Abstract
The cohesin protein complex mediates sister chromatid cohesion to ensure accurate chromosome segregation, and also influences gene transcription in higher eukaryotes. Modest deficits in cohesin function that do not alter chromosome segregation cause significant birth defects. The mechanisms by which cohesin participates in gene regulation have been studied in Drosophila, revealing that it is involved in gene activation by transcriptional enhancers and epigenetic gene silencing by Polycomb group proteins. Recent studies reveal that early DNA replication origins are important for determining which genes associate with cohesin and suggest that cohesin at replication origins is important for establishing both sister chromatid cohesion and enhancer-promoter communication.
Keywords: transcriptional enhancer, gene promoter, sister chromatid cohesion, DNA replication, Polycomb silencing
Cohesin Has Multiple Functions
The roles of the cohesin protein complex increase with organismal complexity. Cohesin mediates sister chromatid cohesion (see Glossary) to ensure accurate chromosome segregation, and also facilitates DNA repair in all eukaryotes [1-3]. In metazoans, cohesin influences gene transcription and development, and minor cohesin deficiencies cause birth defects, such as those displayed in Cornelia de Lange Syndrome (CdLS) [4]. In vertebrates, cohesin interacts with the CTCF (CCCTC-binding factor) protein at the borders of higher-order chromosomal structures called Topologically Associating Domains (TADs) and is crucial for their formation [5].
This article focuses on how cohesin influences gene transcription in the invertebrate Drosophila, in which the relatively small genome simplifies many experiments. TADs occur in Drosophila, but cohesin does not play a key role in their formation [6-10] reducing the likelihood that cohesin’s transcriptional roles involve multiple effects on higher order chromosome structure. Drosophila studies reveal that cohesin supports transcriptional enhancer function and the activities of the PRC1 Polycomb group (PcG) repressive complex at both silenced and active genes. They uncovered a role for DNA replication origins in determining which genes are occupied by cohesin and raise the intriguing possibility that origins may facilitate both sister chromatid cohesion and enhancer-promoter communication, functionally linking genome stability and gene regulation.
Cohesin Structure and Chromosome Binding
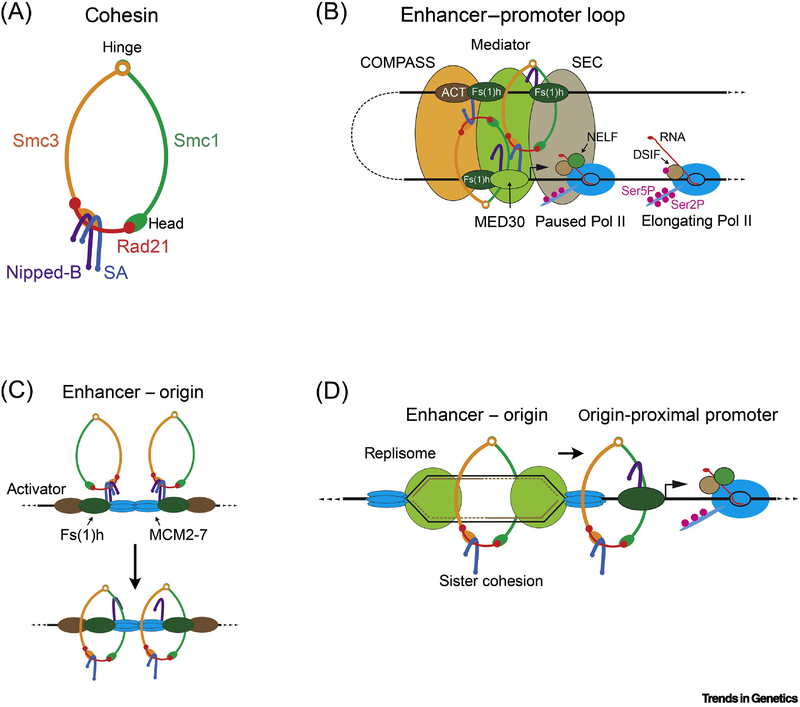
Cohesin is a structural maintenance of chromosome (SMC) complex (Figure 1A). It’s ring structure can topologically encircle DNA and take on multiple conformations. Cohesin contains the Smc1 and Smc3 proteins that fold back on themselves at a hinge, forming rod-like anti-parallel coiled-coils that hold the N and C termini together. The N and C termini form head domains with ABC-type ATPase activity. Smc1 and Smc3 heterodimerize at their hinges. Rad21 interacts with the Smc1 head domain and the Smc3 arm near the head domain. ATP holds the Smc1 and Smc3 head domains together, and without ATP the Smc1 and Smc3 head domains separate but are bridged by Rad21, forming a tripartite ring with a lumen of some 35 by 50 nm (Figure 1A). The SA cohesin subunit interacts with Rad21. The SMC subunit coiled-coil arms are flexible and can interact with each other forming a rod-like structure and can also fold so that the hinge contacts the head domains [11-15].
Figure 1. Models for Cohesin and DNA Replication Origin Roles in Enhancer-Promoter Looping and Sister Chromatid Cohesion.
(A) Schematic of cohesin in the open conformation. (B) Diagram of an enhancer – promoter loop. Activator proteins (ACT) bound to the enhancer recruit the Trr COMPASS histone methylation complex, Mediator, and the SEC (super elongation complex). The Fs(1)h BET domain protein recruits Nipped-B and cohesin to enhancers and linked promoters, and the MED30 subunit of Mediator recruits Nipped-B and cohesin to promoters [38]. Different models for how cohesin facilitates looping have been proposed. In the simplest, a cohesin ring encircles both the enhancer and promoter, similar to the embrace model for sister chromatid cohesion. The diagram depicts alternative ideas, in which cohesin interacts with Mediator and other enhancer and promoter-bound proteins to stabilize looping. Cohesin associates with genes with promoter-proximal paused Pol II [31, 34] which is phosphorylated on the serine 5 residues in the C terminal domain of Rpb1 (pink circles). The NELF and DSIF complexes are required for pausing. The P-TEFb subcomplex of SEC phosphorylates DSIF, NELF and the serine 2 residues in the Rpb1 heptad repeats to induce transition from pausing to transcriptional elongation. (C) Model for recruitment of the MCM2-7 helicase and cohesin to transcriptional enhancers by Fs(1)h and SA [38]. It is envisioned that proteins at enhancers, potentially Fs(1)h, trap MCM2-7 that slides away from the loading sites during origin licensing. This positions origins at enhancers. SA recruits cohesin, allowing Nipped-B to load cohesin topologically at enhancers. (D) Model for establishing sister chromatid cohesion and enhancer-promoter communication [38]. It is theorized that when MCM2-7 unwinds DNA to start replication the new sister chromatids are topologically trapped in cohesin positioned behind the replication fork, passively establishing sister cohesion. Cohesin in front of the fork is pushed to be trapped by proteins present at active promoters, where it participates in stabilizing enhancer-promoter loops.
Cohesin is loaded topologically onto chromosomes by the Nipped-B (Scc2, Mis4, NIPBL) - Mau2 (Scc4) complex, and removed by the Pds5 - Wapl (Rad61) complex. Nipped-B and Pds5 have similar hook-like HEAT repeat structures and compete for binding to Rad21 (Figure 1A). Loading and removal of cohesin from chromosomes involve ATP binding and hydrolysis. Several studies provide mechanistic insights into the cohesin loading and removal, although there is controversy regarding whether the ring opens at the Smc1-Smc3 hinge or the Rad21-Smc3 interface during loading [16-23]. In metazoans, cohesin is loaded immediately after cell division in late telophase-early G1 and is removed from chromosome arms by the Pds5-Wapl complex in prometaphase, and from centromeric regions by the Separase protease at anaphase.
It is unresolved how cohesin mediates sister chromatid cohesion [24, 25]. An embrace model envisions that one cohesin ring encircles both sister chromatids [26] and handcuff models suggest that two cohesin rings, one around each sister, interact with each other [27]. It is also unclear how sister chromatid cohesion is established during S phase. One idea is that the replisome passes through cohesin during DNA replication leaving the two sisters within a cohesin ring, and other models suggest that cohesin is chaperoned from the front of the fork to behind, or that new cohesin is loaded behind the fork.
Cohesin and Transcriptional Elongation
Gene transcription has multiple steps that could be influenced by cohesin. An early idea was that topologically-bound cohesin could alter movement of RNA polymerase II (Pol II) along a gene. The open form of cohesin (Figure 1A) can theoretically accommodate the Pol II holoenzyme, which is on the order of 22 nm in diameter. However, in yeast, cohesin occupies intergenic regions between convergently transcribed genes, suggesting that RNA polymerase pushes cohesin [28, 29]. In contrast, Drosophila cohesin occupies many active genes, with peaks near the transcription start sites and little accumulation between genes, suggesting that Pol II movement does not position cohesin [30]. Although cohesin associates with the full length of the ecdysone receptor gene in Drosophila, cohesin depletion does not alter the rate of transcriptional elongation as measured by nascent RNA synthesis, indicating that cohesin does not hinder Pol II movement [31].
The Drosophila findings contradict in vitro single molecule studies. Transcription causes mammalian cohesin translocation along DNA in vitro, although this translocation is not constrained by nucleosomes or other protein complexes [32]. In contrast, yeast cohesin cannot pass over objects 20 nm in size or larger, and nucleosomes hinder translocation in vitro [33]. A potential explanation for the differences in how Pol II influences cohesin localization and movement in the various systems is that cohesin has different conformations. It may be largely in the open conformation in Drosophila cells, allowing Poll II to pass, while conformations with smaller lumens predominate in vitro and in yeast cells.
Nipped-B and Cohesin Modulate Transition of Paused Pol II into Elongation
Drosophila cohesin associates with genes that have high levels of promoter-proximal paused Pol II [31, 34] (Figure 1B). Pausing requires the NELF (negative elongation factor) and DSIF (DRB sensitivity inducing factor) complexes that interact with Pol II and the nascent RNA transcript (Figure 1B). Paused Pol II is phosphorylated on the serine 5 residues of heptad repeats in the C terminal domain of the Rpb1 Pol II subunit (Figure 1B). Phosphorylation of the serine 2 residues of the heptad repeats and DSIF by the Super Elongation Complex (SEC) and release of NELF is associated with transition into transcriptional elongation (Figure 1B). Despite the strong correlation between cohesin levels and polymerase pausing, cohesin is not required for pausing, and pausing factors are not required for cohesin association [31]. However, depending on the gene, cohesin can inhibit or promote transition of paused Pol II into elongation as measured by total RNA levels and genome-wide PRO-seq (precision run-on sequencing of nascent transcripts) [31, 34].
Nipped-B and cohesin do not significantly influence Pol II recruitment or transcriptional initiation as measured by Pol II ChIP-seq and PRO-seq [34]. Conversely, blocking transcriptional initiation with triptolide, an inhibitor of the TFIIH initiation factor, does not reduce Nipped-B association with promoters [35]. Combined, these studies indicate that cohesin primarily influences gene transcription by facilitating the actions of factors that either promote or inhibit transition of paused Pol II into elongation.
Nipped-B and Cohesin Have Multiple Functions at Transcriptional Enhancers
Transcriptional enhancers bind activator proteins that recruit SEC, Mediator, and the COMPASS histone methylation complex to control transition of paused Pol II into elongation (Figure 1B). Enhancers activate transcription even when they are many kilobases away from a gene, and genetic evidence first indicated that cohesin influences long-range enhancer function [36]. Nipped-B mutations were isolated in a genetic screen for factors that control enhancer-mediated activation of the Drosophila cut and Abdominal-B genes in a dosage-sensitive manner. Subsequent experiments revealed that Nipped-B and cohesin occupy essentially all transcriptional enhancers and many gene promoters in Drosophila cells [30, 34, 37, 38].
Association of cohesin with enhancers and promoters supports the idea that cohesin directly facilitates enhancer-promoter looping (Figure 1B). An early model was that cohesin holds enhancers and promoters together by a mechanism similar to how sister chromatids together, such as the embrace model. It is also now proposed that loop extrusion through the lumen of cohesin can form enhancer-promoter loops in addition to higher order structures such as TADs [39, 40]. However, there are other possible mechanisms, involving protein-protein interactions (Figure 1B) and it remains unsettled how cohesin supports looping.
One potential alternative involves interactions between the Mediator complex and Nipped-B or cohesin subunits (Figure 1B). Mediator occupies all active enhancers and promoters and controls multiple steps in transcription [41]. Substantial evidence indicates that the Nipped-B interacts with Mediator. Nipped-B (NIPBL) and cohesin colocalize with Mediator at enhancers in mouse embryonic stem cells and co-immunoprecipitate from nuclear extracts [42]. MED30 was the only Mediator subunit to interact with Nipped-B in a screen of some 5,000 proteins in Drosophila [43]. MED30 depletion in cultured fly cells reduces Nipped-B occupancy of promoters, consistent with the possibility that MED30 facilitates enhancer-promoter communication through interaction with Nipped-B [38].
Nipped-B might also promote enhancer function by influencing the activity of a COMPASS histone methylation complex (Figure 1B). The Trr COMPASS complex and the mammalian orthologs (MLL/4) monomethylate histone H3 lysine 4 (H3K4me1) at enhancers [44]. Although Trr is an essential protein, the catalytic activity is dispensable for viability and development [45, 46]. In contrast, a hyperactive Trr mutant that increases histone methylation causes adult segmentation defects reminiscent of those caused by Nipped-B overexpression [46, 47]. Nipped-B overexpression stabilizes cohesin chromosome binding, presumably by competitively reducing cohesin interaction with the Pds5 removal factor [37, 47, 48]. Combining the Trr hyperactive mutant with Nipped-B overexpression synergistically increases segmentation defects, indicating that enhancer hypermethylation and cohesin stabilization have similar effects on gene expression and development.
Additional studies will determine if developmental defects with increased Trr and Nipped-B function reflect increased or decreased enhancer activity. For instance, Nipped-B and Pds5 have similar genome-wide effects on mRNA levels, although they have opposing roles in cohesin chromosome binding [37]. The similar effects of proteins with opposing effects on cohesin binding on gene expression suggest that cohesin chromosome-binding dynamics are critical in defining transcription levels.
Many questions remain about how Nipped-B and cohesin facilitate enhancer activity. However, based on the molecular and genetic interactions of Nipped-B with Mediator and COMPASS, it is likely that they influence facets of enhancer function beyond enhancer-promoter looping.
DNA Replication Origins Influence Cohesin Localization
Early DNA replication origins that fire at the start of S phase were mapped genome-wide in multiple Drosophila cell lines [38, 49, 50]. Origins are licensed by binding of ORC (origin recognition complex) to chromosomes, which then recruits Cdc6, Cdt1 and two copies of the MCM2-7 helicase that unwinds DNA to start DNA synthesis at the beginning of S phase [51]. Transcription pushes MCM2-7 away from the loading site before S phase [52]. Cohesin is present at replication origins in both Drosophila and human cells [38, 49, 53].
Recent studies imply that early DNA replication origins dictate which active genes bind Nipped-B and cohesin in Drosophila. Early origins typically occur in transcribed regions of the genome, but meta-origin analysis of Drosophila ChIP-seq data reveals that Nipped-B and cohesin occupancy are more origin-centric than RNA polymerase II [37, 38]. Active genes that are within several kilobases of an origin are more likely to bind cohesin than are genes located farther away.
Strikingly, depletion of the Pds5 or Wapl cohesin removal factors expands the domains of cohesin-occupied genes surrounding origins by several kilobases [37]. Origin-distal genes that normally bind little cohesin bind substantially more. Pds5 depletion causes defects in sister chromatid cohesion, but Wapl depletion does not, indicating that cohesin domain expansion is not caused by a change in sister cohesion. Single molecule DNA fiber assays show that domain expansion also does not reflect a change in the rate of replication fork movement [37].
The current theory is that DNA replication pushes cohesin along the chromosome to be captured by proteins at active genes, and the extent of cohesin spreading is determined by the rate of cohesin removal by the Pds5-Wapl complex in front of the advancing fork. Pds5 or Wapl depletion reduces the rate of cohesin removal, increasing the size of the cohesin domains. Single molecule experiments in Xenopus extracts show that replisomes push cohesin along DNA, and that Pds5 inhibits this movement, although it is unknown if the cohesin conformation is the same in Drosophila cells and Xenopus extracts [54].
Potential Roles of SA and DNA Replication Origins in Sister Chromatid Cohesion and Enhancer-Promoter Communication
Pds5 depletion experiments indicate that Pds5 and the SA cohesin subunit function at origins to establish sister chromatid cohesion. Pds5 depletion does not substantially alter Rad21 cohesin subunit levels at replication origins, but strongly reduces SA, opposite to what is expected for depletion of a cohesin removal factor [37].
A long-standing paradox is that Pds5 is required for both cohesin removal and sister chromatid cohesion. The finding that Pds5 facilitates SA origin association may resolve this paradox. Pds5 forms an alternative stoichiometric complex with the Brca2 DNA repair protein that lacks Wapl [55, 56]. Brca2 depletion increases SA at origins, opposite to the effect of Pds5 depletion [37]. Co-depletion of Pds5 and Brca2 restores origin SA and partially reverses the loss of sister chromatid cohesion caused by depletion of Pds5 alone. Thus, Brca2 antagonizes the roles of Pds5 in both sister chromatid cohesion and SA binding at replication origins, suggesting that Pds5 and SA function at origins to establish sister chromatid cohesion.
SA is present at all enhancers, but not at most active gene promoters [38]. Only 12% of promoters in the Drosophila cells examined are occupied by SA [38]. SA levels are highest at replication origins, implying that the SA-occupied promoters and enhancers are positioned close to origins. Indeed, all SA-occupied promoters and enhancers are replicated at the start of S phase [38].
Enhancers located outside of transcribed regions show higher levels of early DNA synthesis than most, and early origins typically occur within clusters of enhancers [38]. It is theorized that proteins at enhancers trap MCM2-7 being pushed by transcription [52] thereby positioning origins at enhancers (Figure 1C). The enhancer-origin overlap and origin proximity of promoters that bind complete cohesin complexes raise the possibility that DNA replication forks push SA-containing cohesin from enhancers to flanking genes to establish enhancer-promoter communication [38] (Figure 1D).
SA recruits cohesin to origins and enhancers [38] (Figure 1C). SA depletion in cultured cells reduces Nipped-B, Rad21 and Smc1 at origins, enhancers and origin-proximal promoters and increases their levels at origin-distal promoters. This might explain why SA is needed at origins for sister chromatid cohesion. By recruiting cohesin to origins, SA ensures that DNA replication starts where there is a high cohesin density. This could allow the two sister chromatid templates to be trapped within cohesin rings that are behind the fork when MCM2-7 unwinds DNA to initiate replication (Figure 1D). This passive mechanism is consistent with the finding that human NIPBL interacts directly with MCM2-7 [57] and the ability of cohesin to remain bound to chromosomes during replication in the complete absence of the Wapl cohesin removal factor [58]. Distinguishing between a passive mechanism and mechanisms that involve cohesin removal and reloading may be possible by coupling in vitro cohesin loading and DNA replication systems.
The Fs(1)h Protein Facilitates Nipped-B and Cohesin Chromosome Association and Function
The idea that DNA replication facilitates cohesin association with enhancer-targeted genes raises the question of how Nipped-B and cohesin are directed to their targets before replication. Cohesin is removed from chromosomes during mitosis and loaded again in late telophase. One idea is that proteins that remain bound to chromosomes during mitosis act as bookmarks to direct cohesin to the proper genes in G1. A candidate is the Fs(1)h (female sterile homeotic on chromosome 1) protein, which is the sole Drosophila homolog of mammalian BET domain proteins, including BRD4. Mammalian BRD4 binds acetylated histones, remains bound to mitotic chromosomes, and speeds reactivation of gene transcription after cell division, suggesting that it is mitotic bookmark for active genes [59, 60]. ChIP-seq revealed that Fs(1)h associates with chromosomes in a highly origin-centric pattern similar to Nipped-B and cohesin, occupying all enhancers and origin-proximal promoters [38]. The JQ1 small molecule inhibitor globally reduces binding of Fs(1)h to chromosomes and also substantially reduces Nipped-B and Rad21 association with replication origins, enhancers and origin-proximal promoters, with minor effects at origin-distal promoters [38]. This supports the notion that Fs(1)h could direct cohesin association with enhancers and the correct promoters after cell division and before DNA replication.
In vivo evidence from humans and Drosophila further support the idea that BRD4 and Fs(1)h direct NIPBL and Nipped-B function. Individuals with heterozygous BRD4 loss-of-function mutations display growth, physical and intellectual birth defects overlapping those of Cornelia de Lange syndrome (CdLS) caused by heterozygous NIPBL mutations [61]. There are strong genetic interactions between fs(1)h and Nipped-B mutations in Drosophila development [38]. The fs(1)h1 hypomorphic allele permits survival of some 60% of the expected hemizygous fs(1)h1 / Y males. Although heterozygous Nipped-B mutations have no measurable effect on viability [36, 62] they reduce fs(1)h1 / Y male viability several-fold [38]. Nipped-B mutations and fs(1)h1 both suppress segment identity transformations caused by Polycomb (Pc) mutations, and together they synergistically reduce foreleg sex combs [38, 63]. These findings support the idea that Fs(1)h and Nipped-B function closely together in developmental gene regulation in vivo.
Cohesin Influences PRC1 Polycomb Group (PcG) Repressive Complex Activity at Both Silenced and Active Genes
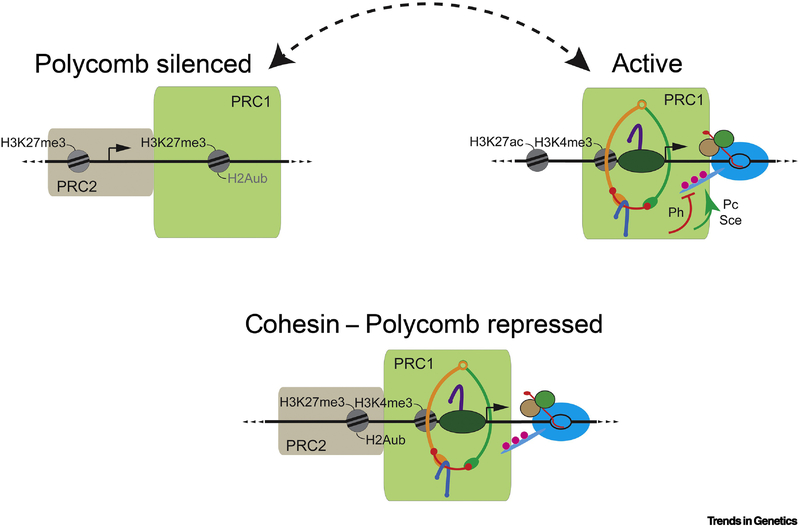
In addition to its roles in enhancer function, cohesin influences gene silencing by Polycomb group (PcG) proteins, and facilitates Polycomb protein association with active genes. The PRC2 complex contains the Enhancer of zeste [E(z)] enzyme that trimethylates histone H3 lysine 27. The H3K27me3 modification covers silenced genes and helps recruit the PRC1 complex with the Sce (Ring1) protein that mono-ubiquitinylates histone H2A. Rad21 (verthandi, vtd) mutations were first isolated as dominant suppressors of haploinsufficient Polycomb (Pc) PRC1 subunit mutant segmental transformations [64] and Nipped-B and other cohesin mutations have similar effects [63, 65]. Genetic antagonism between cohesin and Pc might indicate a dosage-sensitive balance between gene activation by cohesin and silencing by PcG proteins, but further studies revealed that the cohesin-PRC1 relationship is more complex (Figure 2).
Figure 2. Functional Interactions Between Cohesin and the PRC1 Polycomb Complex in Transcriptional Regulation of Silenced and Active Genes.
The upper left shows a Polycomb silenced gene, where the PRC2 Polycomb complex trimethylates histone H3 (H3K27me3) and PRC1 mono-ubiquitinylates histone H2A (H2Aub). H2Aub is not observed at all silenced genes, as indicated by the gray text. The upper right illustrates an active gene promoter where cohesin recruits PRC1 [63]. The Polyhomeotic (Ph) PRC1 subunit inhibits phosphorylation of the C terminal domain of the Pol II Rpb1 subunit, while the Polycomb (Pc) and Sex combs extra (Sce) subunits promote phosphorylation [66]. The opposing effects on serine 5 and serine 2 phosphorylation influences transcriptional elongation and may aid promoter-proximal pausing. The dashed arrow between the silenced and active genes indicates that the PRC1 distribution between active and silenced genes is sensitive to cohesin dosage. Depletion of cohesin increases the PRC1 level at silent genes, which can explain reversal of Pc mutant phenotypes by Nipped-B and cohesin subunit mutations [63] and reduction of H3K27me3 increases PRC1 levels at active genes (unpublished). The bottom diagram illustrates genes that are repressed, but not fully silenced by cohesin and PRC1 [6, 68]. These genes are transcribed at low to moderate levels and marked by H3K27me3 and histone modifications associated with active genes, such as H3K4me3. H2Aub is typically at high levels on these genes. Although rare, these genes encode proteins that regulate transcription, and include the genes encoding the Psc and Su(z)2 PRC1 subunits.
Cohesin is excluded from silenced genes but occupies all the Polycomb Response Elements (PREs) that recruit PcG complexes to silence genes [63, 66]. The role of cohesin at PREs remains unknown, although chromosome conformation capture experiments show that it can support PRE-PRE looping [63] and PRE-PRE interactions can facilitate silencing [67].
In special cases, cohesin cooperates with the PRC1 PcG complex to restrain but not fully silence transcription [6, 68] (Figure 2). These genes have hallmarks associated with silenced genes including H3K27me3 and PRC1 occupancy but are transcribed at low to moderate levels. At these genes, cohesin is not restricted to the PREs, but is present at the promoters and in the gene bodies. These genes also have histone modifications associated with active genes such as H3K4me3, and thus are comparable to bivalent genes in mammalian stem cells. All genes with this pattern encode transcription factors important for development, such as the Enhancer of split and invected-engrailed gene complexes. Strikingly, depletion of either PRC1 or cohesin subunits strongly elevates their expression [6, 68]. Co-depletion of PRC1 and cohesin subunits does not give additive or synergistic increases, indicating that cohesin and PRC1 work together to restrain transcription. It is unclear if these genes are transitioning between silenced and active states, or if this is a stable state that maintains a critical expression level to support development. It is notable, however, that the Psc-Su(z)2 locus encoding PRC1 subunits has this unique state in all cell lines and tissues examined, suggesting that it can be a stable chromatin state [68, 69].
Cohesin directly interacts with PRC1 and recruits PRC1 to active genes, where it influences Pol II activity [63, 66, 70] (Figure 2). Cohesin depletion reduces PRC1 association with active genes with a concomitant increase at silenced genes [63]. This provides an alternative explanation for the dosage-sensitive genetic antagonism between cohesin and PRC1. The idea is that heterozygous Pc mutations cause segmental transformations because of inadequate silencing of genes such as the Abdominal-B and Sex combs reduced HOX genes. The silencing deficiency is alleviated by reducing cohesin because this releases PRC1 from active genes, making more available for silencing. This idea is supported by a dominant-negative Wapl truncation mutant that causes segmental transformations similar to Pc mutants [71]. The mutant Wapl protein stabilizes cohesin binding as measured by in vivo FRAP (fluorescence recovery after photobleaching). The simple interpretation is that stabilizing cohesin at active genes causes more PRC1 to be sequestered, decreasing the amount available for silencing.
PRC1 recruited by cohesin directly impacts transcription of active genes [66]. Depletion of different PRC1 subunits alters phosphorylation of the heptad repeats in Pol II Rpb1 subunit C terminal domain (Figure 2). These include the serine 5 phosphorylation mediated by the TFIIH initiation factor, and the serine 2 phosphorylation mediated by SEC that accompanies transition from pausing to elongation. Different PRC1 subunits have opposite effects on Pol II phosphorylation. Polyhomeotic (Ph) inhibits both serine 5 and serine 2 phosphorylation, and Pc and Sce promote the same modifications. Combined, these activities are predicted to enhance Pol II pausing. Consistent with this idea, PRC1 promotes association of the Spt5 subunit of the DSIF pausing factor with active genes [66]. Many active genes also display changes in elongation and RNA processing detected by 3’ end nascent RNA-seq upon PRC1 subunit depletion [66]. Thus, cohesin influences the transition from pausing to elongation by recruiting PRC1 in addition to supporting enhancer-promoter looping.
The studies on how PRC1 influences transcription of active genes unexpectedly revealed that PRC1 also promotes association of the Spt5 subunit of the DSIF pausing factor with enhancers and PREs, and that depletion of PRC1 subunits increases Pol II phosphorylation and transcription initiation at these regulatory sequences. It is therefore also hypothesized that PRC1 suppresses transcription of cohesin-occupied regulatory sequences in addition to silencing genes.
Concluding Remarks and Future Perspectives
As described above, studies in Drosophila indicate that DNA replication origins are instrumental in determining which active genes are occupied by cohesin, raising new possibilities for how sister chromatid cohesion is established, and the mechanisms by which cohesin supports enhancer-promoter communication. They reveal that cohesin influences transcription of these genes by multiple mechanisms beyond enhancer-promoter looping, including functional interactions with the PRC1 repressive complex at active genes, and in epigenetic gene silencing. Several questions remain open (Outstanding Questions Box). Outside of the roles of cohesin in enhancer-promoter looping, many of the cohesin functions in transcription, and the linkage of these roles to early DNA replication origins discovered in Drosophila have yet to be investigated in mammals, where they could be important for understanding the birth defects associated with mild cohesin deficiencies, and cohesin dysfunction in cancer. The models for establishing sister chromatid cohesion and enhancer–promoter communication suggested by the Drosophila data also remain to be tested and compared to alternative models, including loop extrusion, in Drosophila and mammalian cells.
Outstanding Questions.
1. How many of the cohesin positioning and gene regulation mechanisms observed in Drosophila are conserved in mammals?
A role for cohesin in enhancer-promoter looping is firmly established in mammalian cells, but the potential roles for cohesin in directly influencing the activities of enhancer-binding and Polycomb complexes remain untested. It also remains unknown if cohesin chromosome occupancy is linked to early DNA replication origins.
2. How does cohesin support enhancer-promoter looping?
Drosophila data suggest that DNA replication may facilitate enhancer-promoter communication by pushing cohesin from enhancers to promoters, while the leading current thought is that cohesin brings them together by loop extrusion. There is also the possibility that direct interactions between cohesin and enhancer-binding complexes such as Mediator supports looping.
3. How is sister chromatid cohesion established?
Restriction of SA to origin proximal regions, its recruitment of cohesin to origins, and a correlation between SA levels at origins and sister chromatid cohesion suggest a model in which sister chromatid cohesion is passively established by topological trapping of nascent sister chromatids upon initiation of DNA replication. However, more complex mechanisms such as chaperoning of cohesin around advancing replication forks are possible.
Highlights.
Cohesin participates in gene activation and Polycomb repressive complex function via multiple mechanisms.
Cohesin association with transcriptional enhancers and gene promoters is linked to their proximity to early DNA replication origins.
Cohesin association with early replication origins may be important for establishing both sister chromatid cohesion and enhancer-promoter communication.
Acknowledgements
Research in the author’s laboratory was supported by National Institutes of Health grants GM107814 and GM108872.
Glossary
- Sister chromatid cohesion
The sister chromatids formed by DNA replication in S phase are held together by the cohesin protein complex. The mechanism is currently debated, but requires topological binding of cohesin to DNA.
- Cornelia de Lange syndrome (CdLS)
A genetic birth defect syndrome that displays poor growth, impaired intellectual development and multiple physical abnormalities. It is most frequently caused by dominant loss-of-function mutations in Nipped-B-Like (NIPBL) the human Nipped-B ortholog. Milder forms can be caused by dominant missense or reading frame-preserving mutations affecting cohesin subunits.
- Topologically associating domain (TAD)
higher order chromosomal structure in which sequences within the domain shows high contact frequency with each other and limited contacts with sequences outside the domain. Enhancer-promoter loops typically occur inside TADs.
- Polycomb group (PcG) repressive complex
Protein complexes that mediate epigenetic gene silencing. The main complexes are PRC2, which makes the histone H3 lysine 27 trimethylation (H3K27me3) modification, and PRC1 which makes the histone H2A mono-ubiquitination (H2Aub) mark.
- Replisome
The protein complex containing the DNA polymerase that replicates DNA during S phase.
- Promoter-proximal paused Pol II
RNA polymerase II that initiates transcription and stops several nucleotides downstream of the transcription start site but remains transcriptionally engaged. Pausing requires the NELF (negative elongation factor) and DSIF (DRB-sensitivity inducing factor) protein complexes that interact with Pol II and the nascent RNA transcript.
- Super Elongation Complex (SEC)
A protein complex containing multiple transcriptional elongation factors, including the P-TEFb subcomplex that phosphorylates the serine 2 residues of the heptad repeats in the C terminal domain of the Rpb1 subunit of RNA polymerase II.
- Mediator
A protein complex that interacts with transcriptional activator proteins at enhancers and Pol II at promoters to influence Pol II activity and gene transcription by multiple mechanisms.
- COMPASS
A class of protein complexes that influence gene activation and silencing and have histone methylation activity.
- ORC (origin recognition complex)
The protein complex that binds chromatin to license DNA replication origins in the early G1 phase of the cell cycle.
- MCM2-7
The protein complex with DNA helicase activity that is recruited by ORC and activated to initiate DNA unwinding and replication at the start of the S phase of the cell cycle.
- BET domain proteins
A class of chromatin-binding proteins. BET domains bind to acetylated histone residues.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dorsett D and Merkenschlager M (2013) Cohesin at active genes: a unifying theme for cohesin and gene expression from model organisms to humans. Curr. Opin. Cell Biol 25, 327–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dorsett D and Ström L (2012) The ancient and evolving roles of cohesin in gene expression and DNA repair. Curr. Biol 22, R240–R250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Remeseiro S et al. (2013) Cohesin in development and disease. Development 140, 3715–3718 [DOI] [PubMed] [Google Scholar]
- 4.Deardorff MA et al. (2016) Cornelia de Lange Syndrome In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2019. [PubMed] [Google Scholar]
- 5.Merkenschlager M and Nora EP (2016) CTCF and cohesin in genome folding and transcriptional gene regulation. Annu. Rev. Genomics Hum. Genet 17, 17–43 [DOI] [PubMed] [Google Scholar]
- 6.Schaaf CA et al. (2013) The Drosophila enhancer of split gene complex: architecture and coordinate regulation by notch, cohesin, and polycomb group proteins. G3 (Bethesda) 3, 1785–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramírez F et al. (2018) High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun 9, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szabo Q et al. (2018) TADs are 3D structural units of higher-order chromosome organization in Drosophila. Sci. Adv 4, eaar8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q et al. (2018) Sub-kb Hi-C in D. melanogaster reveals conserved characteristics of TADs between insect and mammalian cells. Nat. Commun 9, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luzhin AV et al. (2019) Quantitative differences in TAD border strength underly the TAD hierarchy in Drosophila chromosomes. J. Cell. Biochem 120, 4494–4503 [DOI] [PubMed] [Google Scholar]
- 11.Hons MT et al. (2016) Topology and structure of an engineered human cohesin complex bound to Pds5B. Nat. Commun 7, 12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulemzina I et al. (2016) A reversible association between Smc coiled coils is regulated by lysine acetylation and is required for cohesin association with the DNA. Mol. Cell 63, 1044–1054 [DOI] [PubMed] [Google Scholar]
- 13.Chavda AP et al. (2017) The torments of the cohesin ring. Nucleus 8, 261–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu X et al. (2018) Suppressor mutation analysis combined with 3D modeling explains cohesin's capacity to hold and release DNA. Proc. Natl. Acad. Sci 115, E4833–E4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bürmann F et al. (2019) A folded conformation of MukBEF and cohesin. Nat. Struct. Mol. Biol 26, 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murayama Y and Uhlmann F (2014) Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature 505, 367–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Çamdere G et al. (2015) The ATPases of cohesin interface with regulators to modulate cohesin-mediated DNA tethering. Elite 4, e11315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murayama Y and Uhlmann F (2015) DNA entry into and exit out of the cohesin ring by an interlocking gate mechanism. Cell 163, 1628–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beckouét F et al. (2016) Releasing activity disengages cohesin's Smc3/Scc1 Interface in a process blocked by acetylation. Mol. Cell 61,563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elbatsh AMO et al. (2016) Cohesin releases DNA through asymmetric ATPase-driven ring opening. Mol. Cell 61, 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu H (2016) Magic acts with the cohesin ring. Mol. Cell 61,489–491 [DOI] [PubMed] [Google Scholar]
- 22.Ouyang Z and Yu H (2017) Releasing the cohesin ring: A rigid scaffold model for opening the DNA exit gate by Pds5 and Wapl. BioEssays 39, 201600207. [DOI] [PubMed] [Google Scholar]
- 23.Petela NJ et al. (2018) Scc2 is a potent activator of cohesin's ATPase that promotes loading by binding Scc1 without Pds5. Mol. Cell 70, 1134–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skibbens RV (2016) Of rings and rods: regulating cohesin entrapment of DNA to generate intra- and intermolecular tethers. PLoS Genet. 12, e1006337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morales C and Losada A (2018) Establishing and dissolving cohesion during the vertebrate cell cycle. Curr. Opin. Cell Biol 52, 51–57 [DOI] [PubMed] [Google Scholar]
- 26.Haering CH et al. (2008) The cohesin ring concatenates sister DNA molecules. Nature 454, 297–301 [DOI] [PubMed] [Google Scholar]
- 27.Robison B et al. (2018) A role for the Smc3 hinge domain in the maintenance of sister chromatid cohesion. Mol. Biol. Cell 29, 339–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glynn EF et al. (2004) Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2, E259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lengronne A et al. (2004) Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430, 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Misulovin Z et al. (2008) Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma 117, 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fay A et al. (2011) Cohesin selectively binds and regulates genes with paused RNA polymerase. Curr. Biol 21, 1624–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davidson IF et al. (2016) Rapid movement and transcriptional re-localization of human cohesin on DNA. EMBO J. 35, 2671–2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stigler J et al. (2016) Single-molecule imaging reveals a collapsed conformational state for DNA-bound cohesin. Cell Rep. 15, 988–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaaf CA et al. (2013) Genome-wide control of RNA polymerase II activity by cohesin. PLoS Genet. 9, e1003382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swain A et al. (2016) Drosophila TDP-43 RNA-binding protein facilitates association of sister chromatid cohesion proteins with genes, enhancers and Polycomb response elements. PLoS Genet. 12, e1006331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rollins RA et al. (1999) Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics 152, 577–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Misulovin Z et al. (2018) Brca2, Pds5 and Wapl differentially control cohesin chromosome association and function. PLoS Genet. 14, e1007225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pherson M et al. (2019) Cohesin occupancy and composition at enhancers and promoters are linked to DNA replication origin proximity in Drosophila. Genome Res. February 22. doi: 10.1101/gr.243832.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hassler M et al. (2018) Towards a unified model of SMC complex function. Curr. Biol 28, R1266–R1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rowley MJ and Corces VG (2018) Organizational principles of 3D genome architecture. Nat. Rev. Genet 19, 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harper TM and Taatjes DJ (2018) The complex structure and function of Mediator. J. Biol. Chem 293, 13778–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kagey MH et al. (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guruharsha KG et al. (2011) A protein complex network of Drosophila melanogaster. Cell 147, 690–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sze CC and Shilatifard A(2016) MLL3/MLL4/COMPASS family on epigenetic regulation of enhancer function and cancer. Cold Spring Harb. Perspect. Med 6, a026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dorighi KM et al. (2017) Mll3 and Mll4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 monomethylation. Mol. Cell 66, 568–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rickels R et al. (2017) Histone H3K4 monomethylation catalyzed by Trr and mammalian COMPASS-like proteins at enhancers is dispensable for development and viability. Nat. Genet 49, 1647–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gause M et al. (2010) Dosage-sensitive regulation of cohesin chromosome binding and dynamics by Nipped-B, Pds5, and Wapl. Mol. Cell. Biol 30, 4940–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ouyang Z et al. (2016) Structural basis and IP6 requirement for Pds5-dependent cohesin dynamics. Mol. Cell 62, 248–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacAlpine HK et al. (2010) Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 20, 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eaton ML et al. (2011) Chromatin signatures of the Drosophila replication program. Genome Res. 21, 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prioleau MN and MacAlpine DM (2016) DNA replication origins-where do we begin? Genes Dev. 30, 1683–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Powell SK et al. (2015) Dynamic loading and redistribution of the Mcm2-7 helicase complex through the cell cycle. EMBO J. 34, 531–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guillou E et al. (2010) Cohesin organizes chromatin loops at DNA replication factories. Genes Dev. 24, 2812–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanke M et al. (2016) Cohesin acetylation and Wapl-Pds5 oppositely regulate translocation of cohesin along DNA. EMBO J. 35, 2686–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brough R et al. (2012) APRIN is a cell cycle specific BRCA2-interacting protein required for genome integrity and a predictor of outcome after chemotherapy in breast cancer. EMBO J. 31, 1160–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kusch T (2015) Brca2-Pds5 complexes mobilize persistent meiotic recombination sites to the nuclear envelope. J. Cell Sci 128, 717–727 [DOI] [PubMed] [Google Scholar]
- 57.Zheng G et al. (2018) MCM2-7-dependent cohesin loading during S phase promotes sister-chromatid cohesion. Elite 7, e33920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rhodes JDP et al. (2017) Cohesin can remain associated with chromosomes during DNA replication. Cell Rep. 20, 2749–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dey A et al. (2000) A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol. Cell. Biol 20, 6537–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao R et al. (2011) Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat. Cell Biol 13, 1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Olley G et al. (2018) BRD4 interacts with NIPBL and BRD4 is mutated in a Cornelia de Lange-like syndrome. Nat. Genet 50, 329–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu Y et al. (2015) Drosophila Nipped-B mutants model Cornelia de Lange syndrome in growth and behavior. PLoS Genet. 11, e1005655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schaaf CA et al. (2013) Cohesin and polycomb proteins functionally interact to control transcription at silenced and active genes. PLoS Genet. 9, e1003560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kennison JA and Tamkun JW (1988) Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc. Natl. Acad. Sci 85, 8136–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hallson G et al. (2008) The Drosophila cohesin subunit Rad21 is a trithorax group (trxG) protein. Proc. Natl. Acad. Sci 105, 12405–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pherson M et al. (2017) Polycomb repressive complex 1 modifies transcription of active genes. Sci. Adv 3, e1700944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kassis JA et al. (2017) Polycomb and trithorax group genes in Drosophila. Genetics 206, 1699–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schaaf CA et al. (2009) Regulation of the Drosophila Enhancer of split and invected-engrailed gene complexes by sister chromatid cohesion proteins. PLoS One 4, e6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park SY et al. (2012) Regulation of Polycomb group genes Psc and Su(z)2 in Drosophila melanogaster. Mech. Dev 128, 536–547 [DOI] [PubMed] [Google Scholar]
- 70.Strübbe G et al. (2011) Polycomb purification by in vivo biotinylation tagging reveals cohesin and Trithorax group proteins as interaction partners. Proc. Natl. Acad. Sci 108, 5572–5577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cunningham MD et al. (2012) Wapl antagonizes cohesin binding and promotes Polycomb-group silencing in Drosophila. Development 139, 4172–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]