Abstract
Background
Split hand/foot malformation (SHFM) is a group of congenital skeletal disorders which may occur either as an isolated abnormality or in syndromic forms with extra-limb manifestations. Chromosomal micro-duplication or micro-triplication involving 17p13.3 region has been described as the most common cause of split hand/foot malformation with long bone deficiency (SHFLD) in several different Caucasian and Asian populations. Gene dosage effect of the extra copies of BHLHA9 gene at this locus has been implicated in the pathogenesis of SHFLD.
Case presentation
The proband was a female child born to non-consanguineous parents. She was referred for genetic evaluation of bilateral asymmetric ectrodactyly involving both hands and right foot along with right tibial hemimelia. The right foot had fixed clubfoot deformity with only 2 toes. The mother had bilateral ectrodactyly involving both hands, but the rest of the upper limbs and both lower limbs were normal. Neither of them had any other congenital malformations or neurodevelopmental abnormalities. Genetic testing for rearrangement of BHLHA9 gene by quantitative polymerase chain reaction confirmed the duplication of the BHLHA9 gene in both the proband and the mother.
Conclusions
We report the first Sri Lankan family with genetic diagnosis of BHLHA9 duplication causing SHFLD. This report along with the previously reported cases corroborate the possible etiopathogenic role of BHLHA9 gene dosage imbalances in SHFM and SHFLD across different populations.
Keywords: Ectrodactyly, Split hand/foot malformation, Split hand/foot malformation with long bone deficiency, 17p13.3 duplication, BHLHA9
Background
Ectrodactyly or split hand/foot malformation (SHFM) is a group of genetic skeletal disorders with variable phenotypes. Complete absence or under-development of the central digits with a cleft in the hands and/or feet is the archetypal phenotype. However, SHFM is clinically heterogeneous, varying from slight shortening of a single central digit to monodactyly in extreme cases. Ectrodactyly may occur as an isolated anomaly affecting only one or more limbs, or in syndromic forms with extra-limb manifestations [1]. To date, single gene pathogenic variants, recurrent genomic duplications and balanced or unbalanced chromosomal rearrangements in several chromosomal loci are reported to be associated with isolated forms of SHFM. Most are inherited in an autosomal dominant pattern. However, incomplete penetrance is a common occurrence in affected families. Several genes in chromosomal loci that are known to regulate embryonic development of the limb buds in animal models are implicated in the genetic etiopathogenesis of the SHFM. One of the most frequently described is the TP63 gene that accounts for about 10% of isolated SHFM [2]. Duplication in the 17p13.3 locus is reported to be associated with SHFM with long bone deficiency (SHFLD), frequently involving the tibia and fibula [3, 4]. The SHFLD critical region in the 17p13.3 locus contains the BHLHA9 (basic helix-loop-helix [BHLH] family member A9) gene. The orthologous gene has been reported to be expressed in the mesenchyme of the developing limb buds in zebrafish and mouse models, thus the BHLHA9 gene is proposed to regulate human limb development [5].
The clinical and genetic heterogeneity of SHFM and SHFLD render the precise diagnosis of the condition challenging, and genetic testing to detect the underlying chromosomal or genetic defect is necessary for confirmation. Herein, we report the case of a mother and daughter with SHFLD with BHLHA9 duplication, which is the first reported Sri Lankan family with a molecular genetic diagnosis of ectrodactyly.
Case presentation
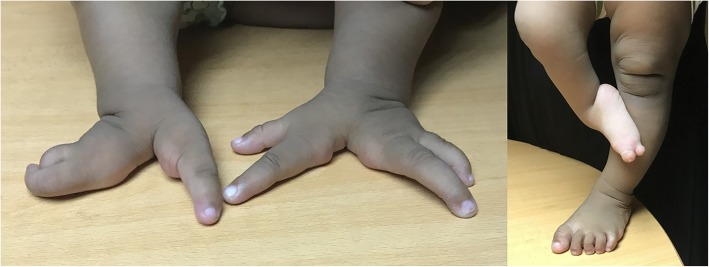
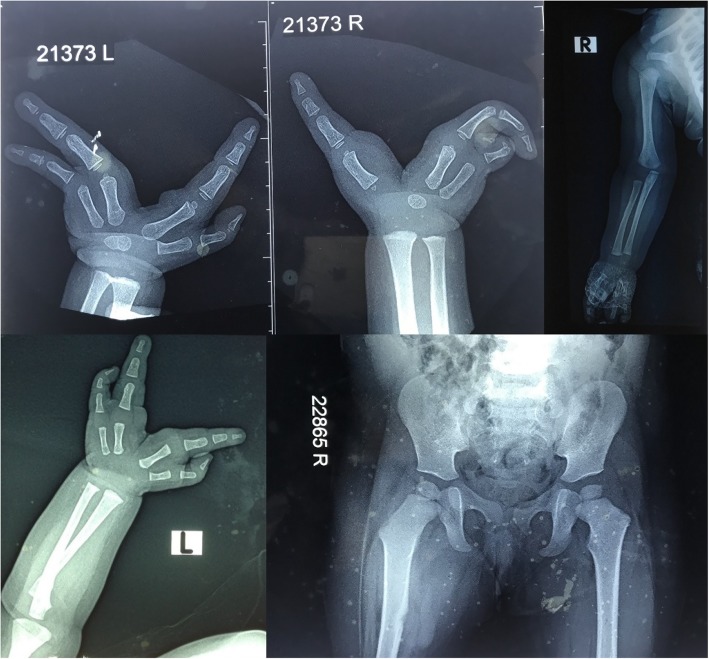
A 4-day-old baby girl was referred from a paediatric tertiary care hospital for the genetic evaluation of bilateral asymmetric ectrodactyly. She is the second child of a non-consanguineous couple; a 25-year-old father and 23-year-old mother. The baby was delivered normally at term following an uncomplicated pregnancy. The birth weight was 2.5 kg and there were no post-natal complications. She had ectrodactyly involving three limbs, with the absence of the third digit on the left hand and the second and third digits on the right hand. The right thumb was clinically normal, but the fourth and fifth digits were malformed. The right foot had fixed clubfoot deformity with only 2 toes (Fig. 1). There was no facial dysmorphism or facial clefts. Radiographs of the upper limbs showed complete absence of the metacarpal bone and the phalanges of the third digit in the left hand and absent metacarpals and phalanges of two digits on the right hand (Fig. 2). During a subsequent evaluation of the proband at the pediatric clinic, right tibial hemimelia was documented in the patient’s medical records by the attending clinician, but the radiological images of the leg were not available for inclusion in this article. Cardiovascular, respiratory and abdominal examination showed no abnormalities. Ultrasonography of the abdomen, brain and bilateral hips were normal.
Fig. 1.
Clinical photographs of the hands and the feet of the proband showing ectrodactyly involving three limbs, with the absence of the third digit on the left hand, absence of 2 digits on the right hand with malformed fingers (except the thumb) and the fixed clubfoot deformity with only 2 toes on the right foot
Fig. 2.
Radiological images of the upper limbs and pelvis of the proband showing complete absence of the metacarpal and the phalanges of the third digit on the left hand and absent metacarpals and phalanges of two digits on the right hand. Rest of the upper limbs and upper femur were radiologically normal
The mother also had bilateral ectrodactyly involving both hands, with the absence of the third digit on the right hand and two digits on the left hand. She had bi-phalangeal fifth digit on the left hand (Fig. 3). She had not previously been investigated for this condition and was otherwise healthy without any remarkable events in the medical and obstetrics history. The first child of the couple who was aged 2 ½ years old at the time of consultation had normal growth and development with no congenital anomalies. There were no other family members or close relatives affected with similar limb deformities or other congenital anomalies.
Fig. 3.

Clinical photographs of the hands of the mother showing bilateral ectrodactyly involving both hands, with the absence of the third digit on the right hand and two digits on the left hand, with bi-phalangeal left 5th digit
Peripheral venous blood samples were obtained from the baby and the mother with informed written consent. Genomic DNA was extracted from the blood samples and quantitative polymerase chain reaction (qPCR) was performed to identify rearrangements of the BHLHA9 gene (Chr17:1173858–1,174,565, hg19)(Ref Seq BHLHA9:NM_001164405). The RPPH1 gene (NR_002312) was used as the gene of reference. The qPCR results showed a BHLHA9/RPPH1 ratio of 1.46 in the baby and 1.50 in the mother confirming the BHLHA9 gene duplication. The unaffected sibling was not available for genetic assessment of her BHLHA9 status. Genetic counseling was offered to the family.
At the time of reporting, the baby was 10 months of age. Her body weight was 7.95 kg (25th centile), length was 72 cm (above 50th centile) and head circumference was 44 cm (between 25th and 50th centile). She had age-appropriate developmental milestones. Hearing and visual assessments were normal. Repeat ultrasonography of the brain and the abdomen showed no abnormalities. 2D-echocardiography showed a structurally and functionally normal heart. She is currently being followed up at the pediatric tertiary care hospital and awaiting reconstructive surgery.
Discussion and conclusions
Ectrodactyly or SHFM is a clinically and genetically heterogeneous group of skeletal disorders with or without extra-limb manifestations. As observed in the present case, the clinical heterogeneity leads to variable expressivity of the malformations among different affected individuals even within the same family [1]. Most of the isolated forms of SHFM; [SHFM1: chromosomal locus 7q21.2, SHFM3: 10q24, SHFM4: 3q28, SHFM5: 2q31] follow the autosomal dominant inheritance pattern. Incomplete penetrance is a common phenomenon in some families. Rare forms of SHFM such as SHFM6 mapped to chromosomal locus 12q13.12 and SHFM2 mapped to chromosomal locus Xq26 have also been described in families that show autosomal recessive and X-linked recessive inheritance patterns, respectively [1, 6].
Duplication at the 17p13.3 locus that was identified in the mother and daughter in the present case is reported to be associated with SHFLD, frequently involving the tibia and rarely other long bones. Some individuals with this duplication are reported to present with isolated SHFM without long bone involvement (Table 1) [3–5, 7–16]. The link between SHFLD and 17p13 duplication was first reported by Lezirovitz et al. in 2008. They identified the chromosomal region 17p13.1–13.3 as the candidate region for SHFLD by linkage analysis and multipoint Lod scores calculations in a Brazilian family with nine affected individuals with SHFM with or without tibial hypoplasia or aplasia [3]. Armour et al., in 2011 investigated 3 families with SHFLD with tibial involvement and narrowed the SHFLD critical region to a 173 kb region at 17p13.3, suggesting BHLHA9 as a possible candidate gene [4]. The critical region was further narrowed to an 11.8 kb region containing only the BHLHA9 gene by Klopocki et al. in 2012, after investigating 17 families with 42 individuals affected with SHFLD. They demonstrated that the expression of BHLHA9 gene in the limb buds of the mouse and zebrafish embryos are restricted to the mesenchyme underlying the apical ectodermal ridge (AER), hence suggesting that BHLHA9 transcription factor regulates the interaction between the AER and the progress zone (PZ) in human limb development and morphogenesis [5].
Table 1.
Studies describing the 17p13.3 duplication/ triplications associated with SHFM or SHFLD
| Reference [No.] | No. of families/ individuals with duplication/ triplication (a) | Limb anomalies of affected individuals | Extra-limb manifestations | Special remarks |
|---|---|---|---|---|
| ezirovitz et al., 2008 [3] |
1 family 9 affected individuals |
SHFM (4), SHFLD [tibial hemimelia] (3), Tibial hemimelia/aplasia only (2) | No |
Autosomal dominant (AD) inheritance/ Variable expressivity (VE)/, Incomplete penetrance (IP), Defined candidate region to 17p13.1 – 17p13.3; ~ 861 kb |
| Armour et al., 2011 [4] |
3 families (?1 with triplicationa) with 12 affected individuals 7 with duplication: 6 affected, 1 unaffected |
Split hand only (3), SHFLD [tibial hypoplasia/aplasia] (9) |
No |
AD inheritance/ VE/ IP Defined critical region of ~ 173 kb, ?BHLHA9 or AC016292 duplication |
| Klopocki et al., 2011 [5] |
17 families (out of 56) 82 with duplication: 42 affected, 40 unaffected |
SHFLD [tibial hemimelia] (18/31) Only SHFM (5) |
No |
AD inheritance/ VE/ IP Sex bias: (Male>Female), affected females with more severe phenotype Defined critical region of ~ 11.8 kb encompassing only BHLHA9 gene |
| Petit et al., 2013 [7] | 2 affected with duplication |
Case 1 SHFLD [R/radial agenesis and hypoplastic R/ulna and L/radial hypoplasia] Case 2 SHFM only |
Case 1 small ASD | Involvement of radius reported for the first time |
| Curry et al., 2013 [8] |
1 affected 14 families in this report (total of 21 families analyzed) with 17p13.3 duplications that includes BHLHA9 |
SHFLD [tibial hemimelia] | Cleft palate, Mild ID | |
| Luk et al., 2014 [9] |
1 affected fetusa Unaffected mothera both with duplication |
Split hands | No | First case with prenatal genetic diagnosis |
| Petit et al., 2014 [10] |
13 families with 42 affected individuals and 19 unaffected obligate carriers; 29 with the duplication (20 affected, 9 unaffected) |
SHFLD (18) | No |
AD inheritance/ VE/ IP Involvement of radius in 2 individuals Femur hypoplasia in one patient Affected males with more severe phenotype |
| Al Kaissi et al., 2014 [11] |
1 affected Father- bilateral partial syndactyly |
SHFM, tibial hemimelia | Sacral hypoplasia, DD, Thrombocyto-penia | |
| Nagata et al., 2014 [12] |
27 families 64 with the duplication/triplication (42/42 affected patients, 22/47 unaffected relatives); 2/1000 Japanese controls positive for duplication |
SHFM (29), SHFLD (11), GWC (2) | NR |
No sex bias SHFLD and GWC more common in triplications |
| Nagata et al., 2015 [13] |
1 affected childa Unaffected mothera both with triplication |
SHFM with tibial aplasia (R)/ hypoplasia (L) and wide R/ distal femoral metaphysis (GWC-like malformation) | No | |
| Cho et al., 2015 [14] |
1 affected fetus with the duplication |
SHFLD, campomelia of R/femur, Bilateral agenesis of fibula, bilateral club feet, oligosyndactyly | No | |
| Fusco et al., 2017 [15] |
3 affected with the duplication |
SHFLD; tibial hypoplasia (2) One with isolated tibial hypoplasia |
One with ASD | |
| Shen et al., 2018 [16] |
1 family with 5 affected individuals, 4 affected fetuses 10 tested (8 individuals, 2 fetuses) 8 with the duplication; 6 affected, 2 unaffected |
SFFM (4) SHFLD (1), bilateral femoral hypoplasia |
No | |
|
Present report 2018 |
1 affected child Affected mother |
SHFM (?with tibial hemimelia) | No |
ASD Atrial septal defect, DD Developmental delay, GWC Goellop Woolfgang Complex, ID Intellectual disability, R Right, L Left
aIndividuals with 17p13.3 triplication
According to GnomAD SV database, the population frequency of BHLHA9 duplication is estimated to be 5 × 10− 5 [17]. Chromosome 17p13.3 duplication encompassing the BHLHA9 gene was reported to be less than 50% penetrant in the OMIM database [18], and Klopocki et al. also reported that only approximately 50% of individuals with the duplication manifest the phenotype [5]. A notable feature is the variable expressivity of the condition even within the same family as observed in the present case [3–5]. In addition, Klopocki et al. observed a sex bias resulting in more affected males than females, with higher risk of unaffected carriers having an affected male offspring compared to an affected female offspring (36% vs. 15%), but more severe phenotypic expression in females when affected [5]. In 2013, Petit et al. reported 2 cases of SHFLD, one with radial agenesis, thus expanding the phenotypic spectrum of SHFLD [7]. The first case of SHFM with a prenatal diagnosis of 17p13.3 triplication was reported in 2014 [9]. In the same year, there were two reports describing the molecular genetic basis of SHFLD in two large cohorts of patients from 13 families in France [10] and 51 families in Japan [12]. They reported the rare but possible involvement of the radius and the femur in patients with SHFLD with BHLHA9 duplication or triplication [10, 12]. These studies suggest that 17p13.3 duplication is the most common cause of SHFLD. The summary of previous studies describing the 17p13.3 duplication associated with SHFM and SHFLD are presented in Table 1.
The BHLHA9 gene in the minimal critical region of 17p13.3 in SHFLD encodes a bHLH transcription factor that is observed to regulate the embryonic limb bud development and morphogenesis in animal experiments. The importance of BHLHA9 gene in the development of limbs in humans was implied by few recent studies that described the association of homozygous missense variants in the BHLHA9 gene with the mesoaxial synostotic syndactyly with phalangeal reduction [19, 20] and with complex camptosynpolydactyly [21]. In a recent study using BHLHA9 knockout mice, Kataoka et al. reported that the BHLHA9 gene is expressed in the early embryonic stages on both the ventral and dorsal surfaces of the PZ. The BHLHA9 knockout mice showed transient aberrant expression of TP63 and FGF8 genes, indicating that the BHLHA9 gene regulates the expression of TP63 and downstream genes [22]. A previous study also using BHLHA9 knockout mice, suggested that the complete deletion of the BHLHA9 causes syndactyly due to reduced apoptosis between digital rays [23], however Kataoka et al. showed the expression of BHLHA9 occurs earlier in the embryo than the interdigital cell apoptosis stage [22]. The BHLHA9 gene is an interesting example of the gene dosage effect; a single gene giving rise to two different phenotypic effects (i.e. syndactyly and ectrodactyly) depending of the gene dosage, however the exact molecular mechanisms of pathogenesis of these conditions still remain unclear.
In the published literature, there are several reports of 17p13.3 duplication involving the BHLHA9 gene region that presented predominantly with developmental delay, intellectual disabilities, autism and occasional brain malformations. Only a small number of these patients presented with subtle hand foot malformations and the limbs were completely normal in most cases [8, 24–27]. These data along with the non-penetrance observed in SHFLD families suggest a complex genetic etiology for SHFLD with the involvement of additional genetic modifiers. Thus, increased BHLHA9 gene dosage by itself is not sufficient to produce the phenotypic effect. Moreover, the duplication breakpoints in patients with SHFLD were observed to occur closer to the BHLHA9 region, compared to patients with 17p13.3 duplication without SHFLD, but with other phenotypic manifestations such as neurodevelopmental abnormalities and facial clefts. This suggests that the disruption of adjacent putative regulatory elements (e.g. in the ABR-TUSC5 region) of the BHLHA9 gene could be an additional contributory factor in the causation of SHFLD [8, 27, 28].
The clinical and genetic heterogeneity of SHFM/SHFLD render the precise diagnosis of this condition challenging, and genetic testing is therefore essential for confirmation. Sowińska-Seidler et al. have proposed a genetic diagnostic flow-chart that would be useful in genetic diagnosis of SHFM. Considering the high frequency of chromosomal rearrangements and genomic micro-duplications among patients with SHFM, genome-wide array comparative genomic hybridization (array CGH) is often considered as the first line of genetic testing. Alternatively, targeted qPCR or multiplex ligation-dependent probe amplification (MLPA) could be used to detect copy number variations associated with SHFM3 and SHFLD. These were shown to have a diagnostic yield of 30–46% [6]. Even with the precise genetic diagnosis, which in itself is a challenge, providing genetic counseling for families with SHFM is a bigger challenging task, due to the reduced penetrance, variable expressivity and the sex bias, which make the risk assessment and the phenotypic prediction a rather daunting task.
Acknowledgements
We would like to thank the family of the proband for their co-operation with this study.
Abbreviations
- AER
Apical ectodermal ridge
- BHLHA9
Basic helix-loop-helix [BHLH] family member A9
- CGH
Comparative genomic hybridization
- MLPA
Multiplex ligation-dependent probe amplification
- PZ
Progress zone
- qPCR
Quantitative polymerase chain reaction
- SHFLD
Split hand/foot malformation with long bone deficiency
- SHFM
Split hand/foot malformation
Authors’ contributions
CSP and NDS obtained the clinical information, collected the literature data and wrote the manuscript. NDS coordinated the study, critically reviewed and edited the manuscript. FE and SM performed genetic evaluation of the proband and the mother. FP coordinated the genetic evaluation, critically reviewed and edited the manuscript. VHWD critically revised the final manuscript for important intellectual content and approved it. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
All data generated in this study are included in this published article.
Ethics approval and consent to participate
Written informed consent was obtained from the parents for the clinical assessment and testing on a consent form approved by the Ethics Review Committee, Faculty of Medicine, University of Colombo.
Consent for publication
Written informed consent was obtained from the parents for the publication of all personal information contained in this case report and accompanying images. A copy of the written consent is available for review by the Editor of this journal.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chamara Sampath Paththinige, Phone: + 94 759251072, Email: paththinige@yahoo.com.
Nirmala Dushyanthi Sirisena, Email: nirmala@anat.cmb.ac.lk.
Fabienne Escande, Email: fabienne.escande@chru-lille.fr.
Sylvie Manouvrier, Email: sylvie.manouvrier@chru-lille.fr.
Florence Petit, Email: florence.petit@chru-lille.fr.
Vajira Harshadeva Weerabaddana Dissanayake, Email: vajira@anat.cmb.ac.lk.
References
- 1.Gurrieri F, Everman DB. Clinical, genetic, and molecular aspects of split-hand/foot malformation: an update. Am J Med Genet A. 2013;161(11):2860–2872. doi: 10.1002/ajmg.a.36239. [DOI] [PubMed] [Google Scholar]
- 2.Duijf PH, van Bokhoven H, Brunner HG. Pathogenesis of split-hand/split-foot malformation. Hum Mol Genet. 2003;12(suppl 1):R51–R60. doi: 10.1093/hmg/ddg090. [DOI] [PubMed] [Google Scholar]
- 3.Lezirovitz K, Maestrelli SR, Cotrim NH, Otto PA, Pearson PL, Mingroni-Netto RC. A novel locus for split-hand/foot malformation associated with tibial hemimelia (SHFLD syndrome) maps to chromosome region 17p13. 1–17p13. 3. Hum Genet. 2008;123(6):625–631. doi: 10.1007/s00439-008-0515-7. [DOI] [PubMed] [Google Scholar]
- 4.Armour CM, Bulman DE, Jarinova O, Rogers RC, Clarkson KB, DuPont BR, et al. 17p13.3 microduplications are associated with split-hand/foot malformation and long-bone deficiency (SHFLD) Eur J Hum Genet. 2011;19(11):1144–1151. doi: 10.1038/ejhg.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klopocki E, Lohan S, Doelken SC, Stricker S, Ockeloen CW, de Aguiar RS, et al. Duplications of BHLHA9 are associated with ectrodactyly and tibia hemimelia inherited in non-Mendelian fashion. J Med Genet. 2012;49(2):119–125. doi: 10.1136/jmedgenet-2011-100409. [DOI] [PubMed] [Google Scholar]
- 6.Sowińska-Seidler A, Socha M, Jamsheer A. Split-hand/foot malformation-molecular cause and implications in genetic counseling. J Appl Genet. 2014;55(1):105–115. doi: 10.1007/s13353-013-0178-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petit F, Andrieux J, Demeer B, Collet LM, Copin H, Boudry-Labis E, et al. Split-hand/foot malformation with long-bone deficiency and BHLHA9 duplication: two cases and expansion of the phenotype to radial agenesis. Eur J Med Genet. 2013;56(2):88–92. doi: 10.1016/j.ejmg.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Curry CJ, Rosenfeld JA, Grant E, Gripp KW, Anderson C, Aylsworth AS, et al. The duplication 17p13. 3 phenotype: analysis of 21 families delineates developmental, behavioral and brain abnormalities, and rare variant phenotypes. Am J Med Genet A. 2013;161(8):1833–1852. doi: 10.1002/ajmg.a.35996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luk HM, Wong VC, Lo IF, Chan KY, Lau ET, Kan AS, et al. A prenatal case of split-hand malformation associated with 17p13. 3 triplication–a dilemma in genetic counseling. Eur J Med Genet. 2014;57(2):81–84. doi: 10.1016/j.ejmg.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Petit F, Jourdain AS, Andrieux J, Baujat G, Baumann C, Beneteau C, et al. Split hand/foot malformation with long-bone deficiency and BHLHA9 duplication: report of 13 new families. Clin Genet. 2014;85(5):464–469. doi: 10.1111/cge.12219. [DOI] [PubMed] [Google Scholar]
- 11.Al Kaissi A, Ganger R, Rötzer KM, Klaushofer K, Grill F. A child with split-hand/foot associated with tibial hemimelia (SHFLD syndrome) and thrombocytopenia maps to chromosome region 17p13. 3. Am J Med Genet A. 2014;164(9):2338–2343. doi: 10.1002/ajmg.a.36614. [DOI] [PubMed] [Google Scholar]
- 12.Nagata E, Haga N, Fujisawa Y, Fukami M, Nishimura G, Ogata T. Femoral-tibial-digital malformations in a boy with the Japanese founder triplication of BHLHA9. Am J Med Genet A. 2015;167(12):3226–3228. doi: 10.1002/ajmg.a.37290. [DOI] [PubMed] [Google Scholar]
- 13.Nagata E, Kano H, Kato F, Yamaguchi R, Nakashima S, Takayama S, et al. Japanese founder duplications/triplications involving BHLHA9 are associated with split-hand/foot malformation with or without long bone deficiency and Gollop-Wolfgang complex. Orphanet J Rare Dis. 2014;9:125. doi: 10.1186/s13023-014-0125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho IA, Park JK, Baek JC, Ha AN, Kang MY, Lee JI, et al. Split hand/foot malformation with long-bone deficiency and BHLHA9 duplication: a prenatal diagnosis report. J Genet Med. 2015;12(2):123–127. doi: 10.5734/JGM.2015.12.2.123. [DOI] [Google Scholar]
- 15.Fusco C, De Nittis P, Alfaiz AA, Pellico MT, Augello B, Malerba N, et al. New Split hand/foot malformation with long bone deficiency familial case. J Pediatr Genet. 2017;6(2):98–102. doi: 10.1055/s-0036-1588029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen Y, Si N, Liu Z, Liu F, Meng X, Zhang Y, et al. 17p13. 3 genomic rearrangement in a Chinese family with split-hand/foot malformation with long bone deficiency: report of a complicated duplication with marked variation in phenotype. Orphanet J Rare Dis. 2018;13:106. doi: 10.1186/s13023-018-0838-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. BioRxiv. 2019:531210.
- 18.Online Mendelian Inheritance in Man, OMIM®. Johns Hopkins University, Baltimore, MD. MIM Number: 612576: 10/03/2013: URL: https://omim.org/.
- 19.Malik S, Percin FE, Bornholdt D, Albrecht B, Percesepe A, Koch MC, et al. Mutations affecting the BHLHA9 DNA-binding domain cause MSSD, mesoaxial synostotic syndactyly with phalangeal reduction, Malik-Percin type. Am J Hum Genet. 2014;95(6):649–659. doi: 10.1016/j.ajhg.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan A, Wang R, Han S, Ahmad W, Zhang X. A novel homozygous missense mutation in BHLHA9 causes mesoaxial synostotic syndactyly with phalangeal reduction in a Pakistani family. Hum Genome Var. 2017;4:17054. doi: 10.1038/hgv.2017.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phadke SR, Kar A, Bhowmik AD, Dalal A. Complex camptosynpolydactyly and mesoaxial synostotic syndactyly with phalangeal reduction are allelic disorders. Am J Med Genet A. 2016;170(6):1622–1625. doi: 10.1002/ajmg.a.37643. [DOI] [PubMed] [Google Scholar]
- 22.Kataoka K, Matsushima T, Ito Y, Sato T, Yokoyama S, Asahara H. Bhlha9 regulates apical ectodermal ridge formation during limb development. J Bone Miner Metab. 2018;36(1):64–72. doi: 10.1007/s00774-017-0820-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schatz O, Langer E, Ben-Arie N. Gene dosage of the transcription factor Fingerin (bHLHA9) affects digit development and links syndactyly to ectrodactyly. Hum Mol Genet. 2014;23(20):5394–5401. doi: 10.1093/hmg/ddu257. [DOI] [PubMed] [Google Scholar]
- 24.Bi W, Sapir T, Shchelochkov OA, Zhang F, Withers MA, Hunter JV, et al. Increased LIS1 expression affects human and mouse brain development. Nat Genet. 2009;41(2):168–177. doi: 10.1038/ng.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruno DL, Anderlid BM, Lindstrand A, van Ravenswaaij-Arts C, Ganesamoorthy D, Lundin J, et al. Further molecular and clinical delineation of co-locating 17p13. 3 microdeletions and microduplications that show distinctive phenotypes. J Med Genet. 2010;47(5):299–311. doi: 10.1136/jmg.2009.069906. [DOI] [PubMed] [Google Scholar]
- 26.Capra V, Mirabelli-Badenier M, Stagnaro M, Rossi A, Tassano E, Gimelli S, et al. Identification of a rare 17p13. 3 duplication including the BHLHA9 and YWHAE genes in a family with developmental delay and behavioural problems. BMC Med Genet. 2012. 10.1186/1471-2350-13-93. [DOI] [PMC free article] [PubMed]
- 27.Ibitoye RM, Roberts J, Goodacre T, Kini U. 17p13. 3 microduplication, a potential novel genetic locus for nonsyndromic bilateral cleft lip and palate. Cleft Palate Craniofac J. 2015;52(3):359–362. doi: 10.1597/13-113. [DOI] [PubMed] [Google Scholar]
- 28.Carter TC, Sicko RJ, Kay DM, Browne ML, Romitti PA, Edmunds ZL, et al. Copy-number variants and candidate gene mutations in isolated split hand/foot malformation. J Hum Genet. 2017;62(10):877–884. doi: 10.1038/jhg.2017.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated in this study are included in this published article.