Abstract
Bitter taste enables the detection of potentially harmful substances and is mediated by bitter taste receptors, TAS2Rs, in vertebrates. Few antagonists and inverse agonists of TAS2Rs have been identified, especially natural compounds. TAS2R16s in humans, apes and Old World monkeys (Catarrhini, Anthropoidea) recognize β-glucoside analogues as specific agonists. Here, we investigated responses of TAS2R16 to β-glucosides in non-anthropoid primates, namely lemurs (Lemuriformes, Strepsirrhini). Salicin acted as an agonist on lemur TAS2R16. Arbutin acted as an agonist in the ring-tailed lemur (Lemur catta) but as an inverse agonist in black lemur (Eulemur macaco) and black-and-white ruffed lemur (Varecia variegata). We identified a strepsirrhine-specific amino acid substitution responsible for the inverse agonism of arbutin. In a food preference test, salicin bitterness was inhibited by arbutin in the black lemur. Structural modelling revealed this locus was important for a rearrangement of the intracellular end of transmembrane helix 7 (TM7). Accordingly, arbutin is the first known natural inverse agonist of TAS2Rs, contributing to our understanding of receptor–ligand interactions and the molecular basis of the unique feeding habit diversification in lemurs. Furthermore, the identification of a causal point mutation suggests that TAS2R can acquire functional changes according to feeding habits and environmental conditions.
Keywords: bitter taste receptor, G-protein-coupled receptor, inverse agonist, lemur, molecular evolution
1. Background
Taste is important for the evaluation of food quality in mammals. Basic tastes are classified as sweet, umami, bitter, salty and sour. Bitter taste is associated with potentially harmful substances in food and thus is essential for survival in mammals. Bitter taste is perceived via bitter taste receptors (TAS2Rs), which are mainly expressed in taste buds on the surface of the tongue and palate epithelium [1,2]. TAS2Rs are G-protein-coupled receptors (GPCRs) and recognize various natural bitter substances as agonists that activate receptors [3,4]. TAS2Rs have various inhibitors (antagonists and inverse agonists). Antagonists have affinity but do not induce the activation of their receptors. Their binding to receptors inhibits the function of agonists. Inverse agonists have the opposite function to that of agonists by decreasing the constitutive activity of receptors. Many antagonists, including natural compounds, have been identified for TAS2Rs, but few inverse agonists (e.g. a synthetic compound, Nα,Nα-bis(carboxymethyl)-l-lysine; BCML to a human TAS2R4 mutant) have been identified [5–10]. Many key amino acid residues for ligand recognition have been identified by homology modelling and site-directed mutagenesis [5,11–19].
Vertebrates have a diverse repertoire of TAS2Rs in the genome, and these are associated with feeding habits and foraging behaviour [20–24]. TAS2R16 is a particularly well-studied TAS2R in terms of cellular biology and population genetics in anthropoids (human, apes and Old World and New World monkeys). β-glucoside analogues, including many natural toxins, are molecules in which sugars are bound to other functional groups via β-glucosidic bonds and are specific agonists of TAS2R16 in humans [3,12,25]. The sensitivity of TAS2R16 to β-glucosides differs among catarrhine primates (human, apes and Old World monkeys) and TAS2R16 of a New World monkey, the common marmoset (Callithrix jacchus), does not respond to β-glucosides [26]. Mouse (Mus musculus), which is closely related to primates, recognizes β-glucosides via TAS2R41 (Tas2r126 in mouse nomenclature), not via TAS2R16 (Tas2r118) [4]. It is not clear when ancestors of the primate lineage obtained the β-glucoside recognition ability observed in catarrhines owing to the lack of studies of TAS2R16 function in non-anthropoid primates (tarsiers and strepsirrhines). In this study, we analysed TAS2R16 responses to β-glucosides in lemurs, belonging to a clade of strepsirrhines, to characterize the evolution of TAS2R16 in primates and to identify the specificity of TAS2R16 in lemurs from the viewpoint of their unique ecology.
Lemurs are endemic to Madagascar and include over a hundred species [27]. These species arose by a unique adaptive radiation in Madagascar [28], which resulted in the dramatic diversification in biological features, including body size, body colour, activity and sociality. In particular, they obtained remarkable variation in feeding habits, including fruits, leaves, plant exudates and insects [27]. Thus, they are a good model for studying the mechanisms underlying the adaptive evolution of taste perception.
In this study, we characterized the functional features and variation in TAS2R16 responses to various β-glucosides in lemur species using cell-based calcium assays. We found that the receptibility of TAS2R16 to β-glucosides is conserved among primates. Furthermore, we found the first natural inverse agonist in non-human animals and identified the critical amino acid residues responsible for its ligand-induced activity using sited-directed mutagenesis and phylogenetic analyses. These findings clarify the functional differentiation of TAS2Rs and molecular mechanism underlying TAS2R–ligand interactions.
2. Results
(a). Different β-glucosides evoked different TAS2R16 responses among lemurs
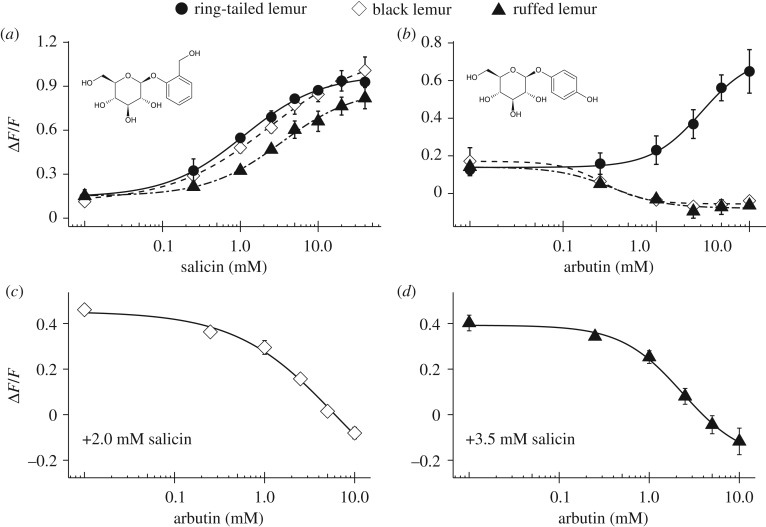
We evaluated the functional features of lemur TAS2R16 using salicin and arbutin. TAS2R16 of three lemurs were activated by salicin (figure 1a). There were significant differences in EC50 values among three lemurs for salicin (ring-tailed lemur: 1.1 ± 0.15 mM; black lemur: 2.3 ± 0.06 mM; ruffed lemur: 3.8 ± 0.68 mM; analysis of variance (ANOVA), p < 0.01). In particular, the EC50 for the ring-tailed lemur was significantly lower than that of the ruffed lemur (Welch's test with Benjamini–Hochberg (BH) correction, p < 0.01), indicating that the sensitivity to salicin is higher in the ring-tailed lemur than in the ruffed lemur. TAS2R16 of tested lemurs showed constitutive activity; IP1 production in the absence of ligands was much greater than that for cells expressing mock or human TAS2R16 (electronic supplementary material, figure S1a).
Figure 1.
Different β-glucosides evoked different responses of TAS2R16 in lemurs. (a,b) HEK293T cells expressing TAS2R16 from each lemur with Gα16/gust44 were stimulated with increasing concentrations of (a) salicin and (b) arbutin. Changes in fluorescence (ΔF/F) upon ligand application were monitored (mean ± s.e.m.). Experiments were performed three to four times independently. (c,d) HEK293T cells expressing TAS2R16 of (c) the black lemur and (d) ruffed lemur with Gα16/gust44 were stimulated with increasing concentrations of arbutin in the presence of (c) 2.0 mM and (d) 3.5 mM salicin. Changes in fluorescence (ΔF/F) upon ligand application were monitored (mean ± s.e.m.). Experiments were performed three to four times independently.
The responses of TAS2R16 to arbutin differed substantially among the three lemurs. TAS2R16 of the ring-tailed lemur was activated by increasing concentrations of arbutin (EC50, ring-tailed lemur: 3.1 ± 0.59 mM). However, TAS2R16 of the black lemur and ruffed lemur were inactivated by increasing concentrations of arbutin (IC50, black lemur: 0.31 ± 0.08 mM; ruffed lemur: 0.52 ± 0.23 mM) (figure 1b). Arbutin inhibited the salicin-mediated activation of TAS2R16 (figure 1c,d) and the constitutive activity of TAS2R16 in the black lemur and ruffed lemur (electronic supplementary material, figure S1b). These results indicate that arbutin acted as an agonist in the ring-tailed lemur and as an inverse agonist in the black lemur and ruffed lemur.
Furthermore, we found that TAS2R16 of the ring-tailed lemur was activated by three synthetic β-glucosides evaluated in this study (β-glucosides #1, #2 and #3). In the black lemur, although β-glucoside #1 partially activated TAS2R16, there was no response to β-glucoside #2 and TAS2R16 was inactivated by β-glucoside #3. In the ruffed lemur, there were no responses to β-glucoside #1, and β-glucosides #2 and #3 inactivated TAS2R16 (electronic supplementary material, figure S2a,b,c). These compounds also showed inhibitory effects on the salicin-mediated activation of TAS2R16 (electronic supplementary material, figure S2d,e).
(b). TAS2R16 function was directly associated with feeding behaviour in the black lemur
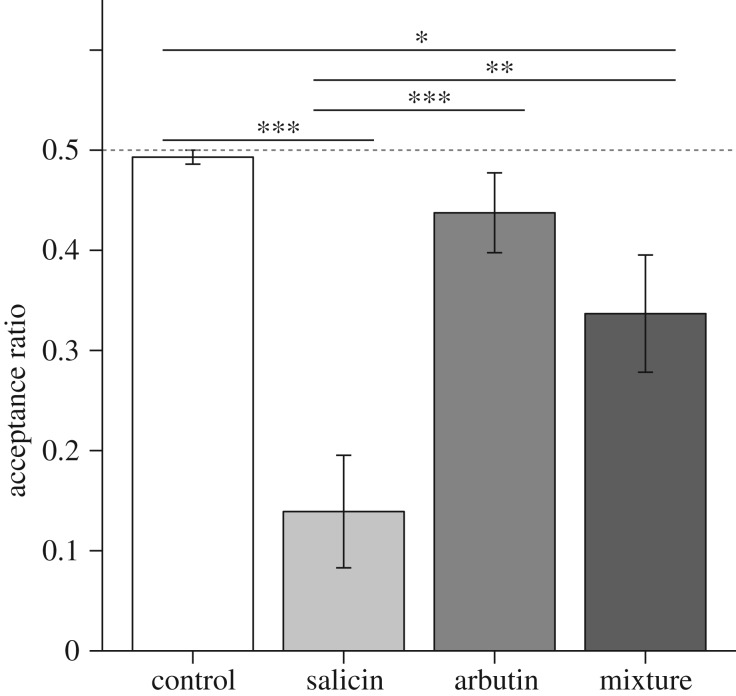
To confirm whether the inverse agonism of TAS2R16 in lemurs reflects their feeding behaviour, we performed a food preference test for β-glucoside sensitivity in a female black lemur (figure 2). The acceptance rate of salicin-treated apple pieces was significantly lower than that of control water-treated pieces (Welch's test with BH correction, p < 0.001). By contrast, the acceptance rate of arbutin-treated food was not different from that of water-treated pieces (p = 0.39). The acceptance rate of the apple pieces treated with a mixture of salicin and arbutin was significantly lower than that of control water-treated pieces (p < 0.05) but significantly greater than that of salicin-treated apple pieces (p < 0.01). This result indicates that arbutin masked the bitterness of salicin at the behavioural level and that TAS2R16 function was directly associated with feeding behaviour.
Figure 2.
Arbutin inhibits salicin bitterness in the black lemur. Water-soaked apple pieces (control) and β-glucoside-soaked pieces were presented to a subject (n = 1) simultaneously. Acceptance ratios for each β-glucoside and the mixture were calculated (mean ± s.e.m.). Behavioural assays (n = 8, respectively) were performed using each β-glucoside. Significant differences were determined by one-way ANOVA followed by Welch's tests with the BH correction (false discovery rate (FDR) q-value, * < 0.05; ** < 0.01; *** < 0.001).
(c). Amino acid residue at position 2827.55 was critical for the activation of lemur TAS2R16 by arbutin
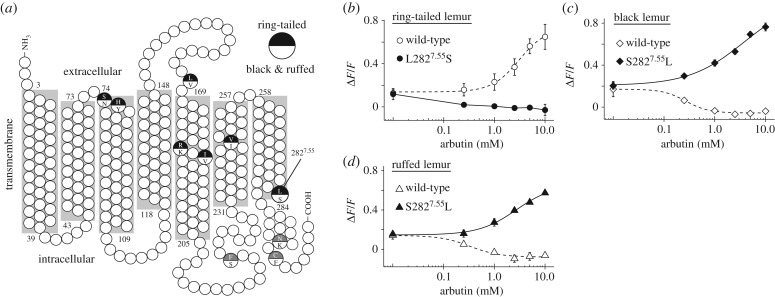
The critical amino acid positions for sensitivity to β-glucosides in TAS2R16 of humans [11] and non-human primates [14] were conserved in lemur TAS2R16 orthologues. TAS2R16 function against arbutin differs between the ring-tailed lemur and the black lemur and ruffed lemur, and we found that ring-tailed lemur TAS2R16 exhibited differences at 10 amino acid residues from those of the black lemur and ruffed lemur (figure 3a; electronic supplementary material, tables S4 and S5).
Figure 3.
Position 2827.55 of TAS2R16 is the critical amino acid residue for arbutin receptibility. (a) The amino acid residue at position 2827.55 was within the 7th transmembrane region. (b–d) Mutant TAS2R16 of (b) the ring-tailed lemur with a Leu-to-Ser substitution at position 2827.55, and of (c) the black lemur and (d) ruffed lemur with a Ser-to-Leu substitution at position 2827.55 were stimulated with increasing concentrations of arbutin. Changes in fluorescence (ΔF/F) were monitored (mean ± s.e.m.). Experiments were performed three to four times independently.
To identify the critical amino acid residues for arbutin recognition, we used site-directed mutagenesis to construct lemur TAS2R16 mutants. Many amino acid residues responsible for ligand specificity in human TAS2R16 are located in the transmembrane and extracellular domains [12]. We generated a three-dimensional model of lemur TAS2R16 using homology modelling (see details in the electronic supplementary material) and selected seven amino acid residues among 10 residues with substitutions in the ring-tailed lemur for site-directed mutagenesis (figure 3a). First, we substituted all seven residues of ring-tailed lemur TAS2R16 and evaluated the responses to arbutin. The 7-site mutant showed a similar response to arbutin to those of wild-type black lemur and ruffed lemur TAS2R16 (electronic supplementary material, figure S3a). Second, we introduced substitutions to combinations of the seven targeted residues in the ring-tailed lemur, measured responses of the mutants to arbutin, and screened the critical residues (electronic supplementary material, figure S3a,b). The mutant with the substitution of one of seven residues (position 2827.55: leucine to serine) showed a similar response to arbutin to those of wild-type black lemur and ruffed lemur TAS2R16 (figure 3b). Furthermore, arbutin also inhibited the salicin-mediated activation of this mutant (electronic supplementary material, figure S4), suggesting that arbutin acted as an inverse agonist on this mutant as well as on wild-type black lemur and ruffed lemur TAS2R16. Third, the responses of reverse substitution mutants of the black lemur and ruffed lemur, which mimicked ring-tailed lemur TAS2R16 (position 2827.55: serine to leucine), to arbutin were measured. The mutants were activated by increasing concentrations of arbutin, similar to wild-type ring-tailed lemur TAS2R16 (figure 3c,d). These results indicate that residue 2827.55 is the most important for inverse agonism of arbutin in lemurs.
(d). Evolutionary history of the amino acid residue at position 2827.55 in TAS2R16
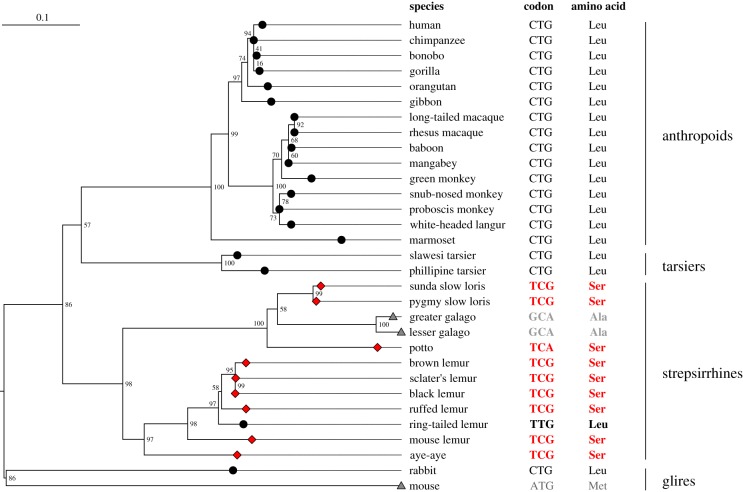
To uncover the evolutionary history of the amino acid residue at position 2827.55 in primate TAS2R16, a phylogenetic tree of primates was reconstructed based on the aligned amino acid sequences of TAS2R16 orthologues. Most strepsirrhines (except for the ring-tailed lemur and galagos) had serine at position 2827.55 and the other mammals had leucine, methionine or alanine (figure 4; electronic supplementary material, table S3), indicating that the ring-tailed lemur specifically acquired the substitution from serine to leucine at position 2827.55, based on parsimony (figure 4). This suggests that TAS2R16 in the last common ancestor of lemurs recognized arbutin as an inverse agonist and possibly exhibited constitutive activity; subsequently, TAS2R16 regained the ability to recognize arbutin as an agonist in the lineage of ring-tailed lemur, as observed in anthropoids.
Figure 4.
Substitution to serine at position 2827.55 is specific to strepsirrhines. Maximum-likelihood tree was reconstructed based on the amino acid sequences of TAS2R16 in primates. The amino acid residue at position 2827.55 in each species is indicated at each tip of the tree (diamonds indicate serine, circles indicate leucine, triangles indicate alanine and methionine). Bootstrap values are expressed at the nodes of the tree. The amino acid residue at position 2827.55 and the corresponding codon of primates are shown next to the species name. (Online version in colour.)
(e). Molecular modelling revealed an additional key residue for the activation of lemur TAS2R16
To identify the structural effect of residue 2827.55, we performed three-dimensional modelling and ligand docking simulations (see details in the electronic supplementary material). Thirteen key residues were identified for salicin and arbutin binding (electronic supplementary material, figure S5b,d). Ten specific residues in the ring-tailed lemur were not included in them. Position 2827.55 was located at the intracellular end of transmembrane helix 7 (TM7) far from the putative ligand binding site. Because position 2627.35 located at TM7 was identified as a key residue for arbutin binding, we examined the effect of the interaction between ligands and residue 2627.35 using site-directed mutagenesis. All F2627.35L mutants (position 2627.35: phenylalanine to leucine) were activated (figure 5; electronic supplementary material, figure S6). This indicates that residue 2627.35 is a key residue for the activation of lemur TAS2R16 and that the effect of residue 2827.55 is dependent on the ligand–TM7 interaction thorough residue 2627.35.
Figure 5.
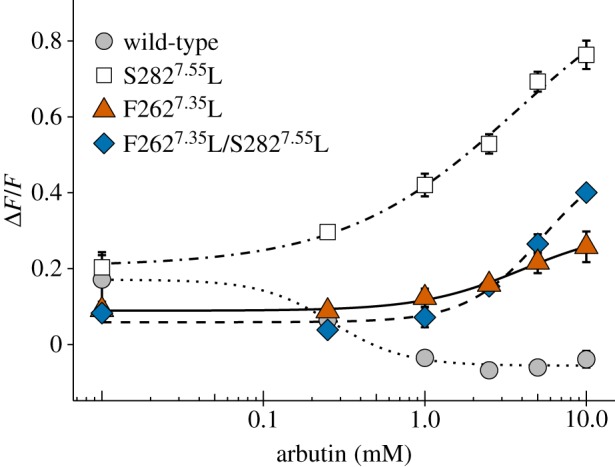
Position 2627.35 is related to the activation of lemur TAS2R16s. HEK293T cells expressing TAS2R16 mutants from the black lemur with Gα16/gust44 were stimulated with increasing concentrations of arbutin. Changes in fluorescence (ΔF/F) upon ligand application were monitored (mean ± s.e.m.). Experiments were performed three to five times independently. (Online version in colour.)
3. Discussion
We analysed the functional features of TAS2R16 in response to β-glucosides in lemurs. A calcium assay and phylogenetic analysis of TAS2R16 indicated that β-glucoside receptibility is conserved among primates and that TAS2R16 in the common ancestor of extant primates is sensitive to β-glucosides. Furthermore, we identified the first natural inverse agonist of TAS2R16 and the key residues responsible for inverse agonist recognition by site-directed mutagenesis.
Anthropoids, including humans, recognize β-glucosides via TAS2R16 [26]. We found that lemurs (strepsirrhines) also recognize β-glucosides via TAS2R16 (figure 1), indicating that β-glucoside-sensitive TAS2R16 was present in the last common ancestor of primates and is conserved in extant primates. Since mice use Tas2r126 (a TAS2R41 orthologue) for β-glucoside recognition, rather than Tas2r118 (a TAS2R16 orthologue) [4], further studies of non-Euarchontoglires mammals are required to characterize the functional evolution of TAS2R16 in mammals. Primates exhibit diverse responses of TAS2R16 to β-glucosides. Humans have high sensitivity to most β-glucoside agonists [25]. Macaques have 10-fold lower sensitivity to β-glucoside agonists in comparison with humans [26]. This study showed that TAS2R16 responses to β-glucosides also vary among lemurs (figure 1; electronic supplementary material, figure S2). Lemur TAS2R16 had constitutive activity (electronic supplementary material, figure S1). While all tested compounds acted as agonists of TAS2R16 in ring-tailed lemur, arbutin acted as an inverse agonist of TAS2R16 in the black lemur and ruffed lemur (figure 1; electronic supplementary material, figure S1).
More than 60 constitutively active wild-type GPCRs have been reported in mammals [29]. Several GPCRs exhibit differences in constitutive activity among species [29]. Several constitutively active mutants have been identified [5,30]. However, we identified the first constitutively active wild-type TAS2R. Most amino acid residues responsible for the constitutive activation of TAS2R are located in the third intracellular loop (ICL3), which plays a critical role in receptor activation [31]. We detected several lemur-specific substitutions in ICL3 of primate TAS2R16s, and these might be responsible for the constitutive activity of TAS2R16 in lemurs. Many constitutively active GPCRs are receptors for neurotransmitters. Thus, future studies should evaluate the role of constitutively active TAS2R in vivo.
Although many inverse agonists have been identified for other GPCRs [29], few inverse agonists for TAS2Rs have been identified (e.g. BCML, an amino acid derivative, for a human TAS2R4 mutant) [5]. Many antagonists have been identified for vertebrate TAS2Rs [5–10]. Arbutin is identified in this study as the first natural inverse agonist. Inverse agonists and antagonists are quite helpful for structural analyses of TAS2Rs because they stabilize the receptor conformation to the inactive state. Crystal structures of many GPCRs have been identified for antagonist- or inverse agonist-bound structures [32]. Since the crystal structures of TAS2Rs have not been determined, most previous studies of TAS2R–ligand interactions are based on models simulated by the structure of other GPCRs (e.g. human β2 adrenergic receptor and bovine rhodopsin) [33–35]. Thus, arbutin may facilitate analyses of the crystal structure of the well-studied bitter taste receptor TAS2R16.
The inverse agonism of arbutin to TAS2R16 was associated with feeding behaviour in the black lemur based on the greater acceptance of apples in a mixture of salicin and arbutin over salicin-treated apples (figure 2). However, the acceptance ratio of the apples in the mixture was slightly lower than that of arbutin-treated apples (p = 0.15) and significantly lower than that of control (water)-treated pieces (p < 0.05), indicating that arbutin partially masks salicin bitterness. Incomplete masking of salicin bitterness by arbutin may be caused by an insufficient amount of arbutin for the complete inhibition of 5 mM salicin or other taste receptor activation. This result suggests that arbutin can mask bitterness derived from specific agonists of TAS2R16, including β-glucosides, which could weaken bitterness in plants containing various β-glucosides. Plants known to contain arbutin, including Ericaceae [36] and Aquifoliaceae [37–39], are distributed worldwide, including in Madagascar [40]. Ilex mitis (Aquifoliaceae), distributed in Madagascar and southern Africa, is consumed by the brown mouse lemur (Microcebus rufus) [41]. Several plants belonging to Ericaceae are eaten by the ring-tailed lemur in Andringitra Massif [42]. Since little is known about the chemical composition of Malagasy plants, further analyses of the repertoire of chemicals in plants eaten by lemurs are required.
Ring-tailed lemur TAS2R16 was activated by all tested β-glucosides, unlike those of black lemur and ruffed lemur (figure 1; electronic supplementary material, figure S2), and exhibited differences at seven amino acid residues in the transmembrane and extracellular regions compared with those of the black lemur and ruffed lemur (figure 3a; electronic supplementary material, tables S4 and S5). We found that position 2827.55, one of the seven residues, was responsible for the activation of lemur TAS2R16 by arbutin using site-directed mutagenesis (figure 3). Except for position 1845.45, these residues are not critical amino acid positions for β-glucoside recognition and signalling in TAS2R16 of humans [11,12] and non-human primates [14]. Position 1845.45 may be related to the activation of lemur TAS2R16 because the orthologous position in human TAS2R16 has been identified as a signalling-related position [12] and its mutants in ring-tailed lemur showed lower responses to arbutin (electronic supplementary material, figure S3). They were also not key residues for ligand binding in the ligand docking simulation (electronic supplementary material, figure S5b,d). However, positions 168 and 2476.55 may have weak effects on β-glucoside binding because they were located within 5 Å from the ligand in our docking simulation.
Position 2827.55 was located at the intracellular end of TM7, far from the putative binding site for salicin and arbutin (electronic supplementary material, figure S5a,c). This region is important for the formation of a G-protein binding pocket during GPCR activation [43]. The intracellular end of TM7 exhibits inward movement during receptor activation in many GPCRs, including rhodopsin, β2 adrenoceptor, μ-opioid receptor and muscarinic acetylcholine receptor [43–47]. Arbutin binding to black lemur and ruffed lemur TAS2R16 may cause an incorrect rearrangement for constructing the G-protein-complementary binding pocket. In lemur TAS2R16, TM7 directly interacts with arbutin at position 2627.35 (figure 5b). This position is a key residue for ligand specificity in human TAS2R16 [12] and, near the side chains, bound to the benzene ring of β-glucosides (electronic supplementary material, figure S5b,d). Using β-glucoside #2 and #3, which have side chains bound to the benzene ring of β-glucosides at the para position, like arbutin, each lemur TAS2R16 showed similar responses to those using arbutin (electronic supplementary material, figure S2). Furthermore, all mutants with F2627.35L (phenylalanine to leucine) were activated by arbutin (figure 5; electronic supplementary material, figure S6). These results indicate that position 2627.35 is a key residue for ligand specificity in lemurs and that the interaction between TM7 and ligands at position 2627.35 has a substantial influence on the rearrangement of the intracellular end of TM7, which includes position 2827.55. Further analyses, including three-dimensional modelling of the G-protein–TAS2R interaction, are required to understand the mechanism underlying this TAS2R–ligand interaction, but these results suggest that TAS2R16 of ring-tailed lemur can easily obtain the active conformation, independent of the structure of the side chains in β-glucoside ligands, and can recognize many β-glucosides as agonists. Ring-tailed lemurs are more omnivorous than the other two lemurs and feed mainly on plants that are poor in nutrients and rich in secondary metabolites, as well as insects [48]. Thus, they might be exposed to a relatively high number of toxins, and the number of compounds with agonist activity may be expanded.
Based on a phylogenetic analysis using TAS2R16 orthologues in placental mammals, we found that strepsirrhines have serine and almost all other mammals have leucine at position 2827.55 (figure 4; electronic supplementary material, table S3), and that strepsirrhines (except for lorises and galagos), tarsiers and almost other non-primate mammals have phenylalanine at position 2627.35 (electronic supplementary material, figure S7 and table S3). These results suggest that the common ancestor of strepsirrhines obtained a substitution at position 2827.55, that lemur TAS2R16 which exhibits inverse agonism by arbutin only has the specific combination of amino acids (F2627.35 and S2827.55), and that arbutin does not activate TAS2R16 in most lemurs and inhibits agonist-dependent activation, and individuals do not perceive arbutin bitterness. The reverse mutation from serine to leucine at position 2827.55 occurred specifically in the ring-tailed lemur linage and recovered the agonism of arbutin (figures 1 and 4).
Our results clarified the general functional characteristics of primate TAS2R16 and the specific features of lemur TAS2R16. β-glucoside-sensitive TAS2R16 is conserved among primates. The responses of TAS2R16 to β-glucosides varied among primates. To determine the precise relationship between feeding habits and TAS2R function, both the range and sensitivity to ligands should be investigated using natural compounds found in plants from the diets of primates. Our discovery of a novel inverse agonist will contribute to analyses of the structure–function relationship, including activation mechanisms, of the well-studied bitter taste receptor TAS2R16.
4. Material and methods
(a). Genomic DNA
Genomic DNAs were isolated from the muscle tissues using the DNAeasy Blood &Tissue Kit (QIAGEN, Hilden, Germany), and from faeces using the QIAamp Fast DNA Stool Mini Kit (QIAGEN) or the NucleoSpin DNA Stool Kit (TaKaRa Bio, Kusatsu, Japan). The sources of genomic DNA are shown in detail in electronic supplementary material, table S1.
(b). Compounds
Bitter compounds were purchased from Sigma-Aldrich (St Louis, MO, USA). Each compound was dissolved in HEPES buffer (130 mM NaCl, 10 mM glucose, 5 mM KCl, 2 mM CaCl2, 1.2 mM MgCl2, 10 mM HEPES, pH 7.4) for the calcium assay, and in water for the behavioural assay.
(c). Sequence determination of TAS2R16
Based on the genome assemblies of the black lemur [49], mouse lemur, greater galago and tarsier [50], primers were designed to amplify TAS2R16 (electronic supplementary material, table S6). Polymerase chain reactions (PCRs) were performed using Tks Gflex DNA Polymerase (TaKaRa Bio) as follows: 1 min of initial denaturation; 30–40 cycles of denaturation at 98°C for 10 s, annealing at a temperature gradient of 50–57°C for 15 s and extension at 68°C for 30 s; and a final extension at 68°C for 5 min. All sequences of PCR products were determined using the BigDye Terminator v. 3.1 Cycle Sequencing Kit and the ABI 3130xl (Applied Biosystems, Foster City, CA, USA).
(d). Construction of expression vectors for TAS2R16 and point mutants
Amplified TAS2R16 was tagged at the N terminus with the first 45 amino acids of rat somatostatin receptor type 3 for cell-surface targeting and at the C terminus with the last 8 amino acids of bovine rhodopsin as an epitope tag [11,26]. The tagged TAS2R16s were sub-cloned into the EcoRV site of the mammalian expression vector pEAK10 using the In-Fusion HD Cloning Kit (Clontech, Fremont, CA, USA). Point mutant vectors were constructed using QuikChange (Agilent Technologies, Santa Clara, CA, USA) and the overlap extension PCR method [51]. All mutations were checked by sequencing.
(e). Cell culture, transfection and calcium assay
Cell culture, transfection and calcium assays were performed as described previously [14,52,53]. The calcium response is expressed as the normalized peak response (F) relative to background fluorescence (F0): ΔF/F (=[F − F0]/F0). ΔF/F values were used for the calculation of dose–response relationships using the drc package in R [54]. Detailed information of calcium assays is described in the electronic supplementary material, methods.
(f). IP1 assay
Because IP3 (inositol triphosphate), the second messenger of TAS2Rs, is quite short-lived, inositol dephosphorylation was stopped at IP1 (inositol phosphate), a metabolite of IP3, by adding LiCl; the concentration of accumulated IP1 was measured as a fluorescence ratio (668 nm/620 nm) by an immunoassay using the IP-One-Gq Kit (Cisbio, Codolet, France). The fluorescence ratio was converted to the IP1 concentration using the standard curve generated from known concentrations of IP1. IP1 production was compared among species and concentrations by Welch's tests with the BH correction. Detailed information is described in the electronic supplementary material, methods.
(g). Behavioural assay
The subject was a female black lemur at the Japan Monkey Centre (Aichi, Japan) who was reared in a group. It was guided into the corridor to the outdoor enclosure for the behavioural assay. Two kinds of apple pieces (15 mm × 8 mm × 8 mm) were used for the food preference test. One was soaked in 100 ml of water (control solution) and another was soaked in the test compound solution (water, 5 mM salicin, 10 mM arbutin and a mixture) overnight before testing. In a trial, the animal was exposed to five water-treated apple pieces and five test compound-treated pieces for 5 min at the same time. The number of pieces eaten was recorded in each trial. Eight trials were performed for each test compound. All trials were recorded using a video camera. The intake rate of apple pieces soaked in the test compound solution (i.e. the acceptance ratio) was calculated as follows: (acceptance ratio) = (number of pieces treated with test compounds eaten)/(total number of pieces eaten). The order of exposure to apple pieces was randomized. Differences in the acceptance ratio among test compounds were assessed by one-way ANOVA, followed by Welch's tests with the BH correction.
(h). Sequence and phylogenetic analysis
Sequences of intact TAS2R16 orthologues were obtained from Hayakawa et al. [21] (Euarchontoglires) and Liu et al. [23] (Laurasiatheria). The genome assemblies of 13 placental mammals were obtained from web sources and are summarized in electronic supplementary material, table S2. TAS2R16 orthologues were identified from the genome assemblies as described previously [21] (see details in the electronic supplementary material). To compare sequences of TAS2R16 orthologues, a multiple sequence alignment was generated based on the amino acid sequences using MAFFT v. 7 [55]. Amino acid residues located in transmembrane domains (TM) were identified by a superscript number following the Ballesteros–Weinstein (BW) numbering method [56]. The residue corresponding to the most conserved residue of class A GPCRs in TM number X is assigned the index X.50, which was identified by X.50 of human TAS2R16 [57,58] and a multiple sequence alignment among human and lemur TAS2R16. A maximum-likelihood tree was reconstructed based on the aligned amino acid sequences using RAxML v. 8.2.10 [59]. The JTT model [60] was used to correct for multiple substitutions. Bootstrap resampling (1000 replicates) was performed [61].
Supplementary Material
Acknowledgements
We thank Dr Mitsugu Araki, Dr Hiroaki Iwata, Dr Biao Ma, Dr Yoko Sasakura and Dr Yasushi Okuno for help with the structural modelling, ligand docking simulations and helpful discussion. We also thank the veterinarians and caretakers, especially Chigusa Tanaka, Yuki Koizumi and Mana Nakakuki, of the Japan Monkey Centre for help with genetic sampling and the behavioural assay; the veterinarians and caretakers, especially Shinichi Kioka and Shu Morikubo, of Ueno Zoological Gardens for help with genetic sampling; Emiko Nishi for help with the cell-based assay; Dr Takashi Ueda, Dr Yoshiro Ishimaru, Dr Takumi Misaka and Dr Keiko Abe for providing the Gα16/gust44 and pEAK10 vectors; Dr Hiroaki Matsunami for providing HEK293T cells; and Dr Takahiro Yamashita for help with the experimental design. We also thank the Proboscis Monkey Functional Genome Consortium (proboscis monkey) and the National Institutes of Health, Dr Richard K. Wilson and The Genome Institute, Washington University School of Medicine (Malayan flying lemur) for making the data of the whole-genome assembly available in the UCSC Genome Browser.
Ethics
The behavioural assay was performed at the Japan Monkey Centre. The study was approved by the Research Ethics Committee of the Japan Monkey Centre (#2017-015) and was performed in accordance with the Ethical Guidelines for Research at the Japan Monkey Centre (1 April 2016) as a collaborative research project with the Japan Monkey Centre (#2017014). The study was also approved by the Animal Welfare and Animal Care Committee of the Primate Research Institute, Kyoto University (#2017-177-05) based on the Guidelines for the Care and Use of Nonhuman Primates of Primates Research Institute, Kyoto University (Version 3, issued in 2010). The use of genetic materials was approved by the Japan Monkey Centre (#2017-018). The study was also performed as a collaborative research project with the Ueno Zoological Gardens (No. 29-100).
Data accessibility
DNA sequences: DDBJ accessions LC414992–LC415000. Original data for the calcium, IP1 and behavioural assays are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.bj20jr7 [62].
Authors' contributions
A.I. designed and conducted experiments, analysed and interpreted data, and wrote the original draft. T.H. conducted experiments, analysed and interpreted data, and wrote and revised the draft. N.S.-H. conducted experiments, analysed and interpreted data, and wrote and revised the draft. H.I. designed experiments and wrote and finalized the draft. All authors approved the final version of the manuscript and agree to be accountable for all aspects of this work.
Competing interests
The authors have no competing interests to report.
Funding
This work was financially supported by the Leading Graduate Program in Primatology and Wildlife Sciences, Kyoto University and KAKENHI grants from the Japan Society for the Promotion of Science (grant nos 18J22288 to A.I., 16K18630 to T.H., 17K15203 to N.S.-H. and 15H02421, 15H05242 and 18H04005 to H.I.), research grant from Kobayashi International Scholarship Foundation, the Terumo Foundation for Life Sciences and Arts and the Umami Manufacturers Association of Japan.
References
- 1.Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJP. 2000. T2Rs function as bitter taste receptors. Cell 100, 703–711. ( 10.1016/S0092-8674(00)80706-0) [DOI] [PubMed] [Google Scholar]
- 2.Chandrashekar J, Hoon MA, Ryba NJP, Zuker CS. 2006. The receptors and cells for mammalian taste. Nature 444, 288–294. ( 10.1038/nature05401) [DOI] [PubMed] [Google Scholar]
- 3.Meyerhof W, Batram C, Kuhn C, Brockhoff A, Chudoba E, Bufe B, Appendino G, Behrens M. 2010. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem. Senses 35, 157–170. ( 10.1093/chemse/bjp092) [DOI] [PubMed] [Google Scholar]
- 4.Lossow K, Hübner S, Roudnitzky N, Slack JP, Pollastro F, Behrens M, Meyerhof W. 2016. Comprehensive analysis of mouse bitter taste receptors reveals different molecular receptive ranges for orthologous receptors in mice and humans. J. Biol. Chem. 291, 15 358–15 377. ( 10.1074/jbc.M116.718544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pydi SP, Sobotkiewicz T, Billakanti R, Bhullar RP, Loewen MC, Chelikani P. 2014. Amino acid derivatives as bitter taste receptor (T2R) blockers. J. Biol. Chem. 289, 25 054–25 066. ( 10.1074/jbc.M114.576975) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pydi SP, Jaggupilli A, Nelson KM, Abrams SR, Bhullar RP, Loewen MC, Chelikani P. 2015. Abscisic acid acts as a blocker of the bitter taste G protein-coupled receptor T2R4. Biochemistry 54, 2622–2631. ( 10.1021/acs.biochem.5b00265) [DOI] [PubMed] [Google Scholar]
- 7.Behrens M, Blank K, Meyerhof W. 2017. Blends of non-caloric sweeteners saccharin and cyclamate show reduced off-taste due to TAS2R bitter receptor inhibition. Cell Chem. Biol. 24, 1199–1204. ( 10.1016/j.chembiol.2017.08.004) [DOI] [PubMed] [Google Scholar]
- 8.Jaggupilli A, Howard R, Upadhyaya JD, Bhullar RP, Chelikani P. 2016. Bitter taste receptors: novel insights into the biochemistry and pharmacology. Int. J. Biochem. Cell Biol. 77, 184–196. ( 10.1016/j.biocel.2016.03.005) [DOI] [PubMed] [Google Scholar]
- 9.Dey B, Kawabata F, Kawabata Y, Yoshida Y, Nishimura S, Tabata S. 2017. Identification of functional bitter taste receptors and their antagonist in chickens. Biochem. Biophys. Res. Commun. 482, 693–699. ( 10.1016/j.bbrc.2016.11.096) [DOI] [PubMed] [Google Scholar]
- 10.Kim MJ, Son HJ, Kim Y, Misaka T, Rhyu M-R. 2015. Umami-bitter interactions: the suppression of bitterness by umami peptides via human bitter taste receptor. Biochem. Biophys. Res. Commun. 456, 586–590. ( 10.1016/j.bbrc.2014.11.114) [DOI] [PubMed] [Google Scholar]
- 11.Sakurai T, et al. 2010. Characterization of the β-D-glucopyranoside binding site of the human bitter taste receptor hTAS2R16. J. Biol. Chem. 285, 28 373–28 378. ( 10.1074/jbc.M110.144444) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas A, Sulli C, Davidson E, Berdougo E, Phillips M, Puffer BA, Paes C, Doranz BJ, Rucker JB. 2017. The bitter taste receptor TAS2R16 achieves high specificity and accommodates diverse glycoside ligands by using a two-faced binding pocket. Sci. Rep. 7, 7753 ( 10.1038/s41598-017-07256-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Born S, Levit A, Niv MY, Meyerhof W, Behrens M. 2013. The human bitter taste receptor TAS2R10 is tailored to accommodate numerous diverse ligands. J. Neurosci. 33, 201–213. ( 10.1523/JNEUROSCI.3248-12.2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai H, et al. 2016. Amino acid residues of bitter taste receptor TAS2R16 that determine sensitivity in primates to β-glycosides. Biophys. Physicobiol. 13, 165–171. ( 10.2142/biophysico.13.0_165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu K, Jaggupilli A, Premnath D, Chelikani P. 2018. Plasticity of the ligand binding pocket in the bitter taste receptor T2R7. Biochim. Biophys. Acta Biomembr. 1860, 991–999. ( 10.1016/j.bbamem.2018.01.014) [DOI] [PubMed] [Google Scholar]
- 16.Nowak S, Di Pizio A, Levit A, Niv MY, Meyerhof W, Behrens M.. 2018. Reengineering the ligand sensitivity of the broadly tuned human bitter taste receptor TAS2R14. Biochim. Biophys. Acta Gen. Subj. 1862, 2162–2173. ( 10.1016/j.bbagen.2018.07.009) [DOI] [PubMed] [Google Scholar]
- 17.Marchiori A, Capece L, Giorgetti A, Gasparini P, Behrens M, Carloni P, Meyerhof W. 2013. Coarse-grained/molecular mechanics of the TAS2R38 bitter taste receptor: experimentally-validated detailed structural prediction of agonist binding. PLoS ONE 8, 1–12. ( 10.1371/journal.pone.0064675) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue AY, Di Pizio A, Levit A, Yarnitzky T, Penn O, Pupko T, Niv MY.. 2018. Independent evolution of strychnine recognition by bitter taste receptor subtypes. Front. Mol. Biosci. 5, 1–14. ( 10.3389/fmolb.2018.00009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Pizio A, Shy N, Behrens M, Meyerhof W, Niv MY.. 2018. Molecular features underlying selectivity in chicken bitter taste receptors. Front. Mol. Biosci. 5, 1–9. ( 10.3389/fmolb.2018.00006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li D, Zhang J. 2014. Diet shapes the evolution of the vertebrate bitter taste receptor gene repertoire. Mol. Biol. Evol. 31, 303–309. ( 10.1093/molbev/mst219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayakawa T, Suzuki-Hashido N, Matsui A, Go Y. 2014. Frequent expansions of the bitter taste receptor gene repertoire during evolution of mammals in the euarchontoglires clade. Mol. Biol. Evol. 31, 2018–2031. ( 10.1093/molbev/msu144) [DOI] [PubMed] [Google Scholar]
- 22.Wang K, Zhao H. 2015. Birds generally carry a small repertoire of bitter taste receptor genes. Genome Biol. Evol. 7, 2705–2715. ( 10.1093/gbe/evv180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z, Liu G, Hailer F, Orozco-terWengel P, Tan X, Tian J, Yan Z, Zhang B, Li M. 2016. Dietary specialization drives multiple independent losses and gains in the bitter taste gene repertoire of Laurasiatherian Mammals. Front. Zool. 13, 1–9. ( 10.1186/s12983-016-0161-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong H, Shang S, Wu X, Chen J, Zhu W, Yan J, Li H, Zhang H. 2017. Genomic evidence of bitter taste in snakes and phylogenetic analysis of bitter taste receptor genes in reptiles. PeerJ 5, e3708 ( 10.7717/peerj.3708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bufe B, Hofmann T, Krautwurst D, Raguse J-D, Meyerhof W. 2002. The human TAS2R16 receptor mediates bitter taste in response to β-glucopyranosides. Nat. Genet. 32, 397–401. ( 10.1038/ng1014) [DOI] [PubMed] [Google Scholar]
- 26.Imai H, Suzuki N, Ishimaru Y, Sakurai T, Yin L, Pan W, Abe K, Misaka T, Hirai H. 2012. Functional diversity of bitter taste receptor TAS2R16 in primates. Biol. Lett. 8, 652–656. ( 10.1098/rsbl.2011.1251) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mittermeier RA, Hawkins F, Louis EE. 2010. Lemurs of Madagascar, 3rd edn Washington, DC: Conservation International. [Google Scholar]
- 28.Ali JR, Huber M. 2010. Mammalian biodiversity on Madagascar controlled by ocean currents. Nature 463, 653–656. ( 10.1038/nature08706) [DOI] [PubMed] [Google Scholar]
- 29.Seifert R, Wenzel-Seifert K. 2002. Constitutive activity of G-proteins-coupled receptors: cause of disease and common property of wild-type receptors. Naunyn. Schmiedebergs. Arch. Pharmacol. 366, 381–416. ( 10.1007/s00210-002-0588-0) [DOI] [PubMed] [Google Scholar]
- 30.Pydi SP, Bhullar RP, Chelikani P. 2012. Constitutively active mutant gives novel insights into the mechanism of bitter taste receptor activation. J. Neurochem. 122, 537–544. ( 10.1111/j.1471-4159.2012.07808.x) [DOI] [PubMed] [Google Scholar]
- 31.Pydi SP, Singh N, Upadhyaya J, Bhullar RP, Chelikani P. 2014. The third intracellular loop plays a critical role in bitter taste receptor activation. Biochim. Biophys. Acta Biomembr. 1838, 231–236. ( 10.1016/j.bbamem.2013.08.009) [DOI] [PubMed] [Google Scholar]
- 32.Xiang J, Chun E, Liu C, Jing L, Al-Sahouri Z, Zhu L, Liu W. 2016. Successful strategies to determine high-resolution structures of GPCRs. Trends Pharmacol. Sci. 37, 1055–1069. ( 10.1016/j.tips.2016.09.009) [DOI] [PubMed] [Google Scholar]
- 33.Brockhoff A, Behrens M, Niv MY, Meyerhof W. 2010. Structural requirements of bitter taste receptor activation. Proc. Natl Acad. Sci. USA 107, 11 110–11 115. ( 10.1073/pnas.0913862107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan J, Abrol R, Trzaskowski B, William AG 3rd. 2012. 3D structure prediction of TAS2R38 bitter receptors bound to agonists phenylthiocarbamide (PTC) and 6-n-propylthiouracil (PROP). J. Chem. Inf. Model. 52, 1875–1885. ( 10.1021/ci300133a) [DOI] [PubMed] [Google Scholar]
- 35.Chen Z, Dong S, Meng F, Liang Y, Zhang S, Sun J. 2018. Insights into the binding of agonist and antagonist to TAS2R16 receptor: a molecular simulation study. Mol. Simul. 44, 322–329. ( 10.1080/08927022.2017.1376325) [DOI] [Google Scholar]
- 36.Pavlović RD, Lakušić B, Došlov-Kokoruš Z, Kovačević N. 2009. Arbutin content and antioxidant activity of some Ericaceae species. Pharmazie 64, 656–659. ( 10.1691/ph.2009.9551) [DOI] [PubMed] [Google Scholar]
- 37.Miura H, Inoue E, Kitamura Y, Sugii M. 1985. Extraction and determination of arbutin in the leaves of Viburnum and Ilex spp. Shoyakugaku Zasshi 39, 181–184. [Google Scholar]
- 38.Choi YH, Sertic S, Kim HK, Wilson EG, Michopoulos F, Lefeber AWM, Erkelens C, Kricun SDP, Verpoorte R. 2005. Classification of Ilex species based on metabolomic fingerprinting using nuclear magnetic resonance and multivariate data analysis. J. Agric. Food Chem. 53, 1237–1245. ( 10.1021/jf0486141) [DOI] [PubMed] [Google Scholar]
- 39.Kim HK, et al. 2010. Metabolic classification of South American Ilex species by NMR-based metabolomics. Phytochemistry 71, 773–784. ( 10.1016/j.phytochem.2010.02.001) [DOI] [PubMed] [Google Scholar]
- 40.Schwery O, Onstein RE, Bouchenak-Khelladi Y, Xing Y, Carter RJ, Linder HP. 2015. As old as the mountains: the radiations of the Ericaceae. New Phytol. 207, 355–367. ( 10.1111/nph.13234) [DOI] [PubMed] [Google Scholar]
- 41.Atsalis S. 1999. Diet of the brown mouse lemur (Microcebus rufus) in Ranomafana National Park, Madagascar. Int. J. Primatol. 20, 193–229. ( 10.1023/A:1020518419038) [DOI] [PubMed] [Google Scholar]
- 42.Goodman SM, Langrand O. 1996. A high mountain population of the ring-tailed lemur Lemur catta on the Andringitra Massif, Madagascar. Oryx 30, 259–268. ( 10.1017/S003060530002175X) [DOI] [Google Scholar]
- 43.Latorraca NR, Venkatakrishnan AJ, Dror RO. 2017. GPCR dynamics: structures in motion. Chem. Rev. 117, 139–155. ( 10.1021/acs.chemrev.6b00177) [DOI] [PubMed] [Google Scholar]
- 44.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauß N, Choe H-W, Hofmann KP, Ernst OP. 2008. Crystal structure of opsin in its G-protein-interacting conformation. Nature 455, 497–502. ( 10.1038/nature07330) [DOI] [PubMed] [Google Scholar]
- 45.Huang W, et al. 2015. Structural insights into μ-opioid receptor activation. Nature 524, 315–321. ( 10.1038/nature14886) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenbaum DM, et al. 2011. Structure and function of an irreversible agonist-β2 adrenoceptor complex. Nature 469, 236–242. ( 10.1038/nature09665) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kruse AC, et al. 2013. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature 504, 101–106. ( 10.1038/nature12735) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simmen B, Sauther ML, Soma T, Rasamimanana H, Sussman RW, Jolly A, Tarnaud L, Hladik A.. 2006. Plant species fed on by Lemur catta in galley forests of southern domain of Madagascar. In Ringtailed lemur biology: Lemur catta in Madagascar (eds Jolly A, Sussman RW, Koyama N, Rasamimanana H), pp. 55–68. Berlin, Germany: Springer. [Google Scholar]
- 49.Meyer WK, Venkat A, Kermany AR, Van De Geijn B, Zhang S, Przeworski M.. 2015. Evolutionary history inferred from the de novo assembly of a nonmodel organism, the blue-eyed black lemur. Mol. Ecol. 24, 4392–4405. ( 10.1111/mec.13327) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindblad-Toh K, et al. 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476–482. ( 10.1038/nature10530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vallejo AN, Pogulis RJ, Pease LR. 2008. PCR mutagenesis by overlap extension and gene SOE. Cold Spring Harb. Protoc. 3, 1–7. ( 10.1101/pdb.prot4861) [DOI] [PubMed] [Google Scholar]
- 52.Purba LHPS, Widayati KA, Tsutsui K, Suzuki-Hashido N, Hayakawa T, Nila S, Suryobroto B, Imai H. 2017. Functional characterization of the TAS2R38 bitter taste receptor for phenylthiocarbamide in colobine monkeys. Biol. Lett. 13, 20160834 ( 10.1098/rsbl.2016.0834) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ueda T, Ugawa S, Yamamura H, Imaizumi Y, Shimada S. 2003. Functional interaction between T2R taste receptors and G-protein α subunits expressed in taste receptor cells. J. Neurosci. 23, 7376–7380. ( 10.1523/JNEUROSCI.23-19-07376.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritz C, Baty F, Streibig JC, Gerhard D. 2015. Dose-response analysis using R. PLoS ONE 10, 1–13. ( 10.1371/journal.pone.0146021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ballesteros JA, Weinstein H. 1995. [19] Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 25, 366–428. ( 10.1016/S1043-9471(05)80049-7) [DOI] [Google Scholar]
- 57.Wiener A, Shudler M, Levit A, Niv MY. 2012. BitterDB: a database of bitter compounds. Nucleic Acids Res. 40, D413–D419. ( 10.1093/nar/gkr755) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dagan-Wiener A, Di Pizio A, Nissim I, Bahia MS, Dubovski N, Margulis E, Niv MY.. 2019. BitterDB: taste ligands and receptors database in 2019. Nucleic Acids Res. 47, D1179–D1185. ( 10.1093/nar/gky974) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. ( 10.1093/bioinformatics/btu033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Bioinformatics. 8, 275–282. ( 10.1093/bioinformatics/8.3.275) [DOI] [PubMed] [Google Scholar]
- 61.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. ( 10.1111/j.1558-5646.1985.tb00420.x) [DOI] [PubMed] [Google Scholar]
- 62.Itoigawa A, Hayakawa T, Suzuki-Hashido N, Imai H. 2019. Data from: A natural point mutation in the bitter taste receptor TAS2R16 causes inverse agonism of arbutin in lemur gestation Dryad Digital Repository. ( 10.5061/dryad.bj20jr7) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Itoigawa A, Hayakawa T, Suzuki-Hashido N, Imai H. 2019. Data from: A natural point mutation in the bitter taste receptor TAS2R16 causes inverse agonism of arbutin in lemur gestation Dryad Digital Repository. ( 10.5061/dryad.bj20jr7) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
DNA sequences: DDBJ accessions LC414992–LC415000. Original data for the calcium, IP1 and behavioural assays are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.bj20jr7 [62].