Abstract
Many marine animals depend upon a larval phase of their life cycle to locate suitable habitat, and larvae use light detection to influence swimming behaviour and dispersal. Light detection is mediated by the opsin genes, which encode light-sensitive transmembrane proteins. Previous studies suggest that r-opsins in the eyes mediate locomotory behaviour in marine protostomes, but few have provided direct evidence through gene mutagenesis. Larvae of the marine annelid Capitella teleta have simple eyespots and are positively phototactic, although the molecular components that mediate this behaviour are unknown. Here, we characterize the spatio-temporal expression of the rhabdomeric opsin genes in C. teleta and show that a single rhabdomeric opsin gene, Ct-r-opsin1, is expressed in the larval photoreceptor cells. To investigate its function, Ct-r-opsin1 was disrupted using CRISPR/CAS9 mutagenesis. Polymerase chain reaction amplification and DNA sequencing demonstrated efficient editing of the Ct-r-opsin1 locus. In addition, the pattern of Ct-r-opsin1 expression in photoreceptor cells was altered. Notably, there was a significant decrease in larval phototaxis, although the eyespot photoreceptor cell and associated pigment cell formed normally and persisted in Ct-r-opsin1-mutant animals. The loss of phototaxis owing to mutations in Ct-r-opsin1 is similar to that observed when the entire photoreceptor and pigment cell are deleted, demonstrating that a single r-opsin gene is sufficient to mediate phototaxis in C. teleta. These results establish the feasibility of gene editing in animals like C. teleta, and extend previous work on the development, evolution and function of the C. teleta visual system. Our study represents one example of disruption of animal behaviour by gene editing through CRISPR/CAS9 mutagenesis, and has broad implications for performing genome editing studies in a wide variety of other understudied animals.
Keywords: opsin, phototaxis, Capitella teleta, annelid, behaviour, genome editing
1. Introduction
Many marine animals have a larval dispersal phase to locate and move towards a suitable habitat. For marine larvae, eyes often provide information about light intensity and direction, and are thought to mediate the positive or negative phototactic responses that are important for both dispersal and settlement [1,2]. The majority of pelagic larvae produced by benthic marine invertebrates have a period of positive photo response [1]. Larvae typically have simple eyes that can be comprised of only two cells: a pigment cell and a photosensory cell [2–5]. The pigment cell shields incoming light, and its close proximity to the photosensory cell is sufficient for detection of the direction of light [3].
Capitella teleta is an annelid worm that burrows in marine sediments and produces a swimming larva as part of its life cycle [6]. Capitella teleta larvae have a pair of eyespots similar to the simple larval eyespots characteristic of many invertebrate larvae, and similar to the prototype pigment-cup eye proposed to represent the ancestral bilaterian condition [3,7]. The larval eyespot in C. teleta is located along the exterior rim of the brain, and is composed of a supporting cell, photosensory cell and pigment cell (figure 1a–d; [8]). The eyespots appear soon after initiation of the larval period, prior to robust swimming [9]. The juvenile photosensory cell appears during late larval stages and temporally coexists with the larval eyespot [10]. During metamorphosis, the larval pigment cell is incorporated into the juvenile eyespot [10], although larval and juvenile photosensory cells appear to be distinct. Capitella teleta larvae exhibit a robust positive phototactic response (electronic supplementary material, figure S1). This behaviour is lost if both photoreceptor and pigment cells are experimentally deleted, demonstrating that phototaxis is mediated by the cerebral eyespots [10,11].
Figure 1.
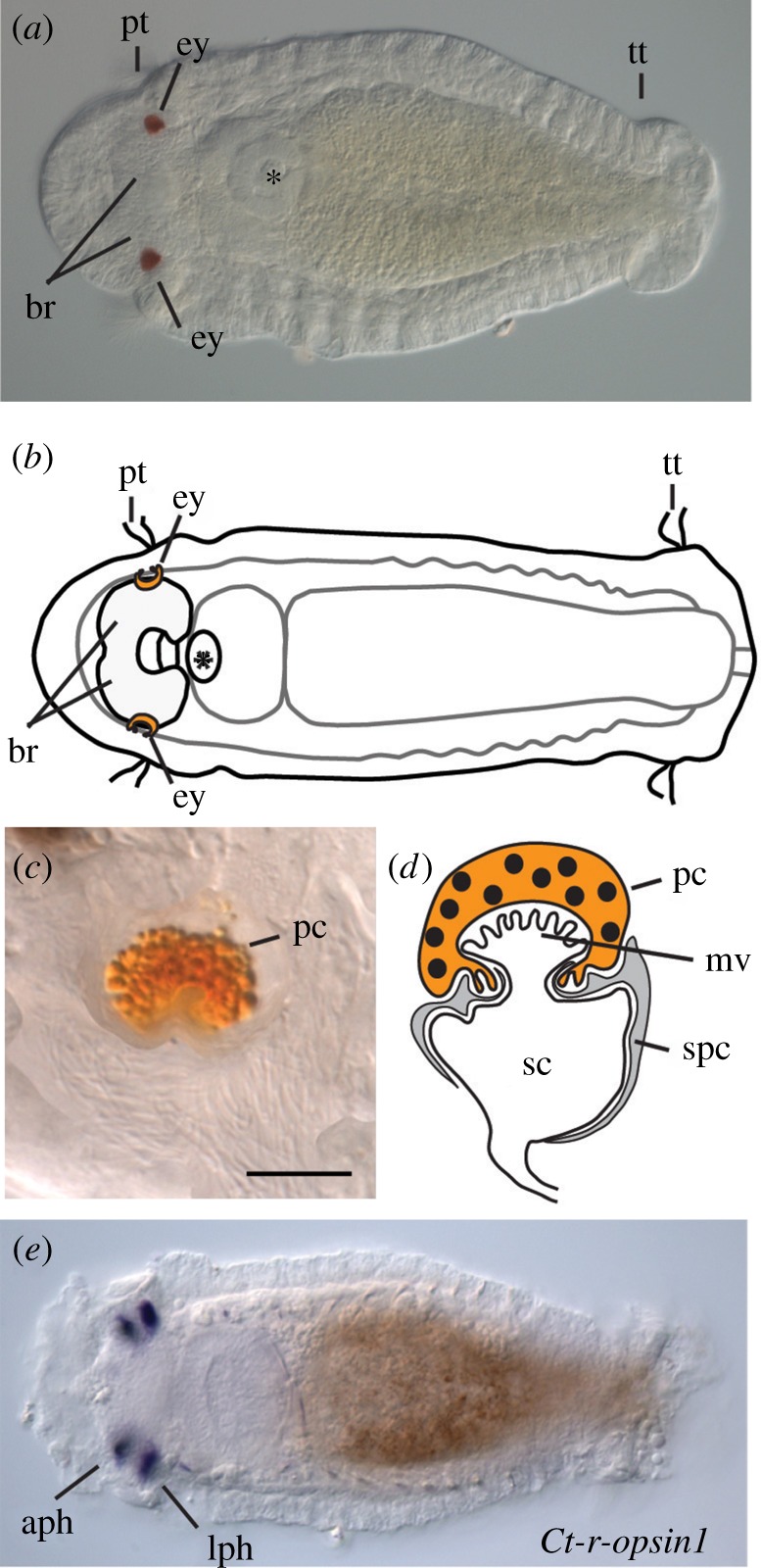
Capitella larval brain and eyespot structure and Ct-r-opsin1 expression. (a) Capitella teleta larva. (b) Schematic of (a). (c) Enlarged image of right larval eye showing pigment granules in the pigment cell. Scale bar, 5 µm. (d) Schematic of larval eye showing sensory, pigment and supporting cells. Adapted from [8]. (e) In situ hybridization showing mRNA expression of Ct-r-opsin1. (a, b and e) are ventral views of a stage 7 larva. Anterior is to the left. aph, adult photoreceptor; br, brain; ey, eye; lph, larval photoreceptor; mv, microvilli; pc, pigment cell; sc, sensory cell; spc, supporting cell; pt, prototroch; tt, telotoch. (Online version in colour.)
Light detection and processing in the photoreceptor cells are mediated by members of the opsin gene family [12], a large monophyletic subclass within the G-protein coupled receptor superfamily [13]. Opsin proteins contain a seven-pass transmembrane domain and a G-protein coupled receptor domain [14]. Different classes of opsin genes are generally associated with distinct photoreceptor cell types, which are distinguished by their apical cell membrane morphology [12]. That is, ciliary photoreceptors express ciliary opsin genes and rhabdomeric photoreceptors express rhabdomeric opsin genes. The cerebral eyes in larval and adult polychaetes typically have rhabdomeric photoreceptor cells, although there are exceptions [3,4,15]. In the last common ancestor of bilaterians, nine classes of opsin genes were thought to have been present [16]. The genome of C. teleta contains nine opsin genes that belong to only two opsin classes: three rhabdomeric opsin (r-opsin) and six neuropsin genes [16,17]. Notably, C. teleta lacks ciliary opsin genes; a similar situation is found in most other lophotrochozoans [16].
Although the evolution and expression of opsin genes has been characterized in many taxa, few studies have demonstrated a functional role for opsin genes in marine larvae. Here, we explore the function of opsin genes in mediating larval phototactic behaviour of C. teleta. The availability of a sequenced genome [18], a comprehensive embryonic fate map [19], and the availability of a breeding laboratory colony make C. teleta a valuable system for studies of development and evolution within the lophotrochozoan clade. We characterize expression of all of the rhabdomeric opsin genes and three neuropsin genes in larvae by in situ hybridization. Using CRISPR/CAS9 mutagenesis, we investigate the function of Ct-r-opsin1, the only opsin gene expressed in the larval photosensory cell. Through direct genotyping, in situ hybridization and behavioural analysis, we demonstrate that Ct-r-opsin1 is sufficient to mediate positive phototaxis. In addition, we establish CRISPR/CAS9 mutagenesis as an efficient method for studies of gene function in C. teleta.
2. Results
(a). Opsin expression in Capitella teleta
We characterized expression of all of the rhabdomeric opsin genes and three of the neuropsin genes present in the C. teleta genome. We analysed these expression patterns during larval development by in situ hybridization (figure 2). Our rationale for focusing on the rhabdomeric opsin genes is that the photoreceptor cells in C. teleta larval eyespots were previously shown to be the rhabdomeric type [8,10], and rhabdomeric opsin genes typically mediate photodetection and vision in protostomes [2,21,22]. One neuropsin gene, Ct-n-opsin1, was undetectable at the stages examined, even with varying conditions (data not shown).
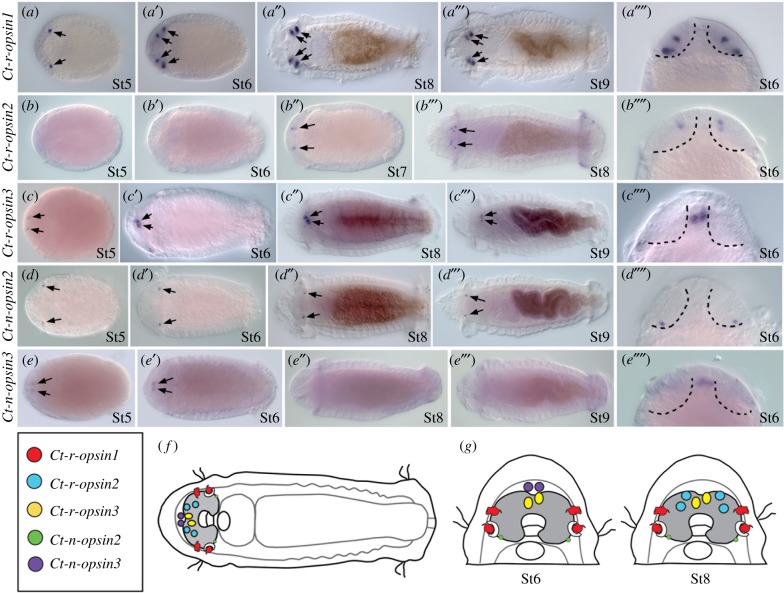
Figure 2.
Expression of opsin genes in the larval brain region and eyespot sensory cells. (a – e′′′′) Expression patterns of opsin genes by in situ hybridization during larval development. Arrows indicate expression. All panels are ventral views. Stages are indicated in bottom right corner and follow the staging system of [20]. (a–e′′′) Anterior is to the left. (a′′′′–e′′′′) Enlarged views of head. Brain lobes are indicated by dotted lines. Anterior is to the top. Brain is shaded in (f) and (g). (a–a′′′′) Ct-r-opsin1, (b–b′′′′) Ct-r-opsin2, (c–c′′′′) Ct-r-opsin3, (d–d′′′′) Ct-n-opsin2, (e–e′′′′) Ct-n-opsin3. (f) Schematic of a stage 7 larva, showing localization of opsin expression domains to the head. Each gene is represented by a distinct colour (see key). (g) Schematics showing magnified views of larval heads (stages 6 and 8) to highlight the spatial relationships among opsin gene expression patterns. (Online version in colour.)
Each opsin gene investigated shows a unique expression pattern, and transcripts of all five genes are restricted to 2–6 cells (figure 2). Ct-r-opsin1 is expressed in both the larval and adult photosensory cells (figures 1e and 2a–a′′′′). The adult photosensory cells are located anterior to the larval eyespots (figure 2a′–a′′′′) [10]. Ct-r-opsin2 is expressed in a small number of cells in the brain region of late larval stages, but is not detectable at early stages (figure 2b–b′′′′). Ct-r-opsin3 is expressed in a pair of medial cells throughout larval stages (figure 2c–c′′′′). Ct-n-opsin2 also shows a stable expression pattern across larval stages and is detected in a pair of lateral cells (figure 2d–d′′′′). Ct-n-opsin3 is only detectable in early larval stages in a pair of cells medial to the brain lobes (figure 2e–e′′′′). Of these, only Ct-r-opsin1 is expressed in the photosensory cell of the eyespot. Ct-r-opsin1 is present as the eyespots form, during the period of larval phototaxis, and in juvenile eyespots (not shown). In summary, transcripts of the five opsin genes are localized to the head, have unique patterns and are closely associated with the brain (figure 2f,g).
(b). CRISPR/CAS9-mediated genome editing of Ct-r-opsin1 is highly efficient
To test the function of Ct-r-opsin1 in C. teleta larvae, we generated a Ct-r-opsin1 mutant using CRISPR/CAS9 gene editing. Ct-r-opsin1 is encoded by three exons (figure 3a). Opsin genes typically have seven transmembrane domains, and they are spread across all three exons in the Ct-r-opsin1 gene. We designed three single guide RNAs (sgRNAs) directed against Ct-r-opsin1, two that target sites in the first exon, and a third that targets a site towards the 5′ end of exon 3 (figure 3a). Fertilized single cell zygotes were microinjected with CAS9 protein/sgRNA complexes containing all three sgRNAs in a single injection cocktail, and F0 stage 7 larvae resulting from these injections were analysed. We examined three different conditions marked by differing ratios of CAS9 : sgRNA (1 : 1, 1.2 : 1, 1.7 : 1) by polymerase chain reaction (PCR) analysis, in situ hybridization, and a phototactic assay (electronic supplementary material, table S1). Additionally, we sequenced DNA extracted from larvae that were injected with the 1.2 : 1 CAS9 : sgRNA ratio.
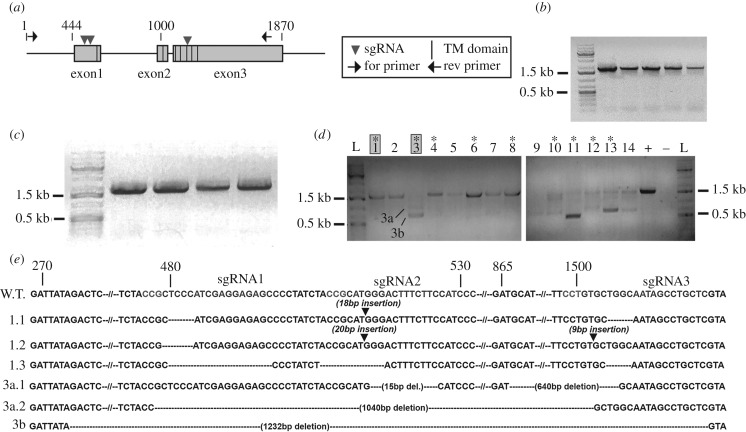
Figure 3.
CRISPR/CAS9-mediated knockout of Ct-r-opsin1. (a) Ct-r-opsin1 genomic locus showing sgRNA target sites (arrowheads), primer binding sites (black arrows), intron-exon structure (grey shaded boxes are exons, intervening black lines are introns), and the seven-transmembrane (TM) domains [11] (black vertical lines within exons). (b,c) Gel electrophoresis showing amplicons of the Ct-r-opsin1 locus from control animals injected with CAS9 protein only (b) or sgRNA only (c). Primer positions are indicated in (a). Each lane represents a PCR product of DNA extracted from an individual larva. (d) Amplicons of the Ct-r-opsin1 locus from 14 individual experimental larvae (1–14) using primers indicated in (a). Larvae resulted from embryos injected with sgRNA/CAS9 complexes. Asterisks indicate lanes from whom bands were cloned and sequenced. Grey shading of lanes 1 and 3 indicates larvae from which sequencing results are shown in figure 3e. For larva 1, the single wild-type-sized band was cloned and multiple clones were sequenced. For larva 3, bands 3a and 3b were cloned separately, and multiple clones from each sequenced. L, ladder. 0.5 kb and 1.5 kb bands of the ladder are marked.+ indicates positive control in which PCR was conducted on gDNA extracted from a wild-type larva, and − indicates the negative (no template) control. (e) Genome sequences showing indels. Wild-type sequence is in the top row (W.T.). Position of target sequences are indicated above sequence. Numbers indicate genomic position within the locus (figure 3a). --//-- represents a large section of sequence that is not shown. Sequences of three individual clones derived from larva 1 are shown as 1.1, 1.2 and 1.3, and sequences of three clones derived from larva 3 are shown as 3a.1, 3a.2 and 3b. Dashes indicate deletions; black arrowheads indicate insertions.
We analysed genome editing events in individual larvae by PCR screening of genomic DNA (figure 3d) and DNA sequencing (figure 3e). CAS9 only and sgRNA only controls displayed an expected amplicon size following PCR analysis (figure 3b and c, respectively). Of the 34 experimental larvae analysed by PCR analysis, 12 larvae had wild-type-sized amplicons (electronic supplementary material, table S1). Figure 3d shows banding patterns of 14 examples from the 34 experimental larvae analysed by PCR analysis. A subset of wild-type and non-wild-type-sized bands from nine experimental individuals were cloned and sequenced (figure 3d, asterisks). All nine larvae had at least one clone with a mutation in the Ct-r-opsin1 gene (total of 53 sequenced clones), and only 3 out of 53 clones displayed a wild-type sequence (electronic supplementary material, table S1). Although bands from four of the sequenced larvae had only wild-type amplicon sizes, only 3 out of 19 clones sequenced from these larvae contained wild-type sequences, indicating that clones appearing to be wild-type by PCR analysis actually contained small indels. Therefore, PCR analysis underestimates the efficiency of CRISPR/CAS9 mutagenesis, because PCR analysis was unable to distinguish small deletions/insertions from wild-type-sized amplicons.
We observed both large and small deletions, and multiple unique cutting events per individual. By analysing single larvae, we could detect the presence of multiple distinct indels (figure 3e). In one example (larva 1), clones isolated from a wild-type-sized amplicon show numerous small indels (figure 3e, clones 1.1, 1.2, 1.3). Sequence 1.1 includes two small deletions at the sites targeted by sgRNA1 (4 bp) and sgRNA3 (4 bp), and there is an 18 bp insertion at the site of sgRNA2 (figure 3e). Therefore, all three guide RNAs caused mutations, and these resulted in frameshifts in the reading frame. Another example comes from larva 3 whose r-opsin1 locus has large-scale deletions (figure 3e; larva 3, clones 3a.1, 3a.2 and 3b). Clone 3a.1 contains two deletions in the sequence targeted by sgRNA2 and sgRNA3, whereas clone 3a.2 contains a 1040 bp deletion spanning the regions targeted by sgRNA1 and sgRNA3. Clone 3b contains a large deletion originating approximately 200 bp 5′ of the sequence targeted by sgRNA1 and extends to the sequence targeted by sgRNA3 (figure 3e). Either a frameshift mutation or large deletion will result in a truncated protein that will probably not localize to the membrane, and therefore be non-functional. Our observations of distinct cutting events within an individual demonstrates the mosaic nature of the genomic mutations.
(c). Effect of CAS9 : sgRNA ratio on efficiency of gene editing
When we varied the molar ratio of CAS9 protein to sgRNA, there were differences in genome editing efficiency (electronic supplementary material, table S1). Because PCR analysis substantially underestimates genome editing events (see the previous section), we ascertained genome editing efficiency by determining the percentage of larvae with a wild-type expression pattern of the Ct-r-opsin1 transcript by in situ hybridization. The 1 : 1 ratio of CAS9 to sgRNA was the least effective. That is, most larvae resulting from zygote injections with a 1 : 1 ratio had wild-type Ct-r-opsin1 expression patterns (84%; electronic supplementary material, table S1). Likewise, a high percentage of larvae resulting from zygotes injected with the sgRNA only or CAS9 only controls displayed wild-type expression (90% and 78%, respectively; electronic supplementary material, table S1, figure 4). By contrast, few larvae resulting from injections with CAS9 : sgRNA ratios of either 1.2 : 1 or 1.7 : 1 displayed wild-type Ct-r-opsin1 expression patterns (6% and 0%, respectively; electronic supplementary material, table S1). In approximately 46% of experimental larvae, Ct-r-opsin1 expression was not detectable (figure 4d). Of the larvae resulting from zygotic injections with the 1.2 : 1 ratio, 8 out of 25 (32%) had no detectable transcript, and of the resulting larvae injected with 1.7 : 1 ratio, 26 out of 49 (53%) had no detectable transcript. Therefore, differences in the ratio between CAS9 and sgRNA in the injectant influenced mutation efficiency, and increasing relative levels of CAS9 produced more robust results.
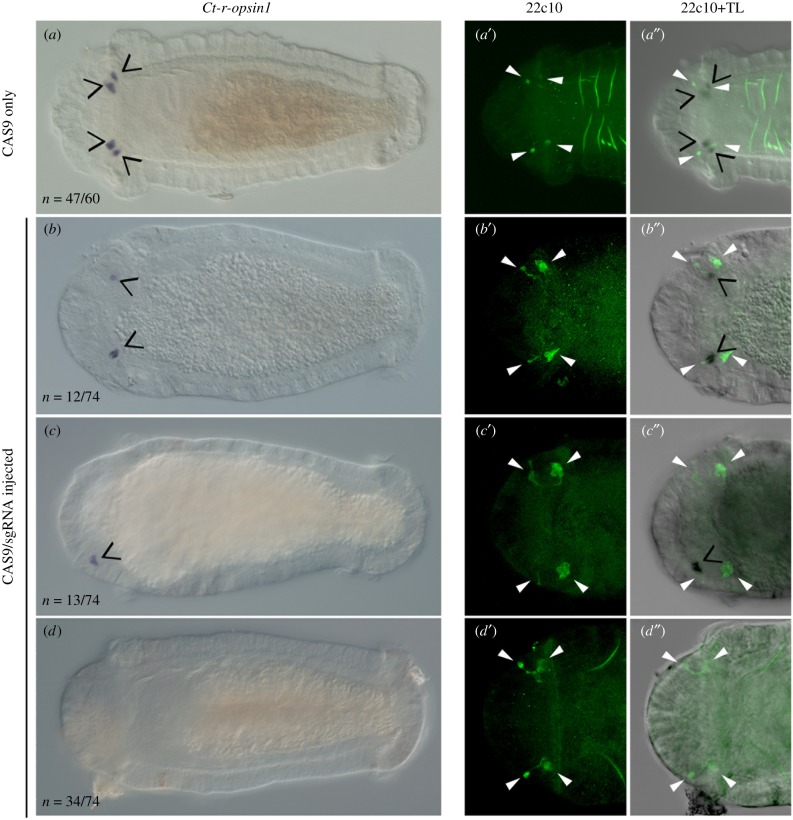
Figure 4.
Expression analysis of Ct-r-opsin1 in mutant larvae. Panels (a–d) show differential interference contrast microscopy images of Ct-r-opsin1 expression. The number of larvae displaying the expression pattern shown over the total number of cases examined is indicated in the bottom left corner. (a) CAS9-only control animals. (b–d) Experimental larvae. Panels (b–d) include larvae injected with the 1.7 : 1 and 1.2 : 1 CAS9 : sgRNA ratio. Panels a′, b′, c′ and d′ are confocal stacks showing 22C10 antibody labelling to visualize sensory cells, and panels a″, b″, c″, d″ show a merge of 22C10 confocal images and transmitted light images. Panels in each row are from a single individual. Open black arrowheads indicate Ct-r-opsin1 expression, and closed white arrowheads indicate sensory cells.
(d). Detection of Ct-r-opsin1 transcript
Multiple distinct expression patterns for Ct-r-opsin1 were recovered from larvae resulting from zygotic injection of the two most effective CAS9 : sgRNA molar ratios (1.2 : 1 and 1.7 : 1) (figure 4a–d). Six expression patterns were observed; the Ct-r-opsin1 transcript was detected in either zero photoreceptors, one photoreceptor, two photoreceptors, three photoreceptors or four photoreceptors. Additionally, in some embryos, although expression was detected in all four photoreceptors, at least one domain was weak relative to the others. This expression pattern was scored as abnormal. We interpret the observed range of expression patterns as a likely sign of mosaicism, or a lack of nonsense-mediated mRNA decay for some, but not all mutations [23].
(e). Normal eyespot formation in Ct-r-opsin1 mutants
We examined whether the eyespots form normally in larvae resulting from CAS9/sgRNA injections into zygotes. The monoclonal antibody 22C10 specifically labels photosensory cells in the juvenile and larval eyes in C. teleta, and co-localizes with Ct-r-opsin1 in the photosensory cells [10,24]. In wild-type larvae, Ct-r-opsin1 mRNA transcript co-localizes with the 22C10 labelling (figure 4a–a′′). In larvae that have disrupted Ct-r-opsin1 expression patterns, all four photosensory cells are present (figure 4b–d′′). Therefore, the photosensory cell of the eyespot develops in the correct location and has axonal processes even in the absence of, or reduction of Ct-r-opsin1 transcript. The pigment cell of the eyespot is also present in larvae resulting from CAS9/sgRNA injections (not shown).
(f). Ct-r-opsin1 knockdown inhibits phototactic behaviour
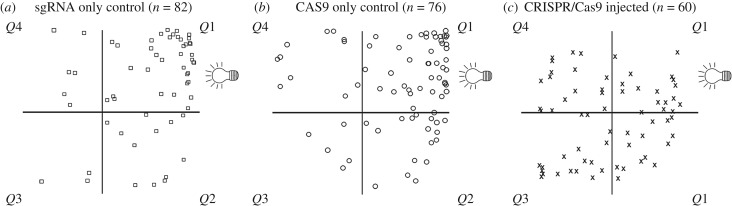
To determine whether Ct-r-opsin1 mediates the phototactic response in C. teleta larvae, previously established phototaxis assays [10] were performed with larvae resulting from zygotes injected with CAS9/sgRNA complexes. Phototaxis assays were performed with 10 larvae at a time, and with only 10 larvae in the cuvette, the larvae swim freely. Each set of 10 larvae was considered as an independent biological replicate. All phototaxis assays were performed with a minimum of five independent replicates per subset of embryos. After a 20 s exposure to a point source of light, the position of each larva was recorded. We observed that both sets of control larvae (CAS9 only and sgRNA only), displayed positive phototaxis as exhibited by displacement towards the light source (figure 5a,b). This behaviour is similar to previous reports of unmanipulated larvae [10]. By contrast, larvae resulting from CAS9/sgRNA injections did not display significant phototaxis (figure 5c). The distribution of larvae in the quadrant closest to the light source (Q1) relative to the other quadrants was higher in the CAS9 only controls compared with larvae resulting from CAS9/sgRNA injections (Fisher's exact test, p = 0.0002; figure 5). The larvae resulting from CAS9/sgRNA injections behaved similarly to larvae in which the photoreceptor and pigment cell were deleted [10]. This indicates that Ct-r-opsin1 expression in the photoreceptor is sufficient to mediate the robust positive phototactic response in C. teleta larvae.
Figure 5.
Loss of phototaxis following CRISPR/Cas9-mediated knockout of Ct-r-opsin1. Schematics represent the area of the glass cuvette used for the phototaxis assay, divided into four quadrants (Q1, Q2, Q3, Q4). Light bulb symbol indicates position of the light source (Q1). Shapes indicate positions of individual larvae 20 s after initial light exposure. Control larvae are indicated by squares (sgRNA only) or circles (CAS9 only), and crosses indicate CAS9/sgRNA injected larvae. The number of animals per quadrant after a 20 s light exposure is (a) n = 44 for Q1 and n = 38 for Q2–4, (b) n = 43 for Q1 and n = 33 for Q2–4, (c) n = 17 for Q1 and n = 43 for Q2–4.
3. Discussion
(a). Comparison of opsin expression patterns
Ct-r-opsin1 is the only r-opsin gene expressed in the photosensory cell of the simple eye in C. teleta larvae. Although not in the eyespots, the two other r-opsins, r-opsin2 and r-opsin3, are also expressed in the cephalic region. Platynereis dumerilii is another annelid whose larva exhibits positive phototaxis, and like in C. teleta, r-opsin is expressed in both larval and adult eyespots [25]. However, unlike the restricted cephalic expression in C. teleta, r-opsin1 and r-opsin3 in P. dumerilii are also expressed in the segmented trunk [26,27]. Furthermore, r-opsin1 and r-opsin3 are expressed in adjacent photoreceptors within the larval eyespot and are co-expressed in the same photoreceptor cells in adult eyes of P. dumerilii [27], a contrast with only a single r-opsin gene expressed in the photosensory cell of the eyespot in C. teleta. There is extraordinary diversity of eye structure, complexity and function in annelids [4]. Studies of opsin gene expression and function are one way to understand this diversity and the evolution of light detection in annelids.
Far less is known about the expression and function of neuropsin genes relative to r-opsins, particularly outside of the vertebrate lineage [16]. Two of the C. teleta neuropsin genes, Ct-n-opsin2 and Ct-n-opsin3, are expressed in a small subset of cells in the brain region. The first described neuropsin, Opn5, is expressed in the brain, spinal cord, eye and testis of mice [28]. Opn5 has a peak sensitivity to ultraviolet light in chicken [29], mice, and humans [30], and in birds, it is thought to function in seasonal reproduction [31]. Although the neuropsin genes are unlikely to be involved in phototaxis, it will be possible to leverage genome editing tools to uncover their function in C. teleta.
(b). Highly efficient CRISPR/CAS9-mediated genome editing in Capitella teleta
The results of this study dramatically improve preliminary attempts at genome editing [6], and demonstrate that CRISPR/CAS9 mediated genome editing is an effective method for generating targeted mutations in C. teleta. Sequence analysis and analysis by in situ hybridization indicate a mutation rate of 94% and 100%, respectively. By contrast, PCR analysis underestimates CRISPR/CAS9-mediated genome editing because it does not detect small indels. Therefore, it is important to carefully choose the detection method for CRISPR/CAS9-induced mutation. Owing to the efficient mutation rate, we can evaluate phenotypes in the F0 generation [32].
We think that our high rate of genome editing was achieved by using three sgRNAs targeted to Ct-r-opsin1 along with injection of CAS9 protein. Injection of three sgRNAs together is highly efficient in generating mutants in other animals [33]. We observed mutations associated with all three sgRNA target sites, and we recovered both large and small deletions. Additionally, microinjecting CAS9 protein has been shown to be substantially more efficient than injecting Cas9 mRNA, and may decrease mosaicism [34,35]. We observed an increased frequency of mutation associated with increasing CAS9 protein levels relative to sgRNA in the injectant. Increasing CAS9 protein concentration may lead to more efficient complex formation in vitro, and in turn, more efficient in vivo genomic editing. Although gene editing was efficient, we did detect mosaicism. This may be explained by DNA editing events that occurred after the zygote cleaved into multiple cells.
(c). Behavioural adaptations to the marine environment
Our results demonstrate that Ct-r-opsin1 is sufficient to mediate positive phototaxis. The Ct-r-opsin1 mutants behaved similarly to animals in which the photoreceptor and pigment cell are experimentally deleted [10]. It is important to note that in our Ct-r-opsin1 mutants, both the pigment cell and photoreceptor cell are present in the correct location, demonstrating that the eyespot forms normally.
One advantage of phototaxis is that it can enhance larval dispersal [39]. Capitella teleta larvae hatch from a brood tube in the sediment and are subsequently free swimming [36]. Positive phototaxis of larvae serves to bring individuals to the ocean surface [12], where they have the potential to be caught in currents that aid in dispersal [37]. Larvae of many polychaetes are positively phototactic for all, or some of their larval life [1]. More broadly, of the benthic marine invertebrates that produce pelagic larvae, the majority of these larvae have a period of positive photo response [1]. These observations emphasize the importance of light and light detection for dispersal of marine larvae to ultimately locate suitable habitat for their subsequent adult benthic life history phase.
Our results represent one of only a few published examples of CRISPR/CAS9-induced mutations causing behavioural changes in animals. In one example, mutation of the receptor for prostaglandin F2∝ prevented the initiation of sexual behaviour in the cichlid fish Astatotilapia burton [38]. In another example, Opsin9 knockout disrupted oocyte maturation-inducing hormone secretion in response to light in the jellyfish Clytia, and prevented maturation of gonads and their subsequent release [39]. CRISPR was also used to knockout orco in Harpengnathos saltator (Indian jumping ant), dramatically affecting social and individual behaviour linked to olfaction [40].
4. Conclusion
Many previous studies have inferred a function for r-opsin in phototaxis of marine protostomes, but few have provided direct demonstration through gene mutagenesis. Our data clearly demonstrate that Ct-r-opsin1 is sufficient to mediate positive phototaxis in C. teleta. Although disruption of Ct-r-opsin1 affects larval behaviour, a morphologically normal sensory neuron of the eyespot forms. This study adds to one of very few examples using CRISPR/CAS9 technology to investigate animal behaviour, and provides mechanistic information of phototaxis in a marine larva. Analysis by genomic sequencing and in situ hybridization show similar high efficiency of the CRISPR/CAS9 system in C. teleta, and analysis of amplicon size by PCR alone is clearly an underestimate of mutation events. To our knowledge, this is the first example of CRISPR/CAS9 mutagenesis in C. teleta, and is among only a few examples in a spiralian. Our use of CRISPR/CAS9 genome editing of the Ct-r-opsin1 gene generates an opportunity to link genotype to phenotype during post-metamorphic stages of the life cycle in future studies. Juvenile and adult worms of C. teleta burrow in the sediment, and we hypothesize that these stages may be negatively phototactic. Studies, such as this, expand the repertoire of functional genomic studies to a wider range of animals, and facilitate our ability to understand the evolution of animal diversity, such as in the case of the extraordinary diversity of eye structure, complexity and function in annelids.
5. Methods
(a). Preparation of single guide RNA and CAS9 protein
There were 19–20 bp target sequences of candidate sgRNAs designed using CRISPRscan (www.crisprscan.org) [41] to target the Ct-r-opsin1 coding sequence. Potential sgRNAs were manually subjected to a BLASTn search of the C. teleta genome (http://genome.jgi.doe.gov/Capca1/Capca1.home.html) to ensure there were no off-target hits. Three candidate sgRNAs targeting Ct-r-opsin1 were selected (sgRNA1: GGAUGGAAGAAAGUCCCAUG; sgRNA2: GGGCUCUCCUCGAUGGGAG; sgRNA3: GAGCAGGCUAUUGCCAGCAC) and custom synthesized by Synthego (www.synthego.com). Lyophilized sgRNAs were diluted in nuclease-free 1x Tris-EDTA (TE) buffer (pH 8.0) to a concentration of 50 µM as a stock solution. Working solutions were created by dilution with nuclease-free water to a concentration of 10 µM. Both stock and working solutions were stored at −20°C. Lyophilized CAS9 protein was purchased from PNAbio (CP01–50), diluted to 2 µg µl−1 with nuclease-free water, and stored as single-use 1 µl aliquots at −80°C. Immediately prior to microinjection, sgRNA and CAS9 protein were mixed, and placed at room temperature for 10 min to enable formation of ribonucleoprotein (RNP) complexes. CAS9/sgRNA RNPs were then mixed with nuclease-free water and a 1 : 10 dilution of 20 mg ml−1 dextran (Texas Red, Molecular Probes™), before loading into needles for microinjection (see ‘Animal husbandry and microinjection' below).
(b). Animal husbandry and microinjection
A laboratory culture of C. teleta was maintained following previously described methods [20]. To obtain zygotes for microinjection, females and males were first separated for 2–5 days, and then combined and checked for the presence of fertilized eggs approximately 10–12 h later. Eggs were dissected from the brood tube in 0.2 µm filtered seawater (FSW). The egg membrane was softened by a 20 s exposure to a freshly prepared 1 : 1 solution of 1 M sucrose : 0.25 M sodium citrate, followed by least three rinses in FSW. Uncleaved embryos were pressure injected using Quartz needles (QF 100–50–10) pulled on a micropipette puller (Sutter Instruments). The needles were filled with the CAS9/sgRNA mixture, a 1 : 10 dilution of 20 mg ml−1 fluorescent dextran (molecular probes) and nuclease-free H20. Injected and uninjected animals from the same brood were raised in FSW plus 60 µg ml−1 penicillin and 50 µg ml−1 streptomycin in separate 35 mm plastic dishes, and compared to determine the health of the brood.
(c). In vitro cleavage assay
To test the ability of CAS9/sgRNA RNPs to cleave Ct-r-opsin1 in vitro, the following components were mixed in a 0.5 ml PCR tube to a total volume of 20 µl: 250 ng of purified Ct-r-opsin1 PCR fragment, 250 ng (approximately 10 pmol) sgRNA, 500 ng CAS9 protein, 2 µl New England Biolabs buffer 3, 2 µl bovine serum albumin (10 mg ml−1). Samples were incubated at 37°C for 1 h. One microlitre RNase was added, and samples incubated for an additional 15 min at 37°C. Next, 1 µl of CAS9 stop solution (30% glycerol, 1% sodium dodecyl sulfate, 250 mM EDTA pH 8.0) was added to dissociate protein from DNA/RNA complex, and DNA fragments resulting from CRISPR/CAS9-induced cleavage were analysed by agarose gel electrophoresis.
(d). Cloning of Capitella teleta opsin genes
Previous analysis identified nine opsin genes in the C. teleta genome (opsin54244, opsin226303, opsin221903, opsin36183, opsin63256, opsin119596, opsin44169, opsin124377 and opsin197851) [17]. Of these, two had previously been cloned (opsin119596, renamed Ct-r-opsin1 (MG225382) and opsin197851, renamed Ct-n-opsin1 (MG710417)). Searches of C. teleta expressed sequence tags (EST) libraries (JGI, Department of Energy, Walnut Creek, CA, USA; [18]) with predicted coding sequences identified opsin44169 (EY644637, renamed Ct-n-opsin3). Because the predicted coding sequence for opsin63256 and opsin36183 were identical, a single pair of primers was designed. Fragments of coding sequence for opsin genes were amplified by PCR from mixed larval stage cDNA, cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA) and sequenced. Primer sequences used were as follows: opsin54244 (F: CCTAACTTCAATCAACACACAGG; R: TTGTCGGAATCGAGGTAAGC), opsin124377 (F: GACTTTAACTCCAGCCATACAGC; R: CAACCGGAGTCTTTTACAGC), opsin63256/36183 (F: TGCTGGTCACGTTACTTTCG; R: ACGATTGGATTCAGACATGC), and opsin44169 (F: GTTAGGGCTTGCAACATGC; R: GAGGAGGCCTTTAACACACC). Sequences of newly cloned, unique gene fragments (those without EST support) were submitted to the National Center for Biotechnology Information (NCBI) as original sequences with the following accession numbers: MG710415 (opsin54244, renamed Ct-r-opsin2), MG710416 (opsin124377, renamed Ct-r-opsin3) and MG710418 (opsin63256/36183, renamed with the single identifier, Ct-n-opsin2). Cloned fragments were used as templates to generate anti-sense RNA probes for in situ hybridization.
(e). Whole mount in situ hybridization
Following fixation in 3.7% paraformaldehyde in FSW overnight at 4°C, larvae were washed in phosphate-buffered saline (PBS), dehydrated through a methanol series to 100% methanol, and stored at −20°C for up to four weeks. Digoxigenin-labelled riboprobes were generated with either the SP6 or T7 MEGAscript kit (Ambion, Inc., Austin, TX, USA) and DIG-11-UTP (Sigma 11209256910). The following riboprobes and working concentrations were used: Ct-r-opsin1, 1047 bp at 0.2 ng µl−1 (SP6 RNA polymerase); Ct-n-opsin1, 865 bp at 1–3 ng µl−1 (T7 RNA polymerase); Ct-r-opsin3, 1176 bp at 1 ng µl−1 (T7); Ct-n-opsin3, 639 bp at 1 ng µl−1 (T7); Ct-n-opsin2, 722 bp at 1 ng µl−1 (SP6) and Ct-r-opsin2, 620 bp at 3 ng µl−1 (T7). Whole-mount in situ hybridization was performed following published protocols [42]. Following hybridization at 65°C for 48–72 h, probes were detected using nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indolyphosphate colour substrate. The reaction was allowed to develop for 30 min −12 h depending upon the probe. Ct-n-opsin1 was not detectable at any stages examined with 1, 2 or 3 ng µl−1 of probe, multiple independent repetitions, or following resynthesis of riboprobe.
(f). Immunohistochemistry
Following in situ hybridization, larvae were washed several times in PBS + 0.1% Triton (PBT), then treated with block solution (PBT + 10% normal goat serum, Sigma G9023) for 45–60 min at room temperature (r.t.). The monoclonal antibody (mAb) 22C10 was diluted 1 : 10 in block solution, and animals were incubated for 2–18 h at 4°C. Animals were washed twice in PBT, followed by four PBT washes of 20–30 min each. Goat anti-mouse-488 secondary antibody (Invitrogen A11001) was diluted 1 : 250 in block solution, and incubated with animals for 2–4 h at r.t., followed by two rinses in PBT, and four PBT washes of 20–30 min each prior to analysis. The mAb 22C10 was deposited to the Developmental Studies Hybridoma Bank by Benzer, S./Colley, N. (DSHB, Department of Biology, University of Iowa, Iowa City, IA, USA).
(g). Microscopy and imaging
Following in situ hybridization, larvae were imaged using an Axioskop 2 motplus compound microscope (Zeiss, Gottingen, Germany), coupled with a SPOT FLEX digital camera (Diagnostic Instruments, Inc., SterlingHeights, MI). Images were captured using SPOT imaging software and analysed using Adobe Photoshop CS6 (v. 13.0). Multiple differential interference contrast microscopy focal planes were merged for some images using Helicon Focus (Helicon Soft Ltd., Kharkov, Ukraine), as noted in figure legends. Following immunohistochemistry, larvae were imaged using a Zeiss LSM 710 confocal microscope (Zeiss, Gottingen, Germany). Z-stack projections were generated using Fiji [43]. All figures were created in Adobe Photoshop CS6 (v. 1.3.0), or Adobe Illustrator CS6 (v. 16.0).
(h). Analysis of CRISPR/CAS9-induced genomic editing
Genomic DNA extraction buffer (0.01 M Tris pH8.0, 0.05 M KCl, 0.3% Tween-20, 0.3% NP-40, 0.001 M EDTA, 0.5 mg ml−1 proteinase K) was freshly prepared and placed on ice. Single larvae were placed on the inside of a lid of a 0.5 ml PCR tube, as much seawater removed as possible, and then 20 µl of extraction buffer was pipetted onto the larva. Tubes were centrifuged briefly to bring larva/buffer to the tube bottom, vortexed, briefly spun again, and then placed at 55°C for 2–3 h. Tubes were vortexed every 30 min during incubation. Next, proteinase K was inactivated by incubation at 98°C for 5 min. PCR was conducted using 5 µl of gDNA as input template with ExTaq DNA polymerase (Takara, RR001A) and Ct-r-opsin1 specific primers (F: 5′ TAACTGGCATGGCATACACG; R: 5′ TTGGATTCCACATAGCAGAGG). Cycling conditions were as follows: initial denaturation, 95°C, 2 min; 35 cycles (95°C, 30 s; 56°C, 30 s; 72°C, 2 min); final extension, 72°C, 5 min. Resulting fragments were analysed by agarose gel electrophoresis.
(i). Phototactic assay and statistical analyses
Phototactic behaviour was assessed using a custom-built chamber based upon the design described in [2]. The chamber consists of a black plastic box, with slits at each end for removable shutters. A diffuser made of sandblasted glass was covered in black electrical tape to restrict light entry to a 5 mm width vertical sliver of glass. Up to 10 larvae were placed in a glass square cuvette (15 × 15 × 4 mm), which was covered on all sides with black electrical tape, aside from a 7.5 mm sliver on one side. A white LED light was positioned to one side of the chamber, and the glass cuvette containing larvae placed within the chamber, orienting the uncovered sliver towards the light source. Larvae were imaged from above by transmitted light that passed through an infrared filter (X-Nite780, LDP LLC). Larvae were dark adapted for at least 1 min, and then the shutter closest to the external light source was removed for 1 min. All behavioural assays were filmed using a xiQ camera, with a frame rate of 90 frames s−1 (MQ042CG-CM; Ximea). Positional information for each larva was recorded 20 s after initial light exposure. The cuvette was divided into four quadrants: quadrant 1 (Q1) nearest the light source, and the remaining quadrants termed quadrants 2, 3 and 4 (Q2–4). Larvae in Q1 at 20 s were scored as ‘near’ the light source (positive phototaxis), and larvae in Q2–4 were added together and scored as ‘far’ from the light source (no phototaxis). Statistical analysis (Fisher's one-tailed exact test) was performed using GraphPad Quick Calcs (http://www.graphpad.com/quickcalcs/).
Supplementary Material
Supplementary Material
Acknowledgements
We thank Alexis Lanza for editing the larval behaviour video. We are grateful for helpful comments on the manuscript from Dr Linlin Zhang and Alexis Lanza. Dr Heather Marlow cloned Ct-r-opsin1 and Ct-n-opsin1 and performed their initial expression analysis.
Data accessibility
Data concerning CRISPR/Cas9 mutagenesis conditions and scoring are provided in the electronic supplementary material, table S1. Sequences of cloned opsin genes have been deposited to NCBI and accession numbers are listed in the Methods. Detailed in situ hybridization protocol is available on the Seaver laboratory website at https://www.whitney.ufl.edu/people/current-research-faculty/elaine-c-seaver-phd/protocols/
Authors' contributions
Conceptualization, E.C.S. and D.M.d.J.; conducted the experiments, E.C.S., D.M.d.J. and S.N.; resources, E.C.S.; writing E.C.S., D.M.d.J. and S.N.; supervision, E.C.S. and D.M.d.J.; funding acquisition, E.C.S. All authors read and approved the final manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was funded by an NSF Research Experience for undergraduates grant no. (DBI-1560356; E.C.S.), and an NSF grant to E.C.S. (IOS1457102).
References
- 1.Thorson G. 1964. Light as an ecological factor in the dispersal and settlement of larvae of marine bottom invertebrates. Ophelia 1, 167–208. ( 10.1080/00785326.1964.10416277) [DOI] [Google Scholar]
- 2.Jekely G, Colombelli J, Hausen H, Guy K, Stelzer E, Nedelec F, Arendt D. 2008. Mechanism of phototaxis in marine zooplankton. Nature 456, 395–399. ( 10.1038/nature07590) [DOI] [PubMed] [Google Scholar]
- 3.Arendt D, Wittbrodt J. 2001. Reconstructing the eyes of urbilateria. Phil. Trans. R. Soc. Lond. B 356, 1545–1563. ( 10.1098/rstb.2001.0971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purschke G, Arendt D, Hausen H, Muller MC. 2006. Photoreceptor cells and eyes in annelida. Arthropod. Struct. Dev. 35, 211–230. ( 10.1016/j.asd.2006.07.005) [DOI] [PubMed] [Google Scholar]
- 5.Purschke G. 2005. Sensory structures in polychaetes. Hydrobiologia 535, 53–78. [Google Scholar]
- 6.Seaver EC. 2016. Annelid models I: Capitella teleta. Curr. Opin Genet. Dev. 39, 35–41. ( 10.1016/j.gde.2016.05.025) [DOI] [PubMed] [Google Scholar]
- 7.Gehring WJ, Ikeo K. 1999. Pax 6 mastering eye morphogenesis and eye evolution. Trends Genet. 15, 371–377. ( 10.1016/S0168-9525(99)01776-X) [DOI] [PubMed] [Google Scholar]
- 8.Rhode B. 1993. Larval and adult eyes in Capitella spec. I (Annelida, Polychaeta). J. Morphol. 217, 327–335. ( 10.1002/jmor.1052170307) [DOI] [PubMed] [Google Scholar]
- 9.Jekely G. 2009. Evolution of phototaxis. Phil. Trans. R. Soc. B 364, 2795–2808. ( 10.1098/rstb.2009.0072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaguchi E, Seaver EC. 2013. The importance of larval eyes in the polychaete Capitella teleta: effects of larval eye deletion on formation of the adult eye. Invert. Biol. 132, 352–367. ( 10.1111/ivb.12034) [DOI] [Google Scholar]
- 11.Butman CA, Grassle JP, Buskey EJ. 1988. Horizontal swimming and gravitational sinking of Capitella sp. I (Annelida: Polychaeta) larvae: implications for settlement. Ophelia 29, 43–57. ( 10.1080/00785326.1988.10430818) [DOI] [Google Scholar]
- 12.Terakita A, Nagata T. 2014. Functional properties of opsins and their contribution to light-sensing physiology. Zool. Sci. 31, 653–659. ( 10.2108/zs140094) [DOI] [PubMed] [Google Scholar]
- 13.Porter ML, Blasic JR, Bok MJ, Cameron EG, Pringle T, Cronin TW, Robinson PR. 2012. Shedding new light on opsin evolution. Proc. R. Soc. B 279, 3–14. ( 10.1098/rspb.2011.1819) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terakita A. 2005. The opsins. Genome Biol. 6, 213 ( 10.1186/gb-2005-6-3-213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verger-Bocquet M. 1992. Polychaeta: sensory structures. In Microscopic anatomy of invertebrates (ed. Harrison W.), pp. 181–196. New York, NY: Wiley-Liss. [Google Scholar]
- 16.Ramirez MD, Pairett AN, Pankey MS, Serb JM, Speiser DI, Swafford AJ, Oakley TH. 2016. The last common ancestor of most Bilaterian animals possessed at least nine opsins. Genome Biol. Evol. 8, 3640–3652. ( 10.1093/gbe/evw135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hering L, Mayer G. 2014. Analysis of the opsin repertoire in the tardigrade Hypsibius dujardini provides insights into the evolution of opsin genes in panarthropoda. Genome Biol. Evol. 6, 2380–2391. ( 10.1093/gbe/evu193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simakov O, et al. 2013. Insights into bilaterian evolution from three spiralian genomes. Nature 493, 526–531. ( 10.1038/nature11696) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer NP, Boyle MJ, Martindale MQ, Seaver EC. 2010. A comprehensive fate map by intracellular injection of identified blastomeres in the marine polychaete Capitella teleta. EvoDevo 1, 8 ( 10.1186/2041-9139-1-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seaver EC, Thamm K, Hill SD. 2005. Growth patterns during segmentation in the two polychaete annelids, Capitella sp. I and Hydroides elegans: comparisons at distinct life history stages. Evol. Dev. 7, 312–326. ( 10.1111/j.1525-142X.2005.05037.x) [DOI] [PubMed] [Google Scholar]
- 21.Fernald RD. 2006. Casting a genetic light on the evolution of eyes. Science 313, 1914–1918. ( 10.1126/science.1127889) [DOI] [PubMed] [Google Scholar]
- 22.Eakin RM. 1979. Evolutionary significance of photoreceptors: in retrospect. Am. Zool. 19, 647–653. ( 10.1093/icb/19.2.647) [DOI] [Google Scholar]
- 23.Popp MW, Maquat LE. 2016. Leveraging rules of nonsense-mediated mRNA decay for genome engineering and personalized medicine. Cell 165, 1319–1322. ( 10.1016/j.cell.2016.05.053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klann M, Seaver EC. 2018. Functional role of pax6 in eye and central nervous system development in the annelid Capitella teleta. bioRxiv. ( 10.1101/481135) [DOI] [PubMed] [Google Scholar]
- 25.Arendt D, Tessmar-Raible K, Snyman H, Dorresteijn AW, Wittbrodt J. 2004. Ciliary photoreceptors with a vertebrate-type opsin in an invertebrate brain. Science 306, 869–871. ( 10.1126/science.1099955) [DOI] [PubMed] [Google Scholar]
- 26.Backfisch B, Rajan VBV, Fischer RM, Lohs C, Arboleda E, Tessmar-Raible K, Raible F. 2013. Stable transgenesis in the marine annelid Platynereis dumerilii sheds new light on photoreceptor evolution. Proc. Natl Acad. Sci. USA 110, 193–198. ( 10.1073/pnas.1209657109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Randel N, Bezares-Calderon LA, Guhmann M, Shahidi R, Jekely G. 2013. Expression dynamics and protein localization of rhabdomeric opsins in Platynereis larvae. Integr. Comp. Biol. 53, 7–16. ( 10.1093/icb/ict046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarttelin EE, Bellingham J, Hankins MW, Foster RG, Lucas RJ. 2003. Neuropsin (Opn5): a novel opsin identified in mammalian neural tissue. FEBS Lett. 554, 410–416. ( 10.1016/S0014-5793(03)01212-2) [DOI] [PubMed] [Google Scholar]
- 29.Yamashita T, Ohuchi H, Tomonari S, Ikeda K, Sakai K, Shichida Y. 2010. Opn5 is a UV-sensitive bistable pigment that couples with Gi subtype of G protein. Proc. Natl Acad. Sci. USA 107, 22 084–22 089. ( 10.1073/pnas.1012498107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kojima D, Mori S, Torii M, Wada A, Morishita R, Fukada Y. 2011. UV-sensitive photoreceptor protein OPN5 in humans and mice. PLoS ONE 6, e26388 ( 10.1371/journal.pone.0026388) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakane Y, Ikegami K, Ono H, Yamamoto N, Yoshida S, Hirunagi K, Ebihara S, Kubo Y, Yoshimura T. 2010. A mammalian neural tissue opsin (Opsin 5) is a deep brain photoreceptor in birds. Proc. Natl Acad. Sci. USA 107, 15 264–15 268. ( 10.1073/pnas.1006393107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haeussler M, Concordet JP. 2016. Genome editing with CRISPR-Cas9: can it get any better? J. Genet. Genomics 43, 239–250. ( 10.1016/j.jgg.2016.04.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sunagawa GA, et al. 2016. Mammalian reverse genetics without crossing reveals Nr3a as a short-sleeper gene. Cell Rep. 14, 662–677. ( 10.1016/j.celrep.2015.12.052) [DOI] [PubMed] [Google Scholar]
- 34.Square T, Romasek M, Jandzik D, Cattell MV, Klymkowsky M, Medeiros DM. 2015. CRISPR/Cas9-mediated mutagenesis in the sea lamprey Petromyzon marinus: a powerful tool for understanding ancestral gene functions in vertebrates. Development 142, 4180–4187. ( 10.1242/dev.125609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Momose T, Concordet JP. 2016. Diving into marine genomics with CRISPR/Cas9 systems. Mar. Genomics 30, 55–65. ( 10.1016/j.margen.2016.10.003) [DOI] [PubMed] [Google Scholar]
- 36.Reish D. 1974. The establishment of laboratory colonies of polychaetous annelids. Thalassia Jugoslavica 10, 181–195. [Google Scholar]
- 37.Randel N, Jekely G. 2016. Phototaxis and the origin of visual eyes. Phil. Trans. R. Soc. B 371, 20150042 ( 10.1098/rstb.2015.0042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Juntti SA, Hilliard AT, Kent KR, Kumar A, Nguyen A, Jimenez MA, Loveland JL, Mourrain P, Fernald RD. 2016. A neural basis for control of cichlid female reproductive behavior by prostaglandin F2a. Curr. Biol. 26, 943–949. ( 10.1016/j.cub.2016.01.067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Artigas GQ, Lapébie P, Leclère L, Takeda N, Deguchi R, Jékely G, Momose T, Houliston E. 2018. A gonad-expressed opsin mediates light-induced spawning in the jellyfish Clytia. Elife 7, e29555 ( 10.7554/eLife.29555) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan H, et al. 2017. An engineered orco mutation produces aberrant social behavior and defective neural development in ants. Cell 170, 736– 747.e739 ( 10.1016/j.cell.2017.06.051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ. 2015. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 12, 982–988. ( 10.1038/nmeth.3543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seaver EC, Kaneshige LM. 2006. Expression of 'segmentation' genes during larval and juvenile development in the polychaetes Capitella sp. I and H. elegans. Dev. Biol. 289, 179–194. ( 10.1016/j.ydbio.2005.10.025) [DOI] [PubMed] [Google Scholar]
- 43.Schindelin J, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. ( 10.1038/nmeth.2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data concerning CRISPR/Cas9 mutagenesis conditions and scoring are provided in the electronic supplementary material, table S1. Sequences of cloned opsin genes have been deposited to NCBI and accession numbers are listed in the Methods. Detailed in situ hybridization protocol is available on the Seaver laboratory website at https://www.whitney.ufl.edu/people/current-research-faculty/elaine-c-seaver-phd/protocols/