with an N, as a result of disturbing the ZF1-stabilizing hydrophobic cluster. Other members of this cluster are L6 in the lead-in sequence, and L28, G27, and L31 in the linker region. RNA binding was also lost with a hydrophilic substitution I63N at C+3 of ZF2. This substitution disturbed the interaction among the core members (hydrophobic portions of K41 and E43, L44 in the lead-in sequence, and P66 beyond the end of the TZF domain), and made the pockets for U4 and A3 unsuitable for binding.
We found that any changes in the length of the C-X5-C and C-X3-H intervals were detrimental to RNA binding. For example, in the C-X5-C intervals, binding was lost when A20 at the C+4 position was deleted, or a residue after it was added (within ZF1), or when S58 was deleted, or a residue was added to the equivalent position within ZF2. Interestingly, residues at these positions (20 and 58) are not very well conserved (Fig. 1, Table 2), and neither of them seems to be involved in forming binding pockets for the bases. However, the simulation model indicates that perturbing the C-X5-C intervals by residue subtraction or addition relaxes the 310 helixes at these intervals, resulting in the inability of the aromatic sidechains of Y18 and Y56 to stack with U9 and U8, and U5 and U4, respectively. Likewise, a decrease or increase within the C-X3-H intervals disrupt the stacking of the F24 and F62 sidechains with their respective bases (U6 and A7, and U2 and A3).
5. The linker region
As shown in Fig. 1A and Table 1, there are several highly conserved residues in the linker region among eukaryotes, including G27, E30, L31, R32, R36, H37, and P38. Besides those belonging to the ZF1 stabilizing hydrophobic cluster mentioned above, E30 also plays a critical role in the stability of ZF1 by interacting with K3 in the first lead-in sequence (Hudson, Martinez-Yamout et al. 2004). A charge reversal or a neutral substitution of E30 rendered TTP unable to bind RNA at all (Table 1). As described above (nuclear localization), substitution mutations of R32 and R36 each decreased binding to RNA (Table 1), more severely when both were mutated (Lai, Perera et al. 2014). H37 is important for the stability of ZF2 (Hudson, Martinez-Yamout et al. 2004), in that a charge reversal or a neutral substitution mutant of H37 severely destabilizes the binding pocket for U2, making RNA binding almost impossible (Table 1). Finally, a P38N substitution reduced RNA binding.
Effect of TZF domain point mutants in mice and S. pombe.
As described above, mutation of any of the eight zinc-coordinating cysteine or histidine residues in the TZF domain prevents the normal interaction between TTP and RNA (Lai, Kennington et al. 2002). We recently decided to address the hypothesis that making a similar mutation in the mouse would mimic the complete knockout (KO) phenotype (Lai, Stumpo et al. 2018). To do this, we used knock-in (KI) technology to mutate a single nucleotide in a codon encoding one of these cysteines (C22 in Fig. 1A, or C116 in mouse TTP, GenBank accession number NP_035886.1). In these mice, the gene encoding TTP, Zfp36, was otherwise intact, so that the expression of TTP remained under the control of its normal locus. We found that the severe inflammatory phenotype of these mice (Fig.5) (Lai, Stumpo et al. 2018) was essentially identical to that described earlier with the conventional KO mice (Taylor, Carballo et al. 1996). We also tested bone marrow derived macrophages and mouse embryo fibroblasts from these mice to answer the question of whether the TTP target mRNAs previously identified from studies of the KO mice behaved similarly in the point mutant KI mice. In both cell types, typical TTP target mRNAs, such as Tnf, Cxcl1, and Cxcl2 mRNAs, were greatly stabilized in one or both of these two cell types. In both cases, the encoded proteins were also hyper-secreted in the cells from the mutant mice. We concluded that both the external inflammatory phenotype of the mice, and the molecular phenotypes seen in the cells, are basically indistinguishable between KO and KI mice and cells. These results suggest, but do not prove incontrovertibly, that all of TTP’s physiological activities derive from its initial, high affinity interaction with its target mRNA binding sites.
Fig. 5. Paw pathology in C129R knock-in mice.
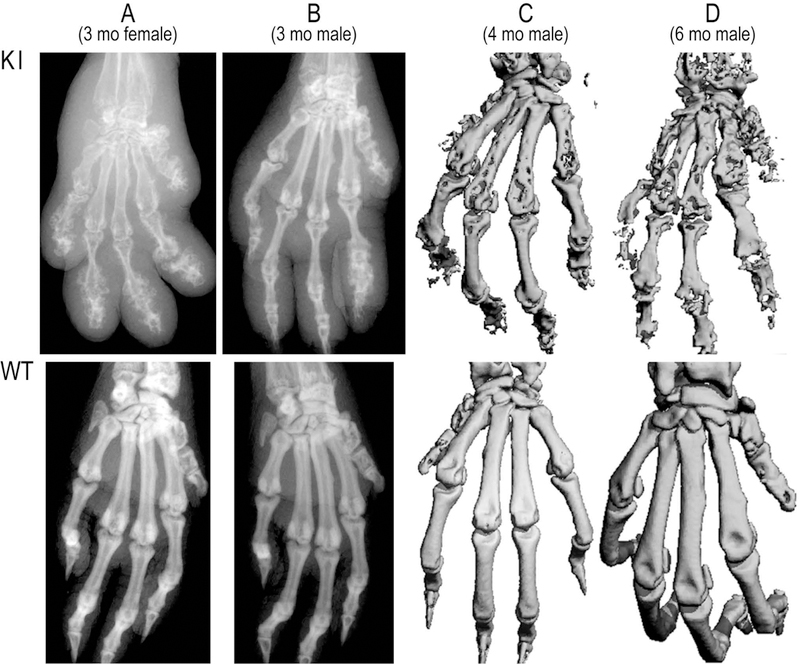
The upper panels show X-rays and micro-CT images from C116R mouse paws, with paws from their WT counterparts shown in the lower panels. A, paw X-rays from female (A) and male (B) mice at 3 months of age. C and D, paw micro-CT images from males at 4 (C) and 6 (D) months of age. Adapted from (Lai, Stumpo et al. 2018), with permission.
We also tested the same general hypothesis in S. pombe, which last shared a common ancestor with mice over a billion years ago. This species expresses only a single protein of the TTP family, Zfs1. In this case, we examined a readily assayable external phenotype, the propensity of the KO cells to flocculate in the presence of calcium (Wells, Huang et al. 2012). We also evaluated the molecular phenotype of the cells, by comparing the increased expression of Zfs1 target transcripts in KO and zinc finger mutant cells (Cuthbertson, Liao et al. 2008, Wells, Huang et al. 2012, Wells, Hicks et al. 2015). As in the case of the mice, both the external, flocculation phenotype, and the molecular phenotype involving increased expression of Zfs1 targets, were essentially identical in the KO and zinc finger mutant cells (Wells, Hicks et al. 2015). Thus, in this species very far removed from mice, the physiological functions of Zfs1 seem to require the presence of an intact mRNA binding domain.
Functional interchangeability of RNA binding domains in S. pombe
As we have discussed above, alignment of the TZF domain from TTP family members from the yeast S. pombe and the three human TTP family members show perfect conservation of the CCCH zinc finger sequences, as well as high conservation of the lead-in sequence, (L/R)YKTE(P/L) (Fig. 1B). However, closer examination revealed significant differences in key residues within the TZF domain, several of which have been shown to play an important role in RNA binding of human TTP (Lai, Perera et al. 2014). For example, a mutation within human TTP that changed arginine at position 1 to leucine, as seen normally in S. pombe and other fungi and some protists, results in a significant decrease in RNA binding (Lai, Perera et al. 2014). In addition, mutation of tyrosine 49 in human TTP to alanine has been shown to dramatically decrease RNA binding and result in the loss of target mRNA regulation (Lai, Perera et al. 2014). Examination of the TTP family member from the pathogenic fungus, Candida guilliermondii, revealed that an alanine naturally occurs at position 49 in this species (XP_001482883.1). Our own resequencing of this species has confirmed this amino acid identity at this site. Despite these variations in what appear to be critical residues, the TZF domains from human, S. pombe, and C. guilliermondii each bound with similar and high affinity to an RNA probe containing the mammalian optimal binding sequence, and solution structure modeling predicted similar binding conformations (Fig. 6) (Wells, Washington et al. 2015).
Fig. 6. Solution structure models of the second zinc finger from the TZF domains from H. sapiens TTP, S. pombe Zfs1, and C. guilliermondii Zfs1.
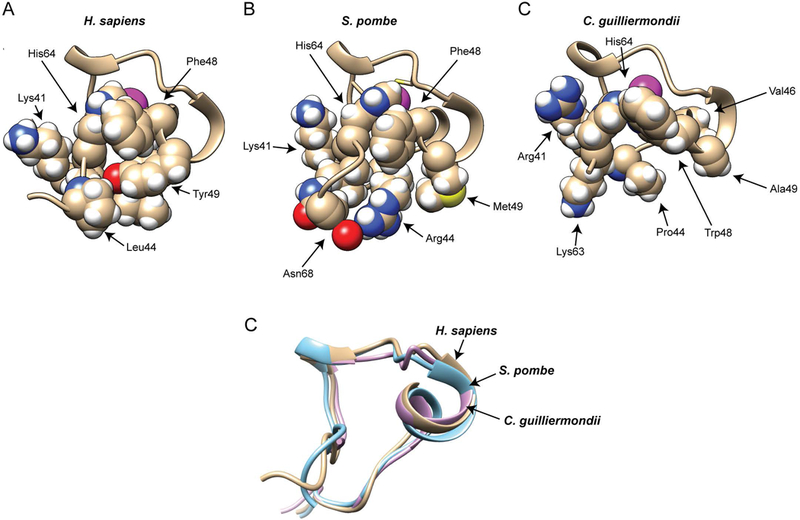
Portions of the solution structure models are shown from the H. sapiens TTP (A), S. pombe Zfs1 (B), and C. guilliermondii Zfs1 (C) TZF domains. Shown are the backbone structures of portions of the second zinc finger from the TZF domains, including selected sidechains. The magenta spheres represent the Zn2+ ions. The peptide backbone ribbon and sidechain carbons are shown in a wheat color, and the atoms of the side chain residues are represented by colored spheres: white, hydrogen; red, oxygen; blue, nitrogen; yellow, sulphur. From (Wells, Hicks et al. 2015), with permission. (D) Superposition of backbone heavy atoms from H. sapiens TTP (in wheat), S. pombe Zfs1 (in cyan), and C. guilliermondii Zfs1 (in magenta) that are shown in (A), (B), and (C).
To address whether the TZF domains from highly divergent species were interchangeable in a live organism, we performed domain swapping experiments in our well-characterized model in S. pombe. In this species, only one family member, Zfs1, is expressed. The loss of Zfs1 in S. pombe results in the up-regulation of several ARE-containing target mRNAs, some of which play roles in cell-cell adhesion (Cuthbertson, Liao et al. 2008, Wells, Huang et al. 2012). This up-regulation of target transcripts and their encoded proteins results in increased calcium-induced flocculation in Zfs1 mutants (Wells, Huang et al. 2012). To determine whether the TZF domain of S. pombe Zfs1 could be replaced with TZF domains from distant species and still complement the Zfs1 KO phenotype, we replaced the endogenous TZF domain with the 64 amino acid domains from human TTP, silkworm Tis11, the TTP family member from the eudicot plant, Chromolaena odorata, and the TTP family member from the pathogenic fungus, C. guilliermondii (Fig. 7A) (Wells, Washington et al. 2015). The TZF domains were cloned into the endogenous Zfs1 locus in S. pombe, utilizing the endogenous promoter and 3’UTR (Fig. 7B, C). Calcium-induced flocculation studies of the complementation strains showed that the strains containing the highly divergent TZF domains responded essentially identically to a wild type strain (Fig. 7D). In addition, mutation of the leucine at position 1 to an arginine in the endogenous Zfs1 protein had no effect on calcium-induced flocculation (Fig. 7D).
Figure 7. Alignment, construction, and expression of TZF domain complementation strains.
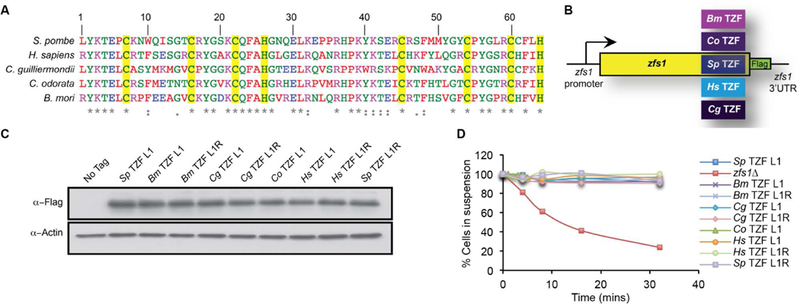
(A) Alignment of TZF domains from the indicated species. Amino acids are color coded according to ClustalW pre-defined colors. The yellow highlight indicates highly conserved CCCH residues. (B) Construct used for swapping the TZF domains from various species into the site of the endogenous S. pombe Zfs1 TZF domain. (Bm = Bombyx mori (silkworm), Cg = Candida guilliermondii, Hs = Homo sapiens, Co = Chromolaena odorata, and Sp = S. pombe). In addition, a 3X Flag tag (Zfs1:Flag) was integrated into the endogenous locus with the endogenous S. pombe zfs1 3’UTR. (C) Western blot analysis of whole cell lysates isolated from the indicated strains and blotted using anti-FLAG antibodies. TZFL and TZFR indicate the amino acid residue at the beginning of the first highly conserved TZF domain lead in sequence, (R/L)YKTEL. (D) Flocculation of the indicated strains was initiated by the addition of CaCl2 and determined using the Helm assay (see (Wells, Huang et al. 2012) for details). The percentage of cells in suspension was measured by optical density. Shown are the means of values from three independent experiments. From (Wells, Hicks et al. 2015), with permission.
To determine whether the TZF domain replacements could complement the mRNA destabilization functions of Zfs1 in S. pombe, we examined the five most up-regulated transcripts previously identified to be Zfs1 targets in this species. The replacement of the endogenous S. pombe Zfs1 TZF domain with the cognate domains from a plant, an insect, a distantly related pathogenic fungus and human TTP, all corrected the molecular phenotype characteristic of Zfs1 deficiency (Fig. 8). In addition, the leucine to arginine mutation at position 1 had no effect on RNA stability in any of the complementation strains (Fig. 8). Taken together, we found that the TZF domains from these distantly related organisms could substitute for the native S. pombe TZF domain, when in the context of the intact protein, as determined by measurement of both target transcript levels and calcium-induced flocculation. These data suggest that, despite the sequence differences discussed above, this domain is functionally interchangeable among these evolutionarily very divergent eukaryotes.
Figure 8. Expression of Zfs1 target transcripts in the TZF domain complementation strains.
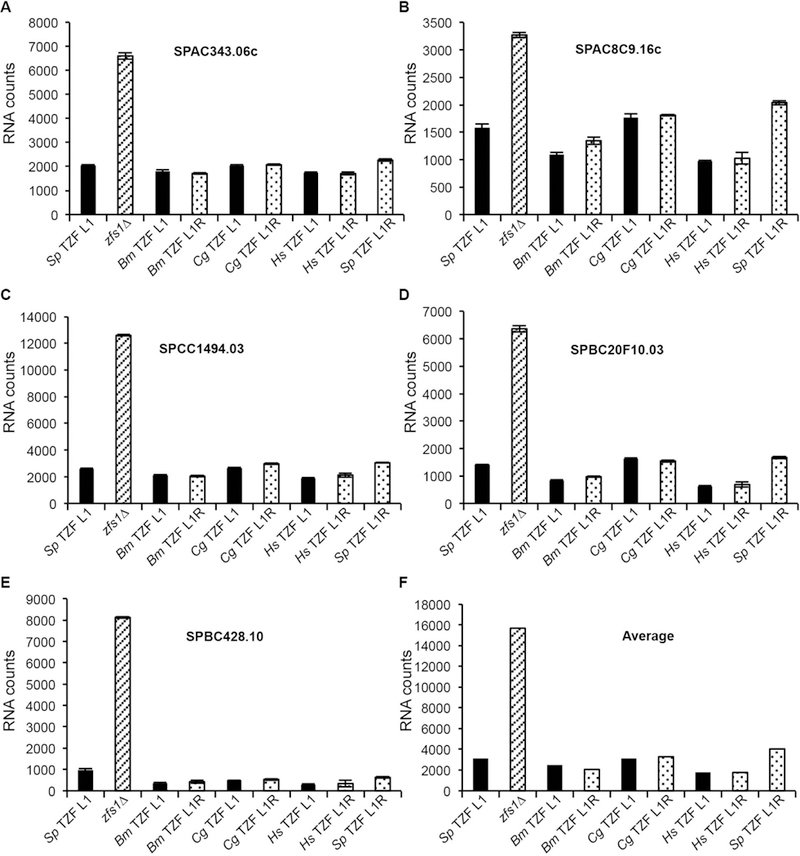
Data are shown from the NanoString analysis of target transcripts in Zfs1:Flag, zfs1Δ, and the complementation strains with the S. pombe TZF domain replaced with the indicated TZF domain described in Figure 7. The NanoString assay measures the abundance of transcripts in an RNA sample, normalized to a collection of housekeeping transcripts (Fortina and Surrey 2008). (A-E) Shown are data from 5 of the 46 potential Zfs1 target transcripts that were analyzed by NanoString. Normalized counts are shown on the y-axis for the indicated transcripts as the mean values from at least four independent isolates, +/− SD. Similar data were obtained with two plant TZF domain replacements (data not shown). (F) Shown are the averages for all transcripts that were increased 2-fold or more in the zfs1Δ strain for the indicated substitution strains. These averages include 17 of the 28 transcripts from the NanoString analysis. From (Wells, Hicks et al. 2015), with permission.
Future directions
We hope in this brief review that we have summarized much that has been learned about this evolutionarily remarkably persistent RNA binding module. Although we know a great deal more than we did when TTP was first discovered, there are many aspects of this that remain to be explored. One of these is the need for additional structures, since much of our discussion has relied on the single NMR structure described in (Hudson, Martinez-Yamout et al. 2004). We think that structures from evolutionarily very distant organisms will be very informative, but it will also be interesting to compare similarities and differences among the structures from the several family members that are expressed in a single organism, such as the three human TTP family members. These will be most useful if they can be determined in dynamic contexts, e.g., with and without RNA, and with the possibilities of regulatory control described above. In addition, although the assumption has been made from RNA binding affinities that target sequences will be conserved among proteins from these very distant organisms, it would be good to have objective data on preferred RNA target sequences, both from proteins with conventional internal spacing, and those few outliers that have gained or lost a residue within the conventional 64 amino acid TZF domain sequence. Even if ideal RNA binding site sequences are conserved in distant lineages, it would be helpful to have careful quantitative measurements of RNA binding affinity to evaluate the influence of some of the amino acid differences that have occurred over the last millennia.
The possibilities of regulation by phosphorylation or other post-translational modifications, and by directly interacting proteins, remain to be explored in detail. For example, we found that members of the mammalian TTP family could interact directly with certain isoforms of the AUF1 protein family, with the binding site being the TZF domain, and we began to explore the effect of this interaction on binding affinity (Kedar, Zucconi et al. 2012). However, much more work needs to be done in terms of identifying such interacting proteins and determining their effects on overall TTP family protein activity.
A major deficiency in our current state of knowledge is that we have been unable to divorce the nuclear import function of the TZF domain from its RNA binding function, since mutations that prevent the former have also destroyed the latter. It may be possible to identify mutants that eliminate the nuclear localization sequence while still maintaining normal RNA binding affinity, but this will probably require some trial and error mutagenesis. In any case, we assume that possible nuclear functions of these shuttling proteins will require the presence of intact RNA binding domains, and it will be fascinating to study these in, for example, mutants that lack functional nuclear export sequences.
Finally, more work needs to be done to identify naturally occurring variants in TZF domains that might lead to disease phenotypes. In man, we assume that homozygous mutations in any of the eight zinc-coordinating amino acids in the three TTP family member proteins would be lethal, but that, based on our mouse studies, heterozygous mutants of these and other severe types should be compatible with life. Resquencing studies have been unhelpful in this regard to date, but the large volume of sequence data being generated now in human populations may well uncover informative mutants in the future.
Acknowledgments
We are grateful to the members of our laboratory for helpful discussions, to Drs. Michael Fessler and Matt Schellenberg for useful comments on the manuscript, and to Dr. Thomas Randall for help with the sequence searches.
Funding Information
Supported by the Intramural Research Program of the NIEHS, NIH.
Footnotes
The authors declare that they have no conflicts of interest.
References.
- Abbehausen C (2019). “Zinc finger domains as therapeutic targets for metal-based compounds - an update.” Metallomics 11(1): 15–28. [DOI] [PubMed] [Google Scholar]
- Baou M, Jewell A and Murphy JJ (2009). “TIS11 family proteins and their roles in posttranscriptional gene regulation.” J Biomed Biotechnol 2009: 634520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshear PJ, Lai WS, Kennington EA, Brewer G, Wilson GM, Guan X and Zhou P (2003). “Characteristics of the interaction of a synthetic human tristetraprolin tandem zinc finger peptide with AU-rich element-containing RNA substrates.” J Biol Chem 278(22): 19947–19955. [DOI] [PubMed] [Google Scholar]
- Blackshear PJ and Perera L (2014). “Phylogenetic distribution and evolution of the linked RNA-binding and NOT1-binding domains in the tristetraprolin family of tandem CCCH zinc finger proteins.” J Interferon Cytokine Res 34(4): 297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer G (1991). “An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro.” Mol Cell Biol 11(5): 2460–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks SA and Blackshear PJ (2013). “Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanisms of action.” Biochim Biophys Acta 1829(6–7): 666–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Deterding LJ and Blackshear PJ (2007). “Phosphorylation site analysis of the anti-inflammatory and mRNA-destabilizing protein tristetraprolin.” Expert Rev Proteomics 4(6): 711–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Deterding LJ, Venable JD, Kennington EA, Yates JR 3rd, Tomer KB and Blackshear PJ (2006). “Identification of the anti-inflammatory protein tristetraprolin as a hyperphosphorylated protein by mass spectrometry and site-directed mutagenesis.” Biochem J 394(Pt 1): 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Dzineku F and Blackshear PJ (2003). “Expression and purification of recombinant tristetraprolin that can bind to tumor necrosis factor-alpha mRNA and serve as a substrate for mitogen-activated protein kinases.” Arch Biochem Biophys 412(1): 106–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E, Cao H, Lai WS, Kennington EA, Campbell D and Blackshear PJ (2001). “Decreased sensitivity of tristetraprolin-deficient cells to p38 inhibitors suggests the involvement of tristetraprolin in the p38 signaling pathway.” J Biol Chem 276(45): 42580–42587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E, Gilkeson GS and Blackshear PJ (1997). “Bone marrow transplantation reproduces the tristetraprolin-deficiency syndrome in recombination activating gene-2 (−/−) mice. Evidence that monocyte/macrophage progenitors may be responsible for TNFalpha overproduction.” J Clin Invest 100(5): 986–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E, Lai WS and Blackshear PJ (1998). “Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin.” Science 281(5379): 1001–1005. [DOI] [PubMed] [Google Scholar]
- Cassandri M, Smirnov A, Novelli F, Pitolli C, Agostini M, Malewicz M, Melino G and Raschella G (2017). “Zinc-finger proteins in health and disease.” Cell Death Discov 3: 17071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Lai WS, Fedic R, Stumpo DJ, Huang W, Li L, Perera L, Brewer BY, Wilson GM, Mason JM and Blackshear PJ (2014). “The Drosophila Tis11 protein and its effects on mRNA expression in flies.” J Biol Chem 289(51): 35042–35060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AR and Dean JL (2016). “The control of inflammation via the phosphorylation and dephosphorylation of tristetraprolin: a tale of two phosphatases.” Biochem Soc Trans 44(5): 1321–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbertson BJ, Liao Y, Birnbaumer L and Blackshear PJ (2008). “Characterization of zfs1 as an mRNA-binding and -destabilizing protein in Schizosaccharomyces pombe.” J Biol Chem 283(5): 2586–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De J, Lai WS, Thorn JM, Goldsworthy SM, Liu X, Blackwell TK and Blackshear PJ (1999). “Identification of four CCCH zinc finger proteins in Xenopus, including a novel vertebrate protein with four zinc fingers and severely restricted expression.” Gene 228(1–2): 133–145. [DOI] [PubMed] [Google Scholar]
- Deveau LM and Massi F (2016). “Three Residues Make an Evolutionary Switch for Folding and RNA-Destabilizing Activity in the TTP Family of Proteins.” ACS Chem Biol 11(2): 435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG and Cowtan K (2010). “Features and development of Coot.” Acta Crystallogr D Biol Crystallogr 66(Pt 4): 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortina P and Surrey S (2008). “Digital mRNA profiling.” Nat Biotechnol 26(3): 293–294. [DOI] [PubMed] [Google Scholar]
- Frederick ED, Ramos SB and Blackshear PJ (2008). “A unique C-terminal repeat domain maintains the cytosolic localization of the placenta-specific tristetraprolin family member ZFP36L3.” J Biol Chem 283(21): 14792–14800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M and Blackshear PJ (2017). “RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins.” Nat Rev Immunol 17(2): 130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingerich TJ, Stumpo DJ, Lai WS, Randall TA, Steppan SJ and Blackshear PJ (2016). “Emergence and evolution of Zfp36l3.” Mol Phylogenet Evol 94(Pt B): 518–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Wang H, Jiang S, Xia J and Jin S (2017). “The Cross-talk between Tristetraprolin and Cytokines in Cancer.” Anticancer Agents Med Chem 17(11): 1477–1486. [DOI] [PubMed] [Google Scholar]
- Gupta G, Bebawy M, Pinto TJA, Chellappan DK, Mishra A and Dua K (2018). “Role of the Tristetraprolin (Zinc Finger Protein 36 Homolog) Gene in Cancer.” Crit Rev Eukaryot Gene Expr 28(3): 217–221. [DOI] [PubMed] [Google Scholar]
- Hitti E, Iakovleva T, Brook M, Deppenmeier S, Gruber AD, Radzioch D, Clark AR, Blackshear PJ, Kotlyarov A and Gaestel M (2006). “Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element.” Mol Cell Biol 26(6): 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson BP, Martinez-Yamout MA, Dyson HJ and Wright PE (2004). “Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d.” Nat Struct Mol Biol 11(3): 257–264. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A and Schulten K (1996). “VMD: visual molecular dynamics.” J Mol Graph 14(1): 33–38, 27–38. [DOI] [PubMed] [Google Scholar]
- Jurrus E, Engel D, Star K, Monson K, Brandi J, Felberg LE, Brookes DH, Wilson L, Chen J, Liles K, Chun M, Li P, Gohara DW, Dolinsky T, Konecny R, Koes DR, Nielsen JE, Head-Gordon T, Geng W, Krasny R, Wei GW, Holst MJ, McCammon JA and Baker NA (2018). “Improvements to the APBS biomolecular solvation software suite.” Protein Sci 27(1): 112–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M and McCammon JA (2002). “Molecular dynamics simulations of biomolecules.” Nat Struct Biol 9(9): 646–652. [DOI] [PubMed] [Google Scholar]
- Kedar VP, Zucconi BE, Wilson GM and Blackshear PJ (2012). “Direct binding of specific AUF1 isoforms to tandem zinc finger domains of tristetraprolin (TTP) family proteins.” J Biol Chem 287(8): 5459–5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A (2010). “The discovery of zinc fingers and their applications in gene regulation and genome manipulation.” Annu Rev Biochem 79: 213–231. [DOI] [PubMed] [Google Scholar]
- Kratochvill F, Machacek C, Vogl C, Ebner F, Sedlyarov V, Gruber AR, Hartweger H, Vielnascher R, Karaghiosoff M, Rulicke T, Muller M, Hofacker I, Lang R and Kovarik P (2011). “Tristetraprolin-driven regulatory circuit controls quality and timing of mRNA decay in inflammation.” Mol Syst Biol 7: 560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WS, Carballo E, Thorn JM, Kennington EA and Blackshear PJ (2000). “Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA.” J Biol Chem 275(23): 17827–17837. [DOI] [PubMed] [Google Scholar]
- Lai WS, Kennington EA and Blackshear PJ (2002). “Interactions of CCCH zinc finger proteins with mRNA: non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs.” J Biol Chem 277(11): 9606–9613. [DOI] [PubMed] [Google Scholar]
- Lai WS, Kennington EA and Blackshear PJ (2003). “Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease.” Mol Cell Biol 23(11): 3798–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WS, Perera L, Hicks SN and Blackshear PJ (2014). “Mutational and structural analysis of the tandem zinc finger domain of tristetraprolin.” J Biol Chem 289(1): 565–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WS, Stumpo DJ, Kennington EA, Burkholder AB, Ward JM, Fargo DL and Blackshear PJ (2013). “Life without TTP: apparent absence of an important anti-inflammatory protein in birds.” Am J Physiol Regul Integr Comp Physiol 305(7): R689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WS, Stumpo DJ, Qiu L, Faccio R and Blackshear PJ (2018). “A Knock-In Tristetraprolin (TTP) Zinc Finger Point Mutation in Mice: Comparison with Complete TTP Deficiency.” Mol Cell Biol 38(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laity JH, Lee BM and Wright PE (2001). “Zinc finger proteins: new insights into structural and functional diversity.” Curr Opin Struct Biol 11(1): 39–46. [DOI] [PubMed] [Google Scholar]
- Maeda K and Akira S (2017). “Regulation of mRNA stability by CCCH-type zinc-finger proteins in immune cells.” Int Immunol 29(4): 149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, Yoshino Y, Morita N and Kaneda N (2002). “Identification of nuclear import and export signals within the structure of the zinc finger protein TIS11.” Biochem Biophys Res Commun 293(4): 1242–1247. [DOI] [PubMed] [Google Scholar]
- Navarro FJ, Chakravarty P and Nurse P (2017). “Phosphorylation of the RNA-binding protein Zfs1 modulates sexual differentiation in fission yeast.” J Cell Sci 130(24): 4144–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Lee TH and Kang TH (2018). “Roles of Tristetraprolin in Tumorigenesis.” Int J Mol Sci 19(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips RS, Ramos SB and Blackshear PJ (2002). “Members of the tristetraprolin family of tandem CCCH zinc finger proteins exhibit CRM1-dependent nucleocytoplasmic shuttling.” J Biol Chem 277(13): 11606–11613. [DOI] [PubMed] [Google Scholar]
- Prabhala P and Ammit AJ (2015). “Tristetraprolin and its role in regulation of airway inflammation.” Mol Pharmacol 87(4): 629–638. [DOI] [PubMed] [Google Scholar]
- Ross CR, Brennan-Laun SE and Wilson GM (2012). “Tristetraprolin: roles in cancer and senescence.” Ageing Res Rev 11(4): 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross EA, Smallie T, Ding Q, O’Neil JD, Cunliffe HE, Tang T, Rosner DR, Klevernic I, Morrice NA, Monaco C, Cunningham AF, Buckley CD, Saklatvala J, Dean JL and Clark AR (2015). “Dominant Suppression of Inflammation via Targeted Mutation of the mRNA Destabilizing Protein Tristetraprolin.” J Immunol 195(1): 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler H and Stoecklin G (2008). “Control of mRNA decay by phosphorylation of tristetraprolin.” Biochem Soc Trans 36(Pt 3): 491–496. [DOI] [PubMed] [Google Scholar]
- Sanduja S, Blanco FF, Young LE, Kaza V and Dixon DA (2012). “The role of tristetraprolin in cancer and inflammation.” Front Biosci (Landmark Ed) 17: 174–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimberg GD, Ok K, Neu HM, Splan KE and Michel SLJ (2017). “Cu(I) Disrupts the Structure and Function of the Nonclassical Zinc Finger Protein Tristetraprolin (TTP).” Inorg Chem 56(12): 6838–6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpo DJ, Broxmeyer HE, Ward T, Cooper S, Hangoc G, Chung YJ, Shelley WC, Richfield EK, Ray MK, Yoder MC, Aplan PD and Blackshear PJ (2009). “Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis.” Blood 114(12): 2401–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpo DJ, Byrd NA, Phillips RS, Ghosh S, Maronpot RR, Castranio T, Meyers EN, Mishina Y and Blackshear PJ (2004). “Chorioallantoic fusion defects and embryonic lethality resulting from disruption of Zfp36L1, a gene encoding a CCCH tandem zinc finger protein of the Tristetraprolin family.” Mol Cell Biol 24(14): 6445–6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaffer MP, Jones AW, Flynn HR, Snijders AP and Nurse P (2018). “Quantitative Phosphoproteomics Reveals the Signaling Dynamics of Cell-Cycle Kinases in the Fission Yeast Schizosaccharomyces pombe.” Cell Rep 24(2): 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes BF and Blackshear PJ (1996). “A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency.” Immunity 4(5): 445–454. [DOI] [PubMed] [Google Scholar]
- Taylor GA, Thompson MJ, Lai WS and Blackshear PJ (1995). “Phosphorylation of tristetraprolin, a potential zinc finger transcription factor, by mitogen stimulation in intact cells and by mitogen-activated protein kinase in vitro.” J Biol Chem 270(22): 13341–13347. [DOI] [PubMed] [Google Scholar]
- Taylor GA, Thompson MJ, Lai WS and Blackshear PJ (1996). “Mitogens stimulate the rapid nuclear to cytosolic translocation of tristetraprolin, a potential zinc-finger transcription factor.” Mol Endocrinol 10(2): 140–146. [DOI] [PubMed] [Google Scholar]
- Wells ML, Hicks SN, Perera L and Blackshear PJ (2015). “Functional equivalence of an evolutionarily conserved RNA binding module.” J Biol Chem 290(40): 24413–24423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells ML, Huang W, Li L, Gerrish KE, Fargo DC, Ozsolak F and Blackshear PJ (2012). “Posttranscriptional regulation of cell-cell interaction protein-encoding transcripts by Zfs1p in Schizosaccharomyces pombe.” Mol Cell Biol 32(20): 4206–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells ML, Perera L and Blackshear PJ (2017). “An Ancient Family of RNA-Binding Proteins: Still Important!” Trends Biochem Sci 42(4): 285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells ML, Washington OL, Hicks SN, Nobile CJ, Hartooni N, Wilson GM, Zucconi BE, Huang W, Li L, Fargo DC and Blackshear PJ (2015). “Post-transcriptional regulation of transcript abundance by a conserved member of the tristetraprolin family in Candida albicans.” Mol Microbiol 95(6): 1036–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington MT, Amann BT, Nathans D and Berg JM (1996). “Metal binding properties and secondary structure of the zinc-binding domain of Nup475.” Proc Natl Acad Sci U S A 93(24): 13754–13759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang EC, Boo SM, Bhattacharya D, Saunders GW, Knoll AH, Fredericq S, Graf L and Yoon HS (2016). “Divergence time estimates and the evolution of major lineages in the florideophyte red algae.” Sci Rep 6: 21361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanocco-Marani T (2010). “TIS11/TTP gene family: it’s never too late for tumor suppressors.” Cell Cycle 9(24): 4771. [DOI] [PubMed] [Google Scholar]


