SUMMARY
Quiescence is a non-proliferative cellular state that is critical to tissue repair and regeneration. Although often described as the “G0 phase”, quiescence is not a single homogeneous state. As cells remain quiescent for longer durations, they move progressively “deeper” and display a reduced sensitivity to growth signals. Deep quiescent cells, unlike senescent cells, can still re-enter the cell cycle under physiological conditions. Mechanisms controlling quiescence depth are poorly understood, representing a currently underappreciated layer of complexity in growth control. Here we show that the activation threshold of a Retinoblastoma (Rb)-E2F network switch controls quiescence depth. Particularly, deeper quiescent cells feature a higher E2F switching threshold and exhibit a delayed traverse through the restriction point (R-point). We further show that different components of the Rb-E2F network can be experimentally perturbed, following computer model predictions, to coarse or fine tune the E2F switching threshold and drive cells into varying quiescence depths.
INTRODUCTION
Cellular quiescence is a “sleep-like” non-proliferative state associated with many cell types in the body (Cheung and Rando, 2013; Coller et al., 2006). It protects cells against stress and toxicity, which is especially important for long-lived cells such as adult stem cells (Cheung and Rando, 2013). Quiescence can revert to proliferation upon physiological growth signals such as serum stimulation; it is thus different from other non-proliferative states such as senescence and terminal differentiation, which are irreversibly arrested under physiological conditions. Inducing quiescent cells to re-enter the cell cycle is fundamental to tissue homeostasis and repair.
Quiescence is often described as the G0 phase; however, it is not a single uniform state. Studies in the ‘70s and ‘80s showed that when fibroblasts and lymphocytes were kept longer under contact inhibition – a quiescence-inducing signal – cells moved progressively “deeper” into quiescence. Deep quiescent cells remained viable and metabolically active (Lemons et al., 2010; Soprano, 1994), but they underwent a longer pre-DNA-synthesis phase when re-entering the cell cycle (Augenlicht and Baserga, 1974; Brooks et al., 1984; Owen et al., 1989; Yanez and O’Farrell, 1989). Deep vs. shallow quiescence is also observed in vivo. In the liver after partial hepatectomy and in the salivary gland after isoproterenol stimulation, cells in older rats took a longer time to initiate DNA synthesis, behaving like deep quiescent cells (Adelman et al., 1972; Bucher, 1963). In contrast, in muscle and neural stem cells, GAlert and “primed” quiescent states were identified respectively, in which cells displayed an accelerated entry into the cell cycle upon stimulation, behaving like shallow quiescent cells (Llorens-Bobadilla et al., 2015; Rodgers et al., 2014). Abnormally deep or shallow quiescent states could conceivably lead to hypo- or hyper-proliferative diseases. Yet, little is known about what controls the depth of quiescence.
Deep quiescence was found correlated with reduced total RNA and protein content, increased chromatin condensation, and decreased transcription activities (Augenlicht and Baserga, 1974; Rossini et al., 1976). “Primed” quiescence and GAlert state, in contrast, were found associated with the upregulation of protein synthesis and mTORC1 activity (Llorens Bobadilla et al., 2015; Rodgers et al., 2014). Deep quiescent cells also exhibit greater expression changes of a transcriptional “quiescence program” than shallow quiescent cells (Coller et al., 2006), suggesting a possible mechanism regulating quiescence depth. Yet, it remains to be determined what gene activities in the quiescence program regulate quiescence depth, apart from those that are affected by quiescence-depth changes.
In this study we sought to develop a mechanistic model to explain and manipulate quiescence depth by focusing on a bistable Rb-E2F network switch (Yao et al., 2008; Yao et al.,2011). The pivotal role of the Rb-E2F pathway in cell proliferation has been well-documented (Attwooll et al., 2004; DeGregori and Johnson, 2006; Frolov and Dyson, 2004; Nevins, 2001; Trimarchi and Lees, 2002). Activator E2Fs (E2F1–3a, E2F for short) are among the few genes that are both necessary and sufficient for quiescence exit: when E2F is knocked out, quiescent cells cannot re-enter the cell cycle (Wu et al., 2001); on the other hand, ectopic E2F expression alone can induce quiescent cells to enter S-phase (Johnson et al., 1993). Previously, we showed that the Rb-E2F pathway network functions as a bistable switch, converting graded and transient serum growth signals into an all-or-none E2F activity, which directly correlates with the all-or-none outcome of quiescence exit (Aguda, 2015; Yao et al., 2008). Here, by coupling modeling and experiments, we show that the minimum serum concentration required to activate the RbE2F bistable switch (“E2F switching threshold” for short) defines quiescence depth. Deeper quiescent cells have a higher E2F switching threshold, require stronger growth stimulation to reenter the cell cycle, and exhibit a delayed commitment to quiescence exit. The E2F switching threshold model provides an integrated framework for both understanding and manipulating quiescence depth. Accordingly, we show that depending on their varying roles in modulating the E2F switching threshold, different components in the Rb-E2F network can be perturbed following model guidance, to serve as coarse- and fine-tuning mechanisms to experimentally create quiescent states with varying depths in the cell.
RESULTS
Characterize cellular features associated with varying quiescence depth
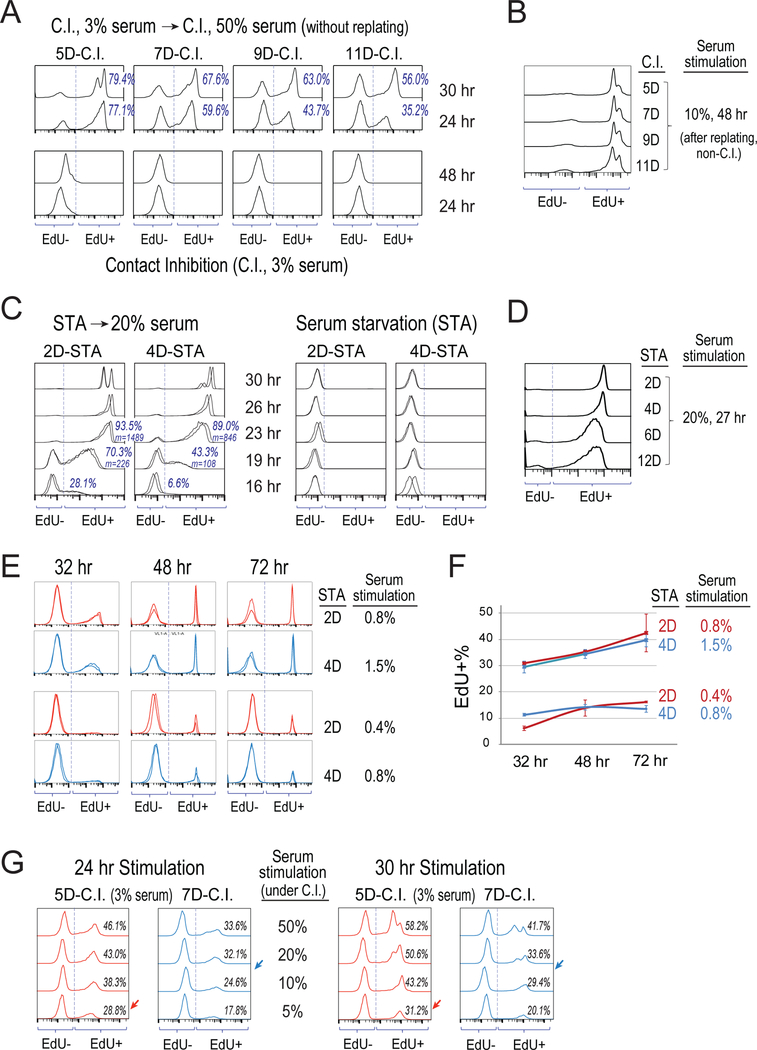
To better understand cellular properties associated with different quiescence depths, we examined the speed and growth-stimulation requirement for cells to exit deep vs. shallow quiescence. First, consistent with earlier findings (Augenlicht and Baserga, 1974; Owen et al., 1989), we observed in our cell model rat embryonic fibroblasts (REF/E23 cells) that under longer-term contact inhibition (C.I.), cells moved into deeper quiescence. As seen in Figure 1A, observed at the indicated time points following serum stimulation, a smaller percentage of cells that were previously under longer-term C.I. (e.g., 11 vs. 5 days, 11D- vs. 5D-C.I. for short) initiated DNA synthesis and thus had positive incorporation of 5-ethynyl-2’-deoxyuridine (EdU). The smaller EdU+% of 11D- vs. 5D-C.I. cells was not due to a subset of cells becoming irreversibly arrested or senescent (as these cells can all proliferate after replated at a non-C.I. condition, Figure 1B); it thus indicated that cells under longer C.I. (11D vs. 5D) were slower to enter S-phase upon stimulation. Consistently, while the majority of 5D-C.I. cells completed S-phase (reaching the maximum EdU intensity) by the 24th hour upon stimulation, 11D-C.I. cells continued to progress through S-phase between the 24th and 30th hour (indicated by the increasing EdU intensity). By the 30th hour, in comparison, many 5D-C.I. cells had already finished division (indicated by the appearance of the 2nd EdU+ peak on the left, Figure 1A).
Figure 1. Measure dynamic features of deep vs. shallow quiescence.
(A-G) S-phase entry following serum stimulation of quiescent cells. Cells were induced to quiescence by contact inhibition (seeded at 3× normal confluency in 3% serum) for 5–11 days (A,B,G) or serum starvation (cultured in 0.02% serum) for 2–12 days (C-F). At time 0, cells were stimulated with higher concentrations of serum as indicated or remained non-stimulated (C.I. control in A, and STA control in C), and with EdU added to the culture medium. Cells were harvested at indicated time points afterwards and measured for their incorporated EdU intensity by flow cytometry. Each histogram represents the distribution of EdU intensity (x-axis) from approximately 10,000 cells, with y-axis = cell number with the height of the (higher) mode normalized to 100%. For serum stimulation of C.I. cells, cells were either stimulated directly while remained at the C.I. condition (A,G) or first replated at a non-C.I. condition prior to serum stimulation (B), as indicated. (A,C) Percentage of EdU+ cells are shown; m = mean EdU intensity of EdU+ cells. Results from duplicate EdU assays are shown in C for each condition. (E,F) Cell division was restricted by low dose of nocodazole at the time of assay (see Methods). Percentages (mean ± s.e.m.) of EdU+ cells calculated in E (duplicates) are shown in F.
Similarly, we observed that deeper quiescence can also be induced by longer-term serum starvation (STA). As seen in Figure 1C, when observed at various time points after serum stimulation of REF/E23 cells previously under STA for 2 or 4 days (2D- or 4D-STA), the EdU+% and mean EdU intensity (m) were both smaller in the 4D-STA cells than the 2D-STA cells, suggesting a delayed entry of S-phase by cells under longer STA. Consistently, the time at which the majority of cells completed S-phase (reaching the maximum EdU intensity) was delayed in 4D-STA cells compared to that in 2D-STA cells (26 hr vs. 23 hr, Figure 1C). This delayed S-phase entry from deep quiescence in REF/E23 cells is also consistent with the previous observation in hematopoietic cells (Lum et al., 2005), where cells under longer-term STA entered S-phase later than those under shorter-term STA (but otherwise had similar rate of S-phase progression). Although being slow in re-entering S-phase, deep quiescent cells under long-term STA (e.g., 12D) are fully reversible and become EdU+ following sufficient serum stimulation (Figure 1D), demonstrating that these cells were not senescent or dead.
Next, we found that deep quiescent cells required stronger growth stimulation to re-enter the cell cycle and initiate DNA synthesis compared to shallow quiescent cells. To drive a similar percentage of cells to exit quiescence (EdU+), serum concentrations required for 4D-STA deep quiescent cells were roughly twice what were required for 2D-STA shallow quiescent cells (Figures 1E and 1F). Correspondingly, upon stimulation with serum at the same concentration (0.8%), fewer 4D-STA cells re-entered the cell cycle as compared to 2D-STA cells (<15% vs. >40%, 72 hr, Figures 1E and 1F). Similarly, to drive cells out of deeper quiescence under C.I., stronger serum stimulation was also required. For example, to drive a similar percentage of 7D- and 5D-C.I. cells to re-enter the cell cycle (EdU+), 10–20% and 5% serum were needed, respectively (arrow pointed, Figure 1G). Consistently, given the same serum stimulation (e.g., 5%), fewer 7D-C.I. cells re-entered the cell cycle as compared to 5D-C.I. cells (Figure 1G).
Put together, we hereby describe quiescence as a heterogeneous cellular state with different depths (Figure 2A). Under either condition, STA and C.I., a) cells move progressively into deeper quiescence under longer-term treatment; b) exiting deeper quiescence requires stronger growth stimulation and longer time; and c) given the same growth stimulation at a non-saturating level, fewer deep quiescent cells exit quiescence than shallow ones.
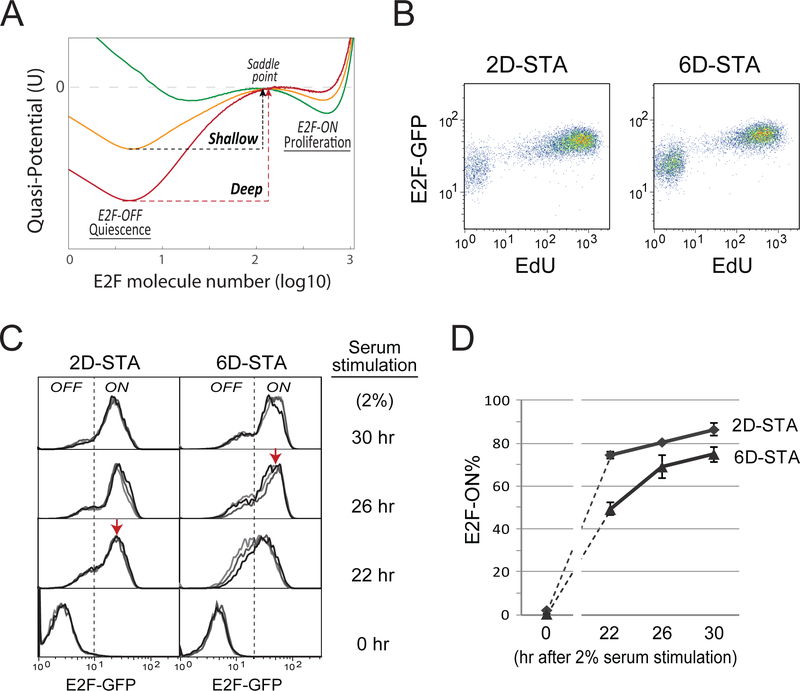
Figure 2. Rb-E2F bistable switch underlies quiescence depth.
(A) E2F quasi-potential landscape and quiescence depth. For each landscape curve, potential values (y-axis) corresponding to given E2F molecule numbers (x-axis) were calculated based on stochastic simulations of the Rb-E2F bistable switch model (see Methods). The three quasipotential curves correspond to three different kI parameter values (kI = 0.15, 0.17, and 0.21 for green, orange, and red curves, respectively; serum concentration = 0.65). The E2F-OFF and E2F-ON states correspond to the E2F molecule numbers at the left and right potential troughs in each curve. Potential barrier for E2F activation ΔP = psaddle− pE2F− (psaddle and PE2F−OFF: the potential peak and trough corresponding to the saddle point and E2F-OFF state, respectively). Deep and shallow quiescent states have relatively higher and lower ΔP. (B) Single-cell correlation between the OFF/ON state of the Rb-E2F switch and cell quiescence/proliferation. REF/E23 cells containing a stably integrated E2F-GFP reporter were serum starved for 2 or 6 days and subsequently stimulated with 20% serum. EdU was added to culture medium at the start of serum stimulation and cells were harvested for EdU assay 26 hours later. Each dot shows the E2F-GFP reporter activity (y-axis) and the incorporated EdU level (x-axis) of a single cell. To recover the GFP signal quenched by the Click-iT EdU reaction, the E2F-GFP activity was measured indirectly using a fluorescein-conjugated GFP antibody (see Methods). (C,D) E2Factivity time courses. (C) Quiescent cells obtained by serum starvation for 2 or 6 days were stimulated with 2% serum. Cells were harvested at indicated time points after serum stimulation and measured for E2F-GFP reporter levels (triplicates). Each histogram represents the E2F-GFP distribution from approximately 10,000 cells (y-axis, the same as Figure 1). Dotted vertical lines indicate the separation between E2F-OFF and E2F-ON cells, which slightly shifted to the right in 6D- vs. 2D-STA cells due to increased auto fluorescence in cells under longer-term serum starvation. Red arrows indicate the fully-ON level of the E2F-GFP reporter. (D) Percentage (mean ± s.e.m.) of E2F-ON cells, calculated from corresponding E2F distributions in C. The dash lines between 0 and 22 hr data points are for the guide of eyes.
Activation threshold of a bistable Rb-E2F network switch underlies quiescence depth
Given the quiescence-depth associated features characterized above, we next probed corresponding control mechanisms. Our earlier work has suggested that the activation of a bistable Rb-E2F network switch marks quiescence exit (Yao et al., 2008; Yao et al., 2011). The E2F-OFF and -ON states correspond to quiescence and proliferation, respectively (Figure 2A). Computationally, deeper quiescence is associated with an increased potential barrier to E2F activation and cell cycle entry and a decreased potential barrier for the reverse transition into quiescence, as shown in the quasi-potential landscape constructed based on stochastic model analysis (Figure 2A) (Ao, 2004; Balazsi et al., 2011; Wang et al., 2006; Xing, 2010; Zhou et al., 2012). We next tested experimentally whether the E2F switching threshold can serve as a metric for quiescence depth. If this is true, understanding what controls the E2F switching threshold could help us reveal what controls quiescence depth.
Indeed, we found that the E2F switching threshold was directly correlated with quiescence depth. Experimentally, we measured the E2F-ON/OFF status using a previously established E2F-GFP system (with an E2F1 promoter-driven destabilized GFP reporter stably integrated into the REF/E23 cell genome, see Methods). First, at the single-cell level, we found that switching E2F from the OFF to ON state was correlated with the exit from both deep and shallow quiescence (6D- and 2D-STA cells, respectively, Figure 2B). Second, consistent with exhibiting delayed DNA replication, deep quiescent cells exhibited delayed E2F activation. As seen in Figure 2C, following the same growth stimulation (2% serum), while shallow quiescent cells (2D-STA) activated E2F to its fully-ON level (indicated by a red arrow) by the 22nd hour, deep quiescent cells (6D-STA) took longer (by the 26th hour) to do so. Third, under the same growth stimulation (2% serum), a smaller percentage of deep vs. shallow quiescent cells was able to switch ON E2F (~73% of 6D-STA cells vs. ~87% of 2D-STA cells, the 30th hour, Figure 2D), consistent with the notion that deep quiescent cells have a higher E2F switching threshold.
Model the control elements of E2F switching threshold and quiescence depth
Given that Rb-E2F bistable switch is an emergent systems property at the network level, we next examined network components that affect its switching threshold. Several cellular factors, including cyclin-dependant kinase (Cdk) 2, Cdk6, and Cdk inhibitor p21, have been found to play critical roles in the cell fate decision between quiescence and proliferation (Laurenti et al., 2015; Overton et al., 2014; Spencer et al., 2013). Consistently, these factors are components of the Rb-E2F network switch and affect its switching dynamics (Yao, 2014; Zhang, 2013).
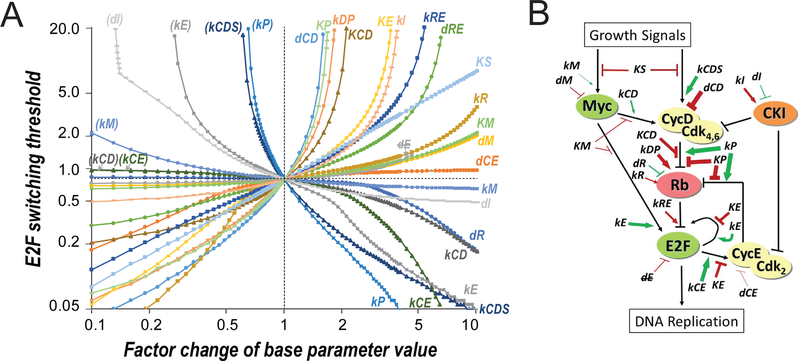
In a previous study, we constructed a simple mathematical model for the Rb-E2F bistable switch (Yao et al., 2008). Here, we first extended this model to include Cdk inhibitors, whose effects were previously lumped inexplicitly into Cdk activities. Based on this extended model (Tables S1 and S2), we simulated the changes of E2F switching threshold resulting from the changes of model parameter values. To this end, for each given parameter change, simulations were run with constant serum inputs ranging from 0 to 20 (%), and the smallest serum input that resulted in an ultrasensitive switch of the steady-state E2F level from OFF to ON (from <0.001 to >0.1 nM, see Methods for details) was considered the E2F switching threshold. We found that 21 out of 26 model parameters – kM, kE, kCD, kCDS, kCE, kR, kI, kDP, kRE, kP, KP, KS, KE, KM, KCD, dM, dCD, dCE, dR, dRE and dI - were “sensitive” in that their variations (increase and/or decrease by a factor of up to 10) resulted in significant modulation of the E2F switching threshold (by a factor of more than 2.5 over the E2F switching threshold in the base model = 0.78, Figure 3A). The steeper the slope of a parameter-sensitivity curve, the larger the modulation of the E2F switching threshold (y-axis, Figure 3A) with a given factor change of the parameter value (x-axis). That is, parameters with steep slopes function as strong “coarse tuning” modulators of the E2F switching threshold and quiescence depth, compared to parameters with low slopes (weak “fine tuning” modulators). The relationship between the 21 sensitive parameters and the Rb-E2F network components (nodes or links) are shown in Figure 3B, together with the relative modulation strength of each parameter (modulator). The 21 modulators can be divided into two groups. Group I contained 13 parameters (labelled in the upper right quadrant, Figure 3A), whose values when increased and decreased caused a higher and lower E2F switching threshold (and thus deep and shallow quiescence), respectively. Group II contained 8 parameters (labelled in the lower right quadrant, Figure 3A), whose values when increased and decreased caused a lower and higher E2F switching threshold (and shallow and deep quiescence), respectively. Furthermore, parameter changes that caused a higher E2F switching threshold (top half, Figure 3A) all resulted in delayed E2F activation upon serum stimulation (Figures S1A and S1B), consistent with the delayed S-phase entry of deep quiescent cells. In summary, our model simulations suggest that many cellular factors can modulate quiescence depth with different efficacies, which are related to different roles of these factors in affecting the E2F switching threshold (as shown in several examples below).
Figure 3. In silico modulators of the E2F switching threshold.
(A) Parameter-sensitivity curves. X-axis = factor change of parameter value. Y-axis = E2F switching threshold (% serum). Both axes are in logarithmic scale. Parameters in parenthesis = repeated parameter labels of those in the lower right quadrant. dE indicates that dE was not considered as a sensitive parameter as increasing its value diminished the separation of E2F-ON and -OFF states (to < 10% of that in the base model) before affecting the E2F switching threshold significantly (i.e., with a factor change > 2.5). Parameter changes resulting in a higher E2F switching threshold (top half) also caused delayed E2F activation upon serum stimulation (see Figures S1A and S1B). (B) Modulators of the E2F switching threshold. Modulators (sensitive parameters) are labelled at their effective positions in the Rb-E2F pathway network. Green and red = increasing the parameter value decreased and increased the E2F switching threshold, respectively. Arrow and bar = increasing the parameter value positively and negatively affected the strength of the pointed node/link, respectively. Thickness of arrow/bar = parameter sensitivity when increasing the parameter value, as determined in the right half of A.
Deep quiescent cells exhibit delayed traverse through the R-point
The modeling result that many cellular factors can affect quiescence depth is consistent with earlier reports that a large transcriptional program is associated with quiescence (Coller et al., 2006; Liu et al., 2007). In a parallel study, we are working to reverse engineer the endogenous quiescence regulatory network by statistical modeling of gene expression changes as cells move into deep quiescence (Fujimaki and Yao, unpublished). Separate from that, in this study we focused on investigating whether the E2F switching threshold model can help us better understand quiescence depth, and particularly, guide us to experimentally manipulate it.
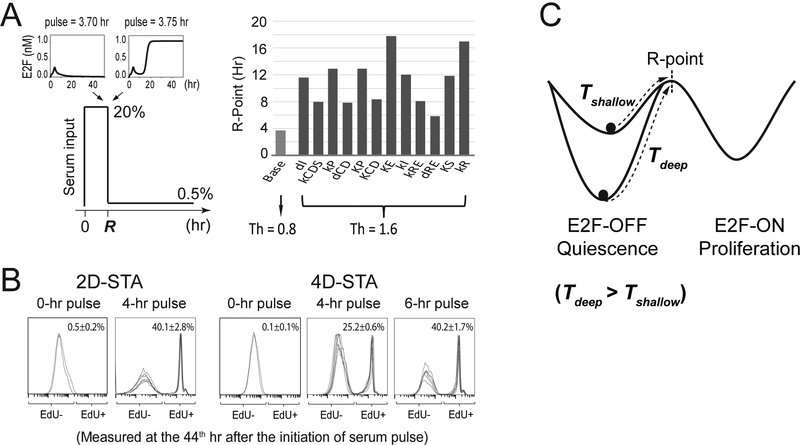
As shown in our earlier work (Yao et al., 2008), once the Rb-E2F bistable switch is activated following a serum pulse, the cell commits to cell cycle entry and passes the R-point; after committing, the cell cycle continues even when serum is removed. In our model simulations, all parameter changes that increased the E2F switching threshold (i.e., into deeper quiescence) increased the minimum serum-pulse duration required for a cell to turn ON the Rb-E2F bistable switch (i.e., the R-point, Figure 4A). This result held true when we considered cell-to-cell variations using more realistic stochastic simulations (Table S3). To experimentally compare the serum-duration requirement for exiting deep vs. shallow quiescence, we applied serum-pulse stimulations where serum starved quiescent cells were treated at a high serum level (20%) for different durations and then switched to a basal serum level (0.3%). The cells that eventually entered the cell cycle following a short serum pulse were identified by positive EdU incorporation. Consistent with the model prediction, we found that a longer serum pulse (6 vs. 4 hours) was required to drive a similar percentage (~40%) of 4D-STA deep vs. 2D-STA shallow quiescent cells to enter the cell cycle (EdU+ by the 44th hour, Figure 4B). Given the same serum pulse (4 hours), a smaller percentage of 4D-STA vs. 2D-STA cells (25.2% vs. 40.1%) were able to enter the cell cycle (Figure 4B). These results suggest that in addition to the previously observed delay in S-phase entry, deep quiescent cells exhibit delayed cell cycle commitment at the R-point upon growth stimulation compared to shallow quiescent cells (Figure 4C).
Figure 4. Deep quiescent cells exhibit delayed passing of the R-point.
(A) Simulated traverse time of the R-point. (Left) Model simulation scheme to determine the time to traverse the R-point (R), which corresponds to the shortest duration of a given serum stimulation (20%) required to activate and sustain the ON-state of the Rb-E2F bistable switch. When the serum duration is shorter than R (upper left inset), the final E2F level will return to the OFF state after the serum input is reduced to a basal level (0.5%); otherwise, the final E2F level will reach and remain at the ON state, even after the serum input is reduced to the same basal level (upper right inset). (Right) Simulated R traverse time (according to the scheme on the left) in the base model (E2F switching threshold Th = 0.8) and with parameter “mutations” that doubled the E2F switching threshold (Th = 1.6). The order of parameters is the same as shown in the top half of Figure 3A. Parameter mutations resulting in Rb-E2F deactivation threshold > 0.5% are not shown. See also Figures S1C and S1D (for deterministic simulation) and Table S3 (for stochastic simulation). (B) Experimentally measured quiescence exit after short serum pulses. Quiescent cells obtained by serum starvation for 2 or 4 days were stimulated with strong serum pulses (20%, at indicated durations), followed by incubation at a basal serum level (0.3%). Cells were harvested at the 44th hour after serum stimulation and measured for EdU incorporation (six replicates in 4- or 6-hr pulse groups and duplicates in 0-hr pulse control groups). Each histogram represents the distribution of EdU intensity from approximately 10,000 cells (y-axis, the same as Figure 1). Cell division was restricted by low dose of nocodazole at the time of assay (see Methods). (C) Time to traverse the R-point is dependent on quiescence depth. Following a given serum stimulation, the time to reach the R-point from deep quiescence (Tdeep) is longer than that from shallow quiescence (Tshallow).
Create deep quiescence experimentally by increasing E2F switching threshold
The ability to control quiescence depth has important implications, e.g., to drive stem cells into deep or shallow reservoirs with desired response rates to signals, or to potentially correct abnormal quiescent states in diseased cells. Therefore, here we asked whether we can experimentally manipulate the depth of quiescent cells by altering the E2F switching threshold. As a proof of principle, we first tested Rb family proteins (Rb for short) and Cdk inhibitors (CKI), which are expressed at high levels in quiescent cells (Cheng et al., 2000; Smith et al., 1996). Rb inhibit E2F; such inhibition is removed when Rb is phosphorylated by cyclin D (CycD)/Cdk4,6 and cyclin E (CycE)/Cdk2, which are in turn repressed by CKI (Figure 3B). In our model, increasing either kR or kI (synthesis rate constants of Rb and CKI, respectively) increased the E2F switching threshold (Figure 3A). Interestingly, increasing kI exhibited a stronger effect than increasing kR in this regard (steeper slope of the kI vs. kR curve, Figure 3A), consistent with that the ratio of unphosphorylated Rb over free form of E2F is more sensitive to enzymatic modifications (by Cyclin/Cdk) than de novo Rb synthesis (Figure S1E). When Cdk is sufficiently active, Rb even with increased synthesis will still be mostly phosphorylated over time, causing only minor changes to the E2F switching threshold. In comparison, an increased level of CKI blocks Cdk activity, leading to inefficient Rb phosphorylation and thus a noticeably higher E2F switching threshold. Consistently, the top parameters showing the highest slope steepness in Figure 3A (i.e., the strongest “coarse tuners”, kCDS, kP, dCD, KP, kDP, and KCD) all directly affect the effective Cyclin/Cdk activities and Rb phosphorylation status.
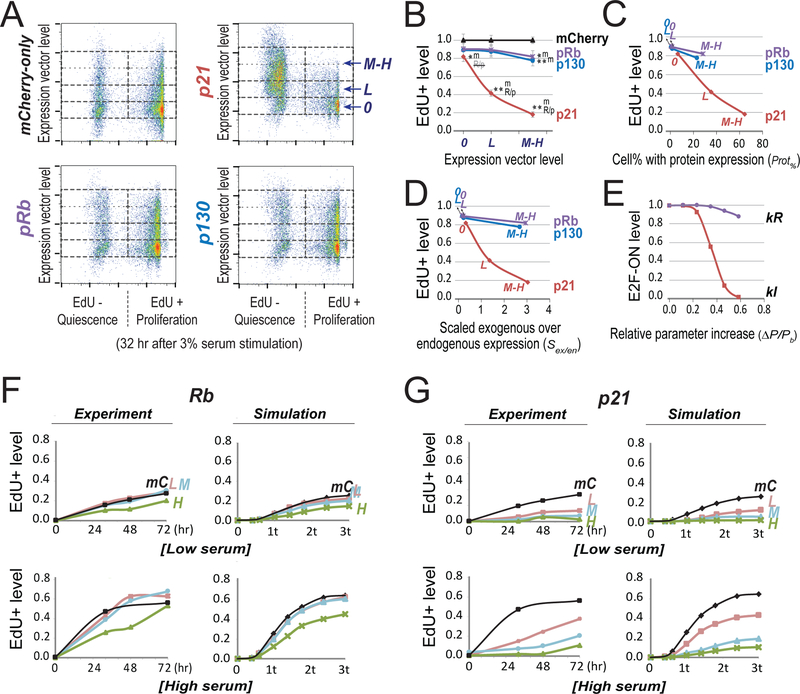
Our experimental results confirmed the model prediction that cells can be driven into deeper quiescence by higher levels of CKI and Rb. Taking advantage of our previously established dose-response-mapping approach (Wong et al., 2011a; Wong et al., 2011b), we examined the changes of quiescence depth resulting from the ectopic expression of p21, a CKI that inhibits both CycD/Cdk4,6 and CycE/Cdk2 (Xiong et al., 1993), and p130 and pRb, two Rb family proteins that play critical roles at quiescence (Smith et al., 1996). Briefly, we transfected REF/E23 cells with expression vectors of p21, pRb, and p130. Transfected cells were brought to quiescence by serum starvation for 2 days. We next stimulated cells with serum (3% BGS), and measured the percentages of cells that were able to exit quiescence (EdU+) under the influence of ectopic p21, pRb, and p130. The introduced expression vector levels (y-axis, Figure 5A) were indicated by the fluorescence intensity associated with a co-transfected mCherry vector in individual cells (as the amounts of two co-transfected vectors were linearly correlated, Figure S2). We found that with a medium-high level of introduced expression vectors (“M-H”, Figures 5A and 5B), p21, pRb, and p130 all led to deeper quiescence as indicated by reduced EdU+ levels upon serum stimulation compared to that of the mCherry-only control.
Figure 5. Experimentally create deep quiescence by increasing E2F switching threshold with Cdk inhibitor p21 and Rb family proteins.
(A) Quiescence exit affected by ectopic expression of p21, pRb and p130. Quiescent cells (2D-STA) containing transfected expression vectors were switched to media containing 3% BGS and EdU, and harvested 32 hours later for EdU incorporation assay. Cell division was restricted by low dose of nocodazole at the time of assay (see Methods). Y-axis = levels of the introduced expression vector in individual cells (indicated by the fluorescence intensity associated with the co-transfected mCherry vector; see also Figure S2). 0, L, and M-H = cell bins of non-transfected, with low and medium-high level of introduced expression vector, respectively. X-axis = EdU-incorporation intensity. (B) Quiescence-exit (EdU+) cell proportion (y-axis) as a function of expression vector level (x-axis). The EdU+ proportion was calculated from six replicate experiments as in A (with the average EdU+% at each expression vector level normalized to that of the mCherry-only control, see Table S4). Single and double star signs (* and **) indicate statistical significance (p < 0.01 and p < 0.001, respectively) in one-sided t-test comparing the data point by the star sign and the text-indicated data point (m, mCherry; R/p, pRb/p130). R/p, the difference from R/p is not statistically significant. Error bar, standard error of the mean. (C,D) Introduced expression vector levels (0, L, M-H) were converted to cell percentages with positive ectopic protein expression (C) and estimated exogenous protein levels (normalized by endogenous expression, D), respectively, as measured by immunoflow cytometry (see Figure S3 for detail). High ectopic p21 expression in quiescence did not cause irreversible arrest (see Figure S4). (E) Simulated quiescence exit affected by parameter changes. X-axis = relative parameter increase (Δ P/Pb = (P-Pb)/Pb, with P = parameter value and Pb = base value). Y-axis = proportion of cells that were able to turn ON the Rb-E2F switch given a parameter change, calculated from 2,000 stochastic simulations. The cell proportion corresponding to the base parameter was normalized to 1.0. Serum input = 1.2 au (au = activation unit; 1 au = 0.78, the E2F switching threshold in the base model). kR and kI, synthesis rate constants of Rb and p21, respectively. (F,G) Time course of quiescence-exit profiles. (Experiment) The EdU+ cell proportions with exogenously expressed pRb (F) or p21 (G) were measured at indicated time points upon serum stimulation of quiescent cells (2D-STA). Low and high serum = 0.8% and 3.0%, respectively. Cell division was restricted by low dose of nocodazole at the time of assay (see Methods). L, M, H: the same as in A; mC = mCherry-only control. (Simulation) E2F-ON cell proportions were calculated at indicated time points (t = model-time unit of 50 hours) upon serum input (low and high serum = 0.96 and 1.01 au, respectively). Each data point reflects the result of 2,000 stochastic simulations. L, M, H for Rb (F): kR = 0.182, 0.184, 0.19, respectively. L, M, H for p21 (G): kI = 0.158, 0.167, 0.171, respectively. mC: kR = 0.18, kI = 0.15.
Furthermore, p21 caused a greater reduction in the EdU+ level than pRb and p130 did with the same level of transfected expression vectors (M-H and L/low, respectively, Figures 5A and 5B). This result was further confirmed when ectopic expression was quantified at the protein level using immunoflow cytometry (Figures S3), and normalized to either the percentage of cells with positive protein expression (Figure 5C) or the mean protein level (Figure 5D, as a ratio over endogenous expression). The experimental observations that p21 was more effective than pRb and p130 in driving cells into deep quiescence were consistent with the stochastic simulation results based on the E2F switching threshold model (Figure 5E), as well as over a separately measured time course (Figures 5F and 5G). Finally, we note that ectopic p21 expression under serum starvation drove cells into deep quiescence but not senescence, as those cells could still proliferate upon strong serum stimulation (Figure S4). Together, these results suggest that as the model predicted, cellular factors such as p21 and Rb can serve as coarse- and fine-tuning factors, respectively, to increase the E2F switching threshold and quiescence depth.
Create shallow quiescence experimentally by reducing E2F switching threshold
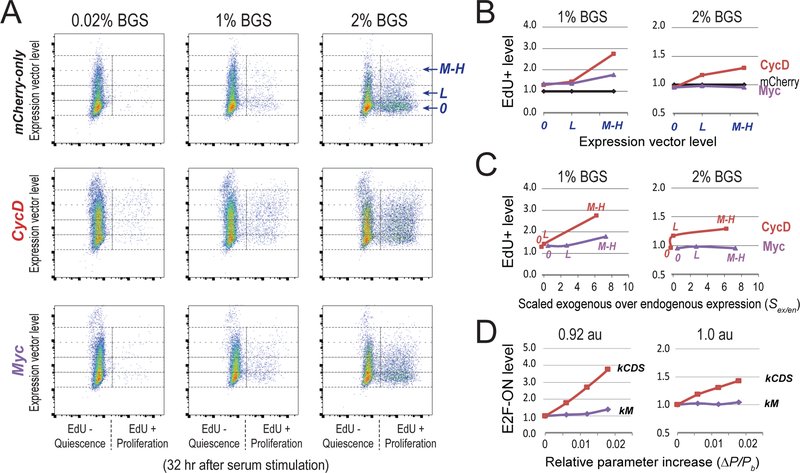
We next asked whether we can drive cells to shallow quiescence by altering the E2F switching threshold. In our model, increasing the synthesis rate constants of CycD and Myc (kCDS and kM, respectively) reduced the E2F switching threshold with high and low efficacies, respectively (steeper slope of the kCDS vs. kM curve, Figure 3A). Experimentally, we tested the influence of CycD and Myc on quiescence exit. We found that as the model predicted, the ectopic expression of CycD and Myc both led to shallow quiescence, and that CycD exhibited a stronger effect than Myc in this regard. As seen in Figures 6A and 6B, upon stimulation with 1% serum, both Myc-and CycD-expressing cells had higher EdU+ levels compared to that of mCherry-only control cells, and this effect was more significant with ectopic expression of CycD than Myc (given the same level of introduced expression vectors). When the serum concentration was increased to 2% (and more cells exited quiescence in the mCherry-only cells, Figure 6A), the weak boosting effect of Myc was buried but CycD was still able to increase the EdU+ level significantly over that of the mCherry control (Figures 6A and 6B).
Figure 6. Experimentally create shallow quiescence by decreasing E2F switching threshold with CycD and Myc.
(A) Quiescence exit affected by ectopic expression of CycD and Myc. Quiescent cells (2D-STA) containing transfected expression vectors were switched to fresh media containing serum at the indicated concentrations and EdU, and harvested 24 hours later for EdU incorporation assay. Y-axis = levels of the introduced expression vector in individual cells (as in Figure 5A). 0, L, and M-H = cell bins of non-transfected, with low and medium-high level of introduced expression vector, respectively. X-axis = EdU-incorporation intensity. (B) Quiescence-exit (EdU+) cell proportion (y-axis) as a function of expression vector level (x-axis). The EdU+ proportion was calculated from A for each expression vector level, and normalized to that of the mCherry-only control. (C) Expression vector levels in B were converted to estimated exogenous protein levels (normalized by endogenous expression) as measured by immunoflow cytometry (see Figure S5 for detail). Labels of 0, L, M-H in each curve correspond to the levels of introduced expression vectors. (D) Model simulated quiescence exit affected by parameter changes. X-axis = relative parameter increase, Y-axis = simulated proportion of cells that were able to turn ON the Rb-E2F switch given a parameter change, as in Figure 5E. The cell proportion corresponding to the base parameter was normalized to 1.0. Serum input = 0.92 and 1.0 au (for 1% and 2% BGS), respectively. kCDS and kM, synthesis rate constants of CycD and Myc, respectively.
The greater effect of CycD vs. Myc in driving quiescent cells to a shallow state was further confirmed when ectopic expression was quantified at the protein level using immunoflow cytometry (Figures S5), and normalized to either the percentage of cells with positive protein expression (Figures S5D and S5G) or the mean protein level (Figure 6C, as a ratio over endogenous expression). This experimental observation was also qualitatively consistent with the simulation results based on the E2F switching threshold model (Figure 6D). This greater efficacy of CycD vs. Myc in creating shallow quiescence is consistent with the findings that Myc alone (without cooperating with E2F, which is inactive at quiescence) is ineffective to activate E2F transcription (Leung et al., 2008) and that CycD counteracts p21 activity which is highly efficient in deepening quiescence (Figure 5). Together, our results suggest that cells can be driven to shallower quiescence by higher expression of CycD and Myc, which serve as coarse- and fine-tuning factors, respectively, to reduce the E2F switching threshold and quiescence depth.
DISCUSSION
The results of this study, particularly the strong model-experiment agreements in both driving cells into deep and shallow quiescence, suggest that our coarse-grained E2F switching threshold model represents an important mechanism controlling quiescence depth. In this regard, our work suggests that the effective Cyclin/Cdk activities and correspondingly the Rb phosphorylation status strongly affect the E2F switching threshold and quiescence depth, as shown in both our modeling analysis (Figure 3A, the top modulators) and experimental tests (Figures 5 and 6, coarse tuners p21 and CycD). These results are consistent with recent findings that Cdk and CKI activities play critical roles in determining the quiescence entry of individual cycling cells (e.g., Cdk2, p21) as well as the heterogeneous cell cycle entry of quiescent cells (e.g., Cdk6, p21) (Laurenti et al., 2015; Overton et al., 2014; Spencer et al., 2013).
As a critical cellular state, quiescence is regulated by complex mechanisms beyond RbE2F, involving other pathways such as Notch-Hes1, p53, autophagy, microRNAs, DNA damage response, and metabolic and stress responses, and is regulated across transcriptional, translational, and epigenetic levels (Cheung and Rando, 2013; Yao, 2014). We propose that together these pathways form a quiescence regulatory network, in which the bistable module, the Rb-E2F switch, provides a necessary cellular mechanism to integrate and convert various graded (analog) growth-stimulating and -inhibitory signals into an all-or-none digital output (E2F-OFF/ON) and cell-fate distinction (quiescence/proliferation) (Figure 2A). Other quiescence regulatory pathways, by impinging on and interacting with the Rb-E2F bistable module, regulate the E2F switching threshold and carry out cellular functions associated with the corresponding quiescence depth (Yao, 2014). In summary, we propose that the metric for deep vs. shallow quiescence in the cell is the E2F switching threshold, a dynamic network property beyond single isolated molecular players. Relatedly, in a parallel RNA-seq analysis we found that for example pRb (but not p21) expression significantly increased as REF/E23 cells move deep into quiescence; yet, pRb unlikely drives deep quiescence alone as many gene clusters were up or down regulated noticeably as well (Fujimaki, Bai, and Yao, unpublished). It is an interesting question what mechanisms differentiate deep quiescence, which is reversible to proliferation, from irreversibly arrested states such as senescence and terminal differentiation. It is possible that when cells move into deeper and deeper quiescence, they eventually become irreversibly arrested when the progressively increasing E2F switching threshold exceeds physiological serum concentration. Consistent with this idea, the expression of Cdk inhibitors (e.g, p16 and p21) is greatly increased under senescence and terminal differentiation (Alcorta et al., 1996; Stein et al., 1999), which leads to a substantially increased E2F switching threshold (Figure 3A, the kI curve). Meanwhile, it has been observed that “irreversibly” arrested cells can become reversible by the forced expression of ectopic Cyclin/Cdks (Latella et al., 2001), which results in a decreased E2F switching threshold (Figure 3A). It has also been recently reported that quiescent fibroblast cells cultured for an extended period (100–150 days) eventually transitioned into senescence (Marthandan et al., 2014). However, accumulated DNA-damage during the long-term culture may cause mutations that were necessary for such a transition (Marthandan et al., 2014). Relatedly, it has been shown that Hes1, a Notch effector, maintains the reversibility of cellular quiescence against senescence (Sang et al., 2008). Therefore, it remains possible that the transition from deep quiescence to senescence involves a certain discrete mechanism(s).
Maintaining appropriate quiescent states and depths is critical to the normal functions of various cell types and tissue homeostasis. Our work in this study helps establish an E2F switching threshold model that can serve as a mechanistic framework to integrate diverse quiescence regulatory activities, and based on which, to guide experimental manipulations of cellular quiescent states. Future studies of single-cell responses to growth signals using quantitative time-lapse analysis with microfluidics will help further better understand quiescence exit heterogeneity, which has been previously demonstrated in the exponential waiting-time behavior before activating the Rb-E2F switch (Lee et al., 2010) and has also been shown related to the cell growth status prior to quiescence entry (Wang et al., 2017). Meanwhile, investigating how the Rb-E2F switch interacts with other quiescence regulatory pathways is crucial for understanding quiescence depth determination in different cell types under different conditions. Further elucidating the nature of quiescence control and heterogeneity will provide the basis for future strategies to regulate the growth responses of quiescent cell reservoirs in vivo and to potentially re-establish normal quiescent states in diseased cells.
Modulating the activation thresholds of cellular network switches has broad implications. In addition to the Rb-E2F bistable switch that regulates cell cycle entry, other bistable and multistable network switches have been recently identified. For example, miR-200/ZEB and RKIP-BACH1 network switches play important roles in epithelial-to-mesenchymal transition and cancer metastasis (Hong et al., 2015; Jolly et al., 2016; Lee et al., 2014; Tian et al., 2013; Zhang et al., 2014). Factors in these networks have been identified to modulate the corresponding response thresholds to cellular and environmental signals, thereby affecting the proportion of cells in different network states. Meanwhile, quiescence-like growth-arrest states in bacterial and mammalian cells are often correlated with persistence or resistance to drug treatment (Pearl Mizrahi et al., 2016; Rotem et al., 2010; Sharma et al., 2010). Identifying the effective modulators of activation/transition thresholds in the underlying regulatory networks may help reduce the stability of these drug-resistant states and enhance therapeutic efficacy.
EXPERIMENTAL PROCEDURES
Cell culture, quiescence entry and exit.
REF/E23 cells were derived from rat embryonic fibroblasts REF52 cells as a single cell clone containing a stably integrated E2F1 promoter-driven destabilized GFP reporter (E2F-GFP reporter for short) as previously described (Yao et al., 2008). Cells were maintained in DMEM (Gibco) supplemented with 10% of bovine growth serum (BGS; HyClone). Cells were regularly passed at a sub-confluent level. For quiescence entry by serum starvation, growing cells were trypsinized and seeded at about 105 cells per well in 6-well cell culture plates, washed twice with DMEM after cell attachment, and cultured in serum-starvation medium (0.02% BGS in DMEM) for 2–12 days. For quiescence entry by contact inhibition, growing cells were trypsinized and seeded at 4×105 cells (3× normal confluency) per well in 12-well cell culture plates and cultured in DMEM containing 3% BGS for 2–6 days. For quiescence exit of serum-starved cells, cells were switched to DMEM containing BGS at indicated concentrations for indicated durations. For quiescence exit of contact-inhibited cells, cells were serum stimulated as above, with or without being replated at a non-contact inhibition condition as indicated.
E2F activity and cell proliferation (EdU incorporation) assays.
To measure E2F activity in individual cells, REF/E23 cells were collected at indicated time points by trypsinization, fixed with 1% formaldehyde, and measured for fluorescence intensity of the E2F-GFP reporter. For the cell proliferation assay, EdU (1 μM) was added to culture medium at the time of serum stimulation. When noted, nocodazole at the lowest effective dose (40 ng ml−1 and 100 ng ml−1 for serum stimulation at non-saturating (≤ 3%) and saturating (≥ 20%) levels, respectively) was applied to restrict cell division while minimizing cell detachment or death. Cells were trypsinized at indicated time points and subjected to Click-iT EdU assay according to the manufacturer’s protocol (Click-iT EdU Alexa Fluor 647 Flow Cytometry Kit, Invitrogen). In the dual GFP-EdU assay, to recover GFP fluorescence quenched by the Click-iT reaction, cells were stained with fluorescein-conjugated GFP antibody (1:300, No. 600–102–215, Rockland) at 4°C overnight. Approximately 10,000 cells from each sample were measured for GFP and/or EdU signal intensity using an LSR II flow cytometer (BD Biosciences), and the data were analyzed using FlowJo software (v. 10.0).
Transfection of expression vectors.
REF/E23 cells were maintained at sub-confluence and transfected with indicated expression vectors using a Neon electroporation system (Invitrogen) following the manufacturer’s instructions. Briefly, cells were electroporated with one 20-millisecond pulse at 1900 volts in a 100 μl Neon tip containing approximately 106 cells and 10 μg of plasmid DNA unless otherwise noted. Expression vectors used in this study all contained the same CMV promoter and essentially the same plasmid backbone (p21-pRc/CMV, a gift from William Kaelin via Addgene; pCDNA3-pRb, a gift from Joseph Nevins; pCMV-p130, from Origene; pCDNA3-FLAG-Myc and pCDNA3-FLAG-CycD1, from GenScript; pCMV-mCherry, a gift from Lingchong You).
Immunoflow cytometry.
After Neon transfection of protein expression vectors, REF/E23 cells were recovered in DMEM containing 10% BGS for 1 day and subsequently switched to serum-starvation medium (0.02% BGS) for 2 days. Typically, cells were harvested by trypsinization, fixed with 4% formaldehyde in DPBS at room temperature (r.t.) for 15 minutes and permeabilized with 0.25% Triton X-100 in DPBS at r.t. for 5 minutes. Cells were then incubated at r.t. for 2 hours with protein-specific primary antibodies (p21(c-19), No. sc-397, Santa Cruz Biotech (SCBT); p130(c-20), No. sc-317, SCBT; pRb(IF8), No. sc-102FITC, SCBT; and for FLAG-Myc and FLAG-CycD1, DYKDDDDK Tag antibody [FITC], No. A01632, GenScript), and subsequently stained at r.t. for 2 hours with a FITC-conjugated secondary antibody (No. sc-2012, SCBT) when necessary and applicable. Antibody-stained cells were measured using an LSR II flow cytometer (BD Biosciences), and the data were analyzed using FlowJo software (v. 10.0).
Model simulations.
To simulate parameter-sensitivity curves, each of the model parameters (Table S2) were systematically “mutated” (increased or decreased from their base values by a factor up to 10, scanned in the logarithmic scale with 100 intervals) using the Parameter Scan function of COPASI (Hoops et al., 2006). Meanwhile, serum input concentration was varied in the range of 0 to 20 (%) with 4000 linear intervals. Simulations were carried out using the deterministic LSODA method in COPASI for a model time course of 1000 hours to ensure that the system reached a steady state. For a given parameter set, the switch of E2F steady state from OFF to ON was determined by passing a cut-off value of [E2F] ≥ 0.1 nM (compared with the mean steady-state values of the E2F-OFF and E2F-ON in the base model, 5.5 ×10−4 and 1.2 nM, respectively). The resultant parameter-sensitivity curve is essentially similar to a two-parameter bifurcation diagram, which shows how a bifurcation point in a one-parameter bifurcation analysis (serum threshold to switch E2F from OFF to ON) changes with variations of another model parameter (Figure S6A, examples generated using Oscill8, http://oscill8.sourceforge.net). Introducing cooperativity (Hill coefficient =1.5) into each Hill function term in the model (Table S1) did not change the qualitative behaviors of the bifurcation diagrams (Figure S6B).
To determine the traverse time of the R-point in a model with base or mutated parameters, a Parameter Scan was run in COPASI for serum-pulse duration (0–30 hours, with 1000 intervals). The amplitude of the serum pulse was 20 (%); after the pulse duration, the serum input was reduced to a basal level within the bistable region (so that if E2F were switched ON, it remained ON at a steady state) (Figure S1D). The minimum pulse duration that switched the E2F steady state from OFF to ON corresponded to the traverse time of the R-point. To determine the deactivation threshold of the Rb-E2F switch (the left boundary of the bistable region) in a model with base or mutated parameters, a Parameter Scan of serum input (0–20 %, with 1000 intervals) was run with the initial condition (model variable values) corresponding to the E2F-ON steady state. The serum input at which the E2F steady state (determined at the 1000th model hour) switched from ON to OFF was considered the deactivation threshold.
To determine the percentage of quiescence-exit cells affected by mutated parameters, stochastic simulations were performed using the Gibson + Bruck method in COPASI. For each indicated value of parameters kR, kI, kM, and kCDS, the E2F-ON percentage was calculated as the percentage of events in 2000 stochastic simulations where the E2F level, following serum stimulation, reached beyond 0.5 nM by the end of a model time course of 500 hours.
Quasi-potential landscape.
To calculate the quasi-potential landscape, simulations were performed based on a stochastic differential equation (SDE) version of the Rb-E2F switch model. To this end, we converted model parameter values and species concentrations in Tables S1 and S2 to molecule numbers, and adopted the chemical Langevin formulation:
where at time t, Xi(t) denotes the molecule number of species i(i = 1,…,n) and X(t) = (X1(t),…,Xn(t)) denotes the system state. The temporal evolution of the system state is calculated based on the rates aj[X(t)] (j = 1,…,M with the corresponding change of molecule number i described in vji. Factors γ and ω represent temporally uncorrelated, statistically independent normal Gaussian noise related to intrinsic and extrinsic noise, respectively, with θ and δ being scaling factors (θ =0.3, δ =25). To generate each curve in the quasi-potential landscape, stochastic simulations were run in 4,000 events (cells) for 3,000 model hours each to get a steady-state histogram of E2F molecule number, which was then fit with a smoothing spline model in Matlab to obtain corresponding probability distribution (pss). The quasi-potential is defined as U = − ln pss+ ln psaddle, where psaddle is the steady-state distribution at the saddle point (Xing, 2010). The resultant quasi-potential landscape corresponds to a one-dimensional projection on the E2F axis of the multidimensional Rb-E2F network system, in which state transitions are affected by environmental signals as well as intrinsic and extrinsic noise.
Statistics.
The indicated p values were obtained using Student’s t test unless otherwise noted (**, p < 0.001; *, p < 0.01).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Rob Brooks, Baltz Aguda, Lingchong You, and Kotaro Fujimaki for critical readings of the manuscript; and Guangbo Liu and Chenglu Chen for technical assistance. This work was supported by grants from the NSF (DMS-1463137, to G.Y. and J.X.; DMS-1418172, to G.Y.), NIH (GM-084905, a T32 fellowship to J.S.K), DARPA (WF911NF-14-1-0395, to G.Y.), and the NSF of China (31500676, to X.W.) and Anhui Province (1508085SQC202, to X.W.).
REFERENCES
- Adelman RC, Stein G, Roth GS, and Englander D (1972). Age-dependent regulation of mammalian DNA synthesis and cell proliferation In vivo. Mechanisms of Ageing and Development 1, 49–59. [Google Scholar]
- Aguda BD (2015). Cell Cycle Control: The Restriction Point. eLS 1, 5. [Google Scholar]
- Alcorta DA, Xiong Y, Phelps D, Hannon G, Beach D, and Barrett JC (1996). Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci U S A 93, 13742–13747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao P (2004). Potential in stochastic differential equations: novel construction. J Phys a-Math Gen 37, L25–L30. [Google Scholar]
- Attwooll C, Lazzerini Denchi E, and Helin K (2004). The E2F family: specific functions and overlapping interests. EMBO J 23, 4709–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augenlicht LH, and Baserga R (1974). Changes in the G0 state of WI-38 fibroblasts at different times after confluence. Exp Cell Res 89, 255–262. [DOI] [PubMed] [Google Scholar]
- Balazsi G, van Oudenaarden A, and Collins JJ (2011). Cellular decision making and biological noise: from microbes to mammals. Cell 144, 910–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks RF, Richmond FN, Riddle PN, and Richmond KM (1984). Apparent heterogeneity in the response of quiescent swiss 3T3 cells to serum growth factors: implications for the transition probability model and parallels with “cellular senescence” and “competence”. J Cell Physiol 121, 341–350. [DOI] [PubMed] [Google Scholar]
- Bucher NL (1963). Regeneration of Mammalian Liver. Int Rev Cytol 15, 245–300. [DOI] [PubMed] [Google Scholar]
- Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes M, and Scadden DT (2000). Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science 287, 1804–1808. [DOI] [PubMed] [Google Scholar]
- Cheung TH, and Rando TA (2013). Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol 14, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller HA, Sang L, and Roberts JM (2006). A New Description of Cellular Quiescence. PLoS Biol 4, e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregori J, and Johnson DG (2006). Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Curr Mol Med 6, 739–748. [DOI] [PubMed] [Google Scholar]
- Frolov MV, and Dyson NJ (2004). Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J Cell Sci 117, 2173–2181. [DOI] [PubMed] [Google Scholar]
- Hong T, Watanabe K, Ta CH, Villarreal-Ponce A, Nie Q, and Dai X (2015). An Ovol2-Zeb1 Mutual Inhibitory Circuit Governs Bidirectional and Multi-step Transition between Epithelial and Mesenchymal States. PLoS Comput Biol 11, e1004569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P, and Kummer U (2006). COPASI--a COmplex PAthway SImulator. Bioinformatics 22, 3067–3074. [DOI] [PubMed] [Google Scholar]
- Johnson DG, Schwarz JK, Cress WD, and Nevins JR (1993). Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature 365, 349–352. [DOI] [PubMed] [Google Scholar]
- Jolly MK, Tripathi SC, Jia D, Mooney SM, Celiktas M, Hanash SM, Mani SA, Pienta KJ, Ben-Jacob E, and Levine H (2016). Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget 7, 27067–27084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latella L, Sacco A, Pajalunga D, Tiainen M, Macera D, D’Angelo M, Felici A, Sacchi A, and Crescenzi M (2001). Reconstitution of cyclin D1-associated kinase activity drives terminally differentiated cells into the cell cycle. Mol Cell Biol 21, 5631–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti E, Frelin C, Xie S, Ferrari R, Dunant CF, Zandi S, Neumann A, Plumb I, Doulatov S, Chen J, et al. (2015). CDK6 levels regulate quiescence exit in human hematopoietic stem cells. Cell Stem Cell 16, 302–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Farquhar KS, Yun J, Frankenberger CA, Bevilacqua E, Yeung K, Kim EJ, Balazsi G, and Rosner MR (2014). Network of mutually repressive metastasis regulators can promote cell heterogeneity and metastatic transitions. Proc Natl Acad Sci U S A 111, E364–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Yao G, Bennett DC, Nevins JR, and You L (2010). Stochastic E2F activation and reconciliation of phenomenological cell-cycle models. PLoS Biol 8, e1000488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemons JMS, Feng X-J, Bennett BD, Legesse-Miller A, Johnson EL, Raitman I, Pollina EA, Rabitz HA, Rabinowitz JD, and Coller HA (2010). Quiescent Fibroblasts Exhibit High Metabolic Activity. PLoS Biol 8, e1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung JY, Ehmann GL, Giangrande PH, and Nevins JR (2008). A role for Myc in facilitating transcription activation by E2F1. Oncogene 27, 4172–4179. [DOI] [PubMed] [Google Scholar]
- Liu H, Adler AS, Segal E, and Chang HY (2007). A transcriptional program mediating entry into cellular quiescence. Plos Genetics 3, 996–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens-Bobadilla E, Zhao S, Baser A, Saiz-Castro G, Zwadlo K, and Martin-Villalba A (2015). Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells that Become Activated upon Brain Injury. Cell Stem Cell 17, 329–340. [DOI] [PubMed] [Google Scholar]
- Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, and Thompson CB (2005).Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 120, 237–248. [DOI] [PubMed] [Google Scholar]
- Marthandan S, Priebe S, Hemmerich P, Klement K, and Diekmann S (2014). Long-term quiescent fibroblast cells transit into senescence. PLoS One 9, e115597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins JR (2001). The Rb/E2F pathway and cancer. Hum Mol Genet 10, 699–03. [DOI] [PubMed] [Google Scholar]
- Overton KW, Spencer SL, Noderer WL, Meyer T, and Wang CL (2014). Basal p21 controls population heterogeneity in cycling and quiescent cell cycle states. Proc Natl Acad Sci U S A 111, E4386–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen TA, Soprano DR, and Soprano KJ (1989). Analysis of the growth factor requirements for stimulation of WI-38 cells after extended periods of density-dependent growth arrest. J Cell Physiol 139, 424–431. [DOI] [PubMed] [Google Scholar]
- Pearl Mizrahi S, Gefen O, Simon I, and Balaban NQ (2016). Persistence to anti-cancer treatments in the stationary to proliferating transition. Cell Cycle 15, 3442–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, King KY, Brett JO, Cromie MJ, Charville GW, Maguire KK, Brunson C, Mastey N, Liu L, Tsai CR, et al. (2014). mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert). Nature 510, 393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossini M, Lin JC, and Baserga R (1976). Effects of prolonged quiescence on neuclei and chromatin of WI-38 fibroblasts. J Cell Physiol 88, 1–11. [DOI] [PubMed] [Google Scholar]
- Rotem E, Loinger A, Ronin I, Levin-Reisman I, Gabay C, Shoresh N, Biham O, and Balaban NQ (2010). Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc Natl Acad Sci U S A 107, 12541–12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang L, Coller HA, and Roberts JM (2008). Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science 321, 1095–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. (2010). A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141, 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EJ, Leone G, DeGregori J, Jakoi L, and Nevins JR (1996). The accumulation of an E2F-p130 transcriptional repressor distinguishes a G0 cell state from a G1 cell state. Mol Cell Biol 16, 6965–6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soprano KJ (1994). WI-38 cell long-term quiescence model system: a valuable tool to study molecular events that regulate growth. J Cell Biochem 54, 405–414. [DOI] [PubMed] [Google Scholar]
- Spencer SL, Cappell SD, Tsai FC, Overton KW, Wang CL, and Meyer T (2013). The Proliferation-Quiescence Decision Is Controlled by a Bifurcation in CDK2 Activity at Mitotic Exit. Cell 155, 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein GH, Drullinger LF, Soulard A, and Dulic V (1999). Differential roles for cyclin-dependent kinase inhibitors p21 and p16 in the mechanisms of senescence and differentiation in human fibroblasts. Mol Cell Biol 19, 2109–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian XJ, Zhang H, and Xing J (2013). Coupled reversible and irreversible bistable switches underlying TGFbeta-induced epithelial to mesenchymal transition. Biophys J 105, 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, and Lees JA (2002). Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol 3, 11–20. [DOI] [PubMed] [Google Scholar]
- Wang J, Huang B, Xia X, and Sun Z (2006). Funneled landscape leads to robustness of cellular networks: MAPK signal transduction. Biophys J 91, L54–56. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang X, Fujimaki K, Mitchell GC, Kwon JS, Della Croce K, Langsdorf C, Zhang HH, and Yao G (2017). Exit from quiescence displays a memory of cell growth and division. Nat Commun 8, 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JV, Yao G, Nevins JR, and You L (2011a). Viral-Mediated Noisy Gene Expression Reveals Biphasic E2f1 Response to MYC. Mol Cell 41, 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JV, Yao G, Nevins JR, and You LC (2011b). Using Noisy Gene Expression Mediated by Engineered Adenovirus to Probe Signaling Dynamics in Mammalian Cells. Methods in Enzymology, Vol 497: Synthetic Biology, Methods for Part/Device Characterization and Chassis Engineering, Pt A 497, 221–237. [DOI] [PubMed] [Google Scholar]
- Wu L, Timmers C, Maiti B, Saavedra HI, Sang L, Chong GT, Nuckolls F, Giangrande P, Wright FA, Field SJ, et al. (2001). The E2F1–3 transcription factors are essential for cellular proliferation. Nature 414, 457–462. [DOI] [PubMed] [Google Scholar]
- Xing J (2010). Mapping between dissipative and Hamiltonian systems. J Phys A: Math Theor 43, 375003. [Google Scholar]
- Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, and Beach D (1993). P21 Is a Universal Inhibitor of Cyclin Kinases. Nature 366, 701–704. [DOI] [PubMed] [Google Scholar]
- Yanez I, and O’Farrell M (1989). Variation in the length of the lag phase following serum restimulation of mouse 3T3 cells. Cell Biol Int Rep 13, 453–462. [DOI] [PubMed] [Google Scholar]
- Yao G (2014). Modelling mammalian cellular quiescence. Interface Focus 4, 20130074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G, Lee TJ, Mori S, Nevins JR, and You L (2008). A bistable Rb-E2F switch underlies the restriction point. Nat Cell Biol 10, 476–482. [DOI] [PubMed] [Google Scholar]
- Yao G, Tan C, West M, Nevins JR, and You L (2011). Origin of bistability underlying mammalian cell cycle entry. Mol Syst Biol 7, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Tian XJ, Zhang H, Teng Y, Li R, Bai F, Elankumaran S, and Xing J (2014). TGF-beta-induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Sci Signal 7, ra91. [DOI] [PubMed] [Google Scholar]
- Zhang T (2013). Phase portraits of the proliferation-quiescence decision. Sci Signal 6, pe37. [DOI] [PubMed] [Google Scholar]
- Zhou JX, Aliyu MD, Aurell E, and Huang S (2012). Quasi-potential landscape in complex multi-stable systems. J R Soc Interface 9, 3539–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.