Summary
Xenograft cell transplantation into immunodeficient mice has become the gold-standard for assessing pre-clinical efficacy of cancer drugs, yet direct visualization of single cell phenotypes is difficult. Here, we report an optically-clear, prkdc−/−, il2rga−/− zebrafish that lack adaptive and natural killer immune cells, can engraft a wide array of human cancers at 37°C, and permit the dynamic visualization of single engrafted cells. For example, photoconversion cell lineage-tracing identified migratory and proliferative cell states in human rhabdomyosarcoma, a pediatric cancer of muscle. Additional experiments identified the preclinical efficacy of combination olaparib PARP-inhibitor and temozolomide DNA-damaging agent as an effective therapy for rhabdomyosarcoma and visualized therapeutic responses using a four-color FUCCI cell cycle fluorescent reporter. These experiments identified that combination treatment arrested rhabdomyosarcoma cells in the G2 cell cycle prior to induction of apoptosis. Finally, patient-derived xenografts could be engrafted into our model, opening new avenues for developing personalized-therapeutic approaches in the future.
Graphical Abstract
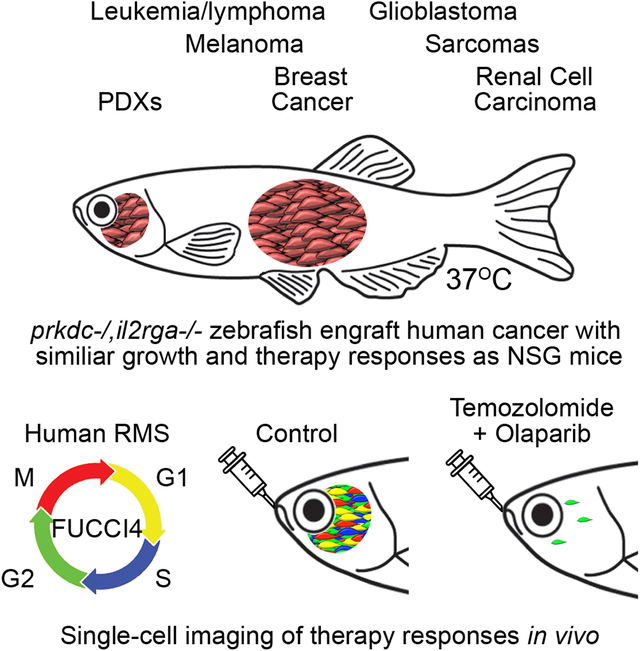
In Brief:
Robust engraftment of human cancers into optically-clear, immune-deficient zebrafish at physiological temperatures enables dynamic visualization of cell biological phenotypes and therapy responses in single cancer cells over time.
Introduction
Cell transplantation has become an important experimental tool to assess normal and malignant cell phenotypes (Goyama et al., 2015). In the setting of cancer, cell transplantation approaches have been used to define mechanisms that drive continued tumor growth, metastasis, and therapy responses (Bakhoum et al., 2018, Stewart et al., 2017, Tian et al., 2017, Das et al., 2016). Using these approaches, cancers arising in genetically engineered mouse models can be readily engrafted into inbred strains of syngeneic mice (Kelly et al., 2007, Bruce and Van Der Gaag, 1963, Curtis et al., 2010). Alternatively, it is also possible to xenograft human cancers into immune deficient mice. Most notably, the current gold-standard for engrafting human cancers is the NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (Ito et al., 2002). Additional immune deficient mouse models have also been created that express human cytokines and facilitate enhanced engraftment of normal blood cell subpopulations as well as acute myeloid leukemias (Rongvaux et al., 2014, Das et al., 2016). Despite the ever-increasing diversity of immune deficient and humanized mouse models, there are inherent limitations. For example, mice are expensive and require large vivarium space - limiting the scale of experimentation. Mice are also furred and imaging through skin is difficult. In fact, single cell imaging modalities largely focus on creating surgical windows and utilizing complex multi-photon imaging modalities to visualize cells in vivo (Ellenbroek and van Rheenen, 2014). Because these windows must be created prior to implantation of tumor, it is not possible to image disseminated cancer cells unbiasedly throughout the whole animal.
To address these issues, others have developed zebrafish xenograft transplantation approaches to engraft human cancers into two-day-old larval fish at a stage prior to the development of the adaptive immune system. Elegant work has shown engraftment of human tumor cells and defined novel mechanisms of metastasis and growth (Fior et al., 2017, Chapman et al., 2014, Welker et al., 2017). Despite these successes, larval fish can only be transplanted with hundreds of cells that often do not contain cancer stem cells that are required for long-term, sustained tumor growth. Moreover, engrafted zebrafish are raised at non-physiological temperatures of ≤ 34°C and thus do not proliferate at similar rates as when grown in mouse or human. Engrafted animals also do not develop histologically similar tumors as those found in humans and engraftment studies are necessarily confined to studies before 10 days of life because fish eventually develop acquired immune responses that kill human cancer cells. Although amazing useful for facile, short-term drug screening, therapy responses in larval fish can only be assessed after submersing the whole fish in water that contains drugs, hence drug penetration and accurate oral uptake are often not known. Finally, submersion therapy cannot achieve accurate drug dosing, optimized drug schedule, and assessment of pharmacodynamics over extended periods beyond one week – all of which are required endpoints for moving pre-clinical animal studies into human clinical trials.
Our group has previously used syngeneic adult zebrafish and partially-immune deficient strains of zebrafish to perform large-scale allogeneic cell transplantation experiments (Blackburn et al., 2012, Tang et al., 2014, Ignatius et al., 2012, Moore et al., 2016a, Smith et al., 2010, Tenente et al., 2014, Moore et al., 2016b). Adult zebrafish have many attributes that make it an ideal cell transplantation model, including high fecundity, low cost, optical clarity and the ability to perform high-throughput drug and tumor evolution studies (Chen et al., 2014, Tang et al., 2014). Yet to date, efficient engraftment of human cancers and tissues into immune deficient zebrafish reared at 37°C has not been rep orted, owing to lack of compound mutant strains that lack T, B, and NK cells and ineffective rearing conditions for highly immune deficient strains. Here, we have generated optically-clear zebrafish that are deficient in prkdc (protein kinase, DNA-activated, catalytic polypeptide) and interleukin-2 receptor gamma a (il2rga). These fish lack T, B, and natural killer cells and robustly engraft human cancer cells that have similar growth kinetics and histopathology as those grown in NSG mice. In addition, we also developed approaches to follow tumor migration and proliferation at single cell resolution in vivo. Using these approaches, we observed important functional diversity in rhabdomyosarcoma cell types that drive proliferation and migration and provide preclinical rationale for combination olaparib PARP inhibitor and the temozolomide DNA damaging agent in pediatric rhabdomyosarcoma. Finally, we established cell transplantation models from patient-derived xenografts (PDXs) for a range of cancer types, opening the model to precision medicine approaches in the future.
Results
Optically clear, prkdc−/−, il2rga−/− zebrafish engraft human cancer
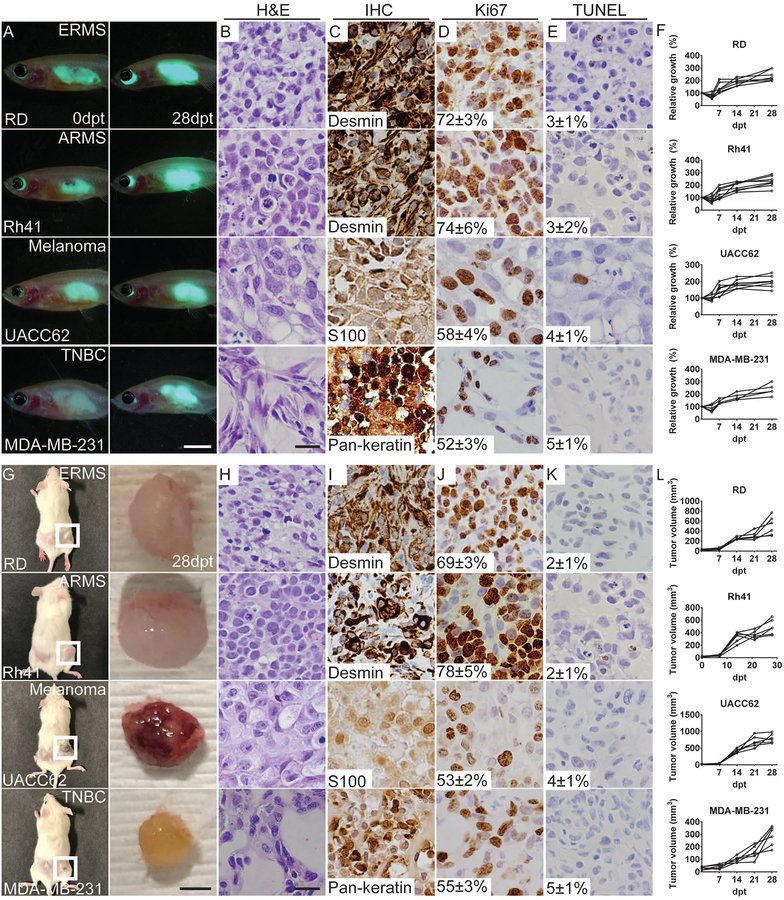
Here, we report an optically-clear zebrafish that is deficient in prkdc and il2rga. These compound mutant fish lack T, B, and NK cells (Tang et al., 2017). Specifically, prkdc−/−, il2rga+/− animals were raised under normal laboratory conditions and incrossed. The resulting progeny were grown under normal laboratory conditions, genotyped by scale resection at 2–3 months of age, and then reared in the presence of antibiotics at 37°C until 6 months of age. As expected, the prkdc−/−, il2rga−/− animals were recovered at expected Mendelian ratios at the time of genotyping (24.8%, n=1,205 of 4850 fish were prkdc−/−, il2rg−/−), with 84.6% survival for fish reared with antibiotics up to 6 months of age (n=22 of 26 animals, Fig. S1A–C). To assess the ability of these animals to engraft human cancers, two-month old prkdc−/−, il2rga−/− zebrafish were transplanted with a panel of lentiviral-GFP expressing human cancer cells (5×105 cells injected intraperitoneally/animal). In total, high engraftment rates were achieved using 16 different fluorescent-labeled cancers that included melanoma, renal cell carcinoma, chronic myeloid leukemia, mantle cell lymphoma, triple-negative breast cancer, and a wide variety of sarcomas (Fig. 1A–F, Fig. S1D–M, and Table S1). Importantly, engraftment persisted for ≥ 28 days with tumors growing substantially over this time with overall morbidity associated with the transplantation procedure being <15% (Fig. S1C). Analysis of engrafted tumors revealed similar histopathology and cell morphological features with each respective cancer type (Fig. 1 and Fig. S1), including expression of diagnostic gene expression markers and pathognomonic gene fusions (Fig. 1C and Fig. S1N–S). Engrafted RMS, melanoma, and breast cancers also had similar rates of proliferation and apoptosis whether grown in prkdc−/−, il2rga−/− zebrafish or NSG mice (Fig. 1D,E,J,K, n=6 mice engrafted/tumor cell line). Together, our results show that the prkdc−/−, il2rga−/− zebrafish robustly engraft a wide array of human cancers at 37°C that grow with similar kinetics and have similar histology as those engrafted into NSG mice.
Figure 1. Human cancers have similar growth kinetics, histology, and rates of proliferation and apoptosis when engrafted into prkdc−/−, il2rga−/− zebrafish or NSG mice.
(A-F) Engraftment of EGFP+ human tumor cells into prkdc−/−, il2rga−/− zebrafish grown at 37°C. Merged fluorescence and brightfield images of whole animals imaged at 0 and 28 days post-transplant (dpt; A). (G-L) Analysis of the same tumors grown in NSG mice for 28 days. Images of engrafted mice shown to left and excised tumors to the right (G). Hematoxylin and eosin stained sections (zebrafish,B; mouse,H) and immunohistochemistry for cell lineage markers (zebrafish,C; mouse,I), Ki67 (zebrafish,D; mouse,J) and TUNEL (zebrafish,E; mouse,K) with the average percentage of positive cells +/− standard deviation noted (n≥3 animals/tumor type). Quantification of relative tumor growth in engrafted prkdc−/−, il2rga−/− zebrafish (F) and NSG mice (L). Human embryonal rhabdomyosarcoma RD (ERMS RD), alveolar rhabdomyosarcoma Rh41 (ARMS Rh41), melanoma UACC62, and triple-negative breast cancer MDA-MB-231. Scale bars equal 0.25 cm (A); 50 μm (B-E, H-K); and 0.5 cm (G). See also Table S1 and Figure S1.
prkdc−/−, il2rga−/− zebrafish engraft patient-derived xenografts
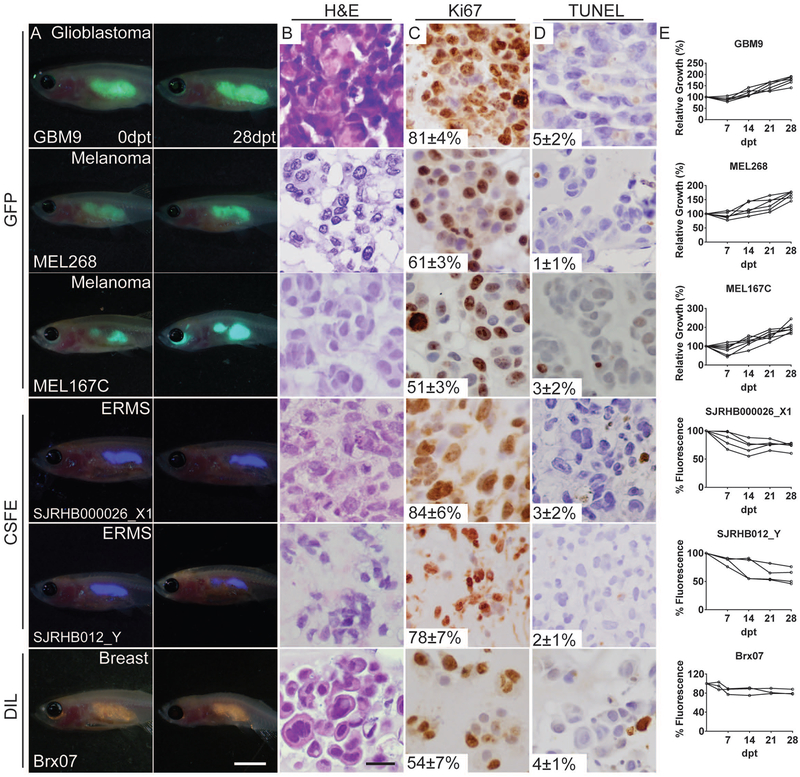
With the long-term goal of using our immune deficient zebrafish as a preclinical model for evaluating patient specific responses to therapy, we next evaluated engraftment of six patient-derived xenografts (PDX) into prkdc−/−, il2rga−/− zebrafish. These included glioblastoma (GBM9, (Tchoghandjian et al., 2012)), two embryonal rhabdomyosarcomas (SJRHB000026_X1 and SJRHB012_Y, (Stewart et al., 2017)), two metastatic BRAFV600E-induced melanomas (MEL167C and MEL268C, (Luo et al., 2014)) and a therapy-resistant ER+/PR+/Her2- lobular breast cancer (BRx-07, (Yu et al., 2014)). These PDXs were created from passaging GBM9 cells under sphere-inducing cell culture conditions, direct passaging from mice (SJRHB000026_X1 and SJRHB012_Y), or following isolation from circulating tumor cells from patients and short-term culture in anchorage-free conditions (MEL167C, MEL268C and BRx-07). As expected based on our cell line engraftment studies, all six PDXs engrafted efficiently into prkdc−/−, il2rga−/− zebrafish for in excess of 28 days, with a subset of fish ultimately succumbing to tumor burden over the course of these experiments (Fig. 2 and Table S1). Tumor cell engraftment was also easily visualized by GFP lentiviral incorporation (GBM9, MEL167C and MEL268C) or by using cytoplasmic dyes (CSFE for SJRHB000026_X1 and SJRHB012_Y ERMS cells and DIL for BRx-07) (Fior et al., 2017).
Figure 2. Prkdc−/−, il2rga−/− zebrafish engraft patient-derived tumors.
Merged fluorescence and brightfield images of prkdc−/−, il2rga−/− zebrafish imaged at 0 and 28 days post-transplant (dpt, A). Hematoxylin and eosin stained sections (B) and immunohistochemistry for Ki67 (C) and TUNEL (D) with the average percentage of positive cells +/− standard deviation noted (n≥3 fish/tumor type). Quantification of relative growth of transplanted cells over time (E). Note that Viafluor- and DiL-labeled cells would not be predicted to increase intensity with time as 100% fluorescence equates to full retention of tumor cells over the experiment. Scale bar equals 0.25 cm (A); 50 μm (B-D).
Visualizing human cancer cell processes at single cell resolution
Cancer is highly heterogeneous and is often comprised of tumor cell types that have specific functions in driving growth, invasion, metastasis, and therapy resistance. Yet, to date, imaging individual human cancer cell behaviors in engrafted mice has been challenging, requiring surgical window construction and complex imaging techniques (Ellenbroek and van Rheenen, 2014). Building on the optical clarity of Casper zebrafish (White et al., 2008), we next refined a method for engrafting human cancer cells into the peri-ocular muscle and dynamically visualizing fluorescently-labeled tumor cells using confocal microscopy (Fig. S2, 5×104 cells/fish). This approach allowed long-term engraftment and assessment of growth for in excess of 21 days post-transplantation (dpt, Fig. S2). Histology, rate of cell proliferation and apoptosis were similar to cancers engrafted by peritoneal injection into prkdc−/−, il2rga−/− zebrafish or those grown in NSG mice (Fig. S2I–Q). Finally, direct quantitation of tumor growth was accomplished by counting individual EGFP+ tumor cells within the engrafted fish (Fig. S2D, H), providing unprecedented access to visualize and quantify cancer cell growth at single cell resolution in vivo.
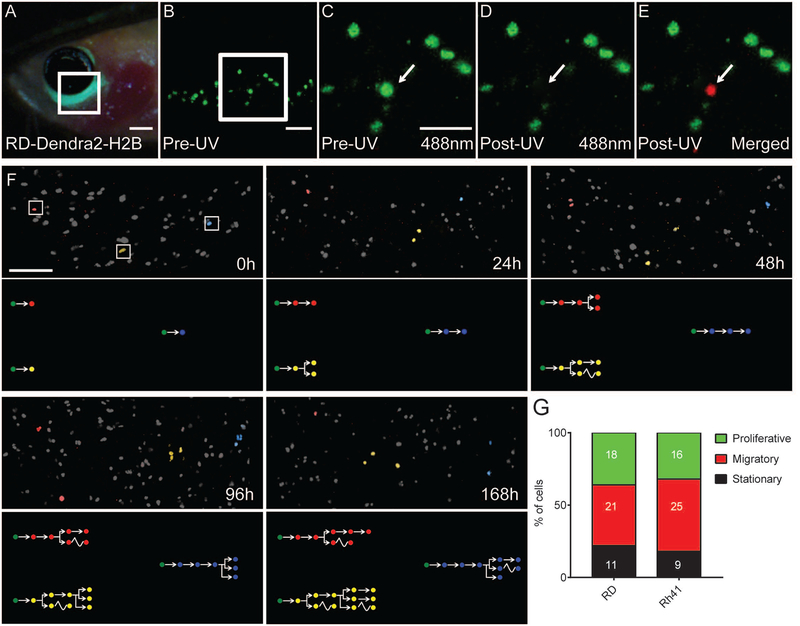
We next optimized approaches to dynamically visualize single cell behavior of cancer cells over extended periods of time in vivo. Our work focused on imaging cell biological differences in rhabdomyosarcoma (RMS), a pediatric soft tissue sarcoma that is comprised of muscle cells arrested at different maturation stages. RMS consists of two major subtypes including embryonal rhabdomyosarcoma (ERMS) that is driven by oncogenic RAS signaling (Langenau et al., 2007, Chen et al., 2013, Shern et al., 2014) and alveolar RMS (ARMS) that is driven by PAX3-FKHR or PAX7-FKHR chromosomal translocations (Barr et al., 1993, Davis et al., 1994). Here, we created human RD ERMS and Rh41 ARMS cells that express a nuclear-localized, photo-convertible H2b-Dendra2 protein (Fig. 3A–E). Following exposure of individual cells to a 405nm UV laser, nuclear-localized H2b-Dendra2 irreversibly switches to a red fluorescent protein that can be detected using a 546nm laser (Chudakov et al., 2007). Using this approach, we individually labeled human RMS cells in vivo and visualized single cell behavior over a course of 7 days (Fig. 3F). In total, 2–3 cells were photo-converted per tumor-bearing fish and then re-imaged at 24, 48, 96 and 168 hours (n=50 cells total per experiment). From this analysis, we observed three functionally distinct RMS cancer cell types that included 1) highly migratory cells, 2) actively proliferating cells and 3) bystander cells that did not divide or move over the 7 days of imaging (Fig. 3G, Fig. S3). This analysis also revealed that proliferative and migratory potential of RMS cells were largely mutually exclusive. Proliferative cells could give rise to migratory cell types, but not vice versa, consistent with our previous findings in zebrafish ERMS (Ignatius et al., 2012). This work is exciting because it identified three phenotypically-distinct human RMS cell types had been presumed based on in vivo zebrafish modeling (Ignatius et al., 2012), but which had not yet been visualized in human RMS in vivo.
Figure 3. Identification of functionally distinct rhabdomyosarcoma cell types using photo-convertible Dendra2-H2b and single cell fate mapping.
(A-E) A prkdc−/−, il2rga−/− zebrafish engrafted with H2b-Dendra2+ RD cells. Cells were engrafted into the peri-ocular muscle and imaged at 7 days post-transplantation. Epifluorescent image of head region (A) and higher magnification confocal images (B, 100X; C-E, 200X magnification). Images were taken before 405nm UV photoconversion (B,C) or immediately post-UV exposure (D-E, merged image E shows excitation using both 488 and 563nm laser). (F) Serially imaging of three photo-converted RD ERMS cells. Cells have been pseudo-colored and tracked at 0, 24, 48, 96 and 168 hours after photoconversion. Cell fates are pictorially depicted within lower panels with migration denoted by wavy lines. (G) Quantification of single cell behaviors in RD ERMS and Rh41 ARMS cells. Number of cells imaged with each phenotype are noted within bar graphs. Scale bar equals 0.1 cm (A); 50 μm (B-E); 200 μm (F). See also Figure S2 and S3.
Preclinical modeling identifies combination olaparib and temozolomide for the treatment of RMS
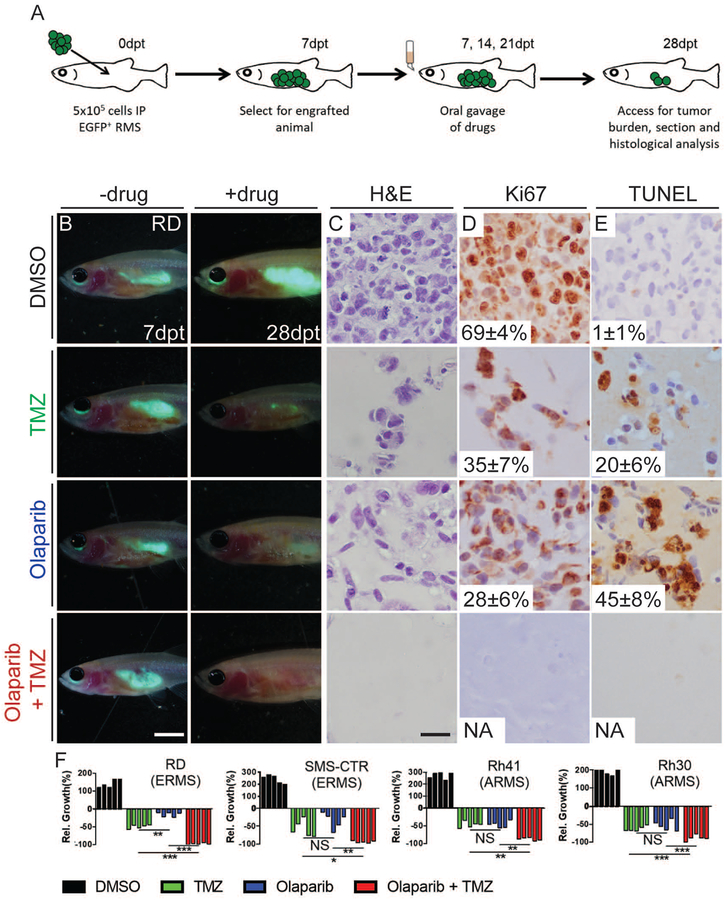
To further establish the prkdc−/−, il2rga−/− zebrafish as a preclinical therapeutic model, we next investigated the hypothesis that the PARP inhibitor olaparib when complexed with temozolomide DNA damaging agent will inhibit RMS growth in vivo. Similar drug combinations curb the growth of human Ewing’s sarcoma in mouse xenograft studies and have rapidly moved into phase II clinical trials for this disease (Garnett et al., 2012, Stewart et al., 2014). Yet, to date, this drug combination has yet to be tested for preclinical efficacy in RMS. To verify that the pharmacokinetics of olaparib and temozolomide are similar in zebrafish when compared with mouse and human, we orally gavaged adult zebrafish with an equivalent dose of 50mg/kg olaparib or 33mg/kg temozolomide which had been previously used in preclinical mouse studies and which achieved a similar dosing as reported in humans (Rajan et al., 2012, Sun et al., 2018, Diez et al., 2010, McMahon et al., 2016, Agarwala and Kirkwood, 2000). Specifically, blood was harvested from 25 animals per time point by cardiac puncture, pooled, and quantified by mass spectrometry. As expected, plasma concentration of olaparib and temozolomide was similar to that found in both mouse preclinical models and human (Fig. S4A,B) (Rajan et al., 2012, Sun et al., 2018, Diez et al., 2010, McMahon et al., 2016, Agarwala and Kirkwood, 2000, Stewart et al., 2014). Next, we assessed the preclinical efficacy of each drug alone or in combination for the ability to curb growth of transplanted GFP-expressing ARMS (Rh30 and Rh41), ERMS (RD and SMS-CTR), and control A673 Ewing’s sarcoma cells into prkdc−/−, il2rga−/− zebrafish. Engrafted fish were photographed after 7 days post transplantation and then orally gavaged with each drug alone or in combination. Animals were dosed three times at 7 day intervals and imaged for effects on tumor growth at 28 days (Fig. 4 and Fig. S4C–I). As expected, single treatment alone had little effect on overall tumor growth while combination treatment led to potent tumor regressions in Ewing’s sarcoma (Fig. S4C–E), similar to preclinical mouse xenograft experiments (Stewart et al., 2014, Gill et al., 2015, Garnett et al., 2012). Remarkably, this drug combination also eliminated engrafted EGFP+ human RMS cells in all four disease models tested (Fig. 4B–F), while only partial responses were observed in single drug treated animals.
Figure 4. Combination treatment of temozolomide and olaparib PARP-inhibitor reduces growth of human RMS cells in engrafted prkdc−/−, il2rga−/− zebrafish.
Schematic of experimental design (A). Merged fluorescence and brightfield images of prkdc−/−, il2rga−/− animals engrafted with EGFP+ RD cells (B). Whole animal imaging of engrafted animal at 7 dpt (prior to drug administration, B, left) and 28 dpt (after three cycles of drug dosing, B, right). Hematoxylin and eosin (C), Ki67 (D), and TUNEL (E) stained sections of fish engrafted with RD RMS cells. The average percentage of positive cells +/− standard deviation is noted (n=5 fish/treatment). Quantification of relative RMS growth following drug administration in RD, SMSCTR, Rh41 and Rh30 (F) cells. *p<0.05, **p<0.01, ***p<0.001, Student’s T-test. Not significant (NS). Scale bar equals 0.25 cm (B) and 50 μm (C-E). Not applicable (NA). See also Figure S4.
To independently verify these results, engrafted animals were sacrificed at the end of the experiment and sectioned to examine histology by hematoxylin and eosin (H&E) staining, cell proliferation by Ki67 antibody staining, and apoptosis using TUNEL. Histological analysis confirmed overall reductions in cellularity in animals treated with single drugs and near complete loss of proliferative cells following three cycles of combination treatment (Fig. 4C–E, Fig. S4G–I). Together, our work has uncovered the remarkable sensitivity of human RMS to combination olaparib and temozolomide using the preclinical prkdc−/−, il2rga−/− zebrafish model.
To extend our findings to more traditional mouse xenograft models, we next assessed single and combination drug treatment on RMS tumors engrafted into NSG mice. Specifically, luciferase-expressing human RD and Rh41 RMS cells were engrafted into the flank muscle of NSG mice. Once tumors reached 300mm3, animals were orally gavaged for five consecutive days with vehicle control, olaparib (50mg/kg), temozolomide (25mg/kg), or drug combination (Drapkin et al., 2018). Tumor growth was assessed weekly by bioluminescence for 28 days. In this experiment, animals that received single drug treatment initially had early tumor responses but regained similar growth rates to vehicle control treated animals by 28 days post-treatment (Fig. S5A–F). By contrast, tumor bearing mice dosed with combination therapy exhibited significant tumor regression by 14 days after treatment, with tumors remaining smaller in size for duration of follow up (Fig. S5A–F). These experiments suggested that combination therapy, but not single drug doses of olaparib and temozolomide, was adequate to retard RMS tumor growth in NSG mice.
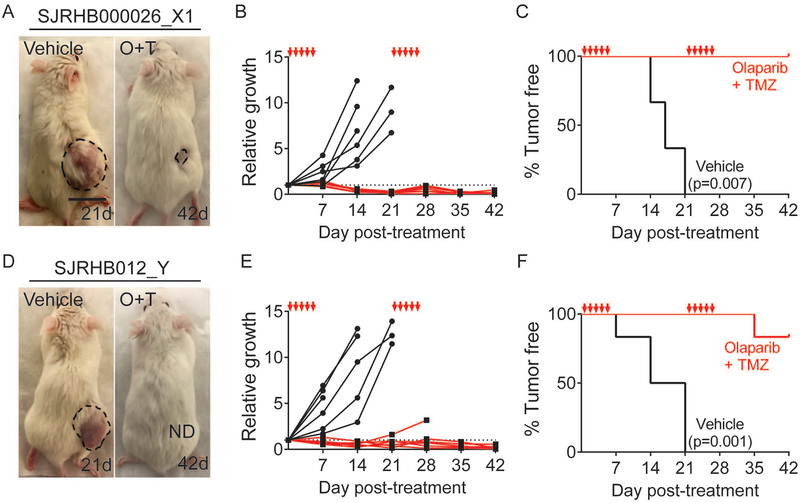
To further verify the effectiveness of combination olaparib and temozolomide on suppressing RMS growth in mouse xenografts, we next extended our studies to a larger cohort of NSG mice and followed drug responses over a longer time. Specifically, luciferase-expressing RD and Rh41 RMS cells were engrafted into NSG mice (N=8/arm). Animals were followed until tumors reached a size of 300mm3, at which time mice were orally gavaged for five consecutive days, allowed to rest for 16 days, and this cycle repeated (Fig. S5G–P). Two animals from each treatment group were sacrificed at 10 days post drug administration to quantify drug effects on Ki67 staining and TUNEL (Fig. S5I,J,N,O). Bioluminescence imaging of luciferase-expressing tumors showed that two cycles of combination drug treatment suppressed RMS growth while tumors continued to grow in vehicle control treated mice (p<0.002 for both RD and Rh41 at 14–35 days post treatment, Student’s T-test). As expected, combination drug treatment resulted in reductions in overall cellularity, reduced Ki67 positive cells and increased numbers of apoptotic TUNEL positive cells (Fig. S5H–J,M–O). Finally, we examined the effectiveness of olaparib and temozolomide in PDX models of ERMS. Specifically, SJRHB000026_X1 and SJRHB012_Y RMS tumors were engrafted into NSG mice (n=6/arm) and assessed for responses to combination therapy as outlined above (Fig. 5). Not unexpectedly, both PDX models exhibited potent tumor regressions following two cycles of drug administration while control treated tumors continued to grow (p≤0.008 at 42d, Fisher’s Exact Test, Fig. 5B,E). Mice engrafted with Human RMS PDXs also had prolonged survival when compared to vehicle control treated mice (SJRHB000026_X1, p=0.007 and SJRHB012_Y, p=0.0001, Mantel-cox log-rank test; Fig. 5C, F). In total, our mouse xenograft experiments using both cell line and PDX models are consistent with our observations reported in zebrafish xenograft studies, strongly supporting the preclinical efficacy of combination olaparib and temozolomide as an effective therapy in human RMS.
Figure 5. Combination treatment of temozolomide and olaparib PARP-inhibitor reduces growth of patient-derived rhabdomyosarcomas grown in NSG mice.
Representative images of mice engrafted with two independent PDX RMS and orally gavaged with vehicle control shown at 21 days post-treatment (21d) or following combination drug treatment at 42 days (42d, A,D). Quantification of relative growth (p=0.001 and p=0.008 at 42d for data shown in B and E, respectively, Fisher’s Exact Test) and Kaplan-Meier survival analysis with p-values denoted within the panels (C,F, n=6 mice/treatment arm). The five days of drug administration are denoted by red arrows in B,C,E and F. See also Figure S5.
Visualizing the pharmacodynamics of drug responses at single cell resolution using zebrafish xenografts
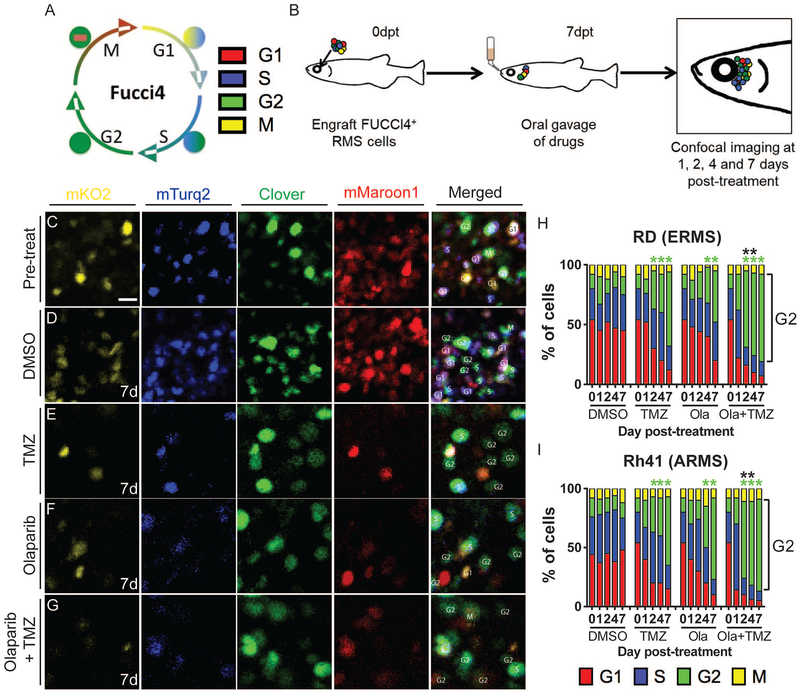
To investigate the effects of the aforementioned drug responses on cell cycle and provide real-time visualization of pharmacodynamic responses to combination therapy in vivo, we next created human RD ERMS and Rh41 ARMS cells that stably express the FUCCI4 cell cycle fluorescent reporter (Bajar et al., 2016). This four-colored reporter emits a combination of turquoise and Kusabira-Orange during G1 phase, turquoise and clover during S phase, clover only during G2 phase and clover and maroon during M phase, thus allowing real time tracking of proliferation and cellular state in live RMS cells at single cell resolution (Fig. 6A). Because fish and mice engraft human tumors with similar proliferation rates, it was not surprising that the overall percentages of RD and Rh41 cells in each cell cycle cell state was the same when grown in either immune deficient zebrafish or NSG mice (Fig. S6). We next assessed the kinetics of therapy responses at single cell resolution by engrafting FUCCI4+ RMS cells into the peri-ocular muscle, allowed engraftment for 7 days, and then orally gavaged fish with either vehicle control, single or combination drug treatment. These fish were then imaged at 1,2,4 and 7 days post-treatment (Fig. 6B, n=100 engrafted cells/time point/drug, n=3 animals analyzed per condition). Using this approach, combination therapy accelerated the time and numbers of G2-arrested RD and Rh41 cells as early as two days post drug treatment when compared with control or single agent treated fish (Fig. 6C–I), suggestive that G2 arrest precedes apoptosis in combination treated animals. Taken together, our work has identified combination olaparib and temozolomide as an effective therapeutic approach in human RMS xenograft studies and established the prkdc−/−, il2rga−/− zebrafish model as an effective in vivo tool for dynamic imaging of pharmacodynamic therapy responses at single cell resolution in vivo.
Figure 6. FUCCI4 cell cycle imaging reveals that combined treatment with olaparib PARP-inhibitor and temozolomide results in rapid G2-cell cycle arrest of human RMS cells engrafted into prkdc−/−, il2rga−/− zebrafish.
Schematic of FUCCI 4-color cell reporter (A) and experimental design (B). Single fluorescent channels or merged images of RD expressing FUCCI4+ RD RMS cells engrafted peri-ocularly into prkdc−/−, il2rga−/− zebrafish (C-G). Merged images to the right denote cell cycle state for individual cells. High magnification images of cells prior to drug treatment (C) and 7 days after receiving control vehicle (D), temozolomide (TMZ,E), olaparib (F), or TMZ + olaparib (G). Quantification of cell cycle effects in RD ERMS (H) and Rh41 ARMS cells (I). Green asterisks denote statistical differences in numbers of G2-arrested cells when compared to vehicle controls and black asterisks denote differences between single olaparib- or single temozolomide-treated animals when compared to combination-treated zebrafish (*p<0.05 by Fisher’s exact test, n=100 cells analyzed per time point, n=3 animals/condition). Scale bar equals 10μm (C-G). See also Figure S6.
Discussion
The Casper prkdc−/−, il2rga−/− immune deficient zebrafish model robustly engrafts a wide array of human cancers, can be reared at 37°C, and permits long-term, single-cell visualization of cancer cell engraftment. Building on the optical clarity of the model, we used a variety of cell labeling approaches to follow cell migration, growth, and therapy responses at single cell resolution in vivo. We envision additional reporters and imaging modalities will allow facile and dynamic visualization of many additional hallmarks of cancer. For example, we have previously used fli1:mCherry fluorescent transgenic zebrafish to visualize the establishment of cancer-associated vascular networks in live animals (Moore et al., 2016b) and refined reporter lines to dynamically visualize RMS cancer stem cells based on myogenic transcriptional changes in vivo (Ignatius et al., 2012, Hayes et al., 2018). Others have used fluorescent reporters to quantify apoptosis in development (To et al., 2016) and to visualize therapy responses in larvae using transcriptional readouts (White et al., 2011, Kaufman et al., 2016). Such approaches when adapted to our model, will be broadly useful for performing drug and genetic screens that modulate a wide array of cancer processes.
The ability of our model to stably engraft cancers from patient-derived xenografts is also particularly exciting. Although engraftment of PDXs from resected and circulating tumor cells is possible using immune deficient mouse models (Luo et al., 2014, Yu et al., 2014), we envision large-scale pre-clinical testing using a patient’s own tumor grown in immune-deficient zebrafish to aid in the stratifying patients into trials for which their tumor is best predicted to respond. When coupled to the inherent advantage of the zebrafish model for large-scale, high-throughput drug screening, it will also be possible to perform combination drug screening using oral-gavage to achieve accurate, clinically-relevant dosing. Our work in the zebrafish also established approaches to assess the pharmacokinetics for drug responses in vivo and to dynamically visualize pharmacodynamics in real-time using adult fish. Like NSG mice, we also envision that these models will also be useful for engraftment of a wider array of tissues including ES, iPSCs, and normal stem cell populations, greatly expanding the types of live cell imaging experiments that can be completed using xenografts in the stem cell and regenerative medicine fields.
Our work also uncovered remarkable functional diversity in human rhabdomyosarcoma cells. For example, using photo-convertible dendra2-H2b cell lineage tracking, we visualized individual cells in vivo and followed their cell fates. We observed three distinct tumor cell behaviors that comprised actively migrating cells or dividing cells that were mutually exclusive, and cells that were bystanders that did little over the seven days of imaging. This observation demonstrated that migratory and proliferative human RMS cells are largely confined to non-overlapping cell subsets, consistent with our earlier findings in zebrafish RMS (Ignatius et al., 2012). These findings are also consistent with intravital imaging studies of mice xenografted with human breast ductal carcinoma that demonstrated the existence of proliferative cancer stem and non-proliferative migratory cell states (Beerling et al., 2016). Melanoma and breast cancers have also been suggested to have distinct and yet oscillating cell states that differ in self-renewal, therapy response, and/or migration (Roesch et al., 2010, Chaffer et al., 2013). Indeed, a subset of rhabdomyosarcoma cells can oscillate between stem cell and differentiated cell states based on elevated NOTCH1 expression (Ignatius et al., 2017), raising the possibility of identifying and imaging these rare cell state changes in human xenografted tumors in the future.
Our preclinical modeling in zebrafish also uncovered combination olaparib PARP inhibitor and temozolomide DNA damaging agent as a potent therapy for the elimination of human RMS tumor cells in vivo. We further demonstrated consistency in results when the same drug combination was tested in mouse xenografts using both RMS cell lines and PDXs. Remarkably, both ERMS and ARMS were responsive to combination treatment, despite transformation by different oncogenic drivers. Even more remarkable, all six models tested showed response to combination therapy, suggesting that our preclinical modeling may have identified a potent drug combination for the treatment of a wide array of RMS. Poly ADP ribose polymerase (PARP) has important roles in repair of single-strand DNA nicks and blocking PARP activity shifts the reliance of cells to repair DNA breaks using the homologous recombination pathway. Thus, tumors deficient in BRCA1, BRCA2 or PALB2 cannot efficiently repair DNA breaks, leading to the death of cells treated with PARP inhibitors (Ray Chaudhuri et al., 2016). Although PARP inhibitors had once been thought to work primarily by blocking enzymatic activity, olaparib has an additional mode of action where it locks PARP in a tight complex with DNA and blocks DNA replication (Murai et al., 2012). These findings suggest a broader utility of olaparib and DNA-damaging agent combination therapy in tumors with intact DNA repair pathways, including RMS. In vitro experiments in other tumor cells have suggested that combination olaparib and temozolomide treatment arrest cells at G2 phase (Murai et al., 2014, Garnett et al., 2012), yet in vivo drug responses and kinetics of combination therapy had not been achieved at single cell resolution in live xenografted animals until this study. Because combination therapy using olaparib and temozolomide is currently being evaluated for efficacy in Phase II trials for Ewing’s sarcoma, our studies provide much-needed preclinical rational for assessing this drug combination in a wider range of pediatric sarcomas, including rhabdomyosarcoma.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources/reagents should be directed to David M. Langenau (dlangenau@mgh.harvard.edu) who is the lead contact for this work. MTA and fish line requests can be made using the following website: https://www.langenaulab.com/fish-orders/order-fish-lines
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animal welfare assurances and human subjects
All animal studies were approved by the Massachusetts General Hospital subcommittee on research animal care under protocols #2011-N-000127 (zebrafish) and #2013N000038 (mouse) and by the Partners human research committee under institutional review board protocol #2009-P-002756. Human tumor samples were harvested under protocol DF/HCC05–300. All immune compromised animals used in this study are kept at BCL2 facilities with regular veterinary checks being peformed. Both fish and mice used in these studies were healthy and were not used in prior studies, ensuring they were drug naïve. All animals used in this study are kept at maximum of 10 animals/4L fish tank and 3mice/cage.
Rearing and husbandry of compound prkdcD3612fs/il2rgaY91fs homozygous zebrafish
prkdcD3612fs/D3612fs,Il2rgaY91fs/+ adult Casper (double mutant for roya9/a9 and nacrew2/w2) (White et al., 2008, Lister et al., 1999) zebrafish were incrossed and double homozygous prkdcD3612fs/D3612fs, Il2rgaY91fs/Y91fs (denoted prkdc−/−, il2rga−/−) animals identified following scale resection and genotyping at 2–3 months of age as previously described (White et al., 2008, Moore et al., 2016a, Tang et al., 2017). Both males and females were used in these studies. Genotyped prkdc−/−, il2rg−/− zebrafish were transferred to autoclaved, sterilized fish water containing penicillin G sodium salt (Sigma, 150 units/ml), streptomycin sulfate salt (Sigma, 100mg/ml) and amphotericin b (Sigma, 2.5μg/ml) for 4–7 days for recovery. After, fish were acclimated to 37°C by incrementally elevating the t emperature over the course of 7 days in the presence of these same antibiotics (32°C on 1–2d, 35°C on 3d, 35.5°C on 4d, 36°C on 5d, 36.5°C on 6d and 37°C on 7d). Following acclimatio n to 37°C, animals were then either held in individual 4L fish tanks containing autoclaved, sterilized fish water supplemented with the same antibiotics outlined above (2L volume/tank) or in tanks housed on a custom built stand-alone 37°C re-circulating system (Aqaurius) and supplemen ted with commercially available fish-grade antibiotics including pimenta racemoa (2.48ml/L, API Pimafix), metronidazole (8.25mg/L, Thomas labs), praziquante (25μg/L, Thomas labs), ciprofloxacin (8.25mg/L, Thomas labs) and penicillin V potassium (8.25mg/L, Thomas labs). For animals raised in 4L fish tanks, water changes were performed every 2–3 days. Fish were fed three times daily with 100mg/animal of spirulina-infused with their cell-line specific culture media mixed at an equal ratio of spirulina, cell-line specific culture media, and FBS. Using these modified husbandry approaches, fish survival was 88.5% by >60 days post-transplant (n=155 of 180 engrafted fish), with fish exhibiting no overt signs of infection after engraftment and exhibiting overall survival rates similar to sham injected controls (Fig. S1A–C). No significant difference in engraftment rates were observed when comparing fish reared in 4L tanks or in our custom-built re-circulating stand alone rack system from Aquarius (Table S1).
Human cell lines, PDXs and authentication
SMS-CTR (male) and Rh30 (male) cells were obtained from Dr. Corinne Linardic (Duke University); UACC62 (sex unknown) and M14 (male) from Dr. David Fisher (MGH); K562 (female), U87 (male) and JeKo-1 (female) from Dr. Marcela Maus (MGH); Rh41 (female), A673 (female), SN12C (male), SK-LMS-1 (female) and HT1080 (male) from Dr. Cyril Benes (MGH); LMS04 (female) and MPNST 14 (sex unknown) from Dr. Jonathan Fletcher (MGH), and the MDA-MB-231 (female) from Dr. Dennis Sgroi (MGH). Remaining cell lines were purchased from AATC. Cell lines were cultured in cell culture media according to ATCC’s recommendation (RD, SMS-CTR, Rh30, UCAA62, U87, A673, SK-LMS-1, LMS04 and MDA-MB-231 in DMEM; Rh41, K562 JeKo-1 and MPNST 14 in RPMI, M14, SN12C and HT1080 in opti-MEM Reduced Serum Medium), supplemented with 10% FBS (Atlanta Biologicals) and 1% PSG (Life Tech), at 5% CO2 and 37°C. Adherent cells were detached and dissociated with 0.025% trypsin for 2 minutes. All human cell lines used in this work were authenticated just prior to use using Small Tandem Repeat profiling using the Whatman FTA sample collection kit (ATCC).
PDX models used in this work included human glioblastoma GBM9 (41-year-old male) (Tchoghandjian et al., 2012); a NOTCH1 mutated recurrent ERMS isolated from the pelvis (SJRHB000026_X1 (4-year-old female), (Stewart et al., 2017)); a p53 mutated recurrent ERMS isolated from the prostate (SJRHB012_Y (17-year-old male), (Stewart et al., 2017)); two metastatic BRAFV600E-induced melanomas (MEL167C (49-year-old male) and MEL268 (age unknown, male), (Luo et al., 2014)) (49-year-old male) and a therapy-resistant HER2+ breast cancer (Brx-07 (age unknown, female), (Yu et al., 2014)). GBM9 cells used for xenograft studies were generated from growing cells in sphere-inducing culture conditions (GBM9) (Neurobasal medium supplemented with 20ng/ml EGF (Life tech), 20ng/ml bFGF (Life tech), 2x B27 (Life tech) and 1% PSG (Life tech), at 5% CO2 and 37°C SJRHB000026_X1 and SJRHB012_Y ERMS from direct passage from mice; and MEL167C, MEL268C and BRx-07 from circulating tumor cells from patients and short-term culture in anchorage-free conditions. GBM9, MEL167C and MEL248C cells were previously infected with GFP lentivirus. SJRHB000026_X1 and SJRHB012_Y were stained with Viafluor SE cell proliferation stain (Biotium) and BRx-07 stained with lipophilic tracer (DIL) to facilitate imaging, both accordingly to manufacturer’s protocol. Selection of PDXs in this study were not influenced by gender of the patient subjects but by availability of samples.
METHODS DETAILS
Lentiviral vectors and creation of stable cell lines
2μg of pLenti-CMV-GFP-puro, pLKO.1-CMV-mKate2-Luc, pLenti-CAG.H2B-Dendra2.W (Addgene #51005), pLL3.7m-Clover-Geminin(1–110)-IRES-mKO2-Cdt(30–120) (Addgene #83841), and pLL3.7m-mTurquoise2-SLBP(18–126)-IRES-H1-mMaroon1 (Addgene #83842) were transfected into HEK293T cells with 2μg pCMV-dR8.91, 0.2 g pVSV-g and TransIT-LT1 reagent (Mirus Bio). Supernatants containing the lentivirus were collected, filtered and added to the cell lines used in this study, in the presence of 4 g/mL polybrene (Millipore). Viral particle containing pLenti-CMV-GFP-puro was added to cell lines in a subset of our studies. Viral particle containing pLKO.1-CMV-mKate2-Luc was added to RD and Rh41 and selected by FACS. Viral particles containing pLenti-CAG.H2B-Dendra2.W was added to RD and Rh41 and selected by FACS. Viral particle containing pLL3.7m-Clover-Geminin(1–110)-IRES-mKO2-Cdt(30–120) and pLL3.7m-mTurquoise2-SLBP(18–126)-IRES-H1-mMaroon1 was added to RD and Rh41 sequentially and selected by FACS.
Human cancer cell transplantation into zebrafish and quantifying tumor growth
Human cancer cell lines were grown to 90% confluence in T75 cell culture flasks, trypsinized if adherent, counted and resuspended at 2.5×107 cells/ml in matrigel (Corning). Clondronate liposomes (Clodrosome) were then added to the injection mix to inhibit early macrophage ingestion of engrafted cells over the first 7 days (Encapsula nanoscience, 1μg/μl). 10–20μl of volume was injected into the peritoneal cavity of recipient fish using 30 1/2 gauge needle (BD, 5×105 cells/fish). For ocular muscle injections, 2μl of cell suspension was injected (5×104 cells/fish). Recipient zebrafish were then raised at 37°C in sterilized fish water containing the aforementioned antibiotics. Epifluorescent whole-animal imaging was completed using the same UV light intensity and camera exposure (Microscope: Olympus MVX10, Camera: Olympus DP74). Tumor volume was determined by quantifying 2D image area using Image J and multiplying by the average GFP fluorescence intensity. These approaches are now standard in the field and are akin to luciferase bioluminescent imaging performed in mouse models (Kaufman et al., 2016, He et al., 2016, Cox et al., 2016, Lobbardi et al., 2017, Yan et al., 2015, Li et al., 2019, Chen et al., 2014). A subset of experiments also utilized direct counting of GFP+ cells using confocal microscopic imaging at 100X (Zeiss LSM710 inverted microscope, see below). For these experiments, maximum projection images were created from 150–200 micron stacks (10 microns per confocal slice) and manually quantified. Recipient fishes were sacrificed when moribund due to large tumor growths or at ≥28dpt, fixed in 4% paraformaldehyde, and sectioned for histological examination as previously described (Tang et al., 2014, Moore et al., 2016b). Similar approaches were used for engraftment of cancer cells labeled with other fluorescent-reporters and PDX tumors.
Single cell imaging of live zebrafish using confocal imaging
Imaging of peri-ocular transplanted cells was performed using an inverted LSM 710 confocal microscope (Zeiss), with Zen software platform (Zeiss) as previously described by Qin et al. (Tang et al., 2016). Engrafted zebrafish were anesthetized using low-dose 0.01% tricaine (western chemical), placed into a 36-mm glass-bottom dish (Thermo Fisher, product #150680), and torso embedded with 1% low-melting point agarose (Thermo Fisher) to stabilize the animal for imaging. To keep the animal under anesthetic during imaging, zebrafish were submerged in 5ml of fish water containing 0.01% tricaine. Animals were imaged for only 2–3 minutes and an experimental temperature of 37°C was maintained usi ng four heat packs (OccuNomix) placed on the imaging stage. Serial Z-stack imaging was carried out using a 10X objective (NA 0.45) achieving an overall 100X magnification. Both single cell fate mapping and FUCCI4 cell cycle imaging studies were carried out in zebrafish that had successfully engrafted tumors for 7 days. Importantly, repeated daily imaging of the engrafted fish using low-dose, 0.01% Tricaine anesthesia for seven days resulted in overall low mortality (n= only 5 of 45 fish died following repeated imaging).
Cell lineage tracing of RD-H2b-Dendra2 and RH41-H2b-Dendra2 cells was carried out by serial Z-stack imaging using 488nm (emission=493–586 nm) and 546nm laser (emission=575–703 nm). Dendra2 photoconversion was then achieved using 405nm laser at 30% laser power delivered over 2 minutes. Subsequent imaging was completed at 24, 48, 96, and 168 hours post-conversion. In the dendra conversion experiments, cells were pseudo colored using ImageJ.
FUCCI4 cell cycle imaging studies imaged mTurq2 (excitation=458 nm, emission=472– 508 nm), clover (excitation=488 nm, emission=493–586 nm), mKo2 (excitation=561 nm, emission=575–703 nm) and mMaroon1 (excitation=633 nm, emission=599–689 nm). Images were manually annotated and counted in ImageJ (n=100 cells per time point analyzed from n≥ 3 animals) and quantified using the Fisher’s Exact Test.
Accessing blood plasma concentration of olaparib and temozolomide in zebrafish
Wild type zebrafish were orally gavaged with either 50mg/kg of olaparib or 33mg/kg temozolomide. 25 animals/time point were sampled at 0, 2, 24, 48 and 96 hours post drug administration and pooled (n=25/time point/drug). Animals were euthanized in ice-water and wiped dry. Blood was collected by puncturing heart of the animal using an EDTA-coated insulin needle (BD bioscience). Zebrafish plasma was obtained as clear supernatant after centrifugation at 1000g for 10min at 4°C. Determina tion of plasma concentration was carried out as previous reported (Sun et al., 2018, Peer et al., 2016). Briefly, stock solution of temozolomide-D3 and olaparib-D4 was dissolved in either acidic methanol acidic methanol made from an ammonium acetate buffer and stabilized with 10% of 1 normal (N) HCL solution or slight warmed methanol. 200ul of temozolomide-D3 or 250ul of olaparib-D4 working solution was added into fish plasma. Liquid-liquid extraction was performed by adding 1ml of ethyl acetate. Supernatant was dried and reconstituted with 100ul of either 10mM ammonium acetate in water with 0.1% formic acid (10mM, pH 3.5)-methanol (20:80, v/v) or 0.1% formic acid in water for temozolomide or olaparib respectively. All mass spectrometry experiments were performed on a Triple TOF 6600 system (AB Sciex) online with a Shimadzu HPLC LC20AD (Shimadzu America) system. Olaparib or temozolomide were separated on an analytical Ascentis express peptide ES-C18 HPLC column, 15cm × 2.1 mm, 2.7um (Sigma-aldrich) preceded by a C18 guard column (2.1 mm ID) (Phenomenex).
Assessing therapeutic responses in zebrafish xenografts
Fluorescent labeled RD, Rh41, SMS-CTR, Rh30 and A673 cells were transplanted intraperitoneally and/or peri-ocularly into recipient fish. Engrafted fish were selected at 7 days post injection and orally gavaged with 10 μl of drug. Working stocks were 1mg/ml olaparib, 0.66mg/ml temozolomide or 0.5mg/ml olaparib and 0.33mg/ml temozolomide (vehicle was 1% DMSO in 1xPBS). Gavage was performed using a Hamilton 22 G needle and 22 G soft-tip catheter tubing (Dang et al., 2016). Drugs were orally administered at 7, 14 and 21 days post-transplant, and recipient fishes imaged as outlined above. At 28 days, animals were fixed in 4% paraformaldehyde, sectioned, and examined histologically. Comparison between groups was performed using ANOVA followed by Student’s T test (n=5 fish/treatment arm).
Mouse xenograft studies
Human tumor cells were embedded into Matrigel at a 1:1 ratio with 2–5×105 cells per 100μl and injected subcutaneously into the flank of 8-week-old female, anaesthetized NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (Charles River Laboratories). For tumors that were unlabeled, growth was monitored using caliper measure. For animals engrafted with luciferase expressing tumor cells, animals were injected with luciferin at 75mg/kg intraperitoneally and imaged weekly (IVIS, PerkinElmer). At the end of the experiment, engrafted recipient mice were sacrificed, tumor extracted, fixed in 4% paraformaldehyde, and sectioned for histological examination as outlined below. These experimental protocols are similar to those previously reported (Yu et al., 2014, Hayes et al., 2018).
A subset of experiments assessed the efficacy of combination olaparib and temozolomide treatment in mice xenografted with luciferase-expressing RD and Rh41 cell lines or unlabeled PDX models (SJRHB000026_X1 and SJRHB012_Y). For all RMS models, cells were engrafted at a dose of 1×106 cells injected subcutaneously into the flank of 8-week old female, NSG mice. When engrafted tumour reached a volume of 300mm3, animals were orally gavaged with vehicle control (1% carboxymethylcellulose or 10% 2-hydroxy-propyl-β-cyclodextrin/PBS), olaparib (50mg/kg), temozolomide (25mg/kg) or combined drugs. Olaparib was administered twice daily, 8 hours apart while temozolomide was administered once a day 4 hours after the first olaparib administration. Animals were treated for five days, allowed to rest for 16 days and this cycle repeated in a subset of experiments. Tumor growth was quantified by either differences in overall luciferase bioluminescent imaging or caliper measure as previously described (Yu et al., 2014, Drapkin et al., 2018, Hayes et al., 2018). Comparison between groups was performed using ANOVA followed by Student’s T test.
Histology, IHC and FISH evaluation
Zebrafish and mice engrafted with human tumors were fixed in 4% paraformaldehyde (PFA), embedded in paraffin and sectioned at 5mm thickness. Sections were stained by hematoxylin and eosin (H&E), IHC or FISH. For immunohistochemistry staining, the primary antibodies were: rabbit monoclonal anti-Ki67 (Abcam), rabbit polyclonal S100 (Leica), mouse monoclonal Pan-keratin (Abcam), rabbit polyclonal Desmin (Abcam), mouse monoclonal anti-myod (Dako), rabbit polyclonal anti-myogenin (Santa Cruz), rabbit monoclonal CD99 (Abcam), TUNEL (Thermo Fisher). Secondary antibodies were Biotinylated Goat Anti-rabbit igG antibody (Vectorlabs) and Biotinylated Horse Anti-mouse antibody (Vectorlabs) and development completed using Vectastain ABC Kit (Vectorlabs). FISH was performed on formalin-fixed paraffin-embedded tumor sections, using LSI EWS1 Dual Color Break Apart Probe (Abbott-Vysis) for A673 xenografts, and Vysis FOXO1 Break Apart Fish Probe for Rh30 and Rh41 xenografts (Abbott-Vysis) respectively, following manufacturer’s protocols.
Cell proliferation and apoptosis were quantified from IHC for Ki67 and TUNEL, respectively. Three tumor-containing fields were selected at random and imaged at 400x using an Olympus BX41 microscope. Quantifications were carried out blinded by NA Iftimia who was given the images without treatment labels and analysis performed using IHC profiler (ImageJ). Samples were analyzed using a fixed threshold for achieving an unbiased, quantitative assessment of the IHC and TUNEL staining within the selected imaged field [26]. Percentage of proliferating (Ki67) and apoptotic (TUNEL) cells was calculated by dividing the number of positively stained cells by the total number of cells counted within the selected fields. n>200 cells analyzed per sample, with less cells being counted for drug treated samples. H&E stained sections were imaged at 400X and quantified as number of cells per unit area.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical details including n-values, p-values, and statistical tests are detailed in the methods, results and figure legends. Data in bar graphs are shown as an absolute number with mean ± SD (standard deviation) noted. ANOVA and Student’s T tests were used to calculate significant differences where indicated. A subset of experiments used the Fisher Exact Test to compare values across two samples. p≤0.05 was considered statistically significant. In all experiments, zebrafish and mice were randomly assigned to experimental groups. All statistical analysis were carried out using Prism 7 (Graphpad).
Supplementary Material
Figure S1. Survival analysis and xenograft engraftment of prkdc−/−, il2rga−/− zebrafish, Related to Figure 1. Kaplan-Meier survival analysis for prkdc−/−, il2rga−/− zebrafish that were genotyped by scale resection at 60–90 days of life and then returned directly to system water without antibiotic treatment and grown at 28.5°C (A) or grown in isolated 4L tanks supplemented with antibiotics and reared at 37°C (B). Kaplan-Mei er survival analysis for prkdc−/−, il2rga−/− fish that were injected with sham control or human cancer cells and grown at 37°C in our recirculating system supplemented with antibiotics (C, n=155 of 181 engrafted animals survived to 60 dpt). *p<0.05 and ***p<0.001, Log-rank test. Not significant (NS). Representative merged fluorescence and brightfield images of EGFP+ human cancers grown in prkdc−/−, il2rga−/− zebrafish (D,I). Images shown at 0 dpt (left, D,I) and 28 dpt (right, D,I), respectively. Hematoxylin and eosin-stained sections of engrafted tumors (E,J). Ki67 immunohistochemistry (F,K), and TUNEL (G,L) with the average percentage of positive cells +/− standard deviation noted (n≥3 fish/tumor type). Quantification of relative growth of EGFP+ human cancer cells successfully engrafted into individual prkdc−/−, il2rga−/− zebrafish (H,M). Tumor type is noted in upper right in leftmost panels D,I and cell line name in lower left. Chronic myeloid leukemia (CML), leiomyosarcoma (LMS), malignant peripheral nerve sheath tumor (MPNST), embryonal rhabdomyosarcoma (ERMS) and alveolar rhabdomyosarcoma (ARMS). Scale bar equals 0.25 cm (D,I); 50 μm (E-G,J-L). Immunohistochemistry for expression of clinical diagnostic makers and pathognomonic gene fusions in xenografted Rh30 ARMS (N,O), Rh41 ARMS (P,Q), and A673 Ewing’s sarcoma (R,S). Diagnostic Fluorescent In situ Hybridization for FKHR (O,Q) or EWS1 (S). Because probes span the known FKHR or EWS1 genomic breakpoints, split break apart denotes translocation. Scale bars equal 50 μm (N,P,R) and 5μm (O,Q,S).
Figure S2. Engraftment of H2b-EGFP+ rhabdomyosarcoma cells into the peri-ocular muscle of prkdc−/−, il2rga−/− zebrafish, Related to Figure 3. (A-H) Live cell imaging and quantitation of RMS tumor cells over time (RD ERMS: A-D; Rh41 ARMS: E-H). Merged brightfield and fluorescent image of the head region of a prkdc−/−, il2rga−/− zebrafish engrafted with EGFP+ tumor cells for 21 days post-transplantation (dpt; A,E). Serial Z-stack confocal image at 0 dpt (B,F) and 21 dpt (C,G). Quantification of EGFP+ cells over time (D,H). (I-Q) Histological analysis of tumors engrafted into the peri-ocular muscle and analyzed at 21 days post-treatment (RD ERMS: I-M; Rh41 ARMS: N-Q). Hematoxylin and eosin stained sections (I,N, low magnification; and J,O high magnification of boxed region). Immunohistochemistry for Ki67 (K,P) and TUNEL (M,Q) with average percent positive cells +/− standard deviation noted (n≥3 fish/tumor). Scale bar equals 0.1 cm (A, E); 50 μm (B, C, F, G), 200 μm (I, N); 50 μm (J-M, O-Q).
Figure S3. Identification of functionally distinct rhabdomyosarcoma cell types using photo-convertible Dendra2-H2b and single cell fate mapping, Related to Figure 3. (A-F) Single channel images of RD-H2b-Dendra2 cells engrafted peri-ocularly into prkdc−/−, il2rga−/− zebrafish, before and after photoconversion, at 0 (A and B), 24 (C), 48 (D), 96 (E), 168 (F) hours respectively. 488 nm imaging (top panels,) and 546 nm imaging (lower panels). Scale bar equals 200 μm (A-F). This is the same animal depicted in Figure 3. Summary of single cell fate mapping of RD-H2b-Dendra2 engrafted cells (B, n= 50 cells followed in 21 animals) and Rh41-H2b-Dendra2 engrafted cells (C, n= 50 cells from n=18 animals). Cell fates are pictorially depicted with migration denoted by wavy lines. Number of cells exhibiting each phenotype are denoted to the left of each fate map rendering.
Figure S4. Combination treatment of temozolomide and olaparib PARP-inhibitor reduces growth of human Ewing’s sarcoma and rhabdomyosarcoma in vivo, Related to Figure 4. Plasma concentration of olaparib (A) or temozolomide (B) when orally gavaged into adult zebrafish (red, n=25 fish/time point) and compared with previously published mouse (blue) and human studies (green). (C) Merged fluorescence and brightfield images of prkdc−/−, il2rga−/− animals engrafted with EGFP+ A673 Ewing’s sarcoma cells. Animals were imaged at 0 and 7 days post-transplant (dpt, prior to drug administration) and at 28 dpt (after three cycles of drug administration). (D) Quantization of relative growth as assessed by fluorescence intensity changes over time. Individual animals are denoted. (E) Waterfall plot showing the same data but depicted by relative growth from day 0 until day 28. ***p<0.001, student’s t-test (n=5). Not significant (NS). Merged fluorescence and brightfield images of prkdc−/−, il2rga−/− animals engrafted with EGFP+ Rh41 cells (F). Whole animal imaging of engrafted animal at 0 and 7 dpt (prior to drug administration, F, left) and 28 dpt (after three cycles of drug dosing, F, right). Hematoxylin and eosin (G), Ki67 (H), and TUNEL (I) stained sections of fish engrafted with Rh41 RMS cells (n≥3 animals/treatment). Scale bar equals 0.25 cm (C,F) and 50 μm (G-I).
Figure S5. Combination olaparib and temozolomide, but not single drug administration, suppresses human RMS growth in NSG mice, Related to Figure 5. Xenograft studies using RD ERMS (A-C, G-K) and Rh41 ARMS (D-F, L-P). Luciferase bioluminescent imaging of RMS cells prior to drug administration (0d) and after specified days of treatment (A,D,G, and L). Waterfall plot quantifying relative RMS growth following drug administration when assessed at 14 day post-drug treatment (B,E). Quantification of relative RMS growth over time with individual animals are denoted (C,F). The five days of drug administration are denoted by red arrows in panel C and F. *p<0.05 by Fisher’s exact test comparing tumor growth in combination treated animals to control or single drug treated mice (n=3 animals/treatment). (G-P) Combination treatment also suppressed RMS growth in a larger cohort of NSG mice. RD ERMS (G-K) and Rh41 ARMS (L-P). Hematoxylin and eosin stained sections (H,M). IHC for Ki67 (I,N) and TUNEL (J,O) with average percent positive cells +/− standard deviation noted. Quantification of relative RMS growth over time with individual animals denoted (K,P). The days of drug administration are denoted by red arrows in panels K and P (p<0.002 for both RD and Rh41 at 14–35 days post treatment, Student’s T-test).
Figure S6. FUCCI4 cell cycle reporter kinetics do not differ between human rhabdomyosarcomas xenografted in prkdc−/−, il2rga−/− zebrafish or NSG mice, Related to Figure 6. Cryostat sections of human RD ERMS grown for 28 days in NSG mice (A) or zebrafish (B) for 28 days. Cryostat sections of human Rh41 ARMS grown for 28 days in NSG mice (C) or zebrafish (D). Single fluorescent channels noted. Merged images to the right denote cell cycle state for individual cells. (E) Schematic of FUCCI 4-color cell reporter used to lentiviral transfect RD and Rh41 RMS cells. Quantification of cell cycle in RD ERMS (F; n=311 cells counted across 3 zebrafish and n=621 cells counted across 6 mice) and Rh41 ARMS (G; n=283 cells counted across 3 zebrafish and n=701 cells counted across 6 mice). Cell states were not significantly different between tumors grown in immune deficient fish or mice when assessed by Student’s T-test (p>0.05). Scale bar equals 10μm (A-D).
Table S1. Engraftment efficiency of human cancers grown in prkdc−/−, il2rga−/− zebrafish, Related to Figure 1 and Figure 2.
Highlights:
prkdc−/−, il2rga−/− zebrafish reared at 37°C engraft a wide array of human cancers
Growth and therapy responses can be dynamically visualized at single cell resolution
Combination olaparib and temozolomide kill xenografted human rhabdomyosarcoma
Engrafting patient-derived cancers opens new avenues for personalized-therapy
Acknowledgments:
We thank Dr. Cyril Benes for helpful discussions and feedback on the work and Florence Lin at the Shriners Hospital for Children – Boston Special Shared Mass Spectrometry Facility for help with mass spectrometry studies. This work is supported by NIH grant R24OD016761 (D.M.L.), R01CA154923 (D.M.L.), R01CA215118, (D.M.L.), R01CA211734 (D.M.L), R01CA1299933, (D.H.), the Liddy Shriver Sarcoma Initiative (D.M.L., J.A.F), the MGH Research Scholars Program (D.M.L.), Shriner’s Hospital for Children (M.N.K), U01CA220323 (N.J.D), HHMI (D.H), National Foundation for Cancer Research (D.H.), Breast Cancer Foundation (D.H.), the Breast Cancer Research Foundation (D.C.S), the Lung Cancer Research Foundation (B.J.D), ASCO Young Investigator Award (B.J.D), the Tosteson & Fund for Medical Discovery Fellowship from MGH (Y.C.), and the Alex’s Lemonade Stand Foundation Young Investigator Award (Y.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests: The General Hospital Corporation has a patent pending on the creation and use of immune compromised zebrafish for engraftment of human cancers. Dr. Langenau is a co-inventor on patent application number USSN 14/903,940. Dr. Haber is on the scientific advisory board for Janssen and co-founder of TorpedoDx. Dr. Lafrate is on the scientific advisory board for Archerdx, Equity and Sanofi.
All data is available in the manuscript or the supplementary materials
References
- Agarwala SS & Kirkwood JM (2000). Temozolomide, a novel alkylating agent with activity in the central nervous system, may improve the treatment of advanced metastatic melanoma. Oncologist, 5, 144–51. [DOI] [PubMed] [Google Scholar]
- Bajar BT, Lam AJ, Badiee RK, Oh YH, Chu J, Zhou XX, Kim N, Kim BB, Chung M, Yablonovitch AL, et al. (2016). Fluorescent indicators for simultaneous reporting of all four cell cycle phases. Nat Methods, 13, 993–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Ngo B, Laughney AM, Cavallo JA, Murphy CJ, Ly P, Shah P, Sriram RK, Watkins TBK, Taunk NK, et al. (2018). Chromosomal instability drives metastasis through a cytosolic DNA response. Nature, 553, 467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr FG, Galili N, Holick J, Biegel JA, Rovera G & Emanuel BS (1993). Rearrangement of the PAX3 paired box gene in the paediatric solid tumour alveolar rhabdomyosarcoma. Nat Genet, 3, 113–7. [DOI] [PubMed] [Google Scholar]
- Beerling E, Seinstra D, de Wit E, Kester L, van der Velden D, Maynard C, Schafer R, van Diest P, Voest E, van Oudenaarden A, et al. (2016). Plasticity between Epithelial and Mesenchymal States Unlinks EMT from Metastasis-Enhancing Stem Cell Capacity. Cell Rep, 14, 2281–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn JS, Liu S, Raiser DM, Martinez SA, Feng H, Meeker ND, Gentry J, Neuberg D, Look AT, Ramaswamy S, et al. (2012). Notch signaling expands a pre-malignant pool of T-cell acute lymphoblastic leukemia clones without affecting leukemia-propagating cell frequency. Leukemia, 26, 2069–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce WR & Van Der Gaag H (1963). A quantitative assay for the number of murine lymphoma cells capable of proliferation in vivo Nature, 199, 79–80. [DOI] [PubMed] [Google Scholar]
- Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F, D’Alessio AC, Young RA & Weinberg RA (2013). Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell, 154, 61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman A, Fernandez del Ama L, Ferguson J, Kamarashev J, Wellbrock C & Hurlstone A (2014). Heterogeneous tumor subpopulations cooperate to drive invasion. Cell Rep, 8, 688–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, DeRan MT, Ignatius MS, Grandinetti KB, Clagg R, McCarthy KM, Lobbardi RM, Brockmann J, Keller C, Wu X, et al. (2014). Glycogen synthase kinase 3 inhibitors induce the canonical WNT/beta-catenin pathway to suppress growth and self-renewal in embryonal rhabdomyosarcoma. Proc Natl Acad Sci U S A, 111, 5349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Stewart E, Shelat AA, Qu C, Bahrami A, Hatley M, Wu G, Bradley C, McEvoy J, Pappo A, et al. (2013). Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer Cell, 24, 710–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudakov DM, Lukyanov S & Lukyanov KA (2007). Tracking intracellular protein movements using photoswitchable fluorescent proteins PS-CFP2 and Dendra2. Nat Protoc, 2, 2024–32. [DOI] [PubMed] [Google Scholar]
- Cox AG, Hwang KL, Brown KK, Evason K, Beltz S, Tsomides A, O’Connor K, Galli GG, Yimlamai D, Chhangawala S, et al. (2016). Yap reprograms glutamine metabolism to increase nucleotide biosynthesis and enable liver growth. Nat Cell Biol, 18, 886–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis SJ, Sinkevicius KW, Li D, Lau AN, Roach RR, Zamponi R, Woolfenden AE, Kirsch DG, Wong KK & Kim CF (2010). Primary tumor genotype is an important determinant in identification of lung cancer propagating cells. Cell Stem Cell, 7, 127–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang M, Henderson RE, Garraway LA & Zon LI (2016). Long-term drug administration in the adult zebrafish using oral gavage for cancer preclinical studies. Dis Model Mech, 9, 811–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Strowig T, Verma R, Koduru S, Hafemann A, Hopf S, Kocoglu MH, Borsotti C, Zhang L, Branagan A, et al. (2016). Microenvironment-dependent growth of preneoplastic and malignant plasma cells in humanized mice. Nat Med, 22, 1351–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ, D’Cruz CM, Lovell MA, Biegel JA & Barr FG (1994). Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res, 54, 2869–72. [PubMed] [Google Scholar]
- Diez BD, Statkevich P, Zhu Y, Abutarif MA, Xuan F, Kantesaria B, Cutler D, Cantillon M, Schwarz M, Pallotta MG, et al. (2010). Evaluation of the exposure equivalence of oral versus intravenous temozolomide. Cancer Chemother Pharmacol, 65, 727–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapkin BJ, George J, Christensen CL, Mino-Kenudson M, Dries R, Sundaresan T, Phat S, Myers DT, Zhong J, Igo P, et al. (2018). Genomic and Functional Fidelity of Small Cell Lung Cancer Patient-Derived Xenografts. Cancer Discov, 8, 600–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenbroek SI & van Rheenen J (2014). Imaging hallmarks of cancer in living mice. Nat Rev Cancer, 14, 406–18. [DOI] [PubMed] [Google Scholar]
- Fior R, Povoa V, Mendes RV, Carvalho T, Gomes A, Figueiredo N & Ferreira MG (2017). Single-cell functional and chemosensitive profiling of combinatorial colorectal therapy in zebrafish xenografts. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, et al. (2012). Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature, 483, 570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SJ, Travers J, Pshenichnaya I, Kogera FA, Barthorpe S, Mironenko T, Richardson L, Benes CH, Stratton MR, McDermott U, et al. (2015). Combinations of PARP Inhibitors with Temozolomide Drive PARP1 Trapping and Apoptosis in Ewing’s Sarcoma. PLoS One, 10, e0140988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyama S, Wunderlich M & Mulloy JC (2015). Xenograft models for normal and malignant stem cells. Blood, 125, 2630–40. [DOI] [PubMed] [Google Scholar]
- Hayes MN, McCarthy K, Jin A, Oliveira ML, Iyer S, Garcia SP, Sindiri S, Gryder B, Motala Z, Nielsen GP, et al. (2018). Vangl2/RhoA Signaling Pathway Regulates Stem Cell Self-Renewal Programs and Growth in Rhabdomyosarcoma. Cell Stem Cell, 22, 414–427.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Mansour MR, Zimmerman MW, Ki DH, Layden HM, Akahane K, Gjini E, de Groh ED, Perez-Atayde AR, Zhu S, et al. (2016). Synergy between loss of NF1 and overexpression of MYCN in neuroblastoma is mediated by the GAP-related domain. Elife, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatius MS, Chen E, Elpek NM, Fuller AZ, Tenente IM, Clagg R, Liu S, Blackburn JS, Linardic CM, Rosenberg AE, et al. (2012). In vivo imaging of tumor-propagating cells, regional tumor heterogeneity, and dynamic cell movements in embryonal rhabdomyosarcoma. Cancer Cell, 21, 680–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatius MS, Hayes MN, Lobbardi R, Chen EY, McCarthy KM, Sreenivas P, Motala Z, Durbin AD, Molodtsov A, Reeder S, et al. (2017). The NOTCH1/SNAIL1/MEF2C Pathway Regulates Growth and Self-Renewal in Embryonal Rhabdomyosarcoma. Cell Rep, 19, 2304–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Hiramatsu H, Kobayashi K, Suzue K, Kawahata M, Hioki K, Ueyama Y, Koyanagi Y, Sugamura K, Tsuji K, et al. (2002). NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood, 100, 3175–82. [DOI] [PubMed] [Google Scholar]
- Kaufman CK, Mosimann C, Fan ZP, Yang S, Thomas AJ, Ablain J, Tan JL, Fogley RD, van Rooijen E, Hagedorn EJ, et al. (2016). A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science, 351, aad2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly PN, Dakic A, Adams JM, Nutt SL & Strasser A (2007). Tumor growth need not be driven by rare cancer stem cells. Science, 317, 337. [DOI] [PubMed] [Google Scholar]
- Langenau DM, Keefe MD, Storer NY, Guyon JR, Kutok JL, Le X, Goessling W, Neuberg DS, Kunkel LM & Zon LI (2007). Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev, 21, 1382–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, He S & Look AT (2019). The MCL1-specific inhibitor S63845 acts synergistically with venetoclax/ABT-199 to induce apoptosis in T-cell acute lymphoblastic leukemia cells. Leukemia, 33, 262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister JA, Robertson CP, Lepage T, Johnson SL & Raible DW (1999). nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development, 126, 3757–67. [DOI] [PubMed] [Google Scholar]
- Lobbardi R, Pinder J, Martinez-Pastor B, Theodorou M, Blackburn JS, Abraham BJ, Namiki Y, Mansour M, Abdelfattah NS, Molodtsov A, et al. (2017). TOX Regulates Growth, DNA Repair, and Genomic Instability in T-cell Acute Lymphoblastic Leukemia. Cancer Discov, 7, 1336–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Mitra D, Sullivan RJ, Wittner BS, Kimura AM, Pan S, Hoang MP, Brannigan BW, Lawrence DP, Flaherty KT, et al. (2014). Isolation and molecular characterization of circulating melanoma cells. Cell Rep, 7, 645–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon M, Frangova TG, Henderson CJ & Wolf CR (2016). Olaparib, Monotherapy or with Ionizing Radiation, Exacerbates DNA Damage in Normal Tissues: Insights from a New p21 Reporter Mouse. Mol Cancer Res, 14, 1195–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore FE, Garcia EG, Lobbardi R, Jain E, Tang Q, Moore JC, Cortes M, Molodtsov A, Kasheta M, Luo CC, et al. (2016a). Single-cell transcriptional analysis of normal, aberrant, and malignant hematopoiesis in zebrafish. J Exp Med, 213, 979–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JC, Tang Q, Yordan NT, Moore FE, Garcia EG, Lobbardi R, Ramakrishnan A, Marvin DL, Anselmo A, Sadreyev RI, et al. (2016b). Single-cell imaging of normal and malignant cell engraftment into optically clear prkdc-null SCID zebrafish. J Exp Med, 213, 2575–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S & Pommier Y (2012). Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res, 72, 5588–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai J, Zhang Y, Morris J, Ji J, Takeda S, Doroshow JH & Pommier Y (2014). Rationale for poly(ADP-ribose) polymerase (PARP) inhibitors in combination therapy with camptothecins or temozolomide based on PARP trapping versus catalytic inhibition. J Pharmacol Exp Ther, 349, 408–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer C, Ronner L, Rodgers L, McCully C, Warren K & Figg W (2016). Quantification of Temozolomide in Nonhuman Primate Fluids by Isocratic Ultra-High Performance Liquid Chromatography-Tandem Mass Spectrometry to Study Brain Tissue Penetration Following Intranasal or Intravenous Delivery. Separations, 3, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan A, Carter CA, Kelly RJ, Gutierrez M, Kummar S, Szabo E, Yancey MA, Ji J, Mannargudi B, Woo S, et al. (2012). A phase I combination study of olaparib with cisplatin and gemcitabine in adults with solid tumors. Clin Cancer Res, 18, 2344–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Callen E, Ding X, Gogola E, Duarte AA, Lee JE, Wong N, Lafarga V, Calvo JA, Panzarino NJ, et al. (2016). Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature, 535, 382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T & Herlyn M (2010). A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell, 141, 583–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongvaux A, Willinger T, Martinek J, Strowig T, Gearty SV, Teichmann LL, Saito Y, Marches F, Halene S, Palucka AK, et al. (2014). Development and function of human innate immune cells in a humanized mouse model. Nat Biotechnol, 32, 364–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, Ambrogio L, Auclair D, Wang J, Song YK, et al. (2014). Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov, 4, 216–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AC, Raimondi AR, Salthouse CD, Ignatius MS, Blackburn JS, Mizgirev IV, Storer NY, de Jong JL, Chen AT, Zhou Y, et al. (2010). High-throughput cell transplantation establishes that tumor-initiating cells are abundant in zebrafish T-cell acute lymphoblastic leukemia. Blood, 115, 3296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E, Federico SM, Chen X, Shelat AA, Bradley C, Gordon B, Karlstrom A, Twarog NR, Clay MR, Bahrami A, et al. (2017). Orthotopic patient-derived xenografts of paediatric solid tumours. Nature, 549, 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart E, Goshorn R, Bradley C, Griffiths LM, Benavente C, Twarog NR, Miller GM, Caufield W, Freeman BB 3rd, Bahrami A, et al. (2014). Targeting the DNA repair pathway in Ewing sarcoma. Cell Rep, 9, 829–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Mikule K, Wang Z, Poon G, Vaidyanathan A, Smith G, Zhang ZY, Hanke J, Ramaswamy S & Wang J (2018). A comparative pharmacokinetic study of PARP inhibitors demonstrates favorable properties for niraparib efficacy in preclinical tumor models. Oncotarget, 9, 37080–37096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Abdelfattah NS, Blackburn JS, Moore JC, Martinez SA, Moore FE, Lobbardi R, Tenente IM, Ignatius MS, Berman JN, et al. (2014). Optimized cell transplantation using adult rag2 mutant zebrafish. Nat Methods, 11, 821–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Iyer S, Lobbardi R, Moore JC, Chen H, Lareau C, Hebert C, Shaw ML, Neftel C, Suva ML, et al. (2017). Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single-cell resolution using RNA sequencing. J Exp Med, 214, 2875–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Moore JC, Ignatius MS, Tenente IM, Hayes MN, Garcia EG, Torres Yordan N, Bourque C, He S, Blackburn JS, et al. (2016). Imaging tumour cell heterogeneity following cell transplantation into optically clear immune-deficient zebrafish. Nat Commun, 7, 10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchoghandjian A, Baeza-Kallee N, Beclin C, Metellus P, Colin C, Ducray F, Adelaide J, Rougon G & Figarella-Branger D (2012). Cortical and subventricular zone glioblastoma-derived stem-like cells display different molecular profiles and differential in vitro and in vivo properties. Ann Surg Oncol, 19 Suppl 3, S608–19. [DOI] [PubMed] [Google Scholar]
- Tenente IM, Tang Q, Moore JC & Langenau DM (2014). Normal and malignant muscle cell transplantation into immune compromised adult zebrafish. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Goldstein A, Wang H, Ching Lo H, Sun Kim I, Welte T, Sheng K, Dobrolecki LE, Zhang X, Putluri N, et al. (2017). Mutual regulation of tumour vessel normalization and immunostimulatory reprogramming. Nature, 544, 250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To TL, Schepis A, Ruiz-Gonzalez R, Zhang Q, Yu D, Dong Z, Coughlin SR & Shu X (2016). Rational Design of a GFP-Based Fluorogenic Caspase Reporter for Imaging Apoptosis In Vivo. Cell Chem Biol, 23, 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker AM, Jaros BD, An M & Beattie CE (2017). Changes in tumor cell heterogeneity after chemotherapy treatment in a xenograft model of glioblastoma. Neuroscience, 356, 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, Langdon E, Tomlinson ML, Mosher J, Kaufman C, et al. (2011). DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature, 471, 518–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RM, Sessa A, Burke C, Bowman T, LeBlanc J, Ceol C, Bourque C, Dovey M, Goessling W, Burns CE, et al. (2008). Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell, 2, 183–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Huo X, Wang S, Feng Y & Gong Z (2015). Stimulation of hepatocarcinogenesis by neutrophils upon induction of oncogenic kras expression in transgenic zebrafish. J Hepatol, 63, 420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, Desai R, Zhu H, Comaills V, Zheng Z, et al. (2014). Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science, 345, 216–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Survival analysis and xenograft engraftment of prkdc−/−, il2rga−/− zebrafish, Related to Figure 1. Kaplan-Meier survival analysis for prkdc−/−, il2rga−/− zebrafish that were genotyped by scale resection at 60–90 days of life and then returned directly to system water without antibiotic treatment and grown at 28.5°C (A) or grown in isolated 4L tanks supplemented with antibiotics and reared at 37°C (B). Kaplan-Mei er survival analysis for prkdc−/−, il2rga−/− fish that were injected with sham control or human cancer cells and grown at 37°C in our recirculating system supplemented with antibiotics (C, n=155 of 181 engrafted animals survived to 60 dpt). *p<0.05 and ***p<0.001, Log-rank test. Not significant (NS). Representative merged fluorescence and brightfield images of EGFP+ human cancers grown in prkdc−/−, il2rga−/− zebrafish (D,I). Images shown at 0 dpt (left, D,I) and 28 dpt (right, D,I), respectively. Hematoxylin and eosin-stained sections of engrafted tumors (E,J). Ki67 immunohistochemistry (F,K), and TUNEL (G,L) with the average percentage of positive cells +/− standard deviation noted (n≥3 fish/tumor type). Quantification of relative growth of EGFP+ human cancer cells successfully engrafted into individual prkdc−/−, il2rga−/− zebrafish (H,M). Tumor type is noted in upper right in leftmost panels D,I and cell line name in lower left. Chronic myeloid leukemia (CML), leiomyosarcoma (LMS), malignant peripheral nerve sheath tumor (MPNST), embryonal rhabdomyosarcoma (ERMS) and alveolar rhabdomyosarcoma (ARMS). Scale bar equals 0.25 cm (D,I); 50 μm (E-G,J-L). Immunohistochemistry for expression of clinical diagnostic makers and pathognomonic gene fusions in xenografted Rh30 ARMS (N,O), Rh41 ARMS (P,Q), and A673 Ewing’s sarcoma (R,S). Diagnostic Fluorescent In situ Hybridization for FKHR (O,Q) or EWS1 (S). Because probes span the known FKHR or EWS1 genomic breakpoints, split break apart denotes translocation. Scale bars equal 50 μm (N,P,R) and 5μm (O,Q,S).
Figure S2. Engraftment of H2b-EGFP+ rhabdomyosarcoma cells into the peri-ocular muscle of prkdc−/−, il2rga−/− zebrafish, Related to Figure 3. (A-H) Live cell imaging and quantitation of RMS tumor cells over time (RD ERMS: A-D; Rh41 ARMS: E-H). Merged brightfield and fluorescent image of the head region of a prkdc−/−, il2rga−/− zebrafish engrafted with EGFP+ tumor cells for 21 days post-transplantation (dpt; A,E). Serial Z-stack confocal image at 0 dpt (B,F) and 21 dpt (C,G). Quantification of EGFP+ cells over time (D,H). (I-Q) Histological analysis of tumors engrafted into the peri-ocular muscle and analyzed at 21 days post-treatment (RD ERMS: I-M; Rh41 ARMS: N-Q). Hematoxylin and eosin stained sections (I,N, low magnification; and J,O high magnification of boxed region). Immunohistochemistry for Ki67 (K,P) and TUNEL (M,Q) with average percent positive cells +/− standard deviation noted (n≥3 fish/tumor). Scale bar equals 0.1 cm (A, E); 50 μm (B, C, F, G), 200 μm (I, N); 50 μm (J-M, O-Q).
Figure S3. Identification of functionally distinct rhabdomyosarcoma cell types using photo-convertible Dendra2-H2b and single cell fate mapping, Related to Figure 3. (A-F) Single channel images of RD-H2b-Dendra2 cells engrafted peri-ocularly into prkdc−/−, il2rga−/− zebrafish, before and after photoconversion, at 0 (A and B), 24 (C), 48 (D), 96 (E), 168 (F) hours respectively. 488 nm imaging (top panels,) and 546 nm imaging (lower panels). Scale bar equals 200 μm (A-F). This is the same animal depicted in Figure 3. Summary of single cell fate mapping of RD-H2b-Dendra2 engrafted cells (B, n= 50 cells followed in 21 animals) and Rh41-H2b-Dendra2 engrafted cells (C, n= 50 cells from n=18 animals). Cell fates are pictorially depicted with migration denoted by wavy lines. Number of cells exhibiting each phenotype are denoted to the left of each fate map rendering.
Figure S4. Combination treatment of temozolomide and olaparib PARP-inhibitor reduces growth of human Ewing’s sarcoma and rhabdomyosarcoma in vivo, Related to Figure 4. Plasma concentration of olaparib (A) or temozolomide (B) when orally gavaged into adult zebrafish (red, n=25 fish/time point) and compared with previously published mouse (blue) and human studies (green). (C) Merged fluorescence and brightfield images of prkdc−/−, il2rga−/− animals engrafted with EGFP+ A673 Ewing’s sarcoma cells. Animals were imaged at 0 and 7 days post-transplant (dpt, prior to drug administration) and at 28 dpt (after three cycles of drug administration). (D) Quantization of relative growth as assessed by fluorescence intensity changes over time. Individual animals are denoted. (E) Waterfall plot showing the same data but depicted by relative growth from day 0 until day 28. ***p<0.001, student’s t-test (n=5). Not significant (NS). Merged fluorescence and brightfield images of prkdc−/−, il2rga−/− animals engrafted with EGFP+ Rh41 cells (F). Whole animal imaging of engrafted animal at 0 and 7 dpt (prior to drug administration, F, left) and 28 dpt (after three cycles of drug dosing, F, right). Hematoxylin and eosin (G), Ki67 (H), and TUNEL (I) stained sections of fish engrafted with Rh41 RMS cells (n≥3 animals/treatment). Scale bar equals 0.25 cm (C,F) and 50 μm (G-I).
Figure S5. Combination olaparib and temozolomide, but not single drug administration, suppresses human RMS growth in NSG mice, Related to Figure 5. Xenograft studies using RD ERMS (A-C, G-K) and Rh41 ARMS (D-F, L-P). Luciferase bioluminescent imaging of RMS cells prior to drug administration (0d) and after specified days of treatment (A,D,G, and L). Waterfall plot quantifying relative RMS growth following drug administration when assessed at 14 day post-drug treatment (B,E). Quantification of relative RMS growth over time with individual animals are denoted (C,F). The five days of drug administration are denoted by red arrows in panel C and F. *p<0.05 by Fisher’s exact test comparing tumor growth in combination treated animals to control or single drug treated mice (n=3 animals/treatment). (G-P) Combination treatment also suppressed RMS growth in a larger cohort of NSG mice. RD ERMS (G-K) and Rh41 ARMS (L-P). Hematoxylin and eosin stained sections (H,M). IHC for Ki67 (I,N) and TUNEL (J,O) with average percent positive cells +/− standard deviation noted. Quantification of relative RMS growth over time with individual animals denoted (K,P). The days of drug administration are denoted by red arrows in panels K and P (p<0.002 for both RD and Rh41 at 14–35 days post treatment, Student’s T-test).
Figure S6. FUCCI4 cell cycle reporter kinetics do not differ between human rhabdomyosarcomas xenografted in prkdc−/−, il2rga−/− zebrafish or NSG mice, Related to Figure 6. Cryostat sections of human RD ERMS grown for 28 days in NSG mice (A) or zebrafish (B) for 28 days. Cryostat sections of human Rh41 ARMS grown for 28 days in NSG mice (C) or zebrafish (D). Single fluorescent channels noted. Merged images to the right denote cell cycle state for individual cells. (E) Schematic of FUCCI 4-color cell reporter used to lentiviral transfect RD and Rh41 RMS cells. Quantification of cell cycle in RD ERMS (F; n=311 cells counted across 3 zebrafish and n=621 cells counted across 6 mice) and Rh41 ARMS (G; n=283 cells counted across 3 zebrafish and n=701 cells counted across 6 mice). Cell states were not significantly different between tumors grown in immune deficient fish or mice when assessed by Student’s T-test (p>0.05). Scale bar equals 10μm (A-D).
Table S1. Engraftment efficiency of human cancers grown in prkdc−/−, il2rga−/− zebrafish, Related to Figure 1 and Figure 2.