Abstract
Chromosome structure in both interphase and M phase cells is strongly influenced by the action of the Cohesin and Condensin protein complexes. The Cohesin complex tethers the identical copies of each chromosome, called sister chromatids, together following DNA replication, and promotes normal interphase chromosome structure and gene expression. In contrast, Condensin is active largely in M phase and promotes the compaction of individual chromosomes. The Xenopus egg extract system provides a uniquely suitable system with which to analyze the functions of both Cohesin and Condensin. Egg extracts, in which the cell cycle state can be manipulated, contain stockpiles of nuclear proteins, including Condensin and Cohesin, sufficient for the assembly of thousands of nuclei per microliter. Egg extract prepared from unfertilized eggs is arrested by the presence of cytostatic factor (CSF) in a state with high levels of M-phase kinase activity, but can be stimulated to enter interphase, in which DNA replication occurs spontaneously.
For cohesion assays, demembranated sperm nuclei are incubated in interphase extract, where they undergo rapid and synchronous DNA replication and cohesion establishment through the recruitment of proteins and other factors (e.g. nucleotides) from the extract. Sister chromatid cohesion is assessed by then driving the extract into M phase by the addition of fresh CSF-arrested extract.
Chromosome condensation occurs spontaneously in M phase extract extracts. Sperm nuclei are therefore added directly to CSF extracts to assay condensation. In the following protocols, we describe basic assays for Cohesin and Condensin function using Xenopus egg extracts (see Figure 1 for schematic overview).
Materials
Reagents
All water for solution preparation is ultrapure (~18.2 MM·cm at 25 °C).
Adenosine triphosphate, disodium salt (ATP) (200 mM in H2O).
Adjust to pH7 with 10M NaOH and store in 10 uL aliquots at −80°C.
Antibody dilution buffer (AbDil) <R>
Antibodies to Xenopus chromosomal proteins of interest
Antibodies to Condensin, topoisomerase II, and CENP-A are ideal.
Biotin-14-dATP (optional) (Invitrogen #19524016)
Chromosome dilution buffer <R>
Chromosome fix solution <R>
Can be stored frozen in aliquots without formaldehyde, adding formaldehyde just prior to use.
CaCl2 (30 mM)
Prepare fresh from 1M stock solution.
Creatine phosphate, also called phosphocreatine (1 M in 10 mM KHPO4 buffer, pH 7)
Prepared from disodium salt, tetrahydrate. Store working stock at −20°C, remainder at −80°C.
Creatine phosphokinase: type 1, from rabbit muscle (5 mg/mL in 10 mM HEPES, pH 7.5, 50 mM NaCl, and 50% glycerol) (Sigma C-3755)
Store in 200μL aliquots at −20C.
Cushion buffer (1xMMR, 40% glycerol)
DAPI (4’,6-diamidino-2-phenylindole dihydrochloride, 1 μg/mL in H2O)
Energy Mix (EM), 35x <R>
Fluorescein-12-dUTP (optional) Invitrogen #C7604
Formaldehyde (~37% commercial stock, ACS grade, VWR International)
Frozen CSF (cytostatic factor)-arrested Xenopus laevis egg extract
Extract can be stored at −80°C in 100 μL aliquots (Gillespie et al. 2012). Alternatively, freshly prepared CSF may also be used. See protocol from Good and Heald in this volume.
Sperm nuclei (stock at >108/mL) (see Hazel and Gatlin, this volume)
Store in 5–10 μL aliquots at −80°C (Tutter and Walter 2006).
MMR = Marc’s Modified Ringers <R>
Mounting medium containing anti-fade reagent <R>
Nail polish to seal coverglass to slide
Quick Fix solution <R>
TBS = Tris-buffered saline <R>
TBS-TX (TBS with 0.1% Triton-X100)
Equipment
Acrylic blocks: 0.5” diameter x ~0.5” long cylinders. Ours were cut from a single acrylic rod (McMaster-Carr # 8528K32) and the ends were polished with fine sandpaper.
Centrifuge equipped to spin Kimax tubes at 6000 rpm (~6600G) in a swinging bucket rotor
We use the Beckman Avanti J25 with either the JS13.1 or JS 7.5 rotor, each equipped with rubber adapters to accommodate the Kimax tubes.
Cover glass: 12 mm round and 18 mm square, both #1 thickness.
Epi-fluorescence microscope for monitoring nuclear assembly.
Preferred configuration would include a 40X phase contrast air objective lens
Moist chamber for immunostaining of samples
We use 15 cm disposable cell culture dishes lines with Parafilm (Bemis NA), covered and placed inside of a glass dish lined with wet paper towels and covered with clear plastic wrap. This is further covered with aluminum foil when using fluorescent reagents.
Spin-down tubes: made from Glass Kimble Kimax-type glass tubes, 15 mL, thick walled (45500–15) with cylindrical acrylic blocks permanently secured in base with ~1 mL Sylgard 184 (two-part silicone elastomer, Dow Corning).
Each set-up also requires an additional loose block, to serve as a chock, on which the cover glass will rest. When the tube is inverted, the chock slides out bringing with it the cover glass. For diagram, see protocols by Wang et al., and Servin and Straight, in this volume).
Water bath at 18–21°C.
Method
Assay for sister chromatid cohesion
- Thaw extract in fingertips and place on ice. Supplement with 1/35th volume EM. Mix by flicking.
- We prefer to work in volumes of 40–100 μL for each sample. Smaller volumes can be affected by evaporative loss, and larger volumes can suffer from inadequate mixing.
- Add nuclei to the desired final concentration. Mix by gently pipetting the mixture up and down 2–3 times with a standard 200 μL pipette tip. This is your nuclear assembly mixture.
- To ensure complete and timely replication keep the concentration of nuclei below 3000/μL. Optional: extracts can be supplemented with 1μM fluorescein-dUTP or biotinylated dATP (both Invitrogen), to identify replicated chromatids during subsequent imaging.
- Stimulate CSF release by adding 1/50th volume of 30 mM CaCl2. Mix by flicking and immediately place in water bath at 18–21°C and start timer. After 45 minutes check nuclear morphology to confirm that the extract is in interphase. To do this, place 3 μL of Quick Fix on a glass slide and add 0.5 μl of the nuclear assembly mixture to the middle of the drop of fix. Cover with 18 mM square coverslip, and observe by epifluorescence microscopy.
- By 45 minutes the nuclei should appear round, and a well-developed nuclear envelope will be evident by phase contrast microscopy. If the extract failed to release from CSF then there will be no nuclear envelope, and the mass of DNA will have a bumpy and uneven edge, indicative of chromosome condensation.
- Allow replication to proceed for 120 minutes.
- At this stage nuclei can become quite large, and sometimes the DNA will have a reticular staining pattern. This is OK.
- Add one volume of CSF extract, freshly thawed immediately before use, supplemented with 1/35th volume energy mix, and warmed to 19°C. Wait an additional 90–120 minutes. Monitor nuclear morphology for entry into M phase again using Quick Fix (as in step 3 above).
- CSF activity is dominant and will drive the interphase extract into M phase, causing condensation of the chromosomes. This will be evident by loss of the nuclear envelope, chromosome condensation, and collapse of chromosomes into discrete clumps.
- During the above incubation, prepare spin-down tubes: Place one loose chock in each spin-down tube, add 4 mL of cushion buffer, and add one clean 12 mm round coverslip.
- Use a Pasteur pipette to push down the coverslip so that it is resting on the loose chock, under the cushion buffer.
- Add one volume of the chromosome assembly mix to 4 volumes of CDil (18–21°C). Wait 15–20 minutes. Fix by the addition of 20 volumes of Chromosome Fix.
- The equivalent of 5 μL of chromosome assembly mix per coverglass provides ample material for morphological analysis.
- After 5 minutes, layer the fixed mixtures onto the cushions in spin-down tubes (see #8 above) and spin at 6000 rpm (6600 g) for 20 minutes.
- The samples can be split to accommodate different downstream analyses. For example, if immunostaining with different antibody combinations is desired, then use a separate spin-down tube for each, dividing samples accordingly.
Aspirate and discard the top third of the liquid from the tube. This will remove diluted extract and minimize background in subsequent immunolocalization experiments.
Pour off and discard the cushion, then invert the tubes to an approximate 45° angle and tap on a clean paper towel until the loose chock begins to slide towards the top of the tube. Collect chock in a gloved hand (avoiding the coverslip!) and use forceps to grab the coverslip and place face up on clean Parafilm. Cover with a generous amount of AbDil (~200 μL). Repeat for all coverslips.
After 3 × 5 minute washes with AbDil, stain with primary and secondary antibodies, and counterstain with DAPI as desired, using standard immunofluorescence procedures. Wash in TBS, and mount the coverslips by inversion on a small (~2 μL) drop of mounting medium using fine tip forceps, let settle for 10 minutes, aspirate liquid from around the coverslip, and seal with nail polish. Store at −20°C and image within 2 weeks.
Analyze cohesion by measuring the distance between sister chromatids. This is most easily done when the chromosome cores are stained with antibodies against Condensin subunits or topoisomerase II.
Troubleshooting
Incomplete DNA replication can occur in poor extracts, and can give an appearance similar to aberrant chromosome cohesion and result in tangled chromatids. Use only excellent CSF extracts prepared from high quality eggs.
Always mix extracts gently, avoiding excess shear force by pipetting slowly, with the pipette tip well clear of the tube bottom.
Spindle-associated forces may enhance the ability to detect defects in sister chromatid cohesion, so we find it best not to add microtubule poisons to the extracts.
Assay for chromosome condensation
To assay condensation, sperm nuclei are added directly to M-phase extract. Following incubation, during which chromatin assembly and chromosome condensation occur, the chromosomes are collected on coverslips for analysis.
Method
-
13.
Thaw CSF extract in fingertips and place on ice. Do not warm above 23°C. Supplement with 1/35th volume EM. Mix by flicking.
-
14.
Add sperm nuclei to a final concentration of <3000 nuclei/μL. Mix by gently pipetting up and down with a standard 200 μL pipette tip.
-
15.Place reaction in water bath at 19–21°C for 90–120 minutes.
- Entry into M phase can be monitored during this time by placing 0.5μL aliquots of reaction mix to 3 μL of Quick Fix as above (step 3). As chromosomes condense and individualize in M phase extract the mass of DNA will have a bumpy and uneven edge and there will be no nuclear envelope.
-
16.During the above incubation, prepare spin-down tubes: Place one loose chock in each spin-down tube, add 4 mL of cushion buffer, and add one clean 12mm round coverslip.
- Use a Pasteur pipette to push down the coverslip so that it is resting on the loose chock, under the cushion buffer.
-
17.
Add one volume of the chromosome assembly mix to 4 volumes of CDil (room temperature). Wait 15–20 minutes. Fix by the addition of 20 volumes of Chromosome Fix. Wait 5 minutes.
-
18.
Layer the samples onto glycerol cushions, spin, and process coverslips for immunofluorescence labeling of chromosomes, as described for cohesion assays above (step 13).
-
19.
The general level of condensation can be determined by measuring the length and width of chromosomes, as well as the loading of condensin proteins onto chromatin. The impact of DNA replication and cohesion establishment on subsequent chromosome condensation can be assessed by analysis of chromosomes that have been “cycled” through interphase extract, as in the cohesion assay protocol (above).
Discussion
The Xenopus egg extract system provides a powerful approach for the in vitro analysis of cohesion and condensation mechanisms. Importantly, the synchrony and rapid cell cycles of the Xenopus embryonic extracts allow us to probe the function of essential conserved proteins in chromosome dynamics, something that is inherently difficult in somatic cells. The assays described here can be used to test the effects of recombinant or mutant proteins on chromosome cohesion and condensation, and to perform depletion-and-rescue experiments. For depletion experiments, freshly thawed CSF extract is incubated with antibody-coated beads prior to CSF release with CaCl2 and the above procedures are followed (see accompanying protocol by Jenness et al. in this volume for depletion protocol). In addition to protein depletion and addition, the extracts can also be treated with pharmacological agents to manipulate DNA replication or enzyme activity, allowing analysis of their impacts on chromosome condensation and cohesion.
The protocols described here represent a synthesis from contributions made by numerous investigators over many years to the study of DNA replication and chromosome dynamics in Xenopus egg extracts. Critical contributions were made by John Newport, Andrew Murray, Tatsuya Hirano, Johannes Walter, and members of their labs, among many others (Hirano et al. 1997; Walter et al. 1998; Losada et al. 1998; Funabiki and Murray 2000; Takahashi et al. 2004). A recent cohesion assay protocol by Shintomi and Hirano is similar to that described here, and should prove useful to others particularly interested in cohesion assays (Shintomi and Hirano 2017). For our lab, the protocol for preparation of frozen CSF-arrested extract as described by Gillespie and Blow (Gillespie et al. 2012) provided a critical technical breakthrough, as it allows us to routinely control cell cycle transitions without the requirement for freshly-prepared extracts, which can be maddeningly inconsistent.
RECIPES
Antibody dilution buffer (AbDil)
| Reagent | Amount to add | Starting concentration |
Final concentration (1X) |
| Tris-HCl, pH 7.4 | 10 mL | 1 M | 20 mM |
| NaCl | 15 mL | 5 M | 150 mM |
| Triton X-100 | 2.5 mL | 20% | 0.1 % |
| BSA | 10 g | powder | 2 % |
| NaN3 | 2.5 mL | 20 % | 0.1 % |
Dissolve and mix the above ingredients in 450 mL of H2O, bring up to 500 mL, filter through 0.2 μm filter, and store at 4°C.
Chromosome dilution buffer (CDil): (Funabiki and Murray 2000)
| Reagent | Amount to add | Starting concentration |
Final concentration (1X) |
| K-HEPES, pH 7.6 | 1 mL | 1 M | 10 mM |
| KCl | 10 mL | 2 M | 200 mM |
| MgCl2 | 0.1 mL | 1 M | 0.5 mM |
| K-EGTA | 0.2 mL | 0.5 M | 0.5 mM |
| Sucrose | 8.57 g | solid | 250 mM |
To prepare 100 mL of CDil, mix and dissolve the reagents listed above in 80 mL of H2O, bring to 100 mL, and store in 10 mL aliquots at −20.
Chromosome fix solution (Funabiki and Murray 2000)
| Reagent | Amount to add | Starting concentration |
Final concentration (1X) |
| MMR | 2 mL | 25 x | 1 x |
| Triton X-100 | 1.25 mL | 20 % | 0.5 % |
| Glycerol | 10 mL | 100 % | 20 % |
| Formaldehyde | 3.65 mL | 37 % | 2.7 % |
Prepare fresh Chromosome fix by mixing the above ingredients and bringing the volume to 50 mL with H2O.
Energy Mix (EM) 35x:
| Reagent | Amount to add | Starting concentration |
Final concentration (35X) |
| ATP | 10 μL | 200 mM | ~67 mM |
| Creatine phosphate | 20 μL | 1 M | ~667 mM |
| Creatine phosphokinase | 0.8 μL | 5 mg/mL | 0.13 mg/mL |
Make fresh 35x EM by mixing the stock solutions as indicated. Keep on ice and discard unused portion at the end of the experiment.
Marc’s Modified Ringers (MMR)
| Reagent |
Amount to add |
Starting concentration |
Final concentration (25X) |
Final concentration (1x) |
| HEPES free acid | 119.2 g | solid | 125 mM | 5 mM |
| NaCl | 584.5 g | solid | 2.5 M | 100 mM |
| KCl | 14.91 g | solid | 50 mM | 2 mM |
| MgCl2 | 20.33 g | solid | 25 mM | 1mM |
| CaCl2 | 29.4 g | solid | 50 mM | 2 mM |
| EDTA, pH8.0 | 20 mL | 0.5 M | 2.5 mM | 0.1 mM |
To prepare a 25x stock, dissolve the above ingredients in 3.8 liters of H2O and adjust with 10 M NaOH to pH 7.8. Bring the volume to 4 L and store at room temperature.
Mounting medium (with anti-fade reagent)
| Reagent |
Amount to add |
Starting concentration | Final concentration |
| Tris-HCl, pH 8.8 | 0.8 mL | 1 M | 20 mM |
| p-phenylenediamine (free base) | 0.20 g | powder | 0.5 % |
| Glycerol | 36 mL | 100 % | 90 % |
Mix the above ingredients together in a 50 ml conical tube and bring to 40 mL with H2O. Dissolve solids by slowly bubbling nitrogen gas through a Pasteur pipette inserted deep into the mixture. Filter thorough a 0.8 μm filter at room temperature (~20°C), and store at −20°C in 1 or 3 mL syringes devoid of air bubbles. Discard when dark brown. NB. This reagent should be handled with gloves and causes stains when spilled.
Quick Fix solution
| Reagent |
Amount to add |
Starting concentration | Final concentration |
| MMR | 80 μL | 25 x | 1 x |
| DAPI | 2 μL | 1 mg/mL | 1 μg/mL |
| Glycerol | 1.2 mL | 100 % | 60 % |
| Formaldehyde | 0.59 mL | 37 % | 11 % |
| H2O | 128 uL |
To make 2 mL of Quick Fix, mix the above ingredients together. Store in 0.5 mL aliquots at −20°C for up to a year.
Tris-buffered saline (TBS)
| Reagent |
Amount to add |
Starting concentration | Final concentration |
| Tris-HCl, pH 7.4 | 20 mL | 1 M | 20 mM |
| NaCl | 30 mL | 5 M | 150 mM |
Mix the above ingredients together and bring to 1 L with H2O.
Figure 1. Schematic illustration of protocols for cohesion and condensation assays.
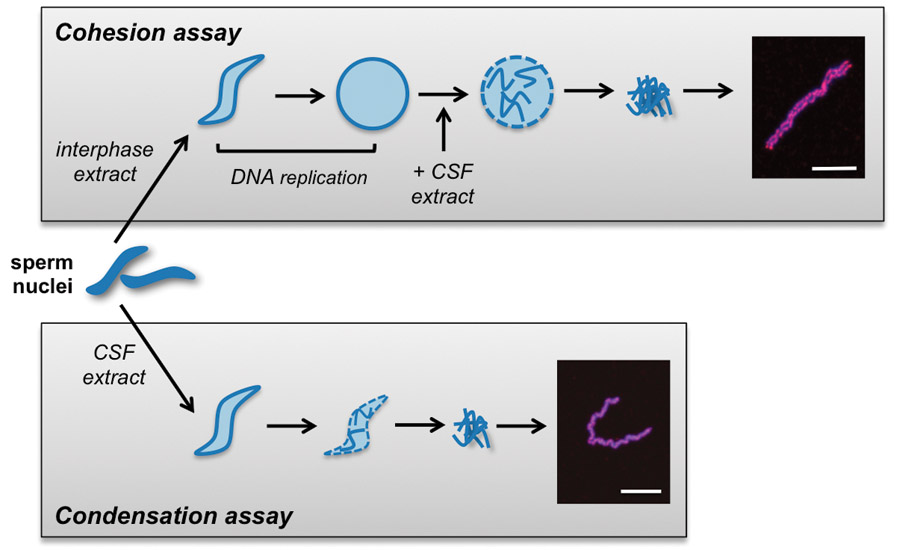
For cohesion assays, in which the tethering together of sister chromatids is analyzed, nuclei are added to interphase extracts to allow DNA replication to occur (top). After DNA replication, the extracts are driven into M phase by the addition of CSF-arrested extract. The resulting condensed chromosomes are collected and analyzed for sister chromatid cohesion by measuring the distance between sister chromatids. In the chromosome condensation assay, demembranated sperm are added directly to M phase-arrested CSF extract, where the haploid genome spontaneously condenses into individual chromatids. In both cases chromosome cores can be identified by immunolocalization of the CapG subunit of Condensin I (red) and the DNA is counterstained with DAPI (blue). Scale bars = 5 μm.
References
- Funabiki H, Murray AW. 2000. The Xenopus Chromokinesin Xkid Is Essential for Metaphase Chromosome Alignment and Must Be Degraded to Allow Anaphase Chromosome Movement. Cell 102: 411–424. [DOI] [PubMed] [Google Scholar]
- Gillespie PJ, Gambus A, Blow JJ. 2012. Preparation and use of Xenopus egg extracts to study DNA replication and chromatin associated proteins. Methods 57: 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Kobayashi R, Hirano M. 1997. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 89: 511–521. [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. 1998. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev 12: 1986–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintomi K, Hirano T. 2017. A Sister Chromatid Cohesion Assay Using Xenopus Egg Extracts. Methods Mol Biol 1515: 3–21. [DOI] [PubMed] [Google Scholar]
- Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. 2004. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol 6: 991–996. [DOI] [PubMed] [Google Scholar]
- Tutter AV, Walter JC. 2006. Chromosomal DNA replication in a soluble cell-free system derived from Xenopus eggs. Methods Mol Biol 322: 121–137. [DOI] [PubMed] [Google Scholar]
- Walter J, Sun L, Newport J. 1998. Regulated chromosomal DNA replication in the absence of a nucleus. Mol Cell 1: 519–529. [DOI] [PubMed] [Google Scholar]


