Abstract
Although we can increasingly measure transcription, chromatin, methylation, etc. at single cell resolution, most assays survey only one aspect of cellular biology. Here we describe sci-CAR, a combinatorial indexing-based co-assay that jointly profiles chromatin accessibility and mRNA in each of thousands of single cells. As a proof-of-concept, we apply sci-CAR to 4,825 cells comprising a time-series of dexamethasone treatment, as well as to 11,296 cells from the adult mouse kidney. With the resulting data, we compare the pseudotemporal dynamics of chromatin accessibility and gene expression, reconstruct the chromatin accessibility profiles of cell types defined by RNA profiles, and link cis-regulatory sites to their target genes on the basis of the covariance of chromatin accessibility and transcription across large numbers of single cells.
One Sentence Summary:
We developed and applied sci-CAR to jointly profile the epigenome and transcriptome of thousands of single cells in systems including cortisol response and whole mouse kidney.
The concurrent profiling of multiple classes of molecules, e.g. RNA and DNA, within single cells has the potential to reveal causal regulatory relationships and to enrich the utility of organism-scale single cell atlases. However, to date, nucleic acid ‘co-assays’ rely on physically isolating each cell, limiting their throughput to a few cells per study (Fig. S1A, Table S1) (1–6).
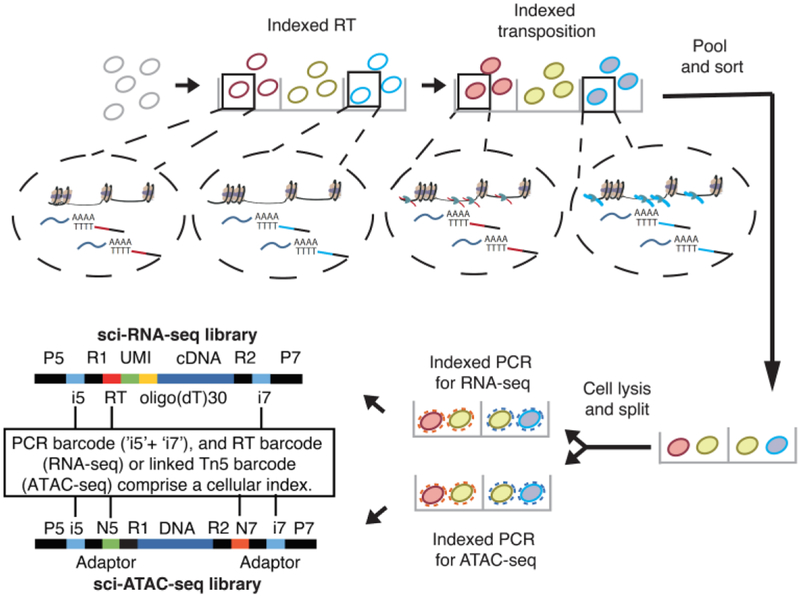
Single-cell combinatorial indexing (“sci”) methods use split-pool barcoding to uniquely label the nucleic acid contents of single cells or nuclei (7–13). Here we describe sci-CAR, which jointly profiles single cell chromatin accessibility and mRNA in a scalable fashion. Sci-CAR effectively combines sci-ATAC-seq and sci-RNA-seq into a single protocol (Fig. 1): (i) Nuclei are extracted, with or without fixation, and distributed to wells. (ii) A first RNA-seq ‘index’ is introduced by in situ reverse transcription (RT) with a poly(T) primer bearing a well-specific barcode and a unique molecular identifier (UMI). (iii) A first ATAC-seq index is introduced by in situ tagmentation with Tn5 transposase bearing a well-specific barcode. (iv) All nuclei are pooled and redistributed by FACS to multiple plates. (v) After second-strand synthesis of cDNA, nuclei in each well are lysed, and the lysate split to RNA and ATAC-dedicated portions. (vi) To provide a second priming site for amplification of 3’ cDNA tags, the RNA-dedicated lysate is subjected to transposition with unindexed Tn5 transposase. 3’ cDNA tags are amplified with primers corresponding to the Tn5 adaptor and RT primer. These primers also bear a well-specific barcode that is the second RNA-seq index. (vii) The ATAC-seq-dedicated lysate is amplified with primers specific to the barcoded Tn5 adaptors from step iii. These primers also bear a well-specific barcode that is the second ATAC-seq index. (viii) Amplicons from RNA-seq and ATAC-seq-dedicated lysates are respectively pooled and sequenced. Each sequence read is associated with two barcodes corresponding to each round of indexing. As with other sci- protocols, most nuclei pass through a unique combination of wells, receiving a unique combination of barcodes that can be used to group reads derived from the same cell. Because the barcodes introduced to RNA-seq and ATAC-seq libraries correspond to specific wells, we can link the mRNA and chromatin accessibility profiles of individual cells.
Fig. 1. sci-CAR workflow.
Key steps outlined in text. RNA-seq: index2 and read1 cover the i5 index, UMI and RT barcode; index1 and read2 cover the i7 index and cDNA fragment. ATAC-seq: read1 and read2 cover genomic DNA sequence. Index 1 and index 2 cover the Tn5 and PCR barcodes.
We applied sci-CAR to a cell culture model of cortisol response, wherein dexamethasone (DEX), a synthetic mimic of cortisol, activates glucocorticoid receptor (GR), which binds to thousands of locations across the genome, altering the expression of hundreds of genes (14–17). We collected lung adenocarcinoma-derived A549 cells after 0, 1 or 3 hrs of 100 nM DEX treatment, and performed a 96 × 576 well sci-CAR experiment. The three timepoints were each represented in 24 wells during the first round of indexing, while the remaining 24 wells contained a mixture of HEK293T (human) and NIH3T3 (mouse) cells (Fig. S1B).
We obtained sci-RNA-seq profiles for 6,093 cells (median 3,809 UMIs) and sci-ATAC-seq profiles for 6,085 cells (median 1,456 unique reads) (Fig. S1C–E). For both data types, reads assigned to the same cell overwhelmingly mapped to one species (Fig. S1F–G). We obtained roughly equivalent UMIs per cell from ‘RNA-only’ plates processed in parallel, albeit at a lower sequencing depth per cell. Aggregated transcriptomes of co-assayed vs. RNA-only plates were well-correlated (r = 0.97–0.98; Fig. S2). In contrast, although co-assayed vs. ‘ATAC-only plates’ were comparable in quality and well-correlated in aggregate (Fig. S3), ATAC-only plates had ~10-fold higher complexity. The lower efficiency of the co-assay for ATAC is likely explained by factors including buffer modifications and our use of only half the lysate.
There were 4,825 cells (70% of either set) for which we recovered both transcriptome and chromatin accessibility data. To confirm that paired profiles truly derived from the same cells, we asked whether cells from mixed human-mouse wells were consistently assigned as human or mouse. Indeed, 1,423/1,425 (99%) of co-assayed cells from those wells were assigned the same species label from both sci-RNA-seq and sci-ATAC-seq profiles (Fig. 2A).
Fig. 2. Joint profiling of chromatin accessibility and transcription in dexamethasone treated A549 cells.
(A) Scatter plot showing the proportion of human reads, out of all reads mapping uniquely to the human or mouse reference genomes, for cells in which both RNA-seq profiles and ATAC-seq profiles were obtained. Only HEK293T (human) and NIH/3T3 (mouse) cells are plotted. (B) t-SNE visualization of A549 cells (RNA-seq) including cells from both sci-CAR and sci-RNA-seq-only plates, colored by DEX treatment time (left) or unsupervised clustering id (right). (C) t-SNE visualization of A549 cells (ATAC-seq) including cells from both sci-CAR and sci-ATAC-seq-only plates, colored by DEX treatment time (left) or unsupervised clustering id (right). (D) t-SNE visualization of A549 cells (ATAC-seq) with linked RNA-seq profiles. If the cell is in cluster 1 (or cluster 2) in both RNA-seq and ATAC-seq, then it is labeled as “Match”, otherwise it is labeled “Discordant”. (E) Distribution of cells from different DEX treatment timepoints in gene expression pseudotime inferred by trajectory analysis. (F) Smoothed line plot showing scaled (with the R function scale) gene expression and promoter accessibility of CKB and ZSWIM6 across pseudotime. Unscaled, unsmoothed data shown in Fig. S5F–G. (G) Smoothed line plot showing the scaled mRNA level and activity change of transcription factors NR3C1 and KLF9 across pseudotime. Unscaled, unsmoothed data shown in Fig. S6D–E.
We next examined the time course of GR activation. DEX treatment of A549 cells increased both transcription and promoter accessibility of markers of GR activation, including NFKBIA, SCNN1A, CKB, PER1 and CDH16 (14, 16) (Fig. S4A–B). Unsupervised clustering or t-SNE visualization of either sci-RNA-seq or sci-ATAC-seq profiles readily separated clusters corresponding to untreated and DEX-treated cells (Fig. 2B–C). Reassuringly, cells from co-assay plates and single-assay plates of either type were intermixed (Figs. S4C).
88% and 93% of co-assayed cells in clusters 1 and 2 of sci-ATAC-seq data were found in corresponding sci-RNA-seq clusters (Fig. S4D–E). Cells with concordant vs. discordant assignments did not significantly differ in read depth (P-value > 0.1, Welch two-sample t-test), but notably fell on the border between clusters 1 and 2 in either t-SNE (Figs. 2D, S4F). While most discordant cells (70%) were from 0 hrs, the remainder tended to derive from 1 hrs rather than 3 hrs (5% of 1 hr vs. 1% of 3 hr cells, P-value = 2.2e-16, Fisher’s Exact Test). Although we cannot rule out that this is due to imperfect clustering, these discordantly assigned cells potentially reflect transitional states in GR activation.
Differential expression (DE) analysis of sci-RNA-seq data revealed significant changes in 2,613 genes (5% FDR) (Table S2). For comparison, a similar analysis with bulk RNA-seq data of DEX treatment in A549 cells at 0 vs. 3 hrs (18) identified 870 DE genes, 536 of which were also DE here. Log2 fold changes were well-correlated between the datasets for DE genes (r = 0.86, Fig. S4G).
Differential accessibility (DA) analysis of sci-ATAC-seq profiles identified significant changes at 4,763 sites (5% FDR) (Table S3). For comparison, a similar analysis of bulk DNase-seq data from DEX-treated A549 cells at 0 vs. 3 hrs (18) identified 672 DA sites, 544 of were also DA here. Log2 fold changes were well-correlated between the datasets for DA sites (rho = 0.68, Fig. S4H).
Of our DA sites, 701 (15%) were promoters, of which 175 overlapped with DE transcripts. Transcripts for genes with DA promoters that were not DE were detected in significantly fewer cells than genes with DA promoters that were DE (median 10% vs. 25%, P-value < 5e-5, unpaired two sample permutation test based on 20,000 simulations), suggesting we may be insufficiently powered to detect DE at many genes with DA promoters. For the 175 genes that are both DA and DE, the log2 fold changes were modestly correlated (rho = 0.63, Fig. S4I), with 130/175 (74%) exhibiting directional concordance (exact two-sided binomial test, P-value = 9e-11).
We ordered cells along a pseudotime trajectory with Monocle (19) based on the top 1,000 DE genes (Fig. S5A). Cells were ordered consistently with the time course (Fig. 2E). Of note, the aforementioned cells from 1 hrs whose cluster assignments were discordant (Figs. 2D, S4F) occurred significantly earlier in pseudotime than cells with concordant assignments (P-value = 3e-5, Wilcoxon rank sum test, Fig. S5B). Of the 2,613 DE genes, 979 (37%) increased and 1,111 (43%) decreased in expression along pseudotime, while 523 (20%) exhibited transient changes (Fig. S5C–D, Tables S2, S4). We exploited the co-assay to examine the dynamics of chromatin accessibility across RNA-defined pseudotime, identifying opening (47%), closing (32%) and transient (21%) DA sites (Fig. S5E, Tables S3, S5). There were eleven genes that showed significant changes in both gene expression and promoter accessibility along pseudotime (5% FDR for both), with well-correlated dynamics (Figs. 2F, S5F–H).
We converted the (cell x site) matrix to a (cell x transcription factor (TF) motif) matrix, simply by counting occurrences of each motif in all accessible sites for each cell (20). The motifs of 91/399 (23%) of expressed TFs were DA across the treatment conditions (5% FDR) (Tables S6–S7). Where ChIP-seq data was available for the same time course (18), we observed consistent dynamics of increasing motif-associated accessibility (Fig. S6A) and TF binding to accessible sites (Fig. S6B). Motif accessibility dynamics across expression-defined pseudotime are summarized in Fig. S6C. The motif of the canonical glucocorticoid receptor NR3C1 was the most activated, even though its expression decreased (Figs. 2G), consistent with its activation by recruitment from the cytosol rather than by increased expression. In contrast, KLF9 is a direct target of GR activation via a feed forward loop (21). Consistent with this, we observe that both its expression and its motif accessibility increase along pseudotime (Fig. 2G, Fig. S6D–E).
Single-cell RNA sequencing studies have recently characterized the transcriptomes of diverse cell types represented in the mammalian kidney (22–24). However, little is known about the epigenetic landscapes that underlie these cell type-specific gene expression programs. To investigate this, we isolated and fixed nuclei from whole kidneys of two 8-week male mice (Fig. S7A). From one sci-CAR experiment, we obtained sci-RNA-seq profiles for 13,893 nuclei (median 1,011 UMIs; Fig. S7B) and sci-ATAC-seq profiles for 13,395 nuclei (median 7,987 unique reads; Fig. S7C). There were 11,296 cells for which we recovered both transcriptome and chromatin accessibility profiles.
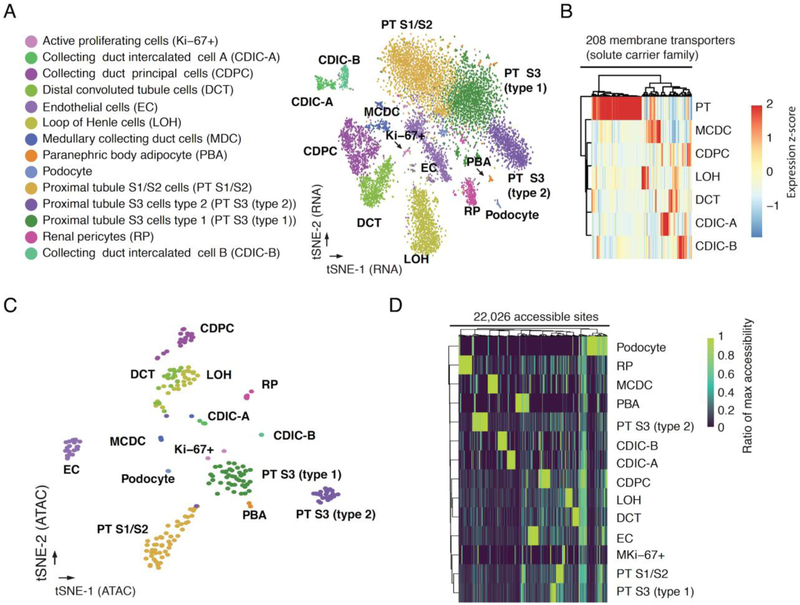
We compared sci-CAR transcriptomes with a recently published single cell RNA-seq dataset of the same tissue generated by Drop-seq (24). After correcting for gene length biases (Drop-seq is biased towards shorter transcripts, and sci-RNA-seq towards longer transcripts) aggregated transcriptomes were reasonably well correlated (r = 0.73, Fig. S7D). Semi-supervised clustering of 10,727 sci-CAR transcriptomes (>500 UMIs) identified 14 groups, ranging in size from 74 (0.7%) to 2,358 (22.0%) cells (Figs. 3A, S7E–F). Established markers identified nearly all cell types (Fig. S8A–B). The expression profiles of proximal tubule cells separate them into three subtypes including S1/S2 cells (Slc5a12+, Gatm+, Alpl+, Slc34a1+), S3 type 1 cells (Slc34a1+, Atp11a+), and S3 type 2 cells (Atp11a+, Rnf24+) (Fig. S8C) (25, 26). The smallest cluster is positive for cell cycle progression markers (Mki67 and Cenpp), and may represent an actively proliferating subpopulation (Fig. S8D) (25, 26). Cell type proportions were well-correlated between replicate kidneys, with the exception of paranephric body adipocytes (1.2% vs. 0.4%), likely due to technical variation in kidney dissection as these reside superficial to the renal fascia (Fig. S7E).
Fig. 3. sci-CAR enables joint profiling of chromatin accessibility and transcription in mouse kidney.
(A) t-SNE visualization of mouse kidney nuclei (RNA-seq). Cell types are assigned based on established marker genes. (B) Heatmap showing the relative expression of genes from the solute carrier group of membrane transport proteins in consensus transcriptomes of each cell type estimated by RNA-seq data from the co-assay. The raw expression data (UMI count matrix) was log-transformed, column centered and scaled (using the R function scale), and the resulting values clamped to [−2, 2]. (C) t-SNE visualization of mouse kidney nuclei (ATAC-seq) after aggregating cells with highly similar transcriptomes (‘pseudocells’), colored by cell types identified from RNA-seq. (D) Heatmap showing the relative chromatin accessibility of cell type-specific sites for each cell type estimated by ATAC-seq data from the co-assay. The raw aggregated ATAC-seq data (read count matrix) was normalized first by the total number of reads for each cell type then by the maximum accessibility score across all cell types.
We identified 8,774 genes that were DE across the 14 cell types (5% FDR), including 1,771 with >2-fold greater expression in the highest vs. second highest cell type (Fig. S9A–B, Tables S8, S9). New marker genes were identified, such as Daam2 for renal pericytes and Calcr for collecting duct intercalated cell B (Fig. S9C–D) (25, 26). We examined expression of solute carrier transporters (SLCs), as these correspond to a principal function of the kidney. 208/345 (60%) of these were DE in subsets of renal tubule cell types, many corresponding to known and potentially novel reabsorption specificities (Figs. 3B, S9E, Table S10).
We compared aggregated sci-CAR chromatin accessibility profiles with published bulk ATAC-seq data on adult mouse kidney (18), and found them to be reasonably well correlated (r = 0.75; Fig. S10A–B). Across all genes, aggregate promoter accessibility correlated with aggregate gene expression (rho = 0.26; Fig. S10C). Nonetheless, a significant challenge for single cell ATAC-seq data, relative to single cell RNA-seq data, is the sparsity of the resulting matrices (8). Thus, our initial efforts to cluster co-assayed cells based solely on their ATAC-seq profiles failed to discover the expected diversity of cell types. We therefore sought to leverage the co-assay aspect of these data to recover the chromatin landscapes of individual cell types.
As a first approach, we simply annotated cell types from transcriptional profiles for ~96% of the 11,296 cells that were successfully co-assayed. We then aggregated ATAC-seq signal for each cell type separately, followed by peak calling (27). As a second approach, we also developed an algorithm to combine the ATAC-seq profiles of cells with highly similar RNA-seq profiles prior to clustering (Fig. S7A). For cells from each RNA-seq-defined cell type, we identified subsets of cells with highly similar expression profiles (a mean of 50 cells assigned to each of 222 ‘pseudo-cells’). We then aggregated the ATAC-seq profiles of each pseudo-cell, and performed t-SNE on these. In contrast with single-cell ATAC-seq data, pseudo-cell chromatin accessibility profiles corresponding to the same cell types clustered together (Fig. 3C). Overall, these analyses illustrate how co-assay data can be leveraged to overcome the relative sparsity of single cell ATAC-seq data and define chromatin accessibility profiles even for closely related cell types.
We identified 22,026 DA sites across the 14 mouse kidney cell types, including 2,096 promoters and 19,930 distal sites (5% FDR; Figs. 3D, S10D–E; Tables S11, S12). In some cases, DA at a gene’s promoter was concordant with DE (Fig. S11A–B), but this was the exception rather than the rule. Out of 2,096 genes with a DA promoter in at least one cell type, 132 genes were also DE (1% FDR) with a >2-fold difference between the first and second ranked cell type. Although promoter accessibility and expression of these genes across cell types are positively correlated (median rho = 0.17), the majority (112/132 or 85%) exhibited maximal promoter accessibility and gene expression in different cell types (Fig. S11C). The relatively weaker correlation compared with what we observed in the A549 dexamethasone time series (rho = 0.63; Fig. S4I) is potentially a consequence of the fact that in the A549 cells, we were comparing changes in promoter accessibility vs. expression, whereas here we are comparing absolute enrichment of accessibility at promoters vs. expression.
We sought to link distal cis-regulatory elements to their target genes based on the covariance of chromatin accessibility and gene expression across large numbers of co-assayed cells. As the sparsity of our single cell profiles makes this challenging, we worked with the aforedescribed 222 pseudo-cells (Fig. S12A). For each gene, we computed correlations between its expression and the adjusted accessibility of all sites within 100 kilobases (kb) of its transcriptional start site (TSS) using LASSO (least absolute shrinkage and selection operator).
Within the top 2,000 DE genes (ranked by q-value), we linked 1,260 distal sites to 321 genes (median 3 sites per gene, out of median 19 sites within 100 kb of TSS tested; Fig. S12BC, Table S13). 44% of sites were linked to the nearest TSS, and 21% to the second nearest TSS (Fig. S12D). Distal site-gene linkages were significantly closer than all possible pairs tested (mean 41 kb for links vs. 48 kb for all pairs tested; P-value < 5e-5, unpaired permutation test based on 20,000 simulations; Fig. S12E).
To evaluate the possibility that the links were artifacts of regularized regression, we permuted the sample IDs of the chromatin accessibility matrix and performed the same analysis. After this permutation, only 4 links were identified (Fig. S12B). To control for correlations between closely located accessible sites in the genome, we separately permuted the peak IDs. This yielded 216 links, or just 17% as many links as without permutation (Fig. S12B).
The 321 genes with linked distal sites were specifically expressed in a variety of cell types (Fig. S12F). For example, the link with the highest correlation is between distal convoluted tubule cell marker gene Slc12a3 and a site 36 kb downstream of its TSS and overlapping its last exon (Fig. S13). The accessibility of this linked site was modestly more specific to distal convoluted tubule cells than the Slc12a3 promoter. In contrast, the accessible site closest to the Slc12a3 promoter (only 216 bp away) was not linked to the Slc12a3 promoter by our approach, nor is its accessibility specific to distal convoluted tubule cells. Similarly, a marker gene for Loop of Henle cells, Slc12a1, is linked to two distal sites (Fig. S14), both of which exhibit accessibility specific to Loop of Henle cells. In contrast, the nearest accessible site (9 kb from the TSS), which was not linked, does not exhibit this specificity.
Links between distal cis-regulatory elements and their target genes can be useful for explaining differential expression across cell types. For example, the cell type-specific expression of Slc6a18, a marker gene for type 2 proximal tubule S3 cells, is not mirrored by cell type-specific promoter accessibility (Fig. S11C). However, from our covariance approach, its TSS is linked to a site 16 kb away whose accessibility is correlated with Slc6a18 expression (Fig. 4A). To quantify the utility of the links between distal cis-regulatory elements and their target genes identified from sci-CAR data, we constructed a linear regression model to predict gene expression differences based on chromatin accessibility at promoters only vs. promoters together with linked distal sites. Including linked distal sites improved predictions by four-fold (P-value < 5e-5, paired permutation test based on 20,000 simulations; Fig. 4B).
Fig. 4. Linking cis-regulatory elements to regulated genes based on covariance in single cell co-assay data.
(A) Top: genome browser plot showing links between accessible distal regulatory sites and the gene Slc6a18. The height corresponds to the correlation coefficient. Bottom: barplots showing the average expression, promoter accessibility and linked site accessibility for cell type-specific marker gene Slc6a18 across different cell types. Gene expression values for each cell were calculated by dividing the raw UMI count by cell-specific size factors. Site accessibilities for each cell were calculated by dividing the raw read count by cell-specific size factors. Error bars represent standard errors of the means. (B) Two linear regression models were built to predict gene expression differences between cell types. The first model predicts changes on the basis of promoter accessibility alone. The second model predicts changes based on the chromatin accessibility of the promoter and distal sites that are linked to it. The boxplot shows the cross-validated r-squared calculated for each gene from the two models.
Our analyses illustrate the advantages of a single cell co-assay over assays that solely profile transcription or chromatin accessibility. Sci-CAR is compatible with fresh or fixed nuclei, and like other sci-seq techniques, can encode multiple samples per experiment. Its throughput can potentially be increased by additional rounds of split-pool indexing (13). With 384 × 384 × 384 sci-CAR, one could potentially co-assay millions of single cells per experiment. A limitation of sci-CAR is the sparsity of the resulting data, particularly with respect to chromatin accessibility. This can potentially be overcome in the future through protocol optimizations, particularly of crosslinking conditions. A second limitation is that although we were able to link distal elements and target genes on the basis of covariance of accessibility and expression, these data remain correlative and involve a minority of DE genes and DA elements.
Notwithstanding these limitations, sci-CAR expands the potential of combinatorial indexing for scalably profiling single cell molecular phenotypes, and may be particularly useful in the context of organism-scale single cell atlases. With further development, we anticipate that additional DNA/RNA co-assays may be realized by simply integrating other sci-seq protocols together with sci-RNA-seq (e.g. methylation + transcripts; chromosome conformation + transcripts; DNA sequence + transcripts) (8–13). A longer-term goal is to adapt single cell combinatorial indexing to span the Central Dogma, such that aspects of DNA, RNA and protein species can be concurrently assayed from each of many single cells.
Supplementary Material
Acknowledgements:
We thank members of the Shendure and Trapnell labs for helpful discussions and feedback, particularly B. Martin, X. Qiu, A. Leith, A. Minkina, Y. Yin, Z. Duan and R. Qiu; as well as R. Hunter, and R. Rualo in the Transgenic Resources Program of University of Washington for their exceptional assistance.
Funding: This work was funded by the Paul G. Allen Frontiers Foundation (Allen Discovery Center grant to JS and CT), grants from the NIH (DP1HG007811 and R01HG006283 to JS; DP2 HD088158 to CT; R35GM124704 to AA), the W. M. Keck Foundation (to CT and JS), the Dale. F. Frey Award for Breakthrough Scientists (to CT), the Alfred P. Sloan Foundation Research Fellowship (to CT), and the Brotman Baty Institute for Precision Medicine. DAC was supported in part by T32HL007828 from the National Heart, Lung, and Blood Institute. JS is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Competing interests: L.C. and F.J.S. declare competing financial interests in the form of stock ownership and paid employment by Illumina, Inc. One or more embodiments of one or more patents and patent applications filed by Illumina may encompass the methods, reagents, and data disclosed in this manuscript.
Data and materials availability: Processed and raw data can be downloaded from NCBI GEO (GSE117089). All methods for making the transposase complexes are described in (7); however, Illumina will provide transposase complexes in response to reasonable requests from the scientific community subject to a material transfer agreement.
Author contributions: J.S. and C.T. designed and supervised the research; J.C. developed technique and performed experiments with assistance from D.C., V.R., R.D., J.M., L.C., F.S. and A.A.; J.C. performed computation analysis with assistance from D.C., V.R., D.A., H.P., A.H. and J.P.; J.S., C.T. and J.C. wrote the paper.
REFERENCES
- 1.Clark SJ et al. , scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun 9, 781 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angermueller C et al. , Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat. Methods 13, 229–232 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hou Y et al. , Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 26, 304–319 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu Y et al. , Simultaneous profiling of transcriptome and DNA methylome from a single cell. Genome Biol. 17, 88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pott S, Simultaneous measurement of chromatin accessibility, DNA methylation, and nucleosome phasing in single cells. Elife. 6 (2017), doi: 10.7554/eLife.23203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo F et al. , Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Res. 27, 967–988 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amini S et al. , Haplotype-resolved whole-genome sequencing by contiguity-preserving transposition and combinatorial indexing. Nat. Genet 46, 1343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cusanovich DA et al. , Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science. 348, 910–914 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramani V et al. , Massively multiplex single-cell Hi-C. Nat. Methods 14, 263–266 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin Y et al. , High-throughput mapping of meiotic crossover and chromosome mis-segregation events in interspecific hybrid mice. bioRxiv (2018), p. 338053. [Google Scholar]
- 11.Vitak SA et al. , Sequencing thousands of single-cell genomes with combinatorial indexing. Nat. Methods 14, 302–308 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulqueen RM et al. , Highly scalable generation of DNA methylation profiles in single cells. Nat. Biotechnol 36, 428–431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao J et al. , Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 357, 661–667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reddy TE et al. , Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 19, 2163–2171 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.John S et al. , Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet 43, 264–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddy TE, Gertz J, Crawford GE, Garabedian MJ, Myers RM, The Hypersensitive Glucocorticoid Response Specifically Regulates Period 1 and Expression of Circadian Genes. Mol. Cell. Biol 32, 3756–3767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vockley CM et al. , Direct GR Binding Sites Potentiate Clusters of TF Binding across the Human Genome. Cell. 166, 1269–1281.e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.ENCODE Project Consortium, An integrated encyclopedia of DNA elements in the human genome. Nature. 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu X et al. , Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buenrostro JD et al. , Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 523, 486–490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chinenov Y, Coppo M, Gupte R, Sacta MA, Rogatsky I, Glucocorticoid receptor coordinates transcription factor-dominated regulatory network in macrophages. BMC Genomics. 15, 656 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L et al. , Transcriptomes of major renal collecting duct cell types in mouse identified by single-cell RNA-seq. Proc. Natl. Acad. Sci. U. S. A 114, E9989–E9998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han X et al. , Mapping the Mouse Cell Atlas by Microwell-Seq. Cell. 172, 1091–1107.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Park J et al. , Comprehensive single cell RNAseq analysis of the kidney reveals novel cell types and unexpected cell plasticity (2017), , doi: 10.1101/203125. [DOI] [Google Scholar]
- 25.Human Protein Atlas, (available at www.proteinatlas.org).
- 26.Uhlen M et al. , A pathology atlas of the human cancer transcriptome. Science. 357 (2017), doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y et al. , Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ, Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cusanovich DA et al. , The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature. 555, 538–542 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobin A et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H et al. , The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pliner HA et al. , Cicero Predicts cis-Regulatory DNA Interactions from Single-Cell Chromatin Accessibility Data. Mol. Cell (2018), doi: 10.1016/j.molcel.2018.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quinlan AR, Hall IM, BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S, Pyl PT, Huber W, HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gentleman R, Carey V, Huber W and Hahne F, genefilter: genefilter: methods for filtering genes from high-throughput experiments (2017). [Google Scholar]
- 36.Grant CE, Bailey TL, Noble WS, FIMO: scanning for occurrences of a given motif. Bioinformatics. 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weirauch MT et al. , Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 158, 1431–1443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolf FA, Angerer P, Theis FJ, SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedman J, Hastie T, Tibshirani R, Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw 33 (2010), doi: 10.18637/jss.v033.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cenpp. The Human Protein Atlas, (available at https://www.proteinatlas.org/ENSG00000188312-CENPP/tissue/kidney#img).
- 42.Pdgfrb. The Human Protein Atlas, (available at https://www.proteinatlas.org/ENSG00000113721-PDGFRB/tissue/kidney#img).
- 43.Slc5a12. The Human Protein Atlas, (available at https://www.proteinatlas.org/ENSG00000148942-SLC5A12/tissue/kidney#img).
- 44.GATM. The Human Protein Atlas, (available at https://www.proteinatlas.org/ENSG00000171766-GATM/tissue/kidney#img).
- 45.Alpl. The Human Protein Atlas, (available at https://www.proteinatlas.org/ENSG00000162551-ALPL/tissue/kidney#img).
- 46.Slc34a1. The Human Protein Atlas, (available at https://www.proteinatlas.org/ENSG00000131183-SLC34A1/tissue/kidney#img).
- 47.Atp11a. The Human Protein Atlas, (available at https://www.proteinatlas.org/ENSG00000068650-ATP11A/tissue/kidney#img).
- 48.Rnf24. The Human Protein Atlas, (available at https://www.proteinatlas.org/ENSG00000101236-RNF24/tissue/kidney#img).
- 49.Mki67. The Human Protein Atlas, (available at https://www.proteinatlas.org/ENSG00000148773-MKI67/tissue/kidney#img).
- 50.Kit. The Human Protein Atlas, (available at https://www.proteinatlas.org/ENSG00000157404-KIT/tissue/kidney#img).
- 51.Daam2. The Human Protein Atlas, (available at https://www.proteinatlas.org/ENSG00000146122-DAAM2/tissue/kidney#img).
- 52.Calcr. The Human Protein Atlas, (available at https://www.proteinatlas.org/ENSG00000004948-CALCR/tissue/kidney#img).
- 53.Zhou X, Wang T, in Current Protocols in Bioinformatics (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.