Abstract
Suppressor of cytokine signaling-3 (SOCS3) is a negative regulator of actions of cytokines and the metabolic hormone leptin. In the hypothalamus, SOCS3 is induced in response to several conditions such as inflammation and high-fat diet feeding, modulates cellular signaling of cytokines and leptin, and mediates the effects of these biological conditions. However, signaling mechanisms controlling hypothalamic Socs3 expression remains to be fully established. To facilitate the identification of molecular pathways of Socs3 induction, we generated a real-time gene expression reporter mouse of Socs3 (Socs3-Luc mice). We successfully detected a remarkable increase in luciferase activity in various tissues of Socs3-Luc mice in response to a peripheral injection of lipopolysaccharide, a potent inducer of inflammation, reflecting expression levels of endogenous Socs3 mRNA. Using ex vivo hypothalamic explants of Socs3-Luc mice, we demonstrate that hypothalamic luciferase activity was significantly elevated in slices stimulated with known inducers of Socs3 such as proinflammatory cytokines IL-6, IL-1β, and TNF-α, lipopolysaccharide, and cAMP-inducing agent forskolin. Using the ex vivo model, we found glycogen synthase kinase-3 (GSK3)β-specific inhibitors to be potent inducers of Socs3. Furthermore, pharmacological inhibitors of β-catenin, a downstream mediator of GSK3β signaling, reduced Socs3 luciferase activity ex vivo. Finally, hypothalamic inhibition of GSK3β hindered leptin-induced phosphorylation of signal transducers and activators of transcription 3 in hypothalamic explants. These results suggest that the Socs3-luciferase mouse is useful for in vivo monitoring of Socs3 gene expression and for ex vivo slice-based screening to identify signaling pathways that control Socs3 in the hypothalamus.
Keywords: Socs3, hypothalamus, luciferase reporter, cytokines, leptin
Suppressor of cytokine signaling-3 (SOCS3) is an inducible inhibitor of cytokine signaling [1, 2]. SOCS3 is induced by various cytokines including IL-6 family cytokines such as leptin and IL-6 [3]. Cytokines stimulate SOCS3 induction primarily via the Janus kinases (JAKs) and the signal transducers and activators of transcriptions (STATs) pathway. Induced SOCS3 inhibits cytokine signaling via the inhibition of JAK activity and targets the receptor complex for proteasomal degradation [4]. Thus, SOCS3 is thought to limit the signaling cascade to prevent an excessive, aberrant response of cytokines.
SOCS3 in the hypothalamus has emerged as a critical regulator of energy balance [4–8]. SOCS3 is significantly induced in the hypothalamus in response to high-fat diet feeding, pregnancy, aging, and long-day photoperiod that promotes positive energy balance [5, 9]. Forced expression of SOCS3 in a distinct population of hypothalamic neurons promoted body weight gain and adiposity [10]. Consistently, induction of the constitutively active form of STAT3 in pro-opiomelanocortin neurons stimulated Socs3 expression and increased body weight [11]. In contrast, brain-specific Socs3 knockout or its haploinsufficiency mice were significantly protected against the development of diet-induced obesity [6, 7]. Furthermore, the range of hypothalamic neuron-specific Socs3 deletion mouse models demonstrates that hypothalamic Socs3 critically regulates body weight and glucose homeostasis [12–14]. Thus, the level of Socs3 expression in the hypothalamus seems to determine body weight and adiposity in vivo.
The effects of hypothalamic Socs3 on energy homeostasis seem to be at least in part mediated through inhibiting leptin actions. Leptin rapidly induces Socs3 expression in the hypothalamus [15]. Increased Socs3 expression leads to the attenuation of cellular signaling of leptin in cultured cells and in animals in a cell-autonomous manner [10, 16, 17]. In contrast, Socs3 deficiency enhances cellular and physiological responses to exogenous leptin [6, 7]. Thus, it is well established that Socs3 is a negative feedback inhibitor of leptin in vivo. Signaling mechanisms mediating leptin-induced Socs3 involve the JAK-STAT3 pathway [15, 18–20]. In addition, recent studies have suggested the existence of a leptin-STAT3-independent mechanism. For example, STAT3 deficiency within leptin receptor (LepR)-expressing hypothalamic cells did not decrease mRNA expression of Socs3 in LepR cells [21]. Along the same line, inactivation of Socs3 in LepR cells displayed a minimum effect on body weight [14]. An increase of Socs3 in LepR neurons does not cause obesity [10]. Clearly, additional work is needed to elucidate signaling networks responsible for Socs3 expression.
Socs3 is also induced by other signaling pathways. Socs3 expression is increased in response to stimulation with inflammatory signals such as Toll-like receptor 4 [22], TNF-α [23], and IL-1β [23] that commonly activate the inhibitor of κ B kinase/nuclear factor κB (NF-κB) pathway [2, 24]. Indeed, activation of the NF-κB pathway is sufficient to induce Socs3 in the hypothalamus and elicit hypothalamic resistance to exogenous leptin [17]. In addition, numerous studies demonstrate that cAMP plays a role to downregulate IL-6 signaling as well as to induce Socs3 in various cell types [25–30]. Importantly, cAMP-elevating agents were reported to diminish cellular sensitivity to exogenous leptin in endothelial cells [30] and in the hypothalamus [31]. These findings raise a possibility that a molecular pathway of Socs3 induction might also interfere with cellular leptin actions. However, the signaling networks that confer the control of Socs3 levels and leptin sensitivity remain to be fully established. Therefore, delineation of molecular pathways responsible for Socs3 induction is of high interest. To facilitate the identification of such molecular pathways, we generated a new Socs3-luciferase reporter mouse (Socs3-Luc) and used hypothalamic explants of Socs3-Luc to search for and characterize a signaling pathway that influences Socs3 expression and cellular leptin action ex vivo.
1. Materials and Methods
A. Generation of Socs3-Luc Mice
A 98.6-kb bacterial artificial chromosome (BAC; bMQ-126K3; Source BioScience) contains the entire Socs3 gene locus. Luciferase2 cDNA was prepared from pGL4.10 (Promega, Madison, WI). The luciferase2 cDNA was introduced into the translation start site of the Socs3 gene in the BAC clone in Escherichia coli cells using BAC recombineering techniques [32]. The recombinant Socs3 luciferase (Socs3-Luc) BAC DNA was prepared by NucleoBond Xtra BAC Kit (Macherey-Nagel, Germany), linearized by PI-SceI digestion, purified with phenol-chloroform, and precipitated with ethanol. Transgenic mice were obtained by injecting the linearized Socs3-luciferase BAC DNA into pronuclei of C57BL/6 mice in the Genetically Engineered Mouse Core at Baylor College of Medicine. Socs3-Luc founders were maintained at a C57BL/6 background. Tail biopsies were used for the genotyping on PCR with primers TL luciferase-F6 5′-CTTCGAGCAGACATGATAAGATAC-3′ and MFSOCS3-F1 5′-TGACGCTCAACGTGAAGAAG-3′ producing the amplicon of 591-bp long. The Institutional Animal Care and Use Committee at Baylor College of Medicine approved all procedures to maintain and use these mice.
B. Protein Extraction and Luciferase Assay
In vitro luciferase activity was quantified using the Luciferase Assay System (Promega, Madison, WI). Tissues were extracted using the 1× Luciferase Cell Culture Lysis Reagent (Promega, Madison, WI), sonicated and vortexed, followed by centrifugation for 10 minutes at 4°C and 15,000 rpm. Supernatant was placed in a new 1.5-mL tube and stored at −80°C. To assess luciferase activity, 4 µL of sample was mixed with 20 µL Luciferase assay reagent, and luminescence was measured using a luminometer (Promega, Madison, WI). The Promega Luc-0-INJ protocol was used to measure luciferase activity in the tissues.
C. In Vivo Imaging
Whole body imaging of luciferase activity in vivo was monitored using an In Vivo MS FX PRO imaging system (Bruker, Billerica, MA). Male and female mice were injected IP with 150 mg/kg body weight of d;-luciferin solution (Promega, Madison, WI) and anesthetized (2.5% isoflurane). Mice were transferred to nose cones within the chamber and imaged 10 minutes after injection of D-luciferin. Exposure time was 10 seconds on the ventral side, and luminescence was quantified using the Bruker imaging software. Bright-field images were also taken of mice with an exposure time of 5 seconds. Tissues, collected from male mice, were also imaged using the In Vivo MS FX PRO. Briefly, male mice were injected IP with 150 mg/kg body weight of D-luciferin solution and dissected 10 minutes later. Tissues were placed in chamber and bright-field, and luminescence images were taken using the same exposure times for in vivo imaging.
D. Organotypic Slice Experiments
Screening of compounds was accomplished using an organotypic slice culture system. Hypothalamic slices were made essentially as described before [31]. Briefly, C57BL/6 mice male and female pups, 8 to 12 days old, were decapitated, and the brains were quickly removed. Hypothalamic tissues were blocked and sectioned in depth of 250 μm on a vibratome (VT1000S, Leica) in chilled Gey’s Balanced Salt Solution (Invitrogen, Carlsbad, CA) enriched with glucose (0.5%) and KCl (30 mM). The coronal slices containing the arcuate nucleus were then placed on Millicell-CM filters (Millipore, pore size 0.4 μm, diameter 30 mm), and maintained at an air-media interface in MEM (Invitrogen, Carlsbad, CA) supplemented with heat inactivated horse serum (25%, Invitrogen, Carlsbad, CA), glucose (32 mM) and GlutaMAX (2 mM, Invitrogen, Carlsbad, CA). Cultures were typically maintained for 10 days in standard medium, which was replaced three times a week. After 10 days, the slices were used for experiments. The following modifications were made for Socs3-luc slices: after brains were sectioned, all sections were cut to the same diameter before being placed on Millicell-CM filters in culture. Slices were maintained 1 to 7 days in standard medium before being transferred to individual cell culture petri dishes (Thermo Fisher Scientific, Waltham, MA) containing low-serum (2% heat inactivated horse serum) phenol-red free MEM supplemented with glucose (32 mM) GlutaMAX (2 mM), 10% HEPES buffer (Invitrogen, Carlsbad, CA), and 0.1 mM d-luciferin. Dishes were sealed with a circular cover glass (Thermo Fisher Scientific, Waltham, MA) using vacuum grease (VWR, Radnor, PA) and placed in a Lumicycle 32 (Actimetrics, Wilmette, IL), a 32-channel luminometer at 37°C to measure luminescence from tissues. Treatments were added 2 days after slices were placed in the Lumicycle. Luciferase activity was analyzed using Lumicycle Analysis software.
E. Reagents
We used the following reagents: lipopolysaccharide (LPS; L4391, Sigma-Aldrich, St. Louis, MO); IL-6 (RMIL6I, Thermo Fisher Scientific, Waltham, MA), IL-1β (I5271, Sigma-Aldrich St. Louis, MO); TNF-α (RMTNFAI, Thermo Fisher Scientific, Waltham, MA); forskolin (#1099, TOCRIS, Bristol, United Kingdom); LY2090314 (SML1438, Sigma-Aldrich); CHIR99021 (SML1046, Sigma-Aldrich St. Louis, MO); methyl 3- 4methylphenyl sulfonyl amino benzoate (MSAB; M60316, Xcess Biosciences, San Diego, CA); and FH535 (S7484, SelleckChem, Houston, TX).
F. Quantitative RT-PCR
Tissues were quickly removed and frozen in liquid nitrogen and kept in −80°C until further processing. Total RNA was extracted using the RNeasy Lipid Tissue Kit (QIAGEN Sciences, Germantown, MD). cDNA was generated by iScript RT Supermix (Bio-Rad Laboratories, Hercules, CA) and used with SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA) for quantitative real-time PCR analysis. Quantitative PCR assays were performed using a CFX384 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA). Normalized mRNA levels were expressed in arbitrary units obtained by dividing the averaged, efficiency-corrected values for sample mRNA expression by that of 18SrRNA expression for each sample. The resulting values were expressed as fold change above average control levels. The primer sequences are as follows: SOCS3 (F-CACCTGGACTCCTATGAGAAAGTG and R-GAGCATCATACTGATCCAGGAACT), PTP1B (F-GGAACAGGTACCGAGATGTCA and R-AGTCATTATCTTCCTGATGCAATT), TCPTP (F-AGGGCTTCCTTCTAAGG and R-GTTTCATCTGCTGCACCTTCTGAG), and 18SrRNA (F- CACGGACAGGATTGACAGATT and R-GCCAGAGTCTCGTTCGTTATC).
G. Immunohistochemistry
For organotypic slices, slices were fixed overnight in 4% paraformaldehyde at 4°C. Then slices were cut out from the membrane and rinsed three times for 10 minutes each in PBS, pH 7.4 and then for 20 minutes in 1% hydrogen peroxide and 1% sodium hydroxide in PBS to quench endogenous peroxidase activity. Following a series of washes with PBS, slices were incubated for 48 to 72 hours at 4°C in pSTAT3 antibodies (1: 3000, 9131, Cell Signaling Technology, Boston, MA) [33] in 3% normal donkey serum (Jackson ImmunoResearch Laboratories, West Grove, PA) with 0.25% Triton X-100 in PBS and 0.02% sodium azide. After washing in PBS, slices were incubated in a biotinylated goat antirabbit antibody IgG (1:1000, BA-1000, Jackson ImmunoResearch Laboratories West Grove, PA) [34] in 3% donkey serum in 0.25 % Triton X-100 in PBS for 1 hour at room temperature. Tissues were then rinsed in PBS and incubated in the avidin-biotin-peroxidase complex kit (1:500, Vectastain Elite ABC kit; Vector Laboratories, Burlingame, CA) for 1 hour. Slices were washed in PBS then reacted in 3,3′-diaminobenzidine (DAB; Sigma-Aldrich St. Louis, MO). Slices were then rinsed in PBS and mounted on slides using Vectashield (Vector Laboratories Burlingame, CA). For fluorescence staining, slices were incubated with Streptavidin, Alexa Fluor™ 488 conjugate (Molecular Probes, Eugene, OR). Immunohistochemical images were analyzed using a bright-field Leica microscope. To measure pSTAT3 signal in the arcuate nucleus, uneven background was eliminated with Adobe Photoshop and then intensity of DAB staining was measured.
H. Statistics
The data are presented as mean ± SEM. Statistical analyses were performed using GraphPad Prism for a two-tailed unpaired Student t test or one- or two-way ANOVA followed by post hoc Tukey, Bonferroni, or Sidak tests. P < 0.05 was considered to be statistically significant.
2. Results
A. Generation and Characterization of Socs3 Luciferase Reporter Mice
To create an in vivo monitoring system for endogenous Socs3 expression in a noninvasive manner, we generated transgenic mice (Socs3-Luc) by engineering a Socs3 BAC such that Luciferase2 gene is driven by Socs3 regulatory elements (Fig. 1A–1C). The firefly luciferase-polyA was inserted into the ATG site of the Socs3 gene in a 93.6-kb BAC containing the entire Socs3 gene locus via homologous recombination, and transgenic mice were generated by pronuclear injection of the modified BAC. Transgenic founders and their offspring were identified by PCR (Fig. 1B). With the use of an in vivo imaging system, we functionally validated Socs3-Luc transgenic mice by monitoring luciferase activity in the reporter mice. In vivo luciferase imaging detected robust luciferase signals in Socs3-Luc but not in wild type (WT) mice (Fig. 1C). Analyzing luciferase activity of tissue lysates from liver and hypothalamus known to express Socs3 we further confirmed that luciferase activity, normalized to the amount of total protein, was specifically detected only in tissues of Socs3-Luc mice (Fig. 1D).
Figure 1.
Generation of Socs3-Luc mice. (A) Schematic of the Socs3-Luc transgene. The full-length luciferase gene was inserted into the ATG codon of the Socs3 gene in a BAC (bMQ126k03). (B) Socs3-Luc mice were identified by PCR analysis. (C) Live imaging of luciferase activity in WT and Socs3-Luc mice. (D) Luciferase enzymatic activity in liver and hypothalamus of WT and Socs3-Luc mice.
Next we examined whether the reporter mice respond to LPS endotoxin [22] that activates a potent, common proinflammatory response and is known to induce Socs3 [2]. We injected LPS into the periphery of Socs3-Luc mice and their littermate controls (WT) and measured luciferase activity using the in vivo imaging system. Total body luciferase activity was markedly stimulated in the Socs3-Luc mice 4 hours after LPS administration, whereas luciferase activity was low in vehicle-injected animals (Fig. 2A). To assess the tissue distribution of luciferase luminescence detected in Socs3-Luc, we dissected internal peripheral organs including small intestine, liver, spleen, kidney, stomach, pancreas, white adipose tissue, brown adipose tissue and heart, at 4 hours after the LPS administration and measured their luciferase activity using in vivo imaging system analysis. Almost all of the tissue samples showed some level of luciferase luminescence induced by LPS (Fig. 2B). We also determined if the Socs3 luciferase reporter recapitulates LPS-induced gene expression pattern of Socs3 mRNA by measuring in vitro biochemical luciferase activity and endogenous Socs3 mRNA in various tissues of Socs3-Luc mice with or without LPS injection. The basal levels of luciferase activities in tissue lysates were detected in all tissues we examined with relatively high in vitro luciferase activity from liver, stomach, muscle, and spleen (Fig. 2C). Upon LPS treatment, the highest induction of luciferase activity was found in intestine, pancreas, kidney, and liver, whereas it was moderate in brain tissues, fat tissues, stomach, and muscle (Fig. 2D). We found a close correlation between luciferase activity and endogenous Socs3 mRNA expression (Fig. 2D and 2E). Endogenous Socs3 mRNA expression was remarkably induced in most of the tissues in an LPS-dependent manner (Fig. 2E). In particular, liver, kidney, stomach, heart, intestine, and pancreas showed highly induced expression of the endogenous Socs3 (Fig. 2E), which closely correlated with the induced expression pattern of the luciferase luminescence. These data thus demonstrate that the Socs3 reporter model faithfully mirrors the tissue distribution of endogenous Socs3 mRNA in vivo.
Figure 2.
Functional validation of Socs3-Luc mice. (A) Luminescence images of Socs3-Luc mice intraperitoneally administered with either saline or 100 ng/kg LPS for 4 h. (B) Bright field images (left) and bioluminescence images (right) of various tissues of Socs3-Luc mice receiving LPS (100 ng/kg for 4 h) or saline. (C and D) In vitro luciferase activity of the indicated tissue lysates extracted from mice treated with either (C) saline or (D) 100 ng/kg LPS for 4 h shown as bioluminescent signal. The same amount of protein (3.5 µg) was used for each reaction. (E) Socs3 mRNA in various tissues of the mice treated with either saline or 100 ng/kg LPS for 4 h. Data are mean ± SEM; n = 3 to 4/per group. *P < 0.05; **P < 0.01; ****P < 0.0001 vs control based on t tests.
B. An Ex Vivo Model to Monitor Hypothalamic Socs3 Expression
Hypothalamic levels of Socs3 crucially regulate energy balance [5, 35]. To explore signaling mechanisms by which hypothalamic Socs3 levels are determined, we used hypothalamic explants of Socs3-Luc mice that allow us to pharmacologically manipulate various signaling cascades and rapidly assess its effect on Socs3 expression. Mediobasal hypothalamus was dissected and placed on a membrane in a dish containing media supplemented with luciferin substrate. Hypothalamic explants were treated with chemicals and peptides, and luminescence was continuously measured in real time with photomultiplier tube detectors during the treatment (Fig. 3A). To confirm that the Socs3 luciferase reporter hypothalamic slice is capable of responding to LPS, we treated slices with LPS and luminescence was continuously measured in real time. As shown in Fig. 3B, slices treated with LPS show a 3.0-fold increase in luciferase activity compared with their PBS controls. Because Socs3 is also produced by proinflammatory cytokines that include IL-6, TNF-α, and IL-1β [22, 23], slices were treated with them to further validate our ex vivo model. As expected, all three of the cytokines tested resulted in an increase in luciferase activity (Fig. 3C–3E). Forskolin, a previously reported inducer of Socs3 in various cells, also displayed a 2.4-fold increase in luciferase activity in hypothalamic explants (Fig. 3F). These results confirm that hypothalamic slices prepared from Socs3-Luc mice can serve as a suitable screening tool for measuring the effects of modulators of Socs3 expression in the hypothalamus.
Figure 3.
Hypothalamic slice cultures as a screening tool for hypothalamic Socs3 modulators. (A) Screening scheme. Hypothalamic slice cultures were made from 8- to 12-d-old Socs3-Luc mice, and luciferase activity of slices was monitored with a Lumicycle 32 following compound treatment. (B–F) Using hypothalamic slice culture, Socs3 luciferase signal was probed ex vivo using known inducers of Socs3. Data are mean ± SEM; n = 3 to 4 per group. **P < 0.01; ****P < 0.0001 vs control based on two-way ANOVA. FSK, forskolin.
C. Inhibitors of Glycogen Synthase Kinase-3β Induces Socs3 Gene Expression in the Hypothalamus
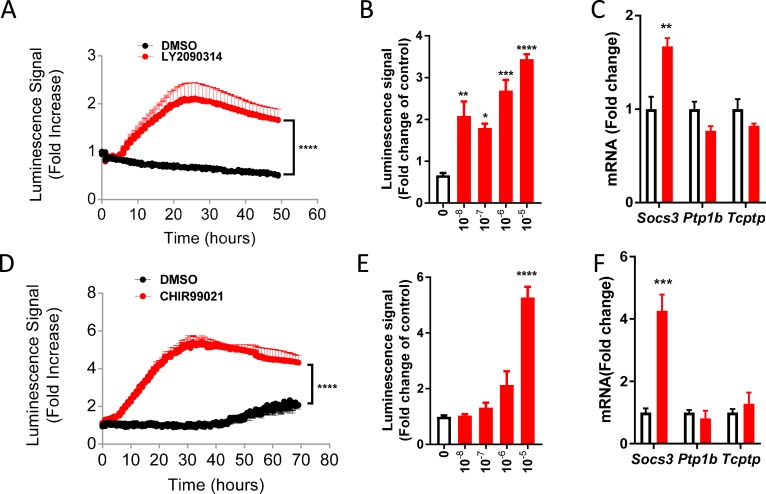
With the use of the ex vivo model to screen compounds for their ability to alter Socs3 expression, we came upon chemicals that target proteins involved in the glycogen synthase kinase-3 (GSK3)β―β-catenin signaling pathway. First, we found that treatment with GSK3 inhibitor LY2090314 [36], a potent and selective small-molecule inhibitor of GSK3α and GSK3β isoforms, induces a 2.1-fold increase in luciferase activity and this occurs in a dose-dependent manner (Fig. 4A and 4B). Consistently, the mRNA level of endogenous Socs3 was elevated in slices treated with LY2090314 (Fig. 4C). Interestingly, we did not detect any increases in other inhibitors of cytokine signaling such as tyrosine protein phosphatases. Hypothalamic induction of Socs3 was also observed in explants treated with CHIR99021 [37], a well-established specific inhibitor for GSK3β with a different kinase selectivity profile than LY2090314 (Fig. 4D and 4F). This increase also occurred in a dose-dependent manner Fig. 4E. These data clearly suggest that pharmacologic inhibition of GSK3β stimulates Socs3 gene expression in the hypothalamus.
Figure 4.
Inhibitors of GSK3β increase Socs3 expression in hypothalamus ex vivo. (A) Luminescence signal observed in Socs3-Luc slices treated with CHIR99021 (10 μM) or dimethyl sulfoxide (DMSO) control. (B) Dose response of CHIR99021 in explants of Socs3 Luc mice. (C) Expression of Socs3, Ptp1b, and Tcptp in hypothalamic slices treated with CHIR99021 (10 μM) or DMSO control for 20 h. (D) Luminescence signal observed in Socs3-Luc slices treated with LY2090314 (10 nM). (E) Dose response of LY2090314 in explants of Socs3-Luc mice. (F) Expression of Socs3, Ptp1b, and Tcptp in hypothalamic slices treated with LY2090314 (10 nM) or DMSO control for 20 h. CHIR99021 and LY2090314, GSK3β inhibitors; Data are mean ± SEM; n = 3 to 4 per group. ****P < 0.0001 for two-way ANOVA (in A and D); *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 for one-way ANOVA followed by Tukey multiple comparisons test (in B and E); ** P < 0.01, ***P < 0.001 vs control based on t tests (in C and F).
D. Inhibitors of β-Catenin Decrease Socs3 Gene Expression in the Hypothalamus
Next, we explored the mechanism by which GSK3β inhibition increased Socs3 transcription. Because GSK3β acts through β-catenin [38], we hypothesized that the action of GSK3β inhibitors in hypothalamus could be mediated through β-catenin. To test this, Socs3-Luc slices were treated with FH535 (50 µM) [39], an inhibitor of β-catenin. This treatment resulted in a decrease in luciferase activity in a dose-dependent manner (Fig. 5A and 5B). Because FH535 is not a selective inhibitor of β-catenin [39], we further tested a more specific β-catenin inhibitor MSAB [40] for its ability to decrease Socs3 luciferase signal. Similar to FH535, treating slices with MSAB significantly decreased Socs3 luciferase activity, and this decrease was also dose-dependent (Fig. 5C and 5D). These data suggest that inhibition of β-Catenin induces Socs3 gene expression.
Figure 5.
Inhibitors of β-catenin, downstream of GSK3β, decrease Socs3 expression. (A) Luminescence signal observed in Socs3-Luc slices treated with FH535 (50 µM) relative to DMSO control. (B) Dose response of FH535 in Socs3-Luc slices. (C) Luminescence signal observed in Socs3-Luc slices treated with MSAB (500 μM) relative to DMSO control. (D) Dose response of MSAB in Socs3-Luc slices. Data are mean ± SEM; n = 3 to 4 per group. ****P < 0.0001 for two-way ANOVA (in A, C, and E); **P < 0.01, ***P < 0.001 vs control for one-way ANOVA followed by Tukey multiple comparisons test (in B and D).
E. A GSK3β Inhibitor Blocks Leptin-Induced STAT3 Phosphorylation in Hypothalamic Slices
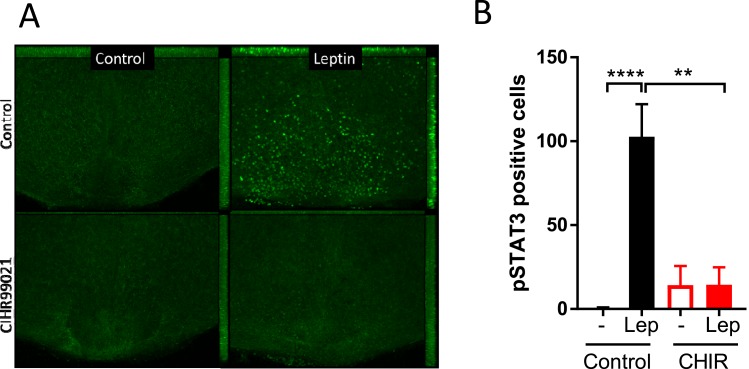
SOCS3 potently limits cellular leptin signaling in the hypothalamus. To examine the functional consequences of GSK3β inhibition, we examined if the treatment desensitizes the hypothalamic cells to leptin using ex vivo hypothalamic slices that recapitulate hypothalamic leptin signaling [31]. Hypothalamic slices were exposed to CHIR99021 (10 µM for 6 hours) followed by leptin stimulation (120 nM for 60 minutes) to examine the effects of GSK3β inhibition. Similar to previous observations, leptin robustly induced phosphorylation of STAT3 in control slices. In contrast, treatment of the slices with both CHIR99021 had a potent inhibitory effect on leptin-induced STAT3 phosphorylation in the hypothalamus (Fig. 6A and 6B). CHIR99021 alone had no effect on pSTAT3 (Fig. 6A and 6B). These data suggest that inhibition of GSK3β impairs hypothalamic LepR signaling.
Figure 6.
The effect of a GSK3β specific inhibitor on hypothalamic leptin signaling. (A) Immunohistochemistry of phosphorylated STAT3 (pSTAT3) in tissues treated with either DMSO or CHIR99021. Slices were pretreated with CHIR99021 (10 μM, 6 h) followed by leptin stimulation (120 nM, 60 min). After fixed with 4% formalin, slices were subjected to immunohistochemistry with anti-pSTAT3 antibodies followed by fluorescence visualization with Alexa Fluor-labeled secondary antibodies. (B) Quantification of hypothalamic pSTAT3. pSTAT3 signal was detected by DAB staining. Values are presented as mean ± SEM. Three separate experiments. **P < 0.01, ****P < 0.0001 for two-way ANOVA followed by Sidak multiple comparisons test.
3. Discussion
SOCS3 plays a critical role in controlling inflammation and metabolism. SOCS3 levels appear to determine cellular sensitivity to cytokines and metabolic hormones, and SOCS3 gene expression is tightly controlled. However, the underlying signaling mechanisms are not fully established. To explore the mechanism of Socs3 induction, we have generated Socs3 reporter mice that allow us to quantitatively monitor changes in Socs3 expression in vivo. We have also developed a hypothalamic slice-based compound screening platform to monitor hypothalamic Socs3 expression. The tool provides an opportunity to identify signaling pathways that control Socs3 expression in hypothalamus ex vivo.
Using the LPS-induced inflammation model that stimulates Socs3 induction in vivo [1, 23, 41], we validated if Socs3-Luc mice could recapitulate the LPS-induced robust induction of Socs3. Socs3 luciferase activity was detected in almost all tissues among those examined after the LPS injection. Importantly, the tissue distribution pattern of LPS-induced luciferase signals closely correlates with that of endogenous Socs3 mRNA. In accordance with the reported studies showing that Socs3 induction emerges only after a few hours following the LPS injection in vivo [42, 43], we found that luciferase signals were observed at 4 hours, but not at 30 minutes after LPS injection in the Socs3-Luc transgenic mice (data not shown). This pattern of luciferase signals was observed in both whole-body mice and individual tissues we examined (small intestine, liver, spleen, kidney, stomach, pancreas, WAT, BAT, and heart). These results are consistent with previous studies that have shown peak expression levels of Socs3 to be around 4 hours in liver and brain tissues [42, 43]. A number of studies have shown that LPS increase SOSC3 rapidly within several minutes in cells such as macrophages in vitro and in vivo. Although the exact reasons for this discrepancy remain unknown, this might be due to difference in the time frame of temporal response to LPS in different tissues. These observations underscore that the Socs3 luciferase reporter model faithfully and robustly reports spatiotemporal dynamics of endogenous Socs3 mRNA in vivo.
To establish a reliable in vitro monitoring system, we first developed a hypothalamic slice culture-based assay that allows real-time monitoring of Socs3 expression using organotypic hypothalamic slices. The organotypic brain slice model is known to partly preserve the in vivo anatomy and function of the hypothalamus [44]. Previous results from the studies by ours and other groups demonstrated that the slice model can genuinely recapitulate in vivo cellular responses to metabolic hormones, anatomically, biochemically, and electrophysiologically [31, 45–48]. In the current study, we show that the ex vivo system reliably reproduces hypothalamic responses to LPS, TNF-α, IL-1β, IL-6, and forskolin, which increase Socs3 mRNA in vivo [22, 23, 26–30]. All of the Socs3 inducers remarkably elevated luciferase activity in Socs3-Luc hypothalamic explants. Thus, the in vivo and ex vivo data collectively suggest that luciferase activity serves as a genuine reporter for monitoring Socs3 expression in real-time, and more importantly, this slice-based imaging model offers a feasible and reliable platform to search for small chemicals for Socs3 expression in vitro.
With the use of the ex vivo monitoring model, we identified several compounds (CHIR99021, LY2090314, FH535, and MSAB) that lead to the modulation of Socs3 expression in the hypothalamus. LY2090314 [36], a potent and selective small-molecule inhibitor of GSK3α and GSK3β isoforms, clearly stimulated Socs3 luciferase activity in the hypothalamic explants, suggesting that inhibition of GSK3 activity results in increased Socs3 gene expression. Further, hypothalamic induction of Socs3 was observed in explants treated with CHIR99021 [37], a structurally distinct specific inhibitor for GSK3β. The effects of LY2090314 and CHIR99021 were observed in the nanomolar or micromolar range, respectively, which are within the effective concentrations established by various cell-based functional assays [36, 49, 50]. GSK3β is known to be involved in several distinct signaling pathways such as cellular signaling mediating insulin, wingless and integration site growth factor, and hedgehog. GSK3β is constitutively active under unstimulated conditions where GSK3β phosphorylates β-catenin, which in turn is subjected to ubiquitin proteasome degradation. In contrast, inhibition of GSK3β by upstream signals such as wingless and integration site growth factor and insulin signaling results in an increase in β-catenin protein levels, which promotes the transcription of its target genes in a T-cell factor/lymphoid enhancer factor-dependent mechanism. Thus, pharmacological inhibition of GSK3β should also lead to activation of β-catenin-dependent gene expression. Ex vivo studies suggest that GSK3β inhibition-induced Socs3 expression is mediated by β-catenin (data not shown), indicating a pathway that links GSK3β and Socs3 via β-catenin in the hypothalamus. The findings in the ex vivo studies presented in this paper provide evidence demonstrating that the GSK3β―β-catenin pathway might act as a potential regulator of Socs3 expression in the hypothalamus.
One potential implication of our results is that GSK3β inhibition may downregulate cellular signaling of leptin via Socs3 induction. Several studies have shown that SOCS3 inhibits cellular actions of leptin by interfering with LepR signaling. Prior studies demonstrated that Socs3 is induced by several distinct signaling pathways that include the JAK-STAT3 pathway, the inhibitor of κ B kinase/NF-κB pathway [2, 17, 24], the endoplasmic reticulum stress-related pathways [51], and the cAMP-related pathways [30, 31]. Notably, all of these pathways limit cellular leptin sensitivity, as well as induce Socs3 in the hypothalamus implicating that the pathways underlying Socs3 induction would inhibit cellular signaling of leptin. Consistent with this notion, our studies indeed demonstrate that a specific inhibitor for GSK3β inhibited leptin-induced STAT3 phosphorylation, while inducing hypothalamic Socs3. The failure of exogenous leptin to modulate key signaling events such as STAT3 phosphorylation within hypothalamic neurons is well documented in various physiological and pathophysiological settings. These phenomena occur in response to high-fat diet feeding, chronic leptin exposure, pregnancy and lactation, age, and seasonal variations in the length of the photoperiod in seasonal animals. Under these conditions, Socs3 is commonly elevated and likely to play a role in suppressing leptin actions. Future studies are warranted to elucidate the role of the GSK3β―β-catenin pathway in Socs3 induction under these conditions.
In addition to leptin actions, SOCS3 commonly suppresses cellular signaling of inflammation. A number of studies have shown that inhibition of GSK3β has the ability to reduce inflammation. For example, GSK3β inhibition protects against LPS-induced endotoxin shock in vivo, decreases proinflammatory cytokine production, and interferes with NF-κB and STAT3 pathways. At the molecular level, GSK3β inhibition was reported to affect the nuclear amounts of transcription factors NF-κB subunit p65 and CREB interacting with the coactivator CBP [52]. GSK3β was also shown to block STAT3 DNA binding activity and STAT3-dependent gene expression [53]. These data suggest the ability of GSK3β inactivation to strongly induce Socs3 and may add another mechanism by which GSK3 inactivation potently suppresses inflammation.
Because Socs3 expression is altered dynamically under various biological conditions such as inflammation, dietary obesity, pregnancy and lactation, seasonal adiposity, intestinal bowel disease, ulcerative colitis, Crohn disease, rheumatoid arthritis, thymocyte differentiation, and atherosclerotic lesions [5, 35, 54–58], this model may also facilitate the functional investigation of SOCS3 in multiple tissues and in diverse physiological and pathophysiological conditions.
In summary, we have created a BAC-transgenic luciferase reporter mouse carrying the Socs3-luciferase reporter in the Socs3 genomic locus. These mice show robust Socs3 activity after administration of LPS in vivo, and hypothalamic explants derived from these animals can be used as a slice-based screening platform to identify signaling networks that alter hypothalamic Socs3 expression.
Acknowledgments
Financial Support: This project was supported by US Department of Agriculture, Agricultural Research Service Grant CRIS 6250-51000-055 (to M.F.); National Institutes of Health (NIH), National Institute of Diabetes and Digestive and Kidney Diseases Grant R01DK104901 (to M.F.); NIH Grant T32HD071839 (to E.L.C.); the Mouse Phenotyping Core at Baylor College of Medicine with funding from the NIH National Human Genome Research Institute (Grant U54 HG006348); the Genetically Engineered Mouse Core supported by National Cancer Institute Grant P30 CA125123; and the National Human Genome Research Institute Grant U42 HG006352.
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- BAC
bacterial artificial chromosome
- DAB
3,3′-diaminobenzidine
- GSK3
glycogen synthase kinase-3
- JAK
Janus kinases
- LepR
leptin receptor
- LPS
lipopolysaccharide
- Luc
Luciferase
- MSAB
methyl 3- 4methylphenyl sulfonyl amino benzoate
- NF-κB
nuclear factor κB
- SOCS3
suppressor of cytokine signaling-3
- STAT3
signal transducers and activators of transcription 3
- WT
wild type
References and Notes
- 1. Croker BA, Krebs DL, Zhang JG, Wormald S, Willson TA, Stanley EG, Robb L, Greenhalgh CJ, Förster I, Clausen BE, Nicola NA, Metcalf D, Hilton DJ, Roberts AW, Alexander WS. SOCS3 negatively regulates IL-6 signaling in vivo. Nat Immunol. 2003;4(6):540–545. [DOI] [PubMed] [Google Scholar]
- 2. Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol. 2007;7(6):454–465. [DOI] [PubMed] [Google Scholar]
- 3. Morris R, Kershaw NJ, Babon JJ. The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. 2018;27(12):1984–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carow B, Rottenberg ME. SOCS3, a major regulator of infection and inflammation. Front Immunol. 2014;5(58):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pedroso JAB, Ramos-Lobo AM, Donato J Jr. SOCS3 as a future target to treat metabolic disorders [published online ahead of print 9 November 2018]. Hormones (Athens). 2018. [DOI] [PubMed] [Google Scholar]
- 6. Mori H, Hanada R, Hanada T, Aki D, Mashima R, Nishinakamura H, Torisu T, Chien KR, Yasukawa H, Yoshimura A. Socs3 deficiency in the brain elevates leptin sensitivity and confers resistance to diet-induced obesity. Nat Med. 2004;10(7):739–743. [DOI] [PubMed] [Google Scholar]
- 7. Howard JK, Cave BJ, Oksanen LJ, Tzameli I, Bjørbaek C, Flier JS. Enhanced leptin sensitivity and attenuation of diet-induced obesity in mice with haploinsufficiency of Socs3. Nat Med. 2004;10(7):734–738. [DOI] [PubMed] [Google Scholar]
- 8. Coppari R, Bjørbæk C. Leptin revisited: its mechanism of action and potential for treating diabetes. Nat Rev Drug Discov. 2012;11(9):692–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Morrison CD. Leptin resistance and the response to positive energy balance. Physiol Behav. 2008;94(5):660–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reed AS, Unger EK, Olofsson LE, Piper ML, Myers MG Jr, Xu AW. Functional role of suppressor of cytokine signaling 3 upregulation in hypothalamic leptin resistance and long-term energy homeostasis. Diabetes. 2010;59(4):894–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ernst MB, Wunderlich CM, Hess S, Paehler M, Mesaros A, Koralov SB, Kleinridders A, Husch A, Münzberg H, Hampel B, Alber J, Kloppenburg P, Brüning JC, Wunderlich FT. Enhanced Stat3 activation in POMC neurons provokes negative feedback inhibition of leptin and insulin signaling in obesity. J Neurosci. 2009;29(37):11582–11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kievit P, Howard JK, Badman MK, Balthasar N, Coppari R, Mori H, Lee CE, Elmquist JK, Yoshimura A, Flier JS. Enhanced leptin sensitivity and improved glucose homeostasis in mice lacking suppressor of cytokine signaling-3 in POMC-expressing cells. Cell Metab. 2006;4(2):123–132. [DOI] [PubMed] [Google Scholar]
- 13. Zhang R, Dhillon H, Yin H, Yoshimura A, Lowell BB, Maratos-Flier E, Flier JS. Selective inactivation of Socs3 in SF1 neurons improves glucose homeostasis without affecting body weight. Endocrinology. 2008;149(11):5654–5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pedroso JA, Buonfiglio DC, Cardinali LI, Furigo IC, Ramos-Lobo AM, Tirapegui J, Elias CF, Donato J Jr. Inactivation of SOCS3 in leptin receptor-expressing cells protects mice from diet-induced insulin resistance but does not prevent obesity. Mol Metab. 2014;3(6):608–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bjørbaek C, Elmquist JK, Frantz JD, Shoelson SE, Flier JS. Identification of SOCS-3 as a potential mediator of central leptin resistance. Mol Cell. 1998;1(4):619–625. [DOI] [PubMed] [Google Scholar]
- 16. Bjørbaek C, El-Haschimi K, Frantz JD, Flier JS. The role of SOCS-3 in leptin signaling and leptin resistance. J Biol Chem. 1999;274(42):30059–30065. [DOI] [PubMed] [Google Scholar]
- 17. Zhang X, Zhang G, Zhang H, Karin M, Bai H, Cai D. Hypothalamic IKKbeta/NF-kappaB and ER stress link overnutrition to energy imbalance and obesity. Cell. 2008;135(1):61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Allison MB, Patterson CM, Krashes MJ, Lowell BB, Myers MG Jr, Olson DP. TRAP-seq defines markers for novel populations of hypothalamic and brainstem LepRb neurons. Mol Metab. 2015;4(4):299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Myers MG Jr, Heymsfield SB, Haft C, Kahn BB, Laughlin M, Leibel RL, Tschöp MH, Yanovski JA. Challenges and opportunities of defining clinical leptin resistance. Cell Metab. 2012;15(2):150–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Allison MB, Pan W, MacKenzie A, Patterson C, Shah K, Barnes T, Cheng W, Rupp A, Olson DP, Myers MG Jr. Defining the transcriptional targets of leptin reveals a role for Atf3 in leptin action. Diabetes. 2018;67(6):1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pan W, Allison MB, Sabatini P, Rupp A, Adams J, Patterson C, Jones JC, Olson DP, Myers MG Jr. Transcriptional and physiological roles for STAT proteins in leptin action. Mol Metab. 2019;22:121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stoiber D, Kovarik P, Cohney S, Johnston JA, Steinlein P, Decker T. Lipopolysaccharide induces in macrophages the synthesis of the suppressor of cytokine signaling 3 and suppresses signal transduction in response to the activating factor IFN-gamma. J Immunol. 1999;163(5):2640–2647. [PubMed] [Google Scholar]
- 23. Starr R, Willson TA, Viney EM, Murray LJ, Rayner JR, Jenkins BJ, Gonda TJ, Alexander WS, Metcalf D, Nicola NA, Hilton DJ. A family of cytokine-inducible inhibitors of signalling. Nature. 1997;387(6636):917–921. [DOI] [PubMed] [Google Scholar]
- 24. Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol. 2007;8(1):49–62. [DOI] [PubMed] [Google Scholar]
- 25. Park ES, Kim H, Suh JM, Park SJ, Kwon OY, Kim YK, Ro HK, Cho BY, Chung J, Shong M. Thyrotropin induces SOCS-1 (suppressor of cytokine signaling-1) and SOCS-3 in FRTL-5 thyroid cells. Mol Endocrinol. 2000;14(3):440–448. [DOI] [PubMed] [Google Scholar]
- 26. Delgado M, Ganea D. Inhibition of IFN-gamma-induced janus kinase-1-STAT1 activation in macrophages by vasoactive intestinal peptide and pituitary adenylate cyclase-activating polypeptide. J Immunol. 2000;165(6):3051–3057. [DOI] [PubMed] [Google Scholar]
- 27. Bousquet C, Chesnokova V, Kariagina A, Ferrand A, Melmed S. cAMP neuropeptide agonists induce pituitary suppressor of cytokine signaling-3: novel negative feedback mechanism for corticotroph cytokine action. Mol Endocrinol. 2001;15(11):1880–1890. [DOI] [PubMed] [Google Scholar]
- 28. Gasperini S, Crepaldi L, Calzetti F, Gatto L, Berlato C, Bazzoni F, Yoshimura A, Cassatella MA. Interleukin-10 and cAMP-elevating agents cooperate to induce suppressor of cytokine signaling-3 via a protein kinase A-independent signal. Eur Cytokine Netw. 2002;13(1):47–53. [PubMed] [Google Scholar]
- 29. Fasshauer M, Klein J, Lossner U, Paschke R. Isoproterenol is a positive regulator of the suppressor of cytokine signaling-3 gene expression in 3T3-L1 adipocytes. J Endocrinol. 2002;175(3):727–733. [DOI] [PubMed] [Google Scholar]
- 30. Sands WA, Woolson HD, Milne GR, Rutherford C, Palmer TM. Exchange protein activated by cyclic AMP (Epac)-mediated induction of suppressor of cytokine signaling 3 (SOCS-3) in vascular endothelial cells. Mol Cell Biol. 2006;26(17):6333–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fukuda M, Williams KW, Gautron L, Elmquist JK. Induction of leptin resistance by activation of cAMP-Epac signaling. Cell Metab. 2011;13(3):331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Balthasar N, Coppari R, McMinn J, Liu SM, Lee CE, Tang V, Kenny CD, McGovern RA, Chua SC Jr, Elmquist JK, Lowell BB. Leptin receptor signaling in POMC neurons is required for normal body weight homeostasis. Neuron. 2004;42(6):983–991. [DOI] [PubMed] [Google Scholar]
- 33. RRID:AB_331586. https://scicrunch.org/resolver/RRID:AB_331586.
- 34. RRID:AB_2313606. https://scicrunch.org/resolver/RRID:AB_2313606.
- 35. Howard JK, Flier JS. Attenuation of leptin and insulin signaling by SOCS proteins. Trends Endocrinol Metab. 2006;17(9):365–371. [DOI] [PubMed] [Google Scholar]
- 36. Atkinson JM, Rank KB, Zeng Y, Capen A, Yadav V, Manro JR, Engler TA, Chedid M. Activating the Wnt/β-catenin pathway for the treatment of melanoma--application of LY2090314, a novel selective inhibitor of glycogen synthase kinase-3. PLoS One. 2015;10(4):e0125028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ring DB, Johnson KW, Henriksen EJ, Nuss JM, Goff D, Kinnick TR, Ma ST, Reeder JW, Samuels I, Slabiak T, Wagman AS, Hammond ME, Harrison SD. Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes. 2003;52(3):588–595. [DOI] [PubMed] [Google Scholar]
- 38. Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci. 2003;116(Pt 7):1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Handeli S, Simon JA. A small-molecule inhibitor of Tcf/beta-catenin signaling down-regulates PPARgamma and PPARdelta activities. Mol Cancer Ther. 2008;7(3):521–529. [DOI] [PubMed] [Google Scholar]
- 40. Hwang S-Y, Deng X, Byun S, Lee C, Lee SJ, Suh H, Zhang J, Kang Q, Zhang T, Westover KD, Mandinova A, Lee SW. Direct targeting of β-catenin by a small molecule stimulates proteasomal degradation and suppresses oncogenic Wnt/β-catenin signaling. Cell Reports. 2016;16(1):28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yasukawa H, Ohishi M, Mori H, Murakami M, Chinen T, Aki D, Hanada T, Takeda K, Akira S, Hoshijima M, Hirano T, Chien KR, Yoshimura A. IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nat Immunol. 2003;4(6):551–556. [DOI] [PubMed] [Google Scholar]
- 42. Lebel E, Vallières L, Rivest S. Selective involvement of interleukin-6 in the transcriptional activation of the suppressor of cytokine signaling-3 in the brain during systemic immune challenges. Endocrinology. 2000;141(10):3749–3763. [DOI] [PubMed] [Google Scholar]
- 43. Yamawaki Y, Kimura H, Hosoi T, Ozawa K. MyD88 plays a key role in LPS-induced Stat3 activation in the hypothalamus. Am J Physiol Regul Integr Comp Physiol. 2010;298(2):R403–R410. [DOI] [PubMed] [Google Scholar]
- 44. Humpel C. Organotypic brain slice cultures: a review. Neuroscience. 2015;305:86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fukuda M, Jones JE, Olson D, Hill J, Lee CE, Gautron L, Choi M, Zigman JM, Lowell BB, Elmquist JK. Monitoring FoxO1 localization in chemically identified neurons. J Neurosci. 2008;28(50):13640–13648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kaneko K, Xu P, Cordonier EL, Chen SS, Ng A, Xu Y, Morozov A, Fukuda M. Neuronal Rap1 regulates energy balance, glucose homeostasis, and leptin actions. Cell Reports. 2016;16(11):3003–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Williams KW, Liu T, Kong X, Fukuda M, Deng Y, Berglund ED, Deng Z, Gao Y, Liu T, Sohn JW, Jia L, Fujikawa T, Kohno D, Scott MM, Lee S, Lee CE, Sun K, Chang Y, Scherer PE, Elmquist JK. Xbp1s in Pomc neurons connects ER stress with energy balance and glucose homeostasis. Cell Metab. 2014;20(3):471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yan J, Mei FC, Cheng H, Lao DH, Hu Y, Wei J, Patrikeev I, Hao D, Stutz SJ, Dineley KT, Motamedi M, Hommel JD, Cunningham KA, Chen J, Cheng X. Enhanced leptin sensitivity, reduced adiposity, and improved glucose homeostasis in mice lacking exchange protein directly activated by cyclic AMP isoform 1. Mol Cell Biol. 2013;33(5):918–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Naujok O, Lentes J, Diekmann U, Davenport C, Lenzen S. Cytotoxicity and activation of the Wnt/beta-catenin pathway in mouse embryonic stem cells treated with four GSK3 inhibitors. BMC Res Notes. 2014;7(1):273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bennett CN, Ross SE, Longo KA, Bajnok L, Hemati N, Johnson KW, Harrison SD, MacDougald OA. Regulation of Wnt signaling during adipogenesis. J Biol Chem. 2002;277(34):30998–31004. [DOI] [PubMed] [Google Scholar]
- 51. Ozcan L, Ergin AS, Lu A, Chung J, Sarkar S, Nie D, Myers MG Jr, Ozcan U. Endoplasmic reticulum stress plays a central role in development of leptin resistance. Cell Metab. 2009;9(1):35–51. [DOI] [PubMed] [Google Scholar]
- 52. Martin M, Rehani K, Jope RS, Michalek SM. Toll-like receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat Immunol. 2005;6(8):777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Beurel E, Jope RS. Differential regulation of STAT family members by glycogen synthase kinase-3. J Biol Chem. 2008;283(32):21934–21944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Münzberg H, Myers MG Jr. Molecular and anatomical determinants of central leptin resistance. Nat Neurosci. 2005;8(5):566–570. [DOI] [PubMed] [Google Scholar]
- 55. Suzuki A, Hanada T, Mitsuyama K, Yoshida T, Kamizono S, Hoshino T, Kubo M, Yamashita A, Okabe M, Takeda K, Akira S, Matsumoto S, Toyonaga A, Sata M, Yoshimura A. CIS3/SOCS3/SSI3 plays a negative regulatory role in STAT3 activation and intestinal inflammation. J Exp Med. 2001;193(4):471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shouda T, Yoshida T, Hanada T, Wakioka T, Oishi M, Miyoshi K, Komiya S, Kosai K, Hanakawa Y, Hashimoto K, Nagata K, Yoshimura A. Induction of the cytokine signal regulator SOCS3/CIS3 as a therapeutic strategy for treating inflammatory arthritis. J Clin Invest. 2001;108(12):1781–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ortiz-Muñoz G, Martin-Ventura JL, Hernandez-Vargas P, Mallavia B, Lopez-Parra V, Lopez-Franco O, Muñoz-Garcia B, Fernandez-Vizarra P, Ortega L, Egido J, Gomez-Guerrero C. Suppressors of cytokine signaling modulate JAK/STAT-mediated cell responses during atherosclerosis. Arterioscler Thromb Vasc Biol. 2009;29(4):525–531. [DOI] [PubMed] [Google Scholar]
- 58. Croom HA, Izon DJ, Chong MM, Curtis DJ, Roberts AW, Kay TW, Hilton DJ, Alexander WS, Starr R. Perturbed thymopoiesis in vitro in the absence of suppressor of cytokine signalling 1 and 3. Mol Immunol. 2008;45(10):2888–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]