Abstract
Background:
Fasciolosis is a shared disease between humans and livestock caused by hepatic trematodes; Fasciola hepatica and F. gigantica. Differentiate between the two species of this genus is essential. High-Resolution Melting (HRM) Analysis represents a new approach to this issue. This method can be performed right after termination of Real-Time PCR. This technique has not been used for identification of adult F. hepatica and F. gigantica genotypes. The aim of this study was to determine Fasciola species by using HRM in isolates taken from Iran, respectively.
Methods:
Ninety-three Fasciola spp. samples were collected from infected slaughtered animals in different regions of Iran, including North West (Ardebil Province) and South East (Zahedan Province) during 2016. Genomic DNA from the samples was extracted using a DNA extraction kit and then after Real-Time PCR amplification, HRM was done.
Results:
Overall, 59 and 34 isolates were identified as F. hepatica and F. gigantica, respectively. The percentages of each species from animals were as follows: sheep (F. hepatica, 80.39% and F. gigantica, 19.61%), cattle (F. hepatica, 42.85% and F. gigantica, 57.15%).
Conclusion:
HRM technique developed in the present study is a powerful, rapid and sensitive technique for epidemiological survey and molecular identification between F. hepatica and F. gigantica.
Keywords: High-resolution melting (HRM), Fasciola hepatica, Fasciola gigantica, CoxI
Introduction
The common liver flukes Fasciola hepatica and F. gigantica are causative agents of fasciolosis: a global zoonotic parasitic disease. Although fasciolosis commonly occurs in liver and biliary ducts, ec-topic fasciolosis may occur in the peritoneal cavity, lungs, subcutaneous tissue, lymph nodes, eye, and other locations (1, 2). Besides causing disease in humans, it can induce mortality and morbidity in sheep and cattle industry (3).
As these flat worms parasitize mollusks and vertebrate hosts, accurate and fast detection of them is of vital importance for facilitating control and prevention strategies of fasciolosis in humans and animals (4). One of the fast, sensitive and specific approach for detection and evolutionary analysis of Fasciola spp. are molecular techniques. Despite existence of many diagnosis tools like microscopy, radiology, ultrasound, CT, MRI, serological and clinical tests for detection of fasciolosis, unsatisfactory amounts of sensitivity and specificity have been obtained in many studies (5). Emerging of molecular methods was a turning point to accurate and precise detection and species identification of parasites. Molecular methods not only offer an appropriate impediment for detection of Fasciola spp. in mammals, but also in detection of infected snails in the epizootiological studies of these flukes (6).
Toward this end, many molecular techniques have been used by researchers that vary from conventional PCR methods like PCR-RFLP, PCR-SSCP and multiplex PCR to quantitative and qualitative real-time PCR (qRT-PCR) technology (7–10).
Indeed real-time PCR has brought the detection of parasitic diseases to another level, with significant sensitivity, easier performance, and no post-PCR operation compared to conventional PCR methods. HRM analysis is a novel method that allows us rapid screening and detection of closely related species in a laboratory (11). The steps in HRM analysis entangle amplification of the wanted region in the presence of a specialized dsDNA binding dye and step by step denaturation of amplicons by increasing the temperature in small increments in order to produce a characteristic melting profile called melting analysis (12). To date, the HRM has mostly been used in molecular studies of parasitic protozoa and rarely for parasitic worms (11,13,14).
The genes targeted for molecular strategies against Fasciola spp. are the ITSI and ITSII of nuclear DNA and COI from mitochondrial DNA. The COI gene fragment introduced as a most variable marker recommended for future analyses (15,3).
In this study, we developed a qualitative real-time PCR method appeared with HRM analysis for the detection of the two Fasciola spp. In samples taken from Ardebil and Zahedan provinces, northwestern and southern Iran, respectively.
The study was conducted as a complementary case to previous studies on genotype (16) and morphological verifications (17).
In this study, we aimed to develop a qualitative real-time PCR method appeared with HRM analysis for the detection of the two Fasciola spp. Samples were taken from Ardebil and Zahedan provinces, northwestern and southern Iran, respectively.
Material and Methods
Sample collection
Ninety-three adult trematodes of Fasciola spp. located within the bile ducts of infected cattle (n=42 samples) and sheep (n=51 samples), were collected from slaughterhouses in two different regions of Iran from Nov 2016 to Jan 2017. The study was conducted in Meshkinshahr City, Ardebil Province, North West and Zabol City, Sistan, and Baluchistan Province, South East, Iran (Fig. 1). The flatworms were extensively washed in physiological saline and preserved in >70° (v/v) ethanol and then frozen at −20 °C until DNA extraction.
Fig. 1:
Sistan and Baluchestan Province (South East) and Ardebil Province (North West), Iran. The Meshkinshahr and Zabol cities are highlighted in yellow
Ethics Committee of Tehran University of Medical Sciences approved the study.
DNA extraction
Before DNA extraction, all individual worms rinsed three times with PBS to remove the ethanol. Genomic DNA (Gdna) was extracted from small portion of apical region of adult trematodes to avoid entry female genitalia that likely to outer sperm. Gdna from individual worms was extracted using a QIAamp DNAeasy, Hilden, Germany, according to the manufacturer’s reference protocols with some modifications. The concentration of the extracted DNA was specified by NanoDrop (Thermo Scientific, Rockford, IL, USA), and after that, the samples were stored at −20 °C for further analysis.
DNA amplification and HRM
The mitochondrial sequences were amplified using specific primers for the COI gene, (forward A.H.S-CO1-F 5′- GGGCATCCTGAGGTTTATGT-3′ and reverse A.H.S-CO1-R 5′- AACATTATCAACCAGGAAAAGACC-3′ design was carried out using the sequences available on GenBank and in continuation. Primers were obtained using the program PRIMER BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) and rechecked with Beacon Designer8.12, PREMIER BIOSOFT software from the consensus sequence obtained by the multiple alignments, with an expected amplicon size of 266 bp for all the sequences.
PCR was performed in 20 Ml final reaction volume containing 10 Ml master mix (Type-it HRM PCR Kit; Qiagen, Hilden, Germany), 5.2 Ml distilled water, 0.4 Ml of each primer, and 4 Ml of template DNA. Enzymatic reaction was performed as follows: the reaction mixture was heated for initial denaturation step at 95 °C for 10 min, followed by 40 cycles of amplification performed at 95 °C for 30 sec for denaturation step, 55 °C for 40 sec, for annealing section, 72 °C for 30 sec related to extension portion, and a final extension step at 72 °C for 6 min after 40 cycles. HRM temperature was raised from 70 °C to 95 °C. During this process, the amplicons obtained from PCR were denatured prior to the development of melting curves in the inflexion point where changes in fluorescence with respect to changes in temperature (Df/Dt) were recorded with a ramp of 0.3 °C/sec (18). Fluorescence dye signaling was measured after each cycle. The kit contained the novel double-stranded DNA-binding fluorescent dye, EvaGreen, and an optimized HRM PCR master mix buffer, consisting of HotStarTaq plus DNA polymerase, Q-Solution, and dNTPs.
We used a positive standard control for F. hepatica and F. gigantica available in the Department of Medical Parasitology and Mycology, School of Public Health, Tehran University of Medical Sciences.
Real-time PCR was carried out in a Mini Opticon real-time PCR detection system (Applied Biosystems Step One Plus Inc., CA, USA). The Real-Time amplification result and Tm analysis were obtained using the Step One PlusTM software ver. 2.3 (Life technologies@). Tm analysis was repeated three times in each run to confirm the repeatability of the Tm assay by estimating the Tm variation within a PCR amplification (intra-assay), and between PCR amplifications (inter-assay). The coefficient of variation (CV) was calculated by dividing the standard deviation (SD) by the arithmetic mean of the measured values of Tm (CV = SD[Le, 2012 #11]/mean value).
Furthermore, to check the uniformity of temperature in the cycler block, a number of samples were re-amplified at different positions of the cycler block during the same amplification cycle. The intra-assay CVs represent the mean CVs of the results obtained from the replications of Fasciola spp.
Results
Real-Time PCR amplification and HRM analysis
In accordance with Real-Time PCR and HRM analysis procedure, among 93 samples containing cattle (n=42 samples) and sheep (n=51 samples) were amplified using partial sequence of COI gene of Fasciola spp., and then HRM was performed. Tm analysis was repeated three times in each run to confirm the repeatability of the Tm assay (Table 1).
Table 1:
Mean Tm, SD, and CV calculated based on intra- and inter-assay of cox1 gene sequence of Fasciola spp.
| Gene | Mean Tm (°C) | SD | Intra-assay CV (%) | Inter-assay CV (%) |
|---|---|---|---|---|
| cox1 | ||||
| F. hepatica | 76°C | 0.14 | 0.05 | 0.12 |
| F. gigantica | 77.1°C | 0.10 | 0.07 | 0.11 |
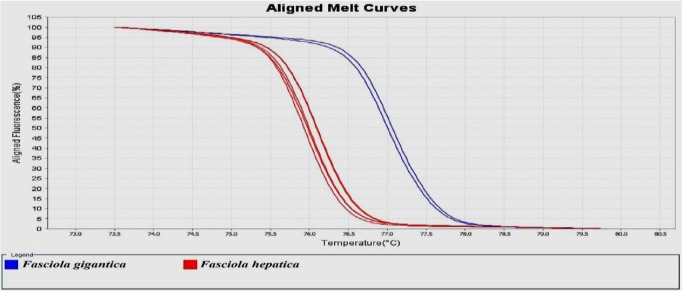
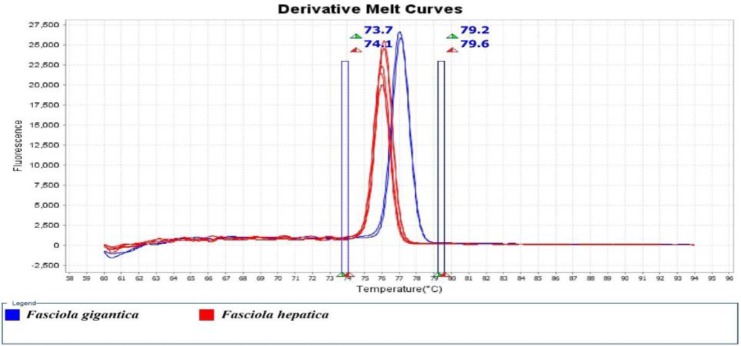
The result of HRM analysis showed that 59 and 34 isolates were identified as F. hepatica and F. gigantica, respectively. The percentages of each species from animals were as follows: Sheep (F. hepatica, 80.39% and F. gigantica, 19.61%), cattle (F. hepatica, 42.85% and F. gigantica, 57.15%). The melting curve and HRM curve analysis of the Fasciola spp. Identified and shown in Figs. 2 and 3. The real-time PCR melting curve results indicated that the mean Tm, SD, CV calculated based on intra- and inter-assay was separated by each genotype of Fasciola spp.
Fig. 2:
HRM based on (EvaGreen) Aligned Melt curves analyses and identified Fasciola spp. In sheep and cattle using COI gene
Fig. 3:
Derivative Melt Curves of the analyzed and identified Fasciola spp. In sheep and cattle using COI gene
Discussion
Following previous studies and because of the importance of fascioliasis in Iran, more comprehensive studies on this parasite and the diagnosis of its species are necessary to achieve this goal (19, 20). In the previous studies, PCR-RFLP technique on ITSI gene was used to determine the species of Fasciola and compare it with the sequencing and morphology methods (16,17).
In the present assay, we successfully developed a real-time PCR and HRM technique on COI gene for rapid, sensitive and precise differentiation of F. hepatica and F. gigantica using only a single pair of primers. The average Tm variation obtained by melting curve analysis was about 1°C. This finding indicates a sufficient ability and reliability of the assay for distinguishing these two parasites.
The real-time PCR and HRM assay often are able to scan even a one SNP, it is likely to be able in significant distinct of the intermediate form of Fasciola spp. in endemic regions. However, this method cannot be a proper substitute for sequencing in terms of aforementioned point.
The results of this study were completely consistent with the previous studies carried out using the PCR-RFLP technique on ITSI gene, sequencing and morphology methods on the Fasciola. isolates of Ardabil Province (16, 17).
Despite the satisfactory results of molecular methods, according to previous studies different sensitivity and specificity have been acquired from variety of DNA based techniques (from conventional to real-time PCR) (15, 21, 22). There are many target genes employed for detection of Fasciola spp. including rRNA gene (ITSI and ITSII) of genomic DNA (22), and NADH and COI of the mitochondrial DNA (17, 19, 23). Several PCRRFLP assays have been described for discrimination of F. hepatica, F. gigantica that targeting the 28S rRNA, ITSI and ITSII (15, 16, 18, 21). In addition, a PCR-SSCP on ITSI region of rRNA was defined for distinction between F. hepatica, F. gigantica and the intermediate species in China (7). A multiplex PCR was designed for simultaneous amplification of mitochondrial DNA and ITS region of Fasciola spp. in mammalian and intermediate hosts (24, 25). Real-time PCR, as a more sensitive molecular approach, has been used in some studies focused on liver fluke (7, 10).
Whenever HRM is incorporated in real-time PCR, this allows us nonstop quantitative detection of the parasites. The present study has been done for the first time in the world while there are some studies used HRM for identification and genotyping of other parasites. For example, the real-time PCR and HRM technique was used for identification of Echinococcus spp. in Isfahan Province of Iran (26, 27). In addition this technique was applied for genotyping of protozoan parasites like Plasmodium spp., Giardia spp. and Cryptosporidium spp. (4, 7, 12, 28).
It has been described as a robust assay for the rapid detection for phylum Platyhelminthes such as Fascioloides magna, Schistosoma spp., Clonorchis sinensis and Opisthorchis viverrini (6, 29).
Finally, this assay needs to be more developed by performing on large sample size in wide geographical regions in the world especially in endemic areas such as Iran. Besides, the efficacy of this technique should be evaluated on discrimination of intermediate species in the region that both of F. hepatica and F. gigantica exist.
The limitations of this study included the small sample size and non-overlapping all geographic regions of Iran. The advantage, however, is that HRM technique was performed for the first time in the world to molecular identification between F. hepatica and F. gigantica.
Conclusion
HRM technique developed in present study is a powerful, rapid and sensitive technique for epidemiological survey and molecular identification between F. hepatica and F. gigantica.
Ethical considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Acknowledgements
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. We are very grateful to Prof. Mehdi Mohebali, Chairman of the Department of Medical Parasitology and Mycology and Prof. Nateghpour, Chief of the National Malaria Diagnostic Laboratory, supported and provided the condition for doing this research.
Footnotes
Conflict of interests
The authors declare that there is no conflict of interests.
References
- 1.Mas-Coma S, Bargues MD, Valero MA. (2005). Fascioliasis and other plant-borne trematode zoonoses. Int J Parasitol, 35(11–12) : 1255–1278. [DOI] [PubMed] [Google Scholar]
- 2.Rokni MB, Lotfy WM, Ashrafi K, Murrell KD. (2014). Neglected Tropical Diseases-Middle East and North Africa. Springer, 59–90. [Google Scholar]
- 3.Simsek S, Utuk AE, Balkaya I. (2011). Molecular differentiation of Turkey cattle isolates of Fasciola hepatica and Fasciola gigantica. Helminthologia, 48, 3–7. [Google Scholar]
- 4.Zhang P, Liu Y, Alsarakibi M, et al. (2012). Application of HRM assays with EvaGreen dye for genotyping Giardia duodenalis zoonotic assemblages. Parasitol Res, 111(5), 2157–2163. [DOI] [PubMed] [Google Scholar]
- 5.Marcilla A, Bargues MD, Mas-Coma S. (2002). A PCR-RFLP assay for the distinction between Fasciola hepatica and Fasciola gigantica. Mol Cell Probes, 16(5):327–333. [DOI] [PubMed] [Google Scholar]
- 6.Cai XQ, Yu HQ, Li R, et al. (2014). Rapid detection and differentiation of Clonorchis sinensis and Opisthorchis viverrini using real-time PCR and high resolution melting analysis. ScientificWorldJournal, 893981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alasaad S, Soriguer RC, Abu-Madi M, et al. (2011). A TaqMan real-time PCR-based assay for the identification of Fasciola spp. Vet Parasitol, 179(1–3), 266–271. [DOI] [PubMed] [Google Scholar]
- 8.Huang W, He B, Wang C, Zhu X. (2004). Characterisation of Fasciola species from Mainland China by ITS-2 ribosomal DNA sequence. Vet Parasitol, 120(1–2), 75–83. [DOI] [PubMed] [Google Scholar]
- 9.Magalhães KG, Passos LKJ, Carvalho OdS. (2004). Detection of Lymnaea columella infection by Fasciola hepatica through Multiplex-PCR. Mem Inst Oswaldo Cruz, 99(4): 421–424. [DOI] [PubMed] [Google Scholar]
- 10.Schweizer G, Meli M, Torgerson PR, et al. (2007). Prevalence of Fasciola hepatica in the intermediate host Lymnaea truncatula detected by real time TaqMan PCR in populations from 70 Swiss farms with cattle husbandry. Vet Parasitol, 150(1–2): 164–169. [DOI] [PubMed] [Google Scholar]
- 11.Ngui R, Lim YA, Chua KH. (2012). Rapid detection and identification of human hookworm infections through high resolution melting (HRM) analysis. PloS One, 7(7):e41996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chua KH, Lim SC, Ng CC, et al. (2015). Development of high resolution melting analysis for the diagnosis of human malaria. Scientific Reports, 5:15671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rojas A, Segev G, Markovics A, Aroch I, Baneth G. (2017). Detection and quantification of Spirocerca lupi by HRM qPCR in fecal samples from dogs with spirocercosis. Parasites Vectors, 10: 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tun S, Ithoi I, Mahmud R, et al. (2015). Detection of helminth eggs and identification of hook-worm species in stray cats, dogs and soil from Klang Valley, Malaysia. PloS One, 10(12): e0142231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rokni MB, Mirhendi H, Mizani A, et al. (2010). Identification and differentiation of Fasciola hepatica and Fasciola gigantica using a simple PCR-restriction enzyme method. Exp Parasitol, 124(2): 209–213. [DOI] [PubMed] [Google Scholar]
- 16.Aryaeipour M, Rouhani S, Bandehpour M, et al. (2014). Genotyping and phylogenetic analysis of Fasciola spp. isolated from sheep and cattle using PCR-RFLP in Ardabil province, northwestern Iran. Iran J Public Health, 43 (10): 1364–71. [PMC free article] [PubMed] [Google Scholar]
- 17.Aryaeipour M, Bozorgomid A, Kazemi B, et al. (2017). Molecular and Morphometrical Characterization of Fasciola Species Isolated from Domestic Ruminants in Ardabil Province, Northwestern Iran. Iran J Public Health, 46(3):318–325. [PMC free article] [PubMed] [Google Scholar]
- 18.Higuera SL, Guhl F, Ramírez JD. (2013). Identification of Trypanosoma cruzi Discrete Typing Units (DTUs) through the implementation of a High-Resolution Melting (HRM) genotyping assay. Parasites Vectors, 6:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rokni MB, Bozorgomid A, Heydarian P, Aryaeipour M. (2018). Molecular Evidence of Human Fasciolosis Due to Fasciola gigantica in Iran: A Case Report. Iran J Public Health, 47(5):750–754. [PMC free article] [PubMed] [Google Scholar]
- 20.Aryaeipour M, Kia EB, Heidari Z, Sayyad T, alaie Z, Rokni MB. (2015). Serological study of Human Fasciolosis in Patients Referring to the School of Public Health, Tehran University of Medical Sciences, Tehran, Iran during 2008–2014. Iran J Parasitol, 10(4):517–522 [PMC free article] [PubMed] [Google Scholar]
- 21.Itagaki T, Tsutsumi K, Sakamoto T, Tsutsumi Y. (1995). Characterization of genetic divergence among species within the genus Fasciola by PCR-SSCP. Japanese J Parasitol, 44, 244–247. [Google Scholar]
- 22.Krämer F, Schnieder T. (1998). Sequence heterogeneity in a repetitive DNA element of Fasciola. Int J Parasitol, 28: 1923–9. [DOI] [PubMed] [Google Scholar]
- 23.Bozorgomid A, Nazari N, Rahimi H, et al. (2016). Molecular Characterization of Animal Fasciola spp. Isolates from Kermanshah, Western Iran. Iran J Public Health, 45(10):1315–1321 [PMC free article] [PubMed] [Google Scholar]
- 24.Le TH, Nguyen KT, Nguyen NT, et al. (2012). Development and evaluation of a single step duplex PCR for simultaneous detection of Fasciola hepatica and Fasciola gigantica (Fasciolidae; Trematoda; Platyhelminthes). J Clin Microbiol, 50(8):2720–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magalhaes KG, Jannotti-Passos LK, Caldeira RL, et al. (2008). Isolation and detection of Fasciola hepatica DNA in Lymnaea viatrix from formalin-fixed and paraffin-embedded tissues through multiplex-PCR. Vet Parasitol, 152(3–4): 333–338. [DOI] [PubMed] [Google Scholar]
- 26.Hosseini-Safa A, Mohag MA, et al. (2016). First report of Tasmanian sheep strain (G2) genotype isolated from Iranian goat using the high resolution melting (HRM) analysis. Gastroenterol Hepatol Bed Bench, 9(Suppl 1):S70–S74. [PMC free article] [PubMed] [Google Scholar]
- 27.Safa AH, Harandi MF, Tajaddini M, et al. (2016). Rapid identification of Echinococcus granulosus and E. canadensis using high-resolution melting (HRM) analysis by focusing on a single nucleotide polymorphism. Jap J Infect Dis, 69(4):300–305. [DOI] [PubMed] [Google Scholar]
- 28.Pangasa A, Jex AR, Campbell BE, et al. (2009). High resolution melting-curve (HRM) analysis for the diagnosis of cryptosporidiosis in humans. Mol Cell Probes, 23(1): 10–5. [DOI] [PubMed] [Google Scholar]
- 29.Radvánský J, Bazsalovicsová E, Králová-Hromadová I, Minárik G, Kádaši L’. (2011). Development of high-resolution melting (HRM) analysis for population studies of Fascioloides magna (Trematoda: Fasciolidae), the giant liver fluke of ruminants. Parasitol Res, 108(1): 201–209. [DOI] [PubMed] [Google Scholar]