Abstract
Shapeshifters, in common mythology, are entities that can undergo multiple physical transformations. As our understanding of G protein-coupled receptors (GPCRs) has accelerated and been refined over the last two decades, we now understand that GPCRs are not static proteins, but rather dynamic structures capable of moving from one posture to the next, and adopting unique functional characteristics at each transition. This model of GPCR dynamics underlies our current understanding of biased agonism—how different ligands to the same receptor can generate different intracellular signals—and constitutive receptor activity, or the level of unbound basal receptor signaling that can be attenuated by inverse agonists. From information derived from related class B receptors, we have recently modeled the structure and molecular dynamics of the full-length pituitary adenylate cyclase activating polypeptide (PACAP, Adcyap1)—selective PAC1 receptor (PAC1R, Adcyap1r1). The class B receptors are different from the class A GPCRs in part from the presence of a large extracellular domain (ECD); the transitions of the ECD along with the dynamics of the transmembrane domains (TMD or 7TM) of the PAC1R describes a series of open- and closed-state conformations that appear to identify the mechanisms for receptor activation. The PAC1R shapeshifts also have the ability of delineating the mechanisms and the design of reagents that may direct biased agonism (or antagonism) for potential therapeutics.
Keywords: PACAP, PAC1 receptor, Molecular dynamics, Markov state modeling, Transition path theory
The more than 800 G protein-coupled receptors (GPCRs) represent the largest family of transmembrane signaling proteins. The family is characterized by heptahelical transmembrane segments linked by three extracellular and three intracellular loops, and from sequence homology and structural specializations, the GPCRs can be divided into five classes: class A (rhodopsin), class B (secretin), class C (glutamate), adhesion and frizzled. The diversity in receptor structure, activation and signaling allows cells to detect a multitude of extracellular instructional cues from neurotransmitters, hormones, paracrine regulators, light, odorants and tastants (Rosenbaum et al. 2009; Wacker et al. 2017; Katritch et al. 2013); hence, GPCRs are critical homeostatic regulators of nearly all physiological systems and are common therapeutic targets for pathophysiological conditions.
The refinements in the cellular, molecular, biochemical and pharmacological studies on GPCRs over the last two decades have vastly changed our perspectives and understandings of GPCR mechanisms. Unlike the “lock and key” concept of receptor mechanism, in which specific ligands can uniquely switch a poised receptor from an inactive “off” state or intermediate transitional state to an active “on” conformation to allow intracellular transducer or scaffolding protein associations for second messenger signaling, the processes are now understood to be more intricate and dynamic to drive the diversity in acute and long-term GPCR responses in many cell types. As well discussed previously (Geppetti et al. 2015; Luttrell et al. 2015), the “lock and key” model of GPCRs did not appear to accommodate several observations on GPCR responses. Signal transduction studies have now shown that many GPCRs can be coupled to multiple intracellular signaling pathways, and yet different ligands can have differential potencies in second messenger responses. The β-adrenergic receptor antagonists carvedilol and propranolol, for example, are actually biased agonists for β-arrestin and ERK activation (Pupo et al. 2016). Isoproterenol is an unbiased β-adrenergic agonist; isoetharine by contrast appears to be biased for ERK activation, whereas salbutamol and albuterol are biased for cAMP signaling. The μ-opioid receptor agonists TRV130 and PZM21 are biased for analgesic Gi activation over ERK signaling, which may have important therapeutic implications (Luttrell et al. 2015; Manglik et al. 2016). Further, endogenous or synthetic ligands can act as inverse agonists to attenuate basal receptor activity; the endogenous agouti-related peptide (AGRP), for example, is an inverse agonist at melanocortin receptors (MC3R/MC4R) to control body weight (Adan and Kas 2003). All of these observations may not be anticipated if the closed and open states of the standard model were equated, respectively, to an activated or inactivated state of a receptor.
From current computational molecular modeling analyses of receptor energy states, the GPCRs are now appreciated not to be static, but to transition fluidly and fleetingly from one conformation to the next, that in aggregate form an intricate ensemble of receptor microstates (Kenakin and Miller 2010; Park 2012; Li et al. 2013; Latorraca et al. 2017). Fine-tuned by the other regulatory influences such as post-translational modifications, accessory proteins and environmental factors, the individual microstates may adopt conformations that can preferentially favor particular signaling events. Specific ligands and allosteric modulators may restrict GPCRs to conformations to favor specific signaling pathways; conversely, in the absence of endogenous ligands, constitutive (basal) GPCR activity from inherently active microstates can be attenuated by inverse agonist regulators that promote GPCRs to inactive conformations. The dynamics of GPCR microstate ensembles provide signaling complexity and diversity, and offer opportunities to develop reagents that amplify desirable GPCR signals and functions, from those that cause maladaptive consequences.
Among the nearly 700 members of the class A receptors, including the β-adrenergic, muscarinic, serotoninergic, dopaminergic and purinergic receptors, the structures of approximately 32 receptors have been solved from crystallography studies (Zhang et al. 2015; Pándy-Szekeres et al. 2018). Perhaps not surprisingly, nearly 40% of all pharmaceutical drugs target class A receptors (Luttrell et al. 2015; Zhang et al. 2015). The structural solutions to the class B receptors, which encompass 15 members including CRH, glucagon, GLP-1, secretin, vasoactive intestinal peptide (VIP) and pituitary adenylate cyclase activating polypeptide (PACAP) receptors, have lagged and the 7TM structures of only 4 members have become available only within the last 5 years (Hollenstein et al. 2013; Siu et al. 2013; Yang et al. 2015; Jazayeri et al. 2016; Jazayeri et al. 2017; Liang et al. 2017; Song et al. 2017; Zhang et al. 2017a; Zhang et al. 2018). Although class B receptors are fewer in number, these receptors are increasingly being recognized as mediating some of the most critical physiological functions, including feeding and energy balance, sensory mechanisms, bone metabolism and stress responses (Harmar et al. 2012; Culhane et al. 2015). Hence, the dysregulation of class B receptor signaling has been implicated in diabetes, obesity, acute and chronic pain including migraine, osteoporosis, and maladaptive stress responses including anxiety, depression, post-traumatic stress disorders (PTSD) and relapse from addiction (Hammack et al. 2009; Stroth and Eiden 2010; Ressler et al. 2011; Roman et al. 2014; Culhane et al. 2015; Hammack and May 2015; Missig et al. 2016; Edvinsson et al. 2018; Miles et al. 2018).
PACAP and PAC1 Receptors
PACAP (ADCYAP1), a member of the VIP/secretin/glucagon family of bioactive peptides, exerts diverse pleiotropic effects in a variety of neurophysiological systems in part upon binding to the class B PAC1 receptor (ADCYAP1R1; Sherwood et al. 2000; Vaudry et al. 2009; Harmar et al. 2012). From the PACAP precursor molecule, the predominant alternative post-translationally processed α-amidated bioactive peptide is the 38-amino acid residue PACAP1–38; the PACAP1–27 bioactive variant is equipotent, but typically 10- to 100-fold less abundant in many tissues (Miyata et al. 1989; Kimura et al. 1990; Arimura et al. 1991; Vaudry et al. 2009). The activation of class B receptors has been suggested to follow a two-domain model (Culhane et al. 2015); the binding of the peptide C-terminus to the large extracellular domain (ECD) of the GPCR facilitates the presentation of the peptide N-terminus to the core of the transmembrane domain (TMD or 7TM) for receptor activation. As with other class B receptor ligands, the C-terminus α-helix of PACAP binds to the ECD of the PAC1 receptor. N-terminal truncation of the PACAP peptide generates PAC1 receptor antagonists; PACAP6–38 competes with the binding of the endogenous ligand, but lacks the necessary N-terminus sequences for PAC1 receptor activation. Previous studies have implicated PACAP/PAC1 receptor signaling in stress-related behavioral disorders, chronic pain and drug addiction relapse, yet approaches to ameliorate the maladaptive PACAP effects have not been developed from a paucity of PAC1 receptor structural and mechanistic information. Hence, as for many class B GPCRs, there are few small molecule compounds that target PACAPergic systems for potential therapeutics.
PACAP binds to three receptor subtypes (Vaudry et al. 2009; Harmar et al. 2012; Blechman and Levkowitz 2013). The PACAP peptides bind selectively to PAC1 receptors; VIP and PACAP bind with near equal high affinity to VPAC1 and VPAC2 receptors (VIPR1 and VIPR2, respectively). Even among class B receptors, the PAC1 receptor is unique in the number of receptor variants from alternative exon usage. PAC1 receptor isoforms arise from the presence or absence of two 84-bp Hip and/or Hop cassettes that encode segments inserted into the 3rd intracellular loop (Spengler et al. 1993; Pisegna and Wank 1996; Vaudry et al. 2009; Harmar et al. 2012; Blechman and Levkowitz 2013). Hence, the PAC1 receptor may be null (neither Hip nor Hop inserts), Hip, Hop1 or HipHop; there may also be shortened variants of Hop1 resulting in Hop2 or HipHop2 inserts. These variants may result in differential receptor interactions with Gαs, Gαq and β-arrestin to impact downstream signaling with respect to potency, efficacy and duration. Also unique to PAC1 receptors and not to other class B GPCRs is the addition of a 21-amino acid segment between the β3 and β4 sheets in the ECD (Pantaloni et al. 1996; Harmar et al. 2012; Blechman and Levkowitz 2013). From polymerase chain reaction (PCR) transcript analyses, nearly all regions of the central nervous system appear to express predominantly both the PAC1null and PAC1Hop1 receptor with the 21-amino acid ECD insert; the null variant may be approximately 2-fold more abundant than the Hop1 isoform. This contrasts with autonomic sympathetic ganglia in which the PAC1Hop1 receptor variant (with the 21-amino acid ECD insert) is the major PAC1 receptor isoform (May and Braas 1995; Braas and May 1999). As the 3rd intracellular loop (ICL3) of GPCRs is a critical determinant of G protein and β-arrestin transducer association and function, the PAC1 receptor variants can add diversity to PAC1 receptor signaling.
PAC1 Receptor Modeling and Microstates
Despite the many physiological roles of the PACAPergic system, there are few pharmacological options for potential therapeutic interventions. Accordingly, using currently available data as templates (Hollenstein et al. 2014; de Graaf et al. 2017; Wootten et al. 2017), we have recently constructed the full-length human PAC1null receptor variant for structural studies and performed microsecond-long molecular dynamic (MD) simulations to delineate PAC1 receptor mechanisms and facilitate rational drug design (Liao et al. 2017). The three-dimensional atomistic homology model of the PAC1 receptor was built from the ECD template from the crystal structure of the “very short” ECD PAC1 receptor variant (PDBID: 3 N94) and the heptahelical TMD template from corticotropin-releasing hormone receptor (CRHR1, PDBID: 4K5Y) and glucagon receptor (GCGR, PDBID: 4L6R). The 21-amino acid ECD sequence (residues 89–109 absent from the ECD PBDID 3 N94 structure) was inserted between the β3 and β4 strands as an extended loop, and the linker region (residues 126–149) was modeled as a helical structure with a coiled N-terminus. To mimic the membrane-bound environment, the 7TM was embedded in the bilayer model of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), while the ECD was fully exposed to the solvent. After energy minimization and 100-ns MD simulation for each system, the most stable model was selected to generate PAC1 receptor states of four different ECD orientations via rotations of the backbone dihedrals in the linker. All of the models, starting with outstretched ECDs, were simulated for 2.0–2.9 μs on the MD-specialized Anton supercomputer. Several stable full-length ligand-free PAC1 receptor models were obtained following 10 μs of total simulation time.
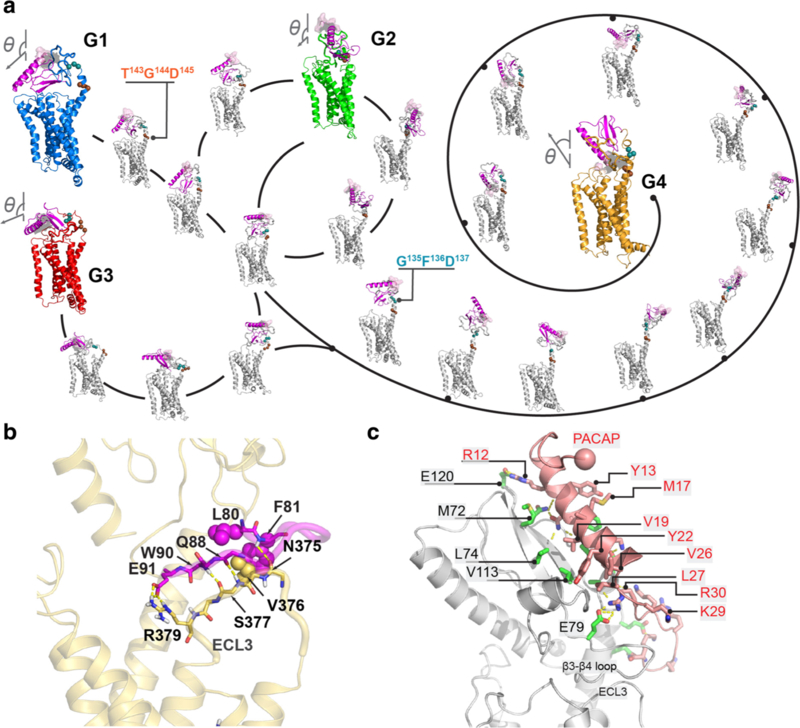
In aggregate with the Markov state model and transition path theory (Weinan and Vanden-Eijnden 2006; Bowman et al. 2009; Prinz et al. 2011), these microsecond-long simulations sampled PAC1 receptor conformational microstates that interconnected three closed (G1–G3) and one open ECD (G4) receptor states (Fig. 1A). Analyses of the transition pathways demonstrated that the conversion paths among the closed states are relatively short (3–5 microstates), while the path to the open state is more remote (11–13 microstates). Thus, the PAC1 receptor open and closed states are likely compartmentalized; the shortest transition times among the closed states are approximately 20–30 μsec, whereas the timescales for open to closed transitions are singularly protracted from 300 to 600 μsec. From these results, we sought to understand the transitional steps for insights into PAC1 receptor activation.
Fig. 1.
PAC1 receptor ensemble of open (G4) and closed (G1–G3) states. (A) Diagram of PAC1R conformational transition by the Markov state model and transition path theory. Vector diagrams (N-to-C of helix 1 in ECD) show the ECD orientations. The ECD N-terminal is show in purple. (B) The “zipper” between residues 80–91 and ECL3 in the open state. Key polar and hydrophobic side chains are shown in sticks and spheres, respectively. (C) Interactions between PACAP1–38 and PAC1R in ECD-open conformation. The sphere indicates the N-terminus of the PACAP peptide. Reprinted with permission from Liao et al. (2017) Sci Rep 7 (1):5427
PAC1 Receptor Extracellular Domain Interactions
To gain a rational understanding of the transitional dynamics for the PAC1 receptor, we first examined the interactions between the ECD and TMD with the hypothesis that similar conformational steps likely underlie PACAP-mediated receptor activation. The ECD of the PAC1 receptor is tethered to the heptahelical TMD via a flexible linker/stalk region (residues 126–149). Shown by MD simulations, the ECD is extremely dynamic in a solvated environment when there are few contacts with the TMD. As described above, the PAC1 receptor is unique from other class B receptors in the presence of a short 21-amino acid loop segment (residues 89–109) between the antiparallel β3 (residues 70–76) and β4 (residues 113–118) sheets of the ECD. For the unbound receptor, the ECD 21-amino acid loop segment contributes to the formation of a backbone zipper with the 3rd extracellular loop (ECL3) through hydrogen bonds at F81-N375ECL3 and Q88-S377ECL3, side-chain salt bridge formation at E91-R379ECL3, and hydrophobic interactions at L80-F81-V376ECL3, thereby supporting and stabilizing the ECD to sustain the PAC1 receptor in an open conformational state (Fig. 1B). This singular distinguishing feature appears key in maintaining the open state of the PAC1 receptor for hundreds of microseconds; indeed, MD simulation of the receptor with the 21-amino acid segment deletion resulted in rapid open to closed state transition within 0.1 μsec, comparable to the transition times observed previously for the glucagon receptor (Yang et al. 2015; Zhang et al. 2018).
The open state of the receptor allows high affinity PACAP1–38 binding to the ECD (Fig. 1C); the C-terminal α-helix formed by PACAP residues 8–27 binds to the ECD groove through hydrophobic (PACAP residues Y13, M17, A18, V19, Y22, L23, V26 and L27 with PAC1R ECD residues M72, L74, V113, V114, F127, P128 and A133) and electrostatic interactions (residue R12PACAP at ECD residue E120PAC1R). This is the prototypic first step in the two-domain receptor activation process; subsequent dynamics of the peptide-bound-ECD would facilitate N-terminal peptide binding to the TMD to initiate mechanisms for intracellular signaling.
PAC1 Receptor Transmembrane Domain Networks
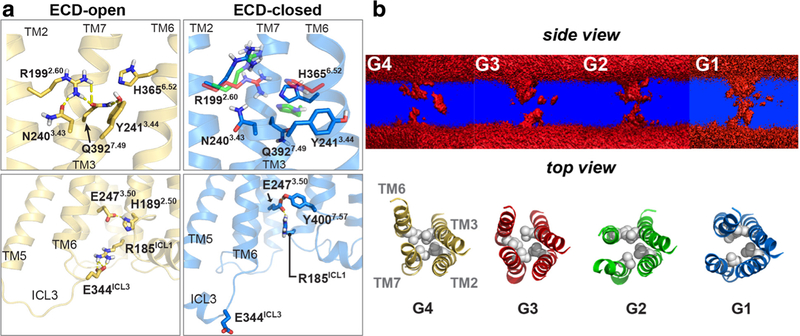
As class B receptors have wider and deeper V-shaped ligand-binding pockets than class A receptors (Hollenstein et al. 2013; Siu et al. 2013), we sought to understand the changes in the TMD core among the G1–G4 transitional states in the context of ionic networks and the creation of a water pathway (Fig. 2). The ECD open receptor (G4) can form several ionic “locks” in the transmembrane (N2403.43-R1992.60-Q3927.49-Y2413.44; Wootten numbering) and intracellular domains (R185ICL1-E2473.50-Y4007.57; Fig. 2A); the transmembrane hydrophobic region (L1922.53, L2443.47, L3586.45 and V3967.53) also constrains the conformations. TM2, TM3, TM6 and TM7 represent the principal boundaries of the receptor core (Fig. 2B). During the open-to-closed transition, the inward shift of TM6 (especially H3656.52) can gradually disrupt the ionic network, resulting in an increase in water density from the G4 (open receptor state) to G1 (closed receptor) transition, i.e., a water pathway through the TMD appears occluded in the open G4 receptor configuration, but most apparent in the closed G1 microensemble state. The water density is less, but still present in the G2, and largely disappears in G3.
Fig. 2.
PAC1 receptor 7TM interactions and water pathway formation. (A) Upper panels: hydrogen-bond network within the 7TM; lower panels: the salt bridge E344ICL3-R185ICL1 in ECD open and closed states. (B) Upper panel, water density cross section (upper panel) in side view of the receptor. Lower panel, top view of receptor core with TM2–TM6 and TM3–TM7 boundaries. The hydrophobic L1922.53, L2443.47, L3586.45 and V3967.53 residues are shown as spheres. An open water pathway is suggested in G1 and G2. Reprinted with permission from Liao et al. (2017) Sci Rep 7 (1):5427
Hence, from these observations, the upright orientation of the ECD and tethering of the ECD loop to ECL3 in the G4 open state likely constrains the movements of TM6 and TM7 in the TMD. In detailed examinations, the occlusion of the water pathway in G3 and G4 is likely a result of the hydrophobic locks formed by L1922.53, L2443.47, L3586.45 and V3967.53, which were identified also in the GCGR and CRHR1 structures (PDBIDs: 4L6R and 4K5Y, respectively). However, when the ECD becomes untethered to ECL3, TM5, TM6 and TM7 are no longer constrained and the transitions from the open pose of G4 to the closed G1 and G2 states result in the creation of a contiguous water passageway in major part from an oblique ~ 2 Å downward shift of TM6. The distances between TM2–TM6, from Cγ–Cγ measurements between L1922.53 and L3586.45, in the closed G1 and G2 configurations are 9.9 and 9.2 Å, respectively, which are wider than those in the G3 or open G4 states (7.7 and 8.8 Å, respectively). Similarly, the Cγ–Cβ distances between L2443.47 and V3967.53 in TM3–TM7 of G1 and G2 are 10.6 and 8.4 Å, respectively, and wider than those in G3 and G4 (8.4 and 6.4 Å, respectively). In short, upon ECD dynamics, the hydrophobic locks within the TMD of the apo PAC1 receptor can be rearranged to create a water pathway during an open-to-closed transition. These effects are distinct from the water pathway mechanism in class A GPCRs that have a tryptophan toggle switch in TM6 to modulate the opening of the pathway and receptor activation (Yuan et al. 2015).
PAC1 Receptor ECD – TMD Network Regulation of Intracellular Loops
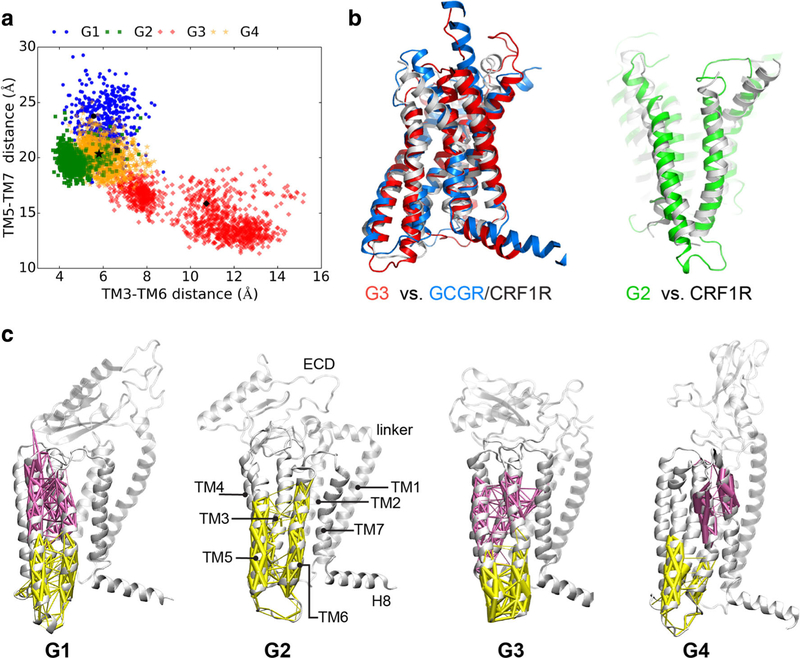
In addition to changes in PAC1 receptor core structure among the transitional states, we also investigated how the dynamics of the PAC1 receptor ECD with the TMD can potentially rearrange the architecture of the intracellular loops to impact the docking of G proteins, β-arrestin or other effector proteins for signaling. Even with very similar initial TMD conformations, the helices in each of the G1–G4 states in the MD simulations can undergo different transitional pathways to reach distinct intracellular domain configurations. Examining the paired distances between intracellular loops (Fig. 3A), the G2 microstates have the most compact TM3–TM6 (4~6 Å) and TM5–TM7 (17~22 Å) distances, while those for G1, G3 and G4 microstates appear more dispersed. For G1, G3 and G4, the intracellular helical distances for TM3–TM6, range 4.5~8 Å, and those for TM5–TM7 are 17~27 Å. Additionally, G3 is capable of segregating within 1.8 μsec into a different conformational microstate group with TM3–TM6 distances of 9~14 Å, and TM5–TM7 distances of 12~17 Å. Interestingly, the intracellular helical arrangement of the G3 microstates is similar to those for the TMD structures for GCGR and CRHR1 (PBDID: 4L6R and 4K5Y), while G2 displays a similar wide V-shaped ligand binding pocket observed in CRHR1 (PBDID: 4K5Y; Fig. 3B). These observations demonstrate convincingly that ECD–TMD interactions have significant roles in determining the helical architecture at the intracellular face of the receptor, and that the different conformational states can be linked together in dynamic networks.
Fig. 3.
PAC1 receptor networks and communities. (A) TM3–TM6 distance (Cβ distance between L251 and L351) is shown as a function of TM5–TM7 distance (Cβ distance between L331 and N404). (B) Structural superpositions of G3 state with GCGR and CRHR1 (PDBIDs: 4L6R and 4K5Y) and G2 state with CRHR1. (C) Community analysis of ECD closed (G1, G2 and G3) and open (G4) states of PAC1 receptor. The community decompositions of TM6 are displayed with weighted edges (thicker edges show greater correlation). Dynamical networks were generated from the last 150 ns of each MD trajectory
To understand such mechanistic effects, the coherent conformational changes were linked together by dynamical network analysis (DNA; Alexander et al. 2010; Black Pyrkosz et al. 2010; Sethi et al. 2009) which has been applied widely to study social networking and traffic control. In dynamical networks, the Cα atoms of amino acid residues are represented as “nodes” and “network communities” are constructed from MD simulation trajectories, which identify the strength of integrations and connections among the nodes and communities (Sethi et al. 2009). Two nodes are interconnected by an edge if the heavy atoms of any two residues are within 4.5 Å for more than 75% of the simulation time. Hence, communities correspond to sets of residues that move in concert with each other, i.e., nodes within one community have stronger connections than between communities.
The community decompositions of TM6 with weighted edges are displayed in Fig. 3C for four ligand-free states of the PAC1 receptor. There are more communities formed among the upper halves of transmembrane helices in the G4 state, reflecting weaker communications in the open apo receptor. Notably in G1, G3 and G4 states, TM6 can be parsed into extracellular (purple) and intracellular (yellow) halves; further, the extracellular half of TM6 is observed to merge into communities containing TM3 or TM7. In the G2 state, in which the ECD interacts with ECL3, the entire TM6 is observed to be in the same community with most of TM5 and the lower half of TM3. As the G2 state and the small molecule antagonist-bound CRHR1 (PDBID: 4K5Y) have similar wide V-shaped ligand-binding pockets (Fig. 3B), the position of the small molecule near the middle of TM5–TM6 of CRHR1 likely plays a similar role in constraining the movements of TM5 and TM6 as a single community. In all of the ligand-free states, TM6 is either separated into two communities, as in G1, G3 and G4, or demonstrates topological dependency with adjacent TM helices (stronger correlations) as in G2. Since all of the correlations and communications are weaker between communities than those within a single community, these ligand-free states may exhibit weak signal propagation from the orthostatic pocket at the extracellular face of the receptor to the intracellular G protein binding site.
PAC1 Versus Glucagon Receptor
The different structural studies of the glucagon receptor have yielded variations in the conformational postures of the receptor. In the original crystallography solution of GCGR with the small molecule antagonist NNC0640 or MK-0893, the ECD was truncated, among other modifications, resulting in a TMD structure with a linker/stalk region structured as an α-helix extension from TM1 (Siu et al. 2013; Jazayeri et al. 2016). Subsequent simulation of this GCGR structure with the ECD attached revealed that the receptor preferred to be in the closed conformation (the ECD folded over the TMD), but that glucagon binding to the ECD, while the GCGR was poised in the open conformation (ECD extended from the TMD), maintained the open receptor state for receptor activation (Yang et al. 2015). Hence, the full-length GCGR in this model appeared to exist in two states, open or closed, dependent on the hinge action of the linker region. However, GCGR crystallography solutions of the full-length receptor (PDBID: 5XEZ) using mAb1 to stabilize the ECD revealed that the linker/ stalk region was more intricate and that instead of an α-helix extension from TM1, the neck could adopt a β-strand configuration for interactions with a β-hairpin of ECL1 to form a β-sheet structure (Zhang et al. 2017a). This structural detail allowed stabilization of the GCGR ECD in an inactive open state. However, more recent structural solutions of the full-length GCGR with the modified glucagon partial agonist NNC1702 revealed that upon peptide binding to the ECD, the linker/stalk region was poised as an α-helix similar to the original solution (PBDID: 4L6R); further, the ECL1 no longer interacted with linker but formed a short α-helix that may help to stabilize peptide–receptor interactions (Zhang et al. 2018). Hence, glucagon peptide binding to the GCGR in this open conformation has been hypothesized to displace the ECL1–linker interaction to facilitate peptide-bound GCGR dynamics for TMD activation. A similar binding pose was revealed in the cryo-EM structure of full-length activated glucagon-like peptide-1 receptor (Zhang et al. 2017b). These recent GCGR structure and mechanism studies closely approximated our current PAC1 receptor ensemble states from MD modeling. As described previously, the 21-amino acid ECD loop segment of the PAC1 receptor could interact with ECL3 and establish a molecular zipper to stabilize PAC1 receptor in the open G4 state. Further, the PAC1 receptor α-helix linker/stalk region could melt into a coil to allow the receptor to adopt the fully closed G3 state. Hence despite some differences in details, the GCGR and PAC1 receptors appear to share mechanistic similarities in maintaining the open receptor states. Whether there are also similarities in ECD transitions and dynamics for bound ligand presentation to the TMD for receptor activation still needs to be determined from peptide agonist and antagonist modeling and simulations.
Shapeshifters in Motion
From computational modeling and molecular dynamics simulations, our recent studies have shown that the PAC1 receptor can undergo a series of transitional conformational changes that in aggregate form an ensemble of microstates. The modeling suggests that the PAC1 receptor appears to preferentially adopt an open conformational state, stabilized by the unique interactions between the ECD 21-amino acid loop segment and the ECL3; disruption of this interaction appears to permit several closed receptor macrostates that rely heavily on the dynamics of the receptor stalk/linker regions. Some of the microstates in the apo receptor likely simulate the activated receptor, conferring a level of basal receptor activity. How PACAP binding to the PAC1 receptor may facilitate or stabilize activated receptor microstates is under study. Further, the sandfly peptide maxadilan is the only PAC1 receptor-specific agonist identified to date (Moro and Lerner 1997); the identification of maxadilan binding to PAC1 receptor orthosteric or allosteric sites and the subsequent changes in receptor conformation may inform the development of specific receptor agonists and antagonist. These may further understanding of receptor conformations and mechanisms driving receptor activation of particular signaling pathways, i.e., Gαs vs Gαq vs. β-arrestin preferring signaling. Increasingly, specific signaling pathways are recognized to mediate particular cellular or physiological responses, while other pathways may facilitate maladaptations leading to disorders. Mu opioid receptor-mediated Gi signaling for example, blocks nociceptive signaling, whereas the concomitant activation of ERK pathways appears to mediate some of the undesirable effects of opioids including tolerance, respiratory depression and constipation (Manglik et al. 2016). Similarly, some of the chronic effects of PAC1 receptor-mediated ERK signaling may be associated with adverse stress-related behaviors and the emotional consequences of chronic pain (Missig et al. 2014; Missig et al. 2016). Understanding how specific ligands or allosteric mediators may channel the PAC1 receptor to specific microstates and signaling pathways may allow the preparation of reagents that favor the benefits of PACAP signaling without engaging those that can lead to maladaptive effects.
Acknowledgements
We thank the UVM REACH grant for the support to this research. Computational resources were provided by Anton (PSC, NIH P41GM103712-S1), Stampede (XSEDE, NSF ACI-1053575), and Vermont Advanced Computing Core (VACC).
Footnotes
Compliance with Ethical Standards
Conflict of Interest The authors declare no competing financial interests.
References
- Adan RA, Kas MJ (2003) Inverse agonism gains weight. Trends Pharmacol Sci 24:315–321 [DOI] [PubMed] [Google Scholar]
- Alexander RW, Eargle J, Luthey-Schulten Z (2010) Experimental and computational determination of tRNA dynamics. FEBS Lett 584: 376–386 [DOI] [PubMed] [Google Scholar]
- Arimura A, Somogyvári-Vigh A, Miyata A, Mizuno K, Coy DH, Kitada C (1991) Tissue distribution of PACAP as determined by RIA: highly abundant in the rat brain and testes. Endocrinology 129: 2787–2789 [DOI] [PubMed] [Google Scholar]
- Black Pyrkosz A, Eargle J, Sethi A, Luthey-Schulten Z (2010) Exit strategies for charged tRNA from GluRS. J Mol Biol 397:1350–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blechman J, Levkowitz G (2013) Alternative splicing of the pituitary adenylate cyclase-activating polypeptide receptor PAC1: mechanisms of fine tuning of brain activity. Front Endocrinol 4:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GR, Huang X, Pande VS (2009) Using generalized ensemble simulations and Markov state models to identify conformational states. Methods 49:197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braas KM, May V (1999) Pituitary adenylate cyclase-activating polypeptides directly stimulate sympathetic neuron neuropeptide Y release through PAC(1) receptor isoform activation of specific intracellular signaling pathways. J Biol Chem 274(39):27702–27710 [DOI] [PubMed] [Google Scholar]
- Culhane KJ, Liu Y, Cai Y, Yan EC (2015) Transmembrane signal transduction by peptide hormones via family B G protein-coupled receptors. Front Pharmacol 6:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf C, Song G, Cao C, Zhao Q, Wang MW, Wu B, Stevens RC (2017) Extending the structural view of class B GPCRs. Trends Biochem Sci 42:946–960 [DOI] [PubMed] [Google Scholar]
- Edvinsson L, Tajti J, Szalárdy L, Vécsei L (2018) PACAP and its role in primary headaches. J Headache Pain 19(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geppetti P, Veldhuis NA, Lieu T, Bunnett NW (2015) G protein-coupled receptors: dynamic machines for signaling pain and itch. Neuron 88: 635–649 [DOI] [PubMed] [Google Scholar]
- Hammack SE, May V (2015) Pituitary adenylate cyclase activating polypeptide in stress-related disorders: data convergence from animal and human studies. Biol Psychiatry 78:167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammack SE, Cheung J, Rhodes KM, Schutz KC, Falls WA, Braas KM, May V (2009) Chronic stress increases pituitary adenylate cyclase-activating peptide (PACAP) and brain-derived neurotrophic factor (BDNF) mRNA expression in the bed nucleus of the stria terminalis (BNST): roles for PACAP in anxiety-like behavior. Psychoneuroendocrinology 34:833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmar AJ, Fahrenkrug J, Gozes I, Laburthe M, May V, Pisegna JR, Vaudry D, Vaudry H, Waschek JA, Said SI (2012) Pharmacology and functions of receptors for vasoactive intestinal peptide and pituitary adenylate cyclase-activating polypeptide: IUPHAR review 1. Br J Pharmacol 166:4–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenstein K, de Graaf C, Bortolato A, Wang MW, Marshall FH, Stevens RC (2014) Insights into the structure of class B GPCRs. Trends Pharmacol Sci 35:12–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenstein K, Kean J, Bortolato A, Cheng RKY, Doré AS, Jazayeri A, Cooke RM, Weir M, Marshall FH (2013) Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature 438:438–443 [DOI] [PubMed] [Google Scholar]
- Jazayeri A, Doré AS, Lamb D, Krishnamurthy H, Southall SM, Baig AH, Bortolato A, Koglin M, Robertson NJ, Errey JC, Andrews SP, Teobald I, Brown AJH, Cooke RM, Weir M, Marshall FH (2016) Extra-helical binding site of a glucagon receptor antagonist. Nature 533:274–277 [DOI] [PubMed] [Google Scholar]
- Jazayeri A, Rappas M, Brown AJH, Kean J, Errey JC, Robertson NJ, Fiez-Vandal C, Andrews SP, Congreve M, Bortolato A, Mason JS, Baig AH, Teobald I, Doré AS, Weir M, Cooke RM, Marshall FH (2017) Crystal structure of the GLP-1 receptor bound to a peptide agonist. Nature 546:254–258 [DOI] [PubMed] [Google Scholar]
- Katritch V, Cherezov V, Stevens RC (2013) Structure-function of the G protein-coupled receptor superfamily. Annu Rev Pharmacol Toxicol 53:531–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenakin T, Miller LJ (2010) Seven transmembrane receptors as shapeshifting proteins: the impact of allosteric modulation and functional selectivity on new drug discovery. Pharmacol Rev 62:265–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura C, Ohkubo S, Ogi K, Hosoya M, Itoh Y, Onda H, Miyata A, Jiang L, Dahl RR, Stibbs HH et al. (1990) A novel peptide which stimulates adenylate cyclase: molecular cloning and characterization of the ovine and human cDNAs. Biochem Biophys Res Commun 166: 81–89 [DOI] [PubMed] [Google Scholar]
- Latorraca NR, Venkatakrishnan AJ, Dror RO (2017) GPCR dynamics: structures in motion. Chem Rev 117(1):139–155 [DOI] [PubMed] [Google Scholar]
- Li J, Jonsson AL, Beuming T, Shelley JC, Voth GA (2013) Ligand-dependent activation and deactivation of the human adenosine a(2A) receptor. J Am Chem Soc 135:8749–8759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang YL, Khoshouei M, Radjainia M, Zhang Y, Glukhova A, Tarrasch J, Thal DM, Furness SGB, Christopoulos G, Coudrat T, Danev R, Baumeister W, Miller LJ, Christopoulos A, Kobilka BK, Wootten D, Skiniotis G, Sexton PM (2017) Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature 546:118–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C, Zhao X, Brewer M, May V, Li J (2017) Conformational transitions of the pituitary adenylate cyclase-activating polypeptide receptor, a human class B GPCR. Sci Rep 7:5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttrell LM, Maudsley S, Bohn LM (2015) Fulfilling the promise of “biased” G protein-coupled receptor agonism. Mol Pharmacol 88: 579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manglik A, Lin H, Aryal DK, McCorvy JD, Dengler D, Corder G, Levit A, Kling RC, Bernat V, Hübner H, Huang XP, Sassano MF, Giguère PM, Löber S, Duan D, Scherrer G, Kobilka BK, Gmeiner P, Roth BL, Shoichet BK (2016) Structure-based discovery of opioid analgesics with reduced side effects. Nature 537:185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May V, Braas KM (1995) Pituitary adenylate cyclase-activating polypeptide (PACAP) regulation of sympathetic neuron neuropeptide Y and catecholamine expression. J Neurochem 65:978–987 [DOI] [PubMed] [Google Scholar]
- Miles OW, Thrailkill EA, Linden AK, May V, Bouton ME, Hammack SE (2018) Pituitary adenylate cyclase-activating peptide in the bed nucleus of the stria terminalis mediates stress-induced reinstatement of cocaine seeking in rats. Neuropsychopharmacology 43:978–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missig G, Roman CW, Vizzard MA, Braas KM, Hammack SE, May V (2014) Parabrachial nucleus (PBn) pituitary adenylate cyclase activating polypeptide (PACAP) signaling in the amygdala: implication for the sensory and behavioral effects of pain. Neuropharmacology 86:38–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missig G, Mei M, Vizzard MA, Braas KM, Waschek JA, Ressler KJ, Hammack SE, May V (2016) Parabrachial PACAP activation of amygdala endosomal ERK signaling regulates the emotional component of pain. Biol Psychiatry (in press):http://www.biologicalpsychiatryjournal.com/article/S0006-3223(0016)32725-32721/fulltext [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata A, Arimura A, Dahl RR, Minamino N, Uehara A, Jiang L, Culler MD, Coy DH (1989) Isolation of a novel 38 residue-hypothalamic polypeptide which stimulates adenylate cyclase in pituitary cells. Biochem Biophys Res Commun 164:567–574 [DOI] [PubMed] [Google Scholar]
- Moro O, Lerner EA (1997) Maxadilan, the vasodilator from sand flies is a specific pituitary adenylate cyclase activating polypeptide type I receptor agonist. J Biol Chem 272:966–970 [DOI] [PubMed] [Google Scholar]
- Pándy-Szekeres G, Munk C, Tsonkov TM, Mordalski S, Harpsøe K, Hauser AS, Bojarski AJ, Gloriam DE (2018) GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res 46:D440–D446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaloni C, Brabet P, Bilanges B, Dumuis A, Houssami S, Spengler D, Bockaert J, Journot L (1996) Alternative splicing in the N-terminal extracellular domain of the pituitary adenylate cyclase-activating polypeptide (PACAP) receptor modulates receptor selectivity and relative potencies of PACAP-27 and PACAP-38 in phospholipase C activation. J Biol Chem 271:22146–22151 [DOI] [PubMed] [Google Scholar]
- Park PS (2012) Ensemble of G protein-coupled receptor active states. Curr Med Chem 19:1146–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisegna JR, Wank SA (1996) Cloning and characterization of signal transduction of four splice variants of the human pituitary adenylate cyclase activating polypeptide receptor. J Biol Chem 271:17267–17274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz JH, Wu H, Sarich M, Keller B, Senne M, Held M, Chodera JD, Schütte C, Noé F (2011) Markov models of molecular kinetics: generation and validation. J Chem Phys 134:174105. [DOI] [PubMed] [Google Scholar]
- Pupo AS, Duarte DA, Lima V, Teixeira LB, Parreiras-E-Silva LT, Costa-Neto CM (2016) Recent updates on GPCR biased agonism. Pharmacol Res 112:49–57 [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Mercer KB, Bradley B, Jovanovic T, Mahan A, Kerley K, Norrholm SD, Kilaru V, Smith AK, Myers AJ, Ramirez M, Engel A, Hammack SE, Toufexis D, Braas KM, Binder EB, May V (2011) Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature 470:492–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman CW, Lezak KR, Hartsock MJ, Falls WA, Braas KM, Howard AB, Hammack SE, May V (2014) PAC1 receptor antagonism in the bed nucleus of the stria terminalis (BNST) attenuates the endocrine and behavioral consequences of chronic stress. Psychoneuroendocrinology 47:151–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum DM, Rasmussen SG, Kobilka BK (2009) The structure and function of G-protein-coupled receptors. Nature 459:356–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi A, Eargle J, Black AA, Luthey-Schulten Z (2009) Dynamical networks in tRNA:protein complexes. Proc Natl Acad Sci U S A 106: 6620–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood NM, Krueckl SL, McRory JE (2000) The origin and function of the pituitary adenylate cyclase-activating polypeptide (PACAP)/ glucagon superfamily. Endocr Rev 21:619–670 [DOI] [PubMed] [Google Scholar]
- Siu FY, He M, de Graaf C, Han GW, Yang D, Zhang Z, Zhou C, Xu Q, Wacker D, Joseph JS, Liu W, Lau J, Cherezov V, Katritch V, Wang MW, Stevens RC (2013) Structure of the human glucagon class B G-protein-coupled receptor. Nature 499(7459):444–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G et al. (2017) Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators. Nature 546:312–315 [DOI] [PubMed] [Google Scholar]
- Spengler D, Waeber C, Pantaloni C, Holsboer F, Bockaert J, Seeburg PH, Journot L (1993) Differential signal transduction by five splice variants of the PACAP receptor. Nature 365:170–175 [DOI] [PubMed] [Google Scholar]
- Stroth N, Eiden LE (2010) Stress hormone synthesis in mouse hypothalamus and adrenal gland triggered by restraint is dependent on pituitary adenylate cyclase-activating polypeptide signaling. Neuroscience 165:1025–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaudry D, Falluel-Morel A, Bourgault S, Basille M, Burel D, Wurtz O et al. (2009) Pituitary adenylate cyclase-activating polypeptide and its receptors: 20 years after the discovery. Pharmacol Rev 61:283–357 [DOI] [PubMed] [Google Scholar]
- Wacker D, Stevens RC, Roth BL (2017) How ligands illuminate GPCR molecular pharmacology. Cell 170:414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinan E, Vanden-Eijnden E (2006) Towards a theory of transition paths. J Stat Phys 123:503–523 [Google Scholar]
- Wootten D, Miller LJ, Koole C, Christopoulos A, Sexton PM (2017) Allostery and biased agonism at class B G protein-coupled receptors. Chem Rev 117:111–138 [DOI] [PubMed] [Google Scholar]
- Yang L, Yang D, de Graaf C, Moeller A, West GM, Dharmarajan V, Wang C, Siu FY, Song G, Reedtz-Runge S, Pascal BD, Wu B, Potter CS, Zhou H, Griffin PR, Carragher B, Yang H, Wang MW, Stevens RC, Jiang H (2015) Conformational states of the full-length glucagon receptor. Nat Commun 6:7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Hu Z, Filipek S, Vogel H (2015) W246(6.48) opens a gate for a continuous intrinsic water pathway during activation of the adenosine A2A receptor. Angew Chem Int Ed Engl 54:556–559 [DOI] [PubMed] [Google Scholar]
- Zhang D, Zhao Q, Wu B (2015) Structural studies of G protein-coupled receptors. Mol Cells 38:836–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H et al. (2018) Structure of the glucagon receptor in complex with a glucagon analogue. Nature 553:106–110 [DOI] [PubMed] [Google Scholar]
- Zhang H et al. (2017a) Structure of the full-length glucagon class B G-protein-coupled receptor. Nature 546:259–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sun B, Feng D, Hu H, Chu M, Qu Q, Tarrasch JT, Li S, Sun Kobilka T, Kobilka BK, Skiniotis G (2017b) Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature 546:248–253 [DOI] [PMC free article] [PubMed] [Google Scholar]