Abstract
The development of physiologically relevant intestinal models fueled by breakthroughs in primary cell-culture methods, has enabled successful recapitulation of key features of intestinal physiology. These advances when paired with engineering methods, for example incorporating chemical gradients or physical forces across the tissues, have yielded ever sophisticated systems enhancing our understanding of the host microbiome impact on human physiology as well as the genesis of intestinal diseases such as inflammatory bowel disease and colon cancer. In this review, we highlight recent advances in the development and usage of primary cell-derived intestinal models incorporating monolayers, organoids, micro-engineered platforms and macrostructured systems as well as our opinion on the expected direction of the field.
Keywords: Intestine, stem cells, in vitro models, organoids, monolayers, organ-on-chips
Current approaches to model intestinal physiology
The small and large intestine, located after the stomach, comprise the lower human gastrointestinal tract and play critical roles in nutrient absorption and housing much of the human microbiome (see Glossary) (Figure 1). In the past decade, model systems have attempted to recapitulate the complex, in vivo intestinal physiology using cell lines derived from intestinal tumors such as Caco-2 cells in place of primary epithelial cells. Advanced organ-on-a-chip systems were created by culturing Caco-2 cells on the geometrically or mechanically-engineered platforms in order to properly mimic the structural and mechanical properties of the human intestine [1–6]. To mimic the mucosal architecture, porous scaffolds were micromolded to villus-like projections on which Caco-2 cells could be cultured [1, 6]. To recapitulate the mechanically dynamic environment, microfluidic systems were developed with fluid flowing both above and below a Caco-2 cell layer growing on a rhythmically stretched flexible surface [2–5]. These systems were designed to mimic the shear forces and contractile motions occurring in the small intestine. Caco-2 cells, along with other tumor cell lines have also been used as surrogate intestinal epithelial cells to probe the interactions between multiple tissue types [7]. These organon-a-chip models incorporating tumor cells offered new capabilities to emulate the structure, function and physiology of the living human intestine not possible with conventional tissue-cultured monolayers. However, as our understanding of these organs progresses, it is clear that these prior tumor model systems fall short in their ability to accurately reflect in vivo physiology as the models do not possess all of the intestinal epithelial subtypes and either lack receptors, transporters, or drug metabolizing enzymes or express the proteins at levels different from that in vivo. Thus in vitro replicas of the intestines that more accurately replicate intestinal physiology are required and will need to utilize primary cells. Accordingly, a suite of platforms employing primary cells in a variety of assay formats, including organoid [96, 97], monolayer, and shaped three-dimensional systems have been developed to recapitulate key properties of these organs. Herein we highlight recent advances of the field, focusing on epithelium derived from primary intestinal epithelial stem cells. Although tumor and immortalized intestinal epithelial cells have been extensively used as in vitro models, they are not covered in this review.
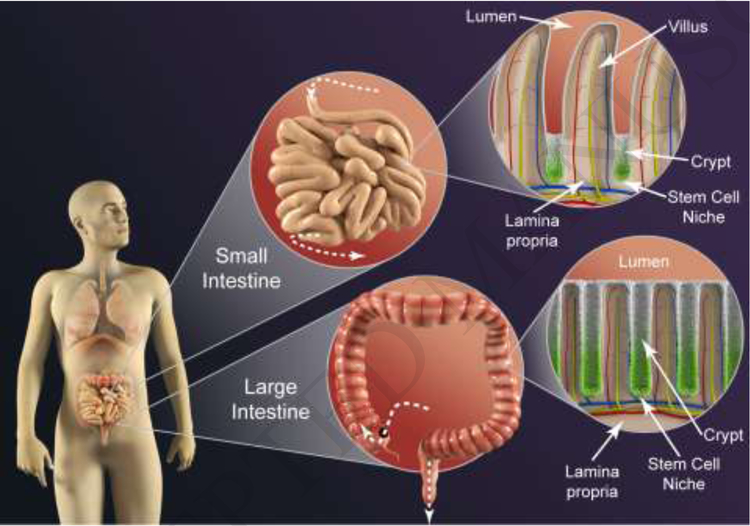
Figure 1. Key Figure. Architectural characteristics of the human intestines.
The small intestinal epithelium hosts an array of repeating crypt/villus units (top right) to maximize absorptive surface area, while the large intestinal epithelium (bottom right) consists solely of crypts. The crypts of both organs harbor proliferative cells and a stem cell niche at each crypt base (green) on non-dividing, differentiated cells along the upper crypt (white), villus and luminal surface.
Intestinal physiology and function
The small intestine secretes water, mucus, enzymes, hormones, and salt to digest and then absorb sugars, amino acids, fatty acids, lipids, and carbohydrates, while the large intestine (colon) absorbs water and salt, and manages waste storage. Both the small and large intestines have critical immunological roles; Peyer’s patches are concentrated areas of lymphoid nodules in the small intestine that ‘sample’ antigens for presentation to the immune system, and the large intestine has a number of innate immune cells such as macrophages and neutrophils. The diverse array of microbes (archaea, bacteria, fungi, and viruses) housed in both intestines play a role in metabolic and endocrine functions in the small and large intestine, and ferment fiber in the large intestine to provide additional nutrients for uptake. To accomplish these vital tasks, the intestines are equipped with specialized architectural features, sophisticated response mechanisms, and discriminatory sensing systems.
Architecturally, micro-scale differences occur prominently in each organ’s epithelial surface to accomplish distinct goals. The epithelium of the small intestine possesses repeating units comprised of a crypt-villus designed to maximize the absorptive surface area. In contrast, the epithelium of the large intestine possesses only crypts, consistent with its role in housing the microbiome and as a waste storage compartment (Table 1).
Table 1.
Characteristics of the Normal Human Intestinal Epithelium
| Characteristic | Small intestine | Colon | Ref |
|---|---|---|---|
| Size | • Diameter: 2–3 cm • Length: 500 cm |
• Diameter: 4–5 cm • Length: 150 cm |
[82] |
| Flow/Transit Time | • Mean transit time: 84 min • Rate: 0.7–3.0 mL/min |
• Mean transit time: 11–14 h • Intermittent flow • Mass movement: ~1 /d |
[83–86] |
| Contractions | Postprandial: • Contraction frequency: 3/s • Contraction amplitude: 20–24 mm Hg Interdigestive: • Contraction Frequency: 0–10/s • Contraction Amplitude: 0–30 mmHg |
Subject to circadian variation • Low-amplitude (<50 mmHg): ~ 61/day • High-amplitude: (>100 mmHg): ~ 5 /day |
[86–88] |
| Epithelial Architecture | • Crypt depth: 132–141 μm • Crypt diameter: 49–50 μm • Villous height: 567–640 μm • Villous diameter: 157–160 μm • Villous/crypt numbers: 1.4–2.3 |
• Crypt depth: 433 μm • Crypt diameter: 74 μm |
[89–91] |
| Cell Lifetime | • Stem cells: long-lived • Absorptive enterocytes: 3–5 d • Paneth cells: 20 d |
• Stem cells: long-lived • Absorptive enterocytes: 3–5 d |
[92–94] |
| Mucus Layer(s) | • One Layer: 50 – 450 μm | • Outer Layer: 300 – 700 μm • One Layer: 50 – 450 μm |
[13–15, 17, 95] |
Contractions and Flow
Contractions mix and propel food and waste through the intestine. The differing properties of these muscular contractions yield significantly different flow rates and transit times through the small and large intestines that are dependent on the specific organ’s function as well as eating/fasting schedules. (Table 1). The contractility of the small intestine is heavily dependent on the time since feeding while the large intestine is subject to a circadian rhythm. Material not absorbed by the small intestine enters the large intestine, where indigestible carbohydrates are fermented by bacteria producing absorbable metabolites or further solidified into feces.
Epithelial Cells
The epithelium acts as a barrier to protect the body from the harsh luminal environment of the digestive tract and its microbes. Additionally, this layer houses and protects the rapidly renewing epithelial stem cells (leucine rich repeat containing G-protein coupled receptor 5 (Lgr5)-positive) within a sheltered microenvironment or niche at the base of the crypts (Table 1, Box 1, Figure I). In contrast, the differentiated cells all of which originate from the stem cells, can be found migrating up the crypts, but are largely located on the villi (small intestine) or the luminal surface (large intestine) [8]. The luminal surface of absorptive cells or enterocytes is covered with microvilli to increase their absorptive area and to enhance uptake of nutrients, vitamins, ions, and water. High-density intercellular contacts such as tight and adherens junctions maintain barrier integrity protecting intestinal tissue and the blood stream from contamination by the luminal contents [9].
Box 1. In Depth Intestinal Physiology.
Tissue Structure
The small and large intestine are each composed of multiple layers: serosa, muscularis externa, submucosa, and mucosa (Figure I). The serosa (or serous membrane) is a smooth tissue that forms the outer membrane of the intestines. Adjacent to the serosa, the muscularis externa or muscular layer propels food stuffs and wastes unidirectionally by peristalsis. The next layer or submucosa supports blood and lymph vessels as well as lymphoid tissue. The mucosa forms the barrier between the lumen of the intestines and the submucosa and is comprised of three sublayers: the epithelium, lamina propria, and muscularis mucosae. The muscularis mucosae performs localized contractions, mixing the intestinal contents while the overlying lamina propria or connective tissue hosts immune cells. Finally, the epithelium lines the intestinal inner surface, interfacing with the luminal contents.
The Stem Cell Niche
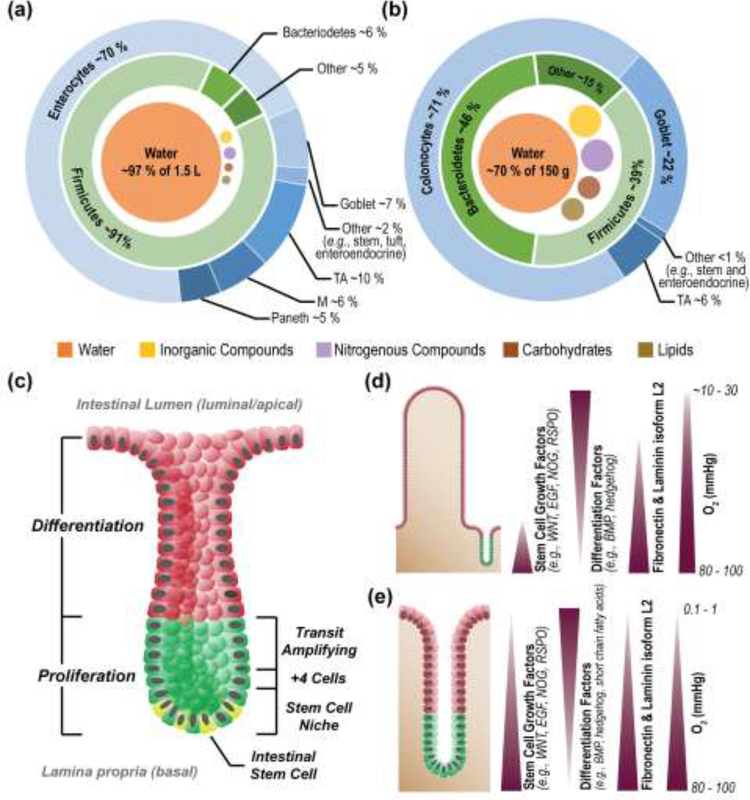
The stem cell niche, provides the necessary factors to support stem cell maintenance and proliferation including Wnts (amalgam of Wingless:integration site, ligand for Frizzled receptors) and R-Spondin (ligand for Lgr5). The major source of these factors is the underlying stromal cells in both the small and large intestine [98, 99]. Paneth cells located within the crypt and adjacent to the stem cells provide additional stem-cell support in the small intestine. Intestinal stem cells in both the large and small intestine give rise to rapidly proliferating or transit amplifying (TA) cells located just above the stem cell niche. As TA cells migrate upward, they give rise to the non-dividing, fully differentiated secretory or absorptive cells lining the intestinal lumen.
Complex Gradients
Stromal cells create high concentrations of growth factors (Wnts and R-Spondins) at the crypt base [98, 99] while mesenchymal cells in the villi and near luminal epithelial cells produce differentiation factors such as bone morphogenic proteins (BMPs) to maintain a high density of enterocytes at the luminal interface [100].
Figure I. Comparison of human small and large intestine.
Luminal contents and epithelial cell types of the small (a) and large (b) intestine [22, 101–105]. Structure of the crypts of the small and large intestines, with major zones and stem cell niche components labelled (c) Chemical gradients across the epithelium of the small and large (d) and large (e) intestine [106]. Images reproduced with permission from the indicated references.
A variety of other specialized cell types are also found in the intestines although in significantly smaller number than the absorptive cells (Box 1, Figure I). Mucus-secreting goblet cells form a thick protective mucus layer covering the intestinal surface. Enteroendocrine cells act as chemosensory cells and regulate appetite satiety and gut motility by secreting a wide range of gut hormones including serotonin, somatostatin, glucose-dependent insulinotropic polypeptide (GIP), glucagon-like peptides (GLPs) in response to chemical stimuli [10]. Tuft cells play a role in defense against protozoa and helminth infections [11]. Microfold cells or M cells are only in the small intestine above Peyer’s patches and they communicate with immune cells by presenting luminal antigens [12].
Mucus Layer
The luminal surfaces of the small and large intestine are protected by a thick blanket of mucus comprised primarily of mucin 2 (Muc2 from goblet cells) (Table 1). The small intestine produces a single mucus layer aiding in resistance to self-digestion, bacteria, and mechanical stresses. In contrast, the large intestinal mucus is composed of two structurally distinct layers, an outer layer hosting bacteria and an inner bacteria-impenetrable layer [13–15]. In both organs, the mucus layer(s) are rapidly replaced. In mice, the mucus layer is renewed every 8–12 h [16] except in the distal colon where it is exchanged on an hourly basis [17].
Supporting Gradients
The orderly migration and differentiation of cells along the crypt/villus axis is maintained by soluble gradients of growth factors, cytokines, microbial products, food metabolites and gases as well as alterations in the physical attributes of the underlying extracellular matrix (ECM) such as stiffness, porosity and receptor contacts (Box 1, Figure I). These gradients are in part generated by the epithelial cells themselves and underlying stromal cells, endothelial cells, and neuronal and immune cells. The circulatory system coupled to luminal content flow and microbial actions set up additional gradients by removal and/or supply of metabolites and gases. Gradients of microbial products such as short chain fatty acids, regulate intestinal physiology by promoting differentiation or suppressing stem cell proliferation [8, 18]. The very steep oxygen gradient across the large intestinal epithelium enables a diverse community of obligate and facultative anaerobes to flourish in the nearly oxygen free lumen of the large intestine while the not-to-distant intestinal stem cells enjoy a normoxic microenvironment [19]. The intervening colonocytes are adapted to live in a hypoxic environment utilizing bacterially-produced butyrate as their primary energy source [8]. Disruption in any of these well-balanced gradients (by injury or inflammation) negatively impacts intestinal homeostasis.
Intestinal Microbiota
The human gut houses 1014 microbes[20] comprised predominantly of bacteria but also including viruses, archaea, and eukarya in the lumen and mucosa (Box 1, Figure I) [21]. The exact species mixture is dynamic and dependent on the luminal environment (nutrients, pH, oxygen, transit times, antibiotic presence, and mechanical stresses) as well as host attributes (age, diet, and disease status). The luminal environment of the human small intestine and large intestine are distinct and optimized for different microbiota. In the small intestine, the lumen is hypoxic, transit time is short (Table 1), and antimicrobial compounds (antimicrobial peptides, bile acids, immunoglobulin A (IgA)) are high. Thus the bacterial composition readily fluctuates over time [22, 23] and oxygen-tolerating facultative anaerobes (like Streptococcus) are dominant in the small intestine, but present at relatively low density, 103-108 cells/g of contents [22, 23] in comparison to the very high bacterial density in the large intestine, These microbes compete with the host cells for the simple sugar molecules produced in the digestive process. In contrast, the long transit time of the large intestine creates a more favorable environment and hence diverse community for the gut microbes. Indeed, the majority of gut microbes including more than 400 bacterial species are located in the large intestine (1011-1012 colony forming units/g feces) [24]. Oxygen[25–27] and nutrients[28] are depleted in the large intestine lumen so that anaerobes ferment the remaining indigestible carbohydrates as an energy source. In turn, these commensal bacteria supply the host with additional calories (up to 10% of host calories) [29], produce required vitamins (B and K) and act to prevent colonization by pathogenic bacteria [30].
In Vitro Intestinal Model Systems Incorporating Primary Epithelium
Long-term, proliferative culture of the human intestinal epithelium in vitro has represented a long-standing and formidable challenge due to the complexity of the interactions between the different cell layers and types within the intestinal mucosa as well as an absence of the factors and microenvironment required for stem cell maintenance. This difficulty was overcome in 2009 when Sato and colleagues identified the key growth factors required to support mouse intestinal stem cells and pioneered the organoid culture model enabling the propagation of intestinal epithelial stem cells in vitro (Box 1 and 2) [31, 32]. The medium composition has been further optimized for culture of intestinal stem cells from humans as well as a variety of other species including, cat, dog, cow, horse, pig, sheep, and chicken [33, 34]. This advance has given rise to a range of in vitro models of the intestine: organoids, monolayers and shaped 3D systems.
Box 2. Sources of Primary Intestinal Cells.
Primary intestinal cells are most often obtained from cadaver samples or biopsied from patients during routine screenings. This tissue can be processed within the laboratory to release crypts containing living intestinal stem cells (ISCs), which thereafter can be expanded. Since these cultures retain their original tissue identity, including factors such as host genetics and disease propensities, biobanks can be established to increase the accessibility of these primary cell lines to wider research audiences and enable population screenings [107]. Concerns remain among the community regarding the ability of these samples to accurately represent human genetic diversity (based on available cadavers or patient accessibility to healthcare), their genetic stability over time (i.e., propensity for acquisition of mutations during long-term culture), and regulations for their acquisition, transfer to others and subsequent use.
While there have been limited attempts to culture full thickness tissue since initial primary culture studies, a moderate level of commercial interest persists today.
Advantages of utilizing full thickness tissue include the presence of a full suite of differentiated epithelial cells as well as an intact lamina propria layer from which interstitial interactions can be studied [76].
Induced pluripotent stem cells (iPSCs) have been proposed as an alternative cell source since they can be readily generated from somatic cells. Given that these can be obtained by far less invasive procedures, such as a simple blood draw, a more genetically diverse population of samples can be utilized, including those from underrepresented populations. Differentiation of iPSCs into the endoderm by activin A, followed by the addition of fibroblast growth factor 2 and Wnt3a has resulted in successful transformation of iPSCs into organoids containing Lgr5+ stem cells [108, 109]. These transformed cells manifest in culture as spheroids that detach from the planar sheets of non-transformed cells, and can be harvested from the medium. A significant drawback has been that the transcriptional profiles of iPSC-derived intestinal cells most closely resemble that of a fetal intestine rather than that of the adult intestine [110]. Until recently, maturation of iPSC-derived stem cells has required in vivo transplantation into living tissue, though maturation in vitro has been enhanced by exposure to IL-2 [111]. While promising, further validations and benchmarking against primary models will be required for widespread adoption of this improved protocol.
Organoids
Intestinal epithelial organoids are cultured from isolated intestinal stem cells or crypts, and possess several general features: a spherical monolayer of cells encasing a lumen of mucus and cellular debris, outwardly extending proliferative regions (i.e., buds) enriched with stem cells, and all differentiated cell types of the intestine (Figure 2). The organoids are typically cultured within Matrigel, a matrix providing the chemical and mechanical cues for intestinal stem cell maintenance and proliferation when combined with specialized media containing Wnt, R-Spondin, Noggin, and epithelial growth factor (EGF) (Box 3). The organoid platform has been the basis of fundamental discoveries regarding stem-cell self-renewal and enabled unique insights between mechanotransduction and stem cell behavior [35]. For example, intestinal stem cells exhibit optimal growth and survival within stiff polyethylene glycol (PEG) hydrogels functionalized with fibronectin, while differentiation and organoid formation proceed optimally on soft PEG hydrogels with laminin coatings [36], reflecting the growth patterns and gradients observed in vivo [18] and providing an avenue for refinement of artificial ECMs [37].
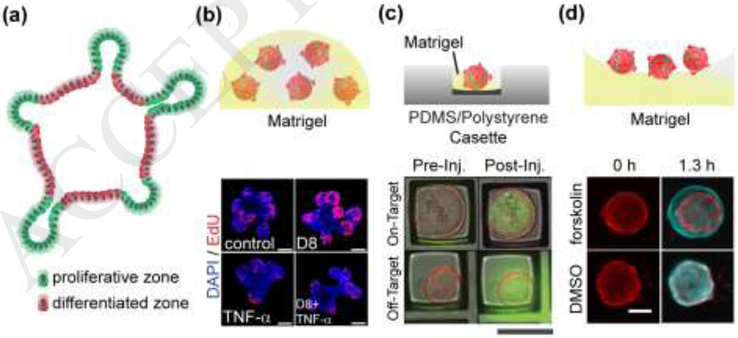
Figure 2. Organoid Systems.
(a) Schematic cross-section through an intestinal organoid. Stem cells (green) are enriched in buds, and differentiated cells (red) are enriched along the luminal aspect. (b) Organoids cultured within Matrigel. The top panel is a schematic of a patty with 5 organoids. The lower panel is a fluorescence image of four different organoids demonstrating that tumor necrosis factor α (TNF-α) decreases EdU incorporation or cell proliferation. Scale bars: 50 μm. Adapted with permission from [42]. (c) Organoids cultured within a microwell. The top panel is a schematic of a side view of a single organoid within a microwell on a microdevice. The lower panel is the top view of an overlaid fluorescence and brightfield image before and after microinjection of two organoids in microwells. The top organoid was successfully injected with a green fluorophore while the lower organoid was not. Scale bars: 200 μm. Reproduced from with permission from [38]. (d) Organoids on a Matrigel surface. The top panel is side-view schematic of 3 organoids on Matrigel. The lower panel is a top view image of two organoids before and after application of forskolin or a DMSO control. Red coloring indicates organoid size prior to stimulus addition while cyan indicates the organoid size after the stimuli. Scale bars: 150 μm. Adapted with permission from [39].
Box 3. Support matrix.
Intestinal epithelial cells require a supporting matrix on which to anchor, migrate, self-renew and differentiate. In vivo intestinal epithelium resides on a supporting basement membrane thought to be comprised primarily of various structural or adhesive proteins such as laminins, collagens, and proteoglycans. In vitro culture systems also supply a supporting matrix most often a hydrogel such as Matrigel or collagen [31, 59]. The matrix influences a wide range of cell behaviors including viability, morphology, tissue organization/architecture, stem cell self-renewal, proliferation and differentiation. The following properties of appear as key considerations when selecting a supporting matrix for an in vitro intestinal model:
Presence of appropriate ligands. Integrins are cell adhesion receptors that bind with ECM ligands including motifs derived from ECM proteins, for example, RGD (arginine-glycine-aspartate), LDV (leucine-aspartate-valine), and GFOGER (glycine-phenylalanine-hydroxyproline-glycine-glutamate-arginine). The ECM ligands are crucial for maintaining cell adhesion, defining the luminal-basal cell polarity, and preventing cell apoptosis (anoikis). Matrigel, collagen, and fibronectin possess the requisite binding sites and have been demonstrated to support the attachment and survival of intestinal epithelial cells [31, 36, 59].
Stiffness. Stiffness is the resistance to deformation in response to an applied force. The intestinal epithelial cells have been shown to be sensitive to matrix stiffness, for example, mouse intestinal epithelial stem cells (ISCs) fail to survive on a hard surface such as polystyrene [59]. Matrix possessing a stiffness on the order of kilopascals (kPa) appears to be ideal for in vitro culture of a self-renewing intestinal epithelium and is to that measured on intact tissue [112]. ISC growth and maintenance is optimal on matrices that are stiffer than that preferred by differentiated intestinal epithelial cells [36].
Porosity. In vivo, intestinal stem cells rely on the growth factors secreted by supporting stromal cells on their basal side and nutrients arriving from the vasculature in the submucosa. Similarly in vitro, a porous matrix with pore sizes larger than the required growth factors and nutrients is best suited for in vitro culture of intestinal stem cells.
A variety of strategies have been developed to facilitate automated and high-throughput analysis of organoids as assay tools (Figure 2). These methods include organoid growth within Matrigel-filled microwells for sorting based on age or phenotype or to act as stable organoid holders for automated microinjection of microbiota [38]. Planar arrays of organoids grown above a Matrigel layer enable fast, reproducible swelling assays to tracking ion and water movement serving as model systems for diseases of transport e.g. cystic fibrosis [39]. Moreover, gradient-forming microdevices have enabled limited control of cell patterning within organoids [40].
The applications for organoids in the basic biomedical sciences have been transformative. A full recitation is well beyond the scope of this review and we provide but a few examples. Intestinal organoids have served as invaluable disease model systems particularly for inflammatory bowel diseases and colon cancer to probe fundamental disease mechanisms and optimize and assess therapeutic compounds [38, 41–43]. Small intestinal organoids are competent to support the full life cycle of Cryptosporidium, a protozoan parasite and a leading cause of diarrhea and global child mortality [44]. Infection of human intestinal organoids with enteroviruses has shifted our understanding of the epithelium, demonstrating its competence to engage in antiviral signaling [45]. Despite the high impact of intestinal organoid culture systems, these constructs fail to recapitulate most of the physiology and architecture of the intestinal epithelium. Further, the organoid morphology with its surrounding hydrogel and enclosed lumen pose fundamental limits on this method’s use as an assay and screening tool.
Monolayer Systems
Culture systems recreating the epithelial cell monolayer covering the intestinal surface are increasingly finding use due to their simplicity, scale up capability, and assay compatibility. When cultured on a porous membrane, monolayer systems also provide access to both the luminal and basal sides of the epithelial as well as the adjacent fluid reservoirs for testing of food components, microorganisms, and bioactive compounds. These systems generate a single layer of intestinal epithelial cells by placing primary cells on a scaffold to which the cells then attach, proliferate and/or differentiate (Figure 3). Utilized scaffolds include nonporous solids, porous membranes, micro-patterned surfaces, and hydrogels to generate fully differentiated monolayers without stem cells, self-renewing monolayers with or without segregation of the stem and differentiated cell types, or proliferative monolayers over a layer of supportive feeder cells.
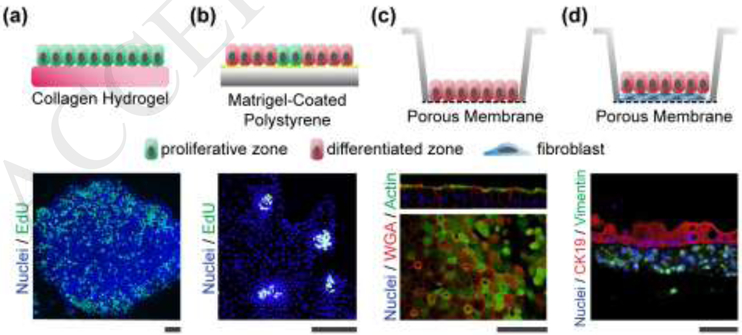
Figure 3. Intestinal monolayer systems.
(a) Self-renewing monolayers. The top panel is a schematic of the side view of the proliferative cells (green) on the hydrogel. The lower panel is a fluorescence image of a patch of cells showing EdU incorporation throughout the monolayer (original image used for illustration purposes). (b) Self-organizing monolayers. The top panel is a schematic of the side view showing the localized region of proliferative cells (green) amongst the larger area of differentiated cells (red). The lower panel is a fluorescence image (top view) of four proliferative zones EdU incorporation localized to these zones. Reproduced with permission from [63]. (c) Differentiated monolayers. The top panel is a schematic of the side view of the differentiated cells (red) on a porous membrane. The lower panel is a fluorescence image of cells showing actin staining which is found in the microvilli covering the differentiated cells, and wheat germ agglutinin (WGA) which binds glycoproteins found in mucus. Reproduced from [70] under a Creative Commons license. (d) Monolayer Co-cultures. The top panel is a schematic of the side view of the differentiated cells (green) overlying the myofibroblasts (blue). The lower panel is a cross section or side view of a fluorescence image demonstrating vimentin-expressing fibroblasts and cytokeratin 19 (CK19)-expressing intestinal epithelial cells. Reproduced from [68] under a Creative Commons license. All scale bars: 50 μm.
Most early monolayer systems generated a short-lived intestinal epithelium largely because the medium composition and scaffolding properties required to maintain stem cells in vitro were unknown [46, 47]. Using extracellular matrix (ECM) coated porous membranes as the scaffold materials, differentiated and nonproliferative epithelial monolayers were readily cultured from a variety of species including murine, porcine and human [48–52]. The monolayers display a characteristic polarized morphology (luminal/basal) and markers of mature intestinal epithelium such as brush border proteins and tight junctions (Figure 3). These monolayers typically form a contiguous, impermeable layer with a transepithelial electrical resistance (TEER) sufficient to support physiological assays such as IgA transcytosis,24 chemokine (CXCL10) secretion [53], inflammatory cytokine (IFN-γ, TNF-α, IL-6, IL-8) synthesis [54, 55], cytotoxicity, barrier function [55, 56], ion transport [52, 57], and hormones production (serotonin, GLP-1, FGF19) [52].
The medium constituents used for self-renewal of organoid stem cells have been adapted to form self-renewing monolayers with stem cells (Figure 3). Selecting a suitable scaffold is also crucial to discourage organoid formation and to enhance cell surface attachment to form a flat layer of cells while still maintaining stem cells [58, 59]. A thick layer of collagen I hydrogel (thickness ≥ 1 mm) supports the formation of self-renewing monolayers with stem cells and all differentiated cell types [59]. A sandwich culture system comprised of a porous membrane (coated with collagen IV) and then overlaid with a collagen I hydrogel also supports the expansion of Lgr5+ stem cells at the sandwich interface but with diminished goblet or enteroendocrine cell presence [60]. Nonporous solid scaffolds such as polydimethylsiloxane (PDMS) only support the self-renewal of stem cells under very specialized conditions [59, 61]. In combination, these observations suggest that stiffness, porosity and ECM contacts of scaffolds synergistically regulate the proliferation of stem cell monolayers [59, 62]. At this time, systems employing thick collagen layers appear best suited for long-term monolayer expansion of stem cell numbers in vitro [59].
Similar to organoids, spontaneous compartmentalization of stem/proliferative cells from differentiated cells is observed in monolayer systems (Figure 3). After mouse large intestine epithelial cells were cultured on a thick collagen layer (1 mm), proliferative cells segregated along the edges of the expanding monolayer, whereas differentiated cells such as goblet cells and colonocytes were located in the center of the monolayer. The reverse self-organization was observed when mouse small intestinal epithelial cells were cultured on a Matrigel coating: proliferative cells were enriched in central regions surrounded by differentiated cells [58, 63]. Self-organization in monolayers can also be precisely controlled by cell culture on a surface with micro-patterned porous regions that enable spatially localized application of growth factors [62]. These self-organized monolayer systems provide simple, robust platforms for assay of chemical gradient impacts on stem and differentiated cells [62, 63].
In vivo, the intestinal epithelium participates in extensive crosstalk with the neighboring environment such as luminal microorganisms, adjacent stroma, and immune cells. Epithelial monolayers provide a simplified tool to study these complex interactions in a controlled manner and co-culture of with fibroblasts, bacteria, viruses, protozoa, neurons and immune cells have all been demonstrated in recent years[64–71]. Placement of fibroblast feeder layers yields a layered tissue suitable for long term epithelial culture or drug absorption and toxicity assays (Figure 3) [64, 68, 72]. Co-culture of epithelium with enteric neurons is in its early stages but will certainly provide novel insights into neuro-intestinal crosstalk [71]. The intestines play a central role in the maturation of the immune system and macrophages co-cultured with monolayers have demonstrated a coordinated epithelial-immune cell response to pathogens [70]. Perhaps one of the most high-value attributes of the monolayer culture systems is the ability to pair them with infectious agents such as noroviruses [65], adenoviruses [66], enterohemorrhagic E. coli [67], and Cryptosporidium parasites [69] to reveal the intricate details of host-pathogen interactions.
Drug absorption, metabolism, and excretion often involve the actions of multiple organs acting in concert and primary intestinal epithelial monolayers coupled to other organs have now replicated complex drug interactions and toxicities observed in humans but not animal models [72–74]. In these systems, the coupling of different organs is accomplished by sequential media transfer between tissues (demonstrated for intestine, liver, kidney proximal tubules, blood-brain barrier, and skeletal muscle) [72], integration of tissues into discrete regions of a single device with connecting fluidic channels (performed for intestine, liver, skin and kidney) [73], or construction of multi-chambered devices with fluid contact via a porous membrane (liver and intestine) [74]. While these systems pose challenges such as the respective scaling of the tissues and a need for a universal media compatible with all organs, their ability to reproduce complex interactions related to drug metabolism and toxicity holds immense promise for their use as complex physiologic systems for drug discovery.
Shaped three-dimensional intestinal systems
Engineered three dimensional systems have the potential to recapitulate intestinal physiology and architecture not possible with organoid or monolayer cultures, for example, modulation of permeability and transport by contractile forces or formation of polarized crypts using chemical gradients. A challenge though is that these shaped three-dimensional systems must accommodate the complex requirements of primary cells with respect to substrate mechanics, adhesion and porosity as well as the growth and differentiation factors needed to support proliferation and proper lineage allocation. This challenge has been addressed using systems falling into one of three general categories (Figure 4): 1) microstructured models, 2) microfluidic systems with mechanical forces, and 3) macrostructured replicas.
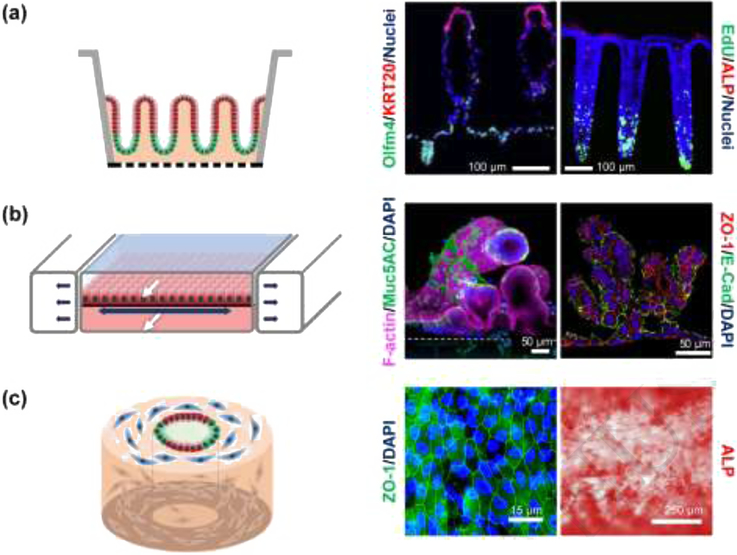
Figure 4. Shaped three-dimensional intestinal systems.
(a) Microstructured systems. Left: Side-view schematic of polarized intestinal crypts with a stem cell niche (green) and differentiated cell zone (red). Middle: Fluorescence image of cross section through an in vitro human small intestine epithelium showing a crypt and two villi. Reproduced with permission from [76]. Right: Fluorescence image of cross section through an in vitro human colon epithelium with three crypts. Stem/proliferative cells are marked by olfactomedin-4 (OLFM4) and differentiated cells by cytokeratin-20 (KRT20). Reproduced from [18] under a Creative Commons license. (b) Microfluidic systems. Left: Side-view schematic of differentiated epithelial cells (red) on a stretchable surface. White arrows mark fluid flow while dark arrows indicate the motion of the stretchable surface. Middle and right: Human small intestinal epithelial cells derived from the organoids of biopsied intestinal tissues (middle), reproduced from [77] under a Creative Commons license, and iPSC derived intestinal epithelial cells (right), reproduced from [78] under a Creative Commons license. Both tissues were grown in microfluidic devices with luminal and basal fluid flow leading to villi formation. (c) Macrostructured replica. Left: Schematic of an angled view of a silk scaffolding with fibroblasts (blue), proliferative epithelial cells (green) and differentiated epithelial cells (red). Middle and right: Fluorescence image of tight junction (ZO-1) staining and alkaline phosphatase (ALP) for human small intestinal cells grown in tubular silk scaffold embedded with myofibroblast. Reproduced from [80] under a Creative Commons license.
Microstructured models recreate the three-dimensional intestinal epithelium by shaping the underlying scaffold so that it replicates the architectural features of the in vivo epithelium (Figure 4, Box 4). A crosslinked collagen hydrogel is molded over a porous membrane mimicking the shape, size and density of crypts in the large intestine or crypts/villi in the small intestine [18, 75, 76]. Remarkably, gradients of growth factors alone are sufficient to polarize the crypts and crypts/villi localizing the stem/proliferative cells to the base of the crypts (forming a stem cell niche) and directing the various differentiated cell types to the luminal surface and villi. The orderly, unidirectional migration and differentiation of cells along the long axis of the crypt recapitulates that observed in vivo. A strength of this system is the facile creation of nearly any chemical gradient across the tissue as demonstrated by the luminal application of short chain fatty acids or basal application of inflammatory cytokines to demonstrate the impact of fermentation products or inflammation, respectively, on stem cell behavior and lineage allocation. To date though, contractile or shear stresses have not been demonstrated for this format.
Box 4. Microfabrication.
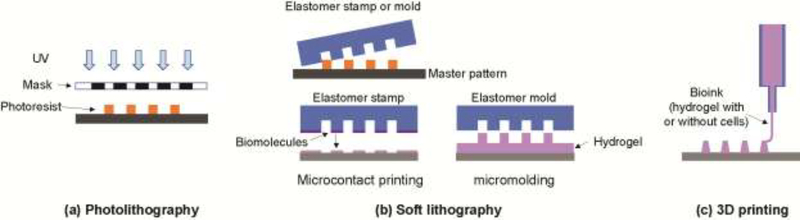
Microfabrication is a construction process to create two or three dimensional shapes with microscale features. Microfabrication was initially developed for semiconductor device manufacturing, but its use has been expanded to the fabrication of any system that requires microscale features including organ on a chip systems. Microfabrication strategies most commonly employed to form organ- or tissues-on-chip include photolithography, soft lithography, and three-dimensional (3D) printing (Figure I).
Photolithography is commonly used to generate shapes or patterns using a photosensitive polymer or photoresist exposed to ultraviolet light through a patterned mask. Polymerization occurs either in the exposed (negative photoresist) or unexposed (positive photoresist) regions followed by removal of nonpolymerized material to reveal the desired features. The resulting device comprised of photoresist pattern can be used as is or transferred to another material such as silicon or metal by deposition or etching processes. While photolithography is a powerful method to create patterns with submicron resolution [113], this method requires significant infrastructure and the range of useable materials is limited.
Soft lithography creates microscale patterns using a stamp or a template molded from a master pattern typically fabricated by photolithography. Elastomers such as PDMS are poured into the master, solidified and then removed to yield a stamp or mold. The stamp or mold is then used as a template to transfer a two or three dimensional pattern to another material with micron resolution [114]. Stamps are typically employed to print two-dimensional patterns of biomolecules onto surfaces (micro-contact printing). Three-dimensional hydrogel shapes are formed by filling a mold loaded followed by gelation and removal from the mold. Elastomeric stamps and molds can typically be used repeatedly, making soft lithography inexpensive (once the master is fabricated) and readily accessible to a broad community.
3D printing creates an object from a digital model generally by placing successive thin layers of material to create the final object. A wide range of 3D printing methods are available for hydrogels, for example, inkjet printing and laser-assisted printing. An advantage of 3D printing is that living cells can be embedded within a shaped hydrogel during the printing process. Inexpensive printers also make this method more widely available than either photo- or soft lithography although the resolution attainable with 3D printing is typically low (tens of microns) [115]. While 3D printing technology is progressing rapidly, the method remains time consuming, and the available printable bio-inks are limited [115, 116].
Figure I. Microfabrication methods.
(a) Photolithography creates patterns by exposing light onto a photoresist film through a patterned mask followed by removal of unpolymerized photoresist. (b) Soft lithography requires a master or starting pattern to form an elastomeric stamp or mold. A stamp transfers a two dimensional pattern of biomolecules (microcontact printing) or a mold forms three dimensional structures from a hydrogel or other material. (c) 3D printing can be used to create structures formed from hydrogels, often with cells embedded.
Microfluidic systems incorporate one or more microchannels above or below the in vivo epithelium and enable facile application of mechanical forces to the epithelium (Figure 4). Stretching and compressive motions can be applied to the cells by manipulating an underlying flexible PDMS membrane. The accompanying fluidic microchannels also enable fluid flows both above and below the epithelial cell layer to apply shear stresses [77, 78]. These systems have recently been adapted for use with primary human duodenal cells by co-culture with intestinal microvascular endothelial cells to recreate both an epithelial luminal compartment as well as a basal vascular zone [77]. In this system, fluid-flow induced shear stresses were sufficient to recreate villi-like protrusions from the epithelial monolayer, suggesting that these forces may play a more dominant role than contractile forces in the formation of small intestinal villi [78]. These formats have been utilized to demonstrate barrier function, nutrient metabolism, and immune responses to inflammatory cytokines showcasing their potential. However, absorption and adsorption of reagents onto the hydrophobic PDMS as well as the need for complex flow systems may ultimately limit device utility. Newer thermoplastics may address some of these limitations [79].
Macrostructured systems possess intestinal features shaped on scales of many millimeters to offer a more intestinal-like macroscale platform relative to that offered by the microfluidic and microstructured systems. The size of these systems offers the potential to investigate long-distance multicellular behavior. Silk-derived tubes support myofibroblasts cultured within the silk matrix itself, while a monolayer of small intestinal cells forms along the inner scaffold surface (Figure 4) [80]. The cell layers collectively respond to the co-culture of E. coli by initiating anti-bacterial gene transcription. Perhaps the most native-like macrostructured tissue is that formed from decellularized human large intestine, which is then repopulated with primary epithelial cells, endothelial cells, and myofibroblasts [81]. Likely due to the native ECM and morphological features, the cells repopulate the scaffolding with formation of crypts, vasculature and stroma closely resembling that of in vivo intestine. These macrosystems offer great potential in identifying organ-level phenomena as well as rare cell behaviors due to their ability to incorporate large numbers of multiple cell types in spatially appropriate arrangements. However, the difficulty in modifying the material properties of the silk scaffold and the challenge in obtaining sufficient primary tissue for decellularization may slow adoption by others.
Concluding Remarks
The ability to culture intestinal epithelial stem cells has enabled the development of in vitro models that recapitulate the phenotypic composition and physiology of the in vivo intestinal epithelium. These models have the potential to offer simple, economical, and flexible experimental platforms with features that neither tumor-cell, organoid, nor animal models can provide. Moreover, micro-engineered intestinal models mimic key physiological attributes such as tissue architecture, chemical gradients, luminal flow, and mechanical motions (peristalsis) not possible with the organoid model system. Unlike animal models, tight control of the microenvironment can be maintained for detailed assay of the impact of a wide range of variables. Ultimately, these models are amenable to the development of personalized and precision medicines for intestinal diseases as the cultured intestinal epithelial cells retain both patient specificity and regional specificity. While the construction and study of these more accurate representations of the intestine have revealed attributes and behaviors of the in vivo epithelium in a manner not possible with prior technologies, many facets of the intestine remain unexplored (see Outstanding Questions), thus presenting new opportunities for future investigation.
Outstanding Questions – (1932/2000 characters including spaces).
How can the movement of luminal liquids or solids in the small and large intestine, and the circadian rhythm in the large intestine, be mimicked in a simplified and robust format in vitro for widespread adoption by end users?
As matrix properties impact cell behavior, how can we standardize the matrix to improve reproducibility, and what are the critical properties required by artificial matrices?
What are the necessary and essential factors to recapitulate the intestinal epithelial cell behaviors in vivo including cell-type ratio, location, migration, growth rate and lifetime for both the stem, proliferative and different cell types in the epithelium?
The in vivo intestinal epithelium is superimposed with an exquisitely structured mucus barrier to protect from bacteria and disease: how can this be reconstructed in vitro, particularly in regards to recreation of the mucus layers of variable density?
How do we recapitulate the complexity of the in vivo microbiota, especially considering the fluctuating composition observed throughout different populations, individuals, and stages of human development?
How do we standardize sources of intestinal tissue and ensure appropriate population diversity?
One goal of microphysiological systems research is to evaluate how organs work in tandem, how do we integrate multiple organ-on-a-chip devices to simulate in vivo physiology?
Intestinal tissues contain various non-epithelial cells including immune cells, endothelial cells, and enteric neurons, as well as epithelial cells. How can we implement all different cell types in the appropriate spatial relation and correct cell ratios to investigate their interactions?
As new platforms increase in complexity with the inclusion of different architectures and multiple cell types, how do we ensure that these systems are reproducible, robust, and easily applied by the biomedical research community?
In light of the rapid progression of this field, there a plethora of hurdles that must be overcome for translation of these platforms to the commercial realm, enabling their widespread use. On the manufacturing end, these include the identification of consistent and reliable sources of human intestinal tissues and epithelial stem cells, as well as extracellular matrices/scaffolding materials, media and growth factors. Metrics and specifications will need to be identified for each of these critical components so that they become more reliable and predictable in their performance. The utilization of standardized protocols for the procurement and differentiation of stem cells, and characterization of the in vitro intestinal epithelium, will accordingly increase experimental reproducibility between laboratories. Once these standards are in place, efforts should be focused on increasing the complexity of in vitro models to recapitulate with high fidelity the intestinal epithelium in terms of phenotypic composition, physiology and function. More realistic models of the intestine can be achieved by incorporation of the full gut microbiome with appropriate mucous layer(s) and gaseous gradients as well as integration with other primary, non-epithelial intestinal cells such as myofibroblasts, enteric neurons, and immune cells. While adoption of state-of-art fabrication methods (e.g. 3D printing) and materials (e.g. synthetic hydrogels) will advance micro-engineered intestinal models, simplicity and ease-of-use should be major considerations to increase practicality and promote acceptance by the end-users.
Highlights.
Breakthroughs in primary cell culture have paved the way for the development of in vitro intestinal models that closely mimic in vivo physiology.
Organoids are the most widely used system for modeling the intestinal epithelium using primary cells, and have recently been applied for studying extracellular matrix-cell interactions, microbiota interfaces, and population screens.
Monolayer culture formats provide a simple and robust approach to generate in vitro intestinal epithelium providing access to both luminal and basal cell sides, and incorporating critical attributes required for screening applications.
Micro-engineered intestinal epithelial models can recapitulate key intestinal physiology including microarchitecture, flow, and peristalsis that are difficult to implement in conventional culture systems, offering more accurate intestine models for preclinical studies.
Acknowledgements.
Supported by the National Institute of Diabetes and Digestive and Kidney Diseases (R01DK109559).
Glossary
- Basal
the outer most side of the epithelium or side closest side to the abdominal cavity.
- Colonocyte
absorptive epithelial cells lining the large intestine (colon) specifically.
- Differentiated cells
non-dividing cells that have developed from stem cells into specialized cells to perform specific functions, like absorption or secretion.
- Enterocyte
absorptive epithelial cells lining the intestines.
- Epithelium
tissue that lines the outer layer of organs, as well as the luminal side of hollow organs.
- Extracellular matrix (ECM)
biological scaffold or support matrix composed of a complex mixture of proteins that support cellular growth, cell adhesion, and differentiation.
- Growth factor
a compound required for normal cellular growth, proliferation, and differentiation (i.e. Wnt, R-Spondin, and Noggin.
- Hydrogel
A polymeric network, derived from synthetic and/or biological constituents, in which water occupies a significant portion of the volume.
- Lamina propria
a thin layer of connective tissue lying beneath the basement membrane of the epithelial cell layer serving as an anchor for the epithelium.
- Lgr5
Leucine-rich G-protein coupled receptor 5, a unique marker for intestinal epithelial stem cells.
- Luminal
Refers to the inner most side of the epithelium facing the interior of the intestine i.e. where food products and bacteria reside. Also referred to as “apical”.
- Mechanotransduction
The translation of external physical and mechanical cues into biochemical signals within the cell to elicit a cellular response.
- Microbiome
all microorganisms in a particular environment such as the human gut, including bacteria, archaea, protists, fungi and viruses.
- Organoid
a multicellular system composed of epithelial cells from one type of organ with a hollow, enclosed lumen and the term used throughout this review rather than the more specialized organ-specific definitions.
- Peristalsis
a series of wave-like muscle contractions that propel the food contents along within the digestive tract.
- Polarized
possessing two or more functions, properties, compositions or shapes in spatially distinct regions. Epithelial cells are polarized into luminal and basal domains. Crypt-villi are polarized along crypt-villi axis with stem cells at the crypt base and differentiated cells in the villi.
- Scaffold
structural support for cell attachment, growth, function and survival.
- Shear stress
force per unit area applied tangentially to a surface. In the intestine, flow moving across the epithelial surface creates a shear stress.
- Stem cells
unspecialized or pluripotent cells that can give rise to more cells of the same type or to other cell types e.g. differentiated cells.
- Transepithelial electrical resistance (TEER)
a measure of ion movement across electrical an under an applied potential. As epithelial barrier function is compromised, more ions move across the cell layer diminishing the resistance to ion movement or TEER.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Costello CM et al. (2014) 3-D intestinal scaffolds for evaluating the therapeutic potential of probiotics. Mol. Pharm 11, 2030–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim HJ et al. (2012) Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 12, 2165–2174 [DOI] [PubMed] [Google Scholar]
- 3.Kim HJ et al. (2016) Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. U. S. A 113, E7–E15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah P et al. (2016) A microfluidics-based in vitro model of the gastrointestinal human– microbe interface. Nature communications 7, 11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trietsch SJ et al. (2017) Membrane-free culture and real-time barrier integrity assessment of perfused intestinal epithelium tubes. Nature communications 8, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu J et al. (2012) In vitro 3D human small intestinal villous model for drug permeability determination. Biotechnol. Bioeng. 109, 2173–2178 [DOI] [PubMed] [Google Scholar]
- 7.Edington CD et al. (2018) Interconnected microphysiological systems for quantitative biology and pharmacology studies. Sci. Rep 8, 4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaiko GE et al. (2016) The colonic crypt protects stem cells from microbiota-derived metabolites. Cell 165, 1708–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lechuga S et al. (2017) Disruption of the epithelial barrier during intestinal inflammation: Quest for new molecules and mechanisms. Biochim. Biophys. Acta 1864, 1183–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Worthington JJ et al. (2018) Enteroendocrine cells-sensory sentinels of the intestinal environment and orchestrators of mucosal immunity. Mucosal Immunol. 11, 3–20 [DOI] [PubMed] [Google Scholar]
- 11.Banerjee A et al. (2018) Interpreting heterogeneity in intestinal tuft cell structure and function. J. Clin. Invest. 128, 1711–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mabbott NA et al. (2013) Microfold (M) cells: important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol. 6, 666–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen A et al. (1985) Adherent and soluble mucus in the stomach and duodenum. Dig. Dis. Sci 30, 55S–62S [DOI] [PubMed] [Google Scholar]
- 14.Pelaseyed T et al. (2015) The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol. Rev 260, 8–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murgia X et al. (2018) The role of mucus on drug transport and its potential to affect therapeutic outcomes. Adv. Drug Del. Rev 124, 82–97 [DOI] [PubMed] [Google Scholar]
- 16.Schneider H et al. (2018) Study of mucin turnover in the small intestine by in vivo labeling. Sci. Rep 8, 5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansson ME (2012) Fast renewal of the distal colonic mucus layers by the surface goblet cells as measured by in vivo labeling of mucin glycoproteins. PLoS One 7, e41009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y et al. (2018) Formation of human colonic crypt array by application of chemical gradients across a shaped epithelial monolayer. Cell. Mol. Gastroenterol. Hepatol 5, 113–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colgan SP et al. (2016) Hypoxia and mucosal inflammation. Annu. Rev. Pathol.: Mech. Dis 11, 77–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bäckhed F et al. (2005) Host-bacterial mutualism in the human intestine. Science 307, 1915–1920 [DOI] [PubMed] [Google Scholar]
- 21.Donaldson GP et al. (2016) Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol 14, 20–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Booijink CC et al. (2010) High temporal and inter individual variation detected in the human ileal microbiota. Environ. Microbiol 12, 3213–3227 [DOI] [PubMed] [Google Scholar]
- 23.Zoetendal EG et al. (2012) The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 6, 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bourlioux P et al. (2003) The intestine and its microflora are partners for the protection of the host: report on the Danone Symposium “The Intelligent Intestine,” helod in Paris, June 14, 2002. Am. J. Clin. Nutr 78, 675–683 [DOI] [PubMed] [Google Scholar]
- 25.Albenberg L et al. (2014) Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 147, 1055–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalantar-Zadeh K et al. (2018) A human pilot trial of ingestible electronic capsules capable of sensing different gases in the gut Nat. Electron. 1, 79–87 [Google Scholar]
- 27.Zheng L et al. (2015) Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A review in the theme: Cellular responses to hypoxia. Am. J. Physiol. Cell Physiol 309, C350–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheithauer TP et al. (2016) Causality of small and large intestinal microbiota in weight regulation and insulin resistance. Mol Metab 5, 759–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McNeil NI (1984) The contribution of the large intestine to energy supplies in man. Am. J. Clin. Nutr 39, 338–342 [DOI] [PubMed] [Google Scholar]
- 30.LeBlanc JG et al. (2013) Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr. Opin. Biotechnol 24, 160–168 [DOI] [PubMed] [Google Scholar]
- 31.Sato T et al. (2009) Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 [DOI] [PubMed] [Google Scholar]
- 32.Ootani A et al. (2009) Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 15, 701–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powell RH et al. (2017) WRN conditioned media is sufficient for in vitro propagation of intestinal organoids from large farm and small companion animals. Biol Open 6, 698–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato T et al. (2011) Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 [DOI] [PubMed] [Google Scholar]
- 35.Fatehullah A et al. (2016) Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 18, 246–254 [DOI] [PubMed] [Google Scholar]
- 36.Gjorevski N et al. (2016) Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560–564 [DOI] [PubMed] [Google Scholar]
- 37.Cruz-Acuña R et al. (2017) Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat. Cell Biol 19, 1326–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williamson IA et al. (2018) A high-throughput organoid microinjection platform to study gastrointestinal microbiota and luminal physiology. Cell. Mol. Gastroenterol. Hepatol 6, 301–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gunasekara DB et al. (2018) Development of arrayed colonic organoids for screening of secretagogues associated with enterotoxins. Anal. Chem 90, 1941–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Attayek PJ et al. (2016) In vitro polarization of colonoids to create an intestinal stem cell compartment. PLoS One 11, e0153795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davoudi Z et al. (2018) Intestinal organoids containing poly(lactic-co-glycolic acid) nanoparticles for the treatment of inflammatory bowel diseases. J. Biomed. Mater. Res., Part A 106, 876–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou Q et al. (2018) Lactobacillus accelerates ISCs regeneration to protect the integrity of intestinal mucosa through activation of STAT3 signaling pathway induced by LPLs secretion of IL-22. Cell Death Differ. DOI: 10.1038/s41418-018-0070-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leibowitz BJ et al. (2018) Targeting p53-dependent stem cell loss for intestinal chemoprotection Sci. Transl. Med 10, eaam7610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heo I et al. (2018) Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat Microbiol 3, 814–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drummond CG et al. (2017) Enteroviruses infect human enteroids and induce antiviral signaling in a cell lineage-specific manner. Proc. Natl. Acad. Sci. U. S. A 114, 1672–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibson PR et al. (1989) Isolation of colonic crypts that maintain structural and metabolic viability in vitro. Gastroenterology 96, 283–291 [DOI] [PubMed] [Google Scholar]
- 47.Whitehead RH et al. (1987) A method for the isolation and culture of human colonic crypts in collagen gels. In Vitro Cell. Dev. Biol 23, 436–442 [DOI] [PubMed] [Google Scholar]
- 48.Moon C et al. (2014) Development of a primary mouse intestinal epithelial cell monolayer culture system to evaluate factors that modulate IgA transcytosis. Mucosal Immunol. 7, 818–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.VanDussen KL et al. (2015) Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 64, 911–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fernando E et al. (2017) A simple, cost-effective method for generating murine colonic 3D enteroids and 2D monolayers for studies of primary epithelial cell function. Am. J. Physiol 313, G467–G475 [DOI] [PubMed] [Google Scholar]
- 51.van der Hee B et al. (2018) Optimized procedures for generating an enhanced, near physiological 2D culture system from porcine intestinal organoids. Stem Cell Res. 28, 165–171 [DOI] [PubMed] [Google Scholar]
- 52.Kozuka K et al. (2017) Development and characterization of a human and mouse intestinal epithelial cell monolayer platform. Stem Cell Rep. 9, 1976–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salem M et al. (2018) P2Y(6) receptors regulate CXCL10 expression and secretion in mouse intestinal epithelial cells. Front. Pharmacol 9, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Graves CL et al. (2014) A method for high purity intestinal epithelial cell culture from adult human and murine tissues for the investigation of innate immune function. J. Immunol. Methods 414, 20–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eaton AD et al. (2017) Primary human polarized small intestinal epithelial barriers respond differently to a hazardous and an innocuous protein. Food Chem. Toxicol 106, 70–77 [DOI] [PubMed] [Google Scholar]
- 56.Bhatt AP et al. (2018) Nonsteroidal anti-inflammatory drug -induced leaky gut modeled using polarized monolayers of primary human intestinal epithelial cells. ACS Infect. Dis. 4, 46–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yin JY et al. (2018) Molecular basis and differentiation-associated alterations of anion secretion in human duodenal enteroid monolayers. Cell. Mol. Gastroenterol. Hepatol 5, 591–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y et al. (2018) Monolayer culture of intestinal epithelium sustains Lgr5+ intestinal stem cells. Cell Discovery 4, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y et al. (2017) Self-renewing monolayer of primary colonic or rectal epithelial cells. Cell. Mol. Gastroenterol. Hepatol 4, 165–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tong Z et al. (2018) Towards a defined ECM and small molecule based monolayer culture system for the expansion of mouse and human intestinal stem cells. Biomaterials 154, 60–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scott A et al. (2016) Long-term renewable human intestinal epithelial stem cells as monolayers: A potential for clinical use. J. Pediatr. Surg 51, 995–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim R et al. (2018) Formation of arrays of planar, murine, intestinal crypts possessing a stem/proliferative cell compartment and differentiated cell zone. Lab Chip 18, 2202–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thorne CA et al. (2018) Enteroid Monolayers Reveal an Autonomous WNT and BMP Circuit Controlling Intestinal Epithelial Growth and Organization. Dev. Cell 44, 624–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ayehunie S et al. (2018) Human primary cell-based organotypic microtissues for modeling small intestinal drug absorption. Pharm. Res 35, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ettayebi K et al. (2016) Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holly MK et al. (2018) Adenovirus infection of human enteroids reveals interferon sensitivity and preferential infection of goblet cells. J. Virol 92, DOI: 10.1128/JVI.00250-00218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.In J et al. (2016) Enterohemorrhagic Escherichia coli reduces mucus and intermicrovillar bridges in human stem cell-derived colonoids. Cell. Mol. Gastroenterol. Hepatol 2, 48–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Madden LR et al. (2018) Bioprinted 3D primary human intestinal tissues model aspects of native physiology and ADME/Tox functions. iScience 2, 156–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mirhashemi ME et al. (2018) Transcriptome analysis of pig intestinal cell monolayers infected with Cryptosporidium parvum asexual stages. Parasites Vectors 11, 176–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Noel G et al. (2017) A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci. Rep 7, 45270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Puzan M et al. (2018) Enteric nervous system regulation of intestinal stem cell differentiation and epithelial monolayer function. Sci. Rep 8, 6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vernetti L et al. (2017) Functional coupling of human microphysiology systems: intestine, liver, kidney proximal tubule, blood-brain barrier and skeletal muscle. Sci. Rep 7, 42296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maschmeyer I et al. (2015) A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 15, 2688–2699 [DOI] [PubMed] [Google Scholar]
- 74.Chen HJ et al. (2018) A pumpless body-on-a-chip model using a primary culture of human intestinal cells and a 3D culture of liver cells. Lab Chip 18, 2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Y et al. (2017) In vitro generation of mouse colon crypts. ACS Biomater. Sci. Eng 3, 2502–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Y et al. (2017) A microengineered collagen scaffold for generating a polarized cryptvillus architecture of human small intestinal epithelium. Biomaterials 128, 44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kasendra M et al. (2018) Development of a primary human small intestine-on-a-chip using biopsy-derived organoids. Sci. Rep. 8, 2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Workman MJ et al. (2017) Enhanced utilization of induced pluripotent stem cell-derived human intestinal organoids using microengineered chips. Cell. Mol. Gastroenterol. Hepatol. 5, 669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hosic S et al. (2018) Rapid prototyping of a multilayer microphysiological system for primary human intestinal epithelial culture. bioRxiv DOI: 10.1101/400721400721 [DOI] [Google Scholar]
- 80.Chen Y et al. (2017) In vitro enteroid-derived three-dimensional tissue model of human small intestinal epithelium with innate immune responses. PLoS One 12, e0187880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen HJ et al. (2016) A recellularized human colon model identifies cancer driver genes. Nat. Biotechnol. 34, 845–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Helander HF et al. (2014) Surface area of the digestive tract - revisited. Scand. J. Gastroenterol 49, 681–689 [DOI] [PubMed] [Google Scholar]
- 83.Arhan P et al. (1981) Segmental colonic transit time. Dis. Colon Rectum 24, 625–629 [DOI] [PubMed] [Google Scholar]
- 84.Kerlin P et al. (1982) Relationship of motility to flow of contents in the human small intestine. Gastroenterology 82, 701–706 [PubMed] [Google Scholar]
- 85.Kim SK (1968) Small intestine transit time in the normal small bowel study. Am. J. Roentgenol. Radium Ther 104, 522–524 [DOI] [PubMed] [Google Scholar]
- 86.Narducci F et al. (1987) Twenty four hour manometric recording of colonic motor activity in healthy man. Gut 28, 17–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bassotti G et al. (2001) Low-amplitude propagated contractile waves: a relevant propulsive mechanism of human colon. Dig. Liver Dis 33, 36–40 [DOI] [PubMed] [Google Scholar]
- 88.Seidl H et al. (2012) Comparison of small-bowel motility of the human jejunum and ileum. Neurogastroenterol. Motil 24, e373–e380 [DOI] [PubMed] [Google Scholar]
- 89.Bijlsma PB et al. (1995) Differential in vivo and in vitro intestinal permeability to lactulose and mannitol in animals and humans: a hypothesis. Gastroenterology 108, 687–696 [DOI] [PubMed] [Google Scholar]
- 90.Halm DR et al. (2000) Secretagogue response of goblet cells and columnar cells in human colonic crypts. Am. J. Physiol.-Cell Physiol 277, C212–C233 [DOI] [PubMed] [Google Scholar]
- 91.Trbojevic-Stankovic JB et al. (2010) Morphometric study of healthy jejunal and ileal mucosa in adult and aged subjects. Histol. Histopathol. 25, 153–158 [DOI] [PubMed] [Google Scholar]
- 92.Cheng H et al. (1974) Origin differentiation and renewal of the four main epithelial cell types in the mouse small intestine v. Unitarian theory of the origin of the four epithelial cell types. Dev. Dyn 141, 537–561 [DOI] [PubMed] [Google Scholar]
- 93.Darwich AS et al. (2014) Meta-analysis of the turnover of intestinal epithelia in preclinical animal species and humans. Drug Metab. Dispos 42, 2016–2022 [DOI] [PubMed] [Google Scholar]
- 94.Sanderson IR et al. (2000) Development of the Gastrointestinal Tract. PMPH-USA
- 95.Atuma C et al. (2001) The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. American journal of physiology. Gastrointestinal and liver physiology 280, G922–929 [DOI] [PubMed] [Google Scholar]
- 96.Spence JR (2017) Taming the wild west of organoids, enteroids, and mini-guts. Cell. Mol. Gastroenterol. Hepatol 5, 159–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stelzner M et al. (2012) A nomenclature for intestinal in vitro cultures. Am. J. Physiol 302, G1359–G1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Greicius G et al. (2018) PDGFRα+ pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc. Natl. Acad. Sci. U. S. A, E3173–E3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shoshkes-Carmel M et al. (2018) Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature 557, 242–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Haramis A-PG et al. (2004) De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science 303, 1684–1686 [DOI] [PubMed] [Google Scholar]
- 101.Claesson MJ et al. (2011) Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. U. S. A 108 Suppl 1, 4586–4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.De Filippo C et al. (2010) Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U. S. A 107, 14691–14696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fujio-Vejar S et al. (2017) The gut microbiota of healthy Chilean subjects reveals a high abundance of the phylum Verrucomicrobia. Front. Microbiol 8, 1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gerbe F et al. (2012) The intestinal epithelium tuft cells: specification and function. Cell. Mol. Life Sci. 69, 2907–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kasai C et al. (2015) Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol 15, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang Y et al. (2018) Bioengineered systems and designer matrices that recapitulate the intestinal stem cell niche. Cell. Mol. Gastroenterol. Hepatol 5, 440–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van de Wetering M et al. (2015) Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Iwao T et al. (2014) Differentiation of human induced pluripotent stem cells into functional enterocyte-like cells using a simple method. Drug Metab. Pharmacokinet 29, 44–51 [DOI] [PubMed] [Google Scholar]
- 109.Spence JR et al. (2011) Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Finkbeiner SR et al. (2015) Transcriptome-wide analysis reveals hallmarks of human intestine development and maturation in vitro and in vivo. Stem Cell Rep. 4, 1140–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jung KB et al. (2018) Interleukin-2 induces the in vitro maturation of human pluripotent stem cell-derived intestinal organoids. Nature Communications 9, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Roeder R et al. (1999) Compliance, elastic modulus, and burst pressure of small-intestine submucosa (SIS), small diameter vascular grafts. J. Biomed. Mater. Res., Part A 47, 65–70 [DOI] [PubMed] [Google Scholar]
- 113.Pimpin A et al. (2012) Review on micro- and nanolithography techniques and their applications. Eng. J 16, 37–56 [Google Scholar]
- 114.Whitesides GM et al. (2001) Soft lithography in biology and biochemistry. Annu. Rev. Biomed. Eng 3, 335–373 [DOI] [PubMed] [Google Scholar]
- 115.Mandrycky C et al. (2016) 3D bioprinting for engineering complex tissues. Biotechnol. Adv 34, 422–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sears NA et al. (2016) A review of three-dimensional printing in tissue engineering. Tissue Eng., Part B 22, 298–310 [DOI] [PubMed] [Google Scholar]