Abstract
The olfactory bulb (OB) ratio is the size of the OB relative to the cerebral hemisphere, and is used to estimate the proportion of the forebrain devoted to smell. In birds, OB ratio correlates with the number of olfactory receptor (OR) genes and therefore has been used as a proxy for olfactory acuity. By coupling OB ratios with known OR gene repertoires in birds, we infer minimum repertoire sizes for extinct taxa, including non-avian dinosaurs, using phylogenetic modelling, ancestral state reconstruction and comparative genomics. We highlight a shift in the scaling of OB ratio to body size along the lineage leading to modern birds, demonstrating variable OR repertoires present in different dinosaur and crown-bird lineages, with varying factors potentially influencing sensory evolution in theropods. We investigate the ancestral sensory space available to extinct taxa, highlighting potential adaptations to ecological niches. Through combining morphological and genomic data, we show that, while genetic information for extinct taxa is forever lost, it is potentially feasible to investigate evolutionary trajectories in extinct genomes.
Keywords: olfactory bulb, dinosaur, olfaction, avian evolution, theropod evolution
1. Introduction
With more than 10 000 species, birds are the most diverse extant tetrapod clade [1], displaying highly specialized ecological and sensory niche adaptations. Originating within theropod dinosaurs, birds have undergone numerous morphological [2] and genomic [3] transformations. Adaptation to novel ecological niches often requires changes in sensory perception [4,5]. Of these, olfaction is a crucial sensory system, through which many vertebrates interact with their environment. Birds have been shown to use smell to forage, navigate, avoid predators and recognize conspecific individuals [6–9].
Olfaction is governed by a family of G-protein coupled receptors, known as olfactory receptors (ORs), found in the olfactory epithelium of the nasal passage. Each nasal cilium in the olfactory epithelium expresses a single OR, and extends a neuron connecting to the olfactory bulb (OB) in the forebrain. When an odorant molecule binds to the expressed OR, the OB is signalled via this neuron, which constitutes the sensation of smell. At the genomic level, OR proteins are encoded by single-exon OR genes, which have evolved through complex interactions of gene duplication and selection [10]. OR genes fall into two major classes: Class I, which principally binds aquatic-borne odorant molecules and Class II, binding air-borne odorants. Within these classes, OR proteins are further divided into monophyletic gene subfamilies based on shared sequence identity [5]. The number and proportion of functional and non-functional OR genes varies greatly across vertebrates, with genes in several related gene families gaining or losing function across phylogeny [10–12].
A gross correlation between reliance on a sensory system and the proportion of total brain size devoted to that sense has long been observed [13]: e.g. the large optic lobes of actinopterygian fishes [14] or the large OBs of mammals [15]. Because the OB receives neuronal extensions from the nasal cilia, bulb size must correlate, to some extent, with the number of cilia in the nasal epithelium, and general olfactory acuity should be represented by relative OB size. Because each cilium expresses only one OR protein, some of the variation in OB size may reflect the number of functionally expressed OR genes or previously functioning, but currently pseudogenized, genes. The OB ratio is defined as the greatest diameter of the OB divided by the greatest diameter of the cerebral hemisphere [16,17], and has been used as a proxy for the proportion of the forebrain devoted to olfaction. Extant birds exhibit a large disparity in OB ratios, and a correlation between OB ratio and OR repertoire size has been observed [8,17,18]. It is possible that this correlation also existed in extinct theropod dinosaurs.
Increasing availability of sequenced vertebrate genomes permits inference of genomic information about ancestral taxa, such as gene composition and chromosome number [19–24]. Here, we combine morphological (OB ratios for extant and fossil taxa) and genomic data (genomes for extant sauropsid taxa) to estimate the number of OR genes in extinct theropod dinosaurs using a combination of ancestral state reconstruction, conserved sequence identity and phylogenetically informed linear modelling. We determine that the minimum ancestral OR repertoire size for Dinosauria would have been between 360 and 500 genes, and that tyrannosaurus probably possessed highly expanded OR repertoires. Using shared sequence identity across extant taxa, we explore the potential odorant space that extinct taxa may have experienced. This study represents an initial step at combining genomic information with morphometrics to infer aspects of the ecology of long extinct taxa.
2. Material and methods
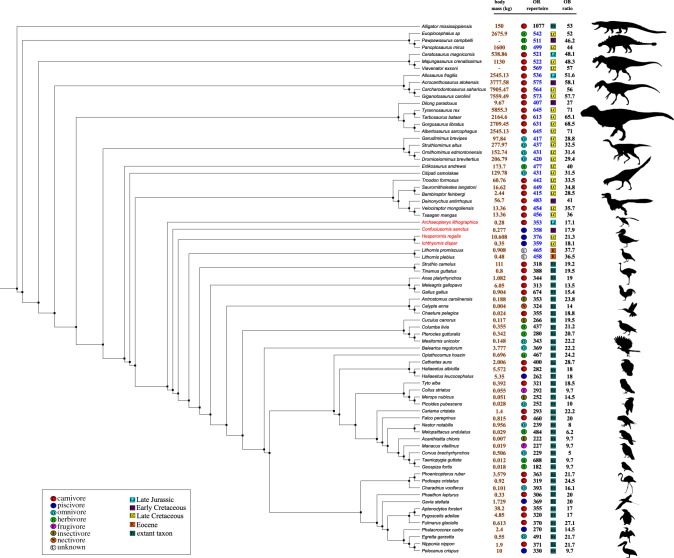
The total OR repertoire size (counts of functional and non-functional genes) for extant birds were taken from Khan et al. [18]. OR gene sequences for birds and non-avian sauropsid taxa were downloaded from RefSeq and augmented with genes mined from raw genome files (electronic supplementary material). We obtained OB ratios for 42 extant and two extinct avian species, four extinct stem birds, Alligator mississippiensis, and 25 non-avian theropod and three ornithiscian dinosaurs (electronic supplementary material, table S1) [17,18,25–28]. Where multiple OB ratios were available, values were averaged. All OB ratios are expressed as percentages. Body mass and diet data were collated from Lislevand et al. [29], the Encyclopedia of Life [30] and Animal Diversity Web ([31], accessed 2018; electronic supplementary material, table S1). We investigated the correlation of OB ratio with total OR repertoire and individual gene family sizes using phylogenetically corrected correlation. Correlations of OB ratios with trait data were done using phylogenetic generalized least squares (PGLS), using a composite phylogeny for all 76 taxa (electronic supplementary material; figure 1).
Figure 1.
Species phylogeny. Olfactory receptor bulb ratios were analysed across a total of 76 taxa. OR gene repertoires (both functional and non-functional) from extant birds were used to infer repertoire sizes for extinct species (blue). Additional life-history data were analysed for evolutionary correlations. Species within the clade Avialae are labelled in red. (Online version in colour.)
We estimated ancestral OB ratios at internal nodes using maximum likelihood (ML), phylogenetic independent contrasts (PIC) and generalized least squares (GLS), with an all-taxon phylogeny and a tree limited to extant birds (with and without outlier taxa, see the electronic supplementary material). Additionally, PGLS was used to model OR gene repertoire size as a function of OB ratio and predict OR gene numbers across all extinct taxa. See the electronic supplementary material for further details.
3. Results
(a). Olfactory bulb and olfactory receptor repertoires
The distribution of OB ratios observed in extant birds is significantly smaller than that of non-avian dinosaurs (t-test, p ≤ 0.001). OB ratios in extant birds range from 5.0 (American crow, Corvus brachyrhynchos) to 28.7 (turkey vulture, Cathartes aura). OR gene repertoires range from 182 (medium ground finch, Geospiza fortis) to 688 (zebra finch, Taeniopygia guttata). The smallest OB ratio among non-avian dinosaurs is observed in Archaeopteryx lithographica (17.1), whereas Albertosaurus sarcophagus and Tyrannosaurus rex possess OB ratios of 71.0 (electronic supplementary material, table S1).
There is a significant, positive correlation between OB ratio and OR repertoire size in extant birds (phylogenetically corrected: p = 0.021, r = 0.428; uncorrected: p ≤ 0.001, r = 0.651; electronic supplementary material table S2). We also observed significant correlations between OB ratio and several OR gene families (electronic supplementary material, table S2). Modelling OB ratios as a function of diet, while accounting for phylogeny, recovers piscivory as significantly correlated with OB ratio (p = 0.005), with the mean and range of OB ratios being smaller in piscivores (electronic supplementary material, figure S1).
The correlation between body mass and bulb ratio was significant when accounting for phylogeny across (i) all taxa (p = 0.031, r = 0.529; electronic supplementary material, figure S2), (ii) non-avian dinosaurs (p < 0.001, r = 0.836), and (iii) non-avian dinosaurs + stem birds (p < 0.001, r = 0.77). However, the correlations are not significant when considering all of Avialae (p = 0.719, r = 0.160) or restricting the analysis solely to crown Aves (p = 0.564, r = 0.359). Finally, there is no significant correlation between body mass and repertoire size among extant birds (all taxa: p = 0.334, r = −0.152, electronic supplementary material, figure S3; removing the three anomalous outlier taxa: p = 0.751, r = −0.052).
(b). Ancestral olfactory bulb and repertoire ratios
Estimates for the ancestral sauropsid OB ratio based on extant data (ML: 32.09, GLS: 25.49, PIC: 25.49) and OR repertoire size in the ancestral genome (ML: 488, GLS: 497, PIC: 497) were remarkably consistent across methods, both with and without outlier taxa (electronic supplementary material, figures S4, S5 and tables S3, S4). No significant differences were found among the distributions of reconstructed OB ratios among the different methods. Upon addition of fossil data, OB ratio estimates were 31.82, 43.21 and 43.21 for ML, GLS and PIC, respectively. Species estimates are given in the electronic supplementary material, figures S6–S8 and table S5.
(c). Estimates of dinosaur olfactory receptor repertoire size
PGLS modelling of OR repertoire size using OB ratio data from all extant birds resulted in
Using this, OR repertoire sizes for extinct taxa were estimated from OB ratios (electronic supplementary material, table S1; figure 1). Fossil OR estimates ranged from approximately 344 (Hesperornis regalis) to approximately 645 (Tyrannosaurus rex, Albertosaurus sarcophagus). Inclusion of Alligator mississippiensis in the modelling produces similar reconstructions of OR repertoire size (electronic supplementary material, table S6).
(d). Inferring minimum repertoire size using genomic data
A total of 2975 sauropsid and 5899 avian ORs gene sequences were collated, with 1321 representative sauropsid ORs chosen after clustering (electronic supplementary material, tables S7 and S8). Using sequence identity between sauropsids and birds, 363 orthologous OR genes (87 mined and 276 collated from other studies) were present in both sauropsid and avian taxa (65% shared identity). This gives a minimum estimate of the number of OR genes present in the most recent common ancestor of all Sauropsida, as this taxon also possessed an unknown number of OR pseudogenes, free from the constraints of selection. Class I ORs show a propensity for hydrophilic ligands [32], and are associated with aquatic odorants, whereas Class II ORs have a tendency to bind hydrophobic odorants, and are considered terrestrial ‘air-borne’ ORs [12]. The number of shared OR genes within the aquatic odorant Class I (α) subfamily was estimated at 47 ORs. Only a single receptor was found shared across sauropsids and birds in the Class I (β) subfamily OR55. The terrestrial odorant Class II (γ) had 315 shared OR genes (table 1). Based on their presence in modern birds and alligators, it is likely that at least one copy of each of these genes existed in all dinosaur taxa.
Table 1.
OR genes showing at least 65% shared identity between sauropsids and birds were considered present in extinct dinosaur taxa. (The number of inferred OR genes per family are displayed.)
| class | family | number of estimated OR genes |
|---|---|---|
| Class I (α,β) | OR 51 | 23 |
| OR 52 | 24 | |
| OR 55 | 1 | |
| class total | 48 | |
| Class II (γ) | OR 1/3/7 | 2 |
| OR 2/13 | 30 | |
| OR 4 | 38 | |
| OR 5/8/9 | 72 | |
| OR 6 | 58 | |
| OR 10 | 82 | |
| OR 11 | 3 | |
| OR 12 | 18 | |
| OR 14 | 12 | |
| class total | 315 |
4. Discussion
(a). Olfactory bulb and diet
Across all niches, only piscivory showed a significant association with OB ratio, after correcting for phylogeny. Previous studies have found ‘semi-aquatic’ habitat [9] or piscivorous diet [18] to be associated with larger OB ratio and OR repertoire size in birds. Given the similarity of OB size in piscivorous birds across a vast time-span, it is possible that the odorant space associated with aquatic habitats may constrain the OR repertoire evolution and, therefore, relative OB size, as has been observed in semi-aquatic mammals [5]. The average number of OR genes (functional and non-functional) in the ‘water-borne’ odorant gene families across all four extant piscvores was higher compared to all other diets (OR 51: piscivore: 21, other diets: 15; OR52: piscivores: 18, other diets: 15), suggesting an expansion of one receptor class (Class I) over the other (Class II) for this dietary niche. A similar pattern may have been present in the repertoires of Hesperornis, Lithornis and Ichthyornis.
(b). Body mass and olfactory bulb scaling
As Steiger et al. [8] observed, OB ratio in extant birds correlates significantly with the number of OR genes. OB ratio is also positively correlated with body mass across all taxa. However, this correlation with body mass is a function of the non-avian dinosaurs: no such relationship is observed within crown clade Aves (electronic supplementary material, figure S2). Stem birds represent an ambiguous case, given their position and low sample size, fitting equally with Aves or non-avian dinosaurs. Lower OB ratios are observed across Avialae [25,33], and there is no relationship observed between body mass and OR repertoire size among birds, with modern birds having a roughly constant OR repertoire relative to body size. Modern birds are generally smaller than their non-avian dinosaur relatives. Interestingly, this pattern implies that, despite a severely reduced OR repertoire relative to other sauropsids and mammals, modern birds have larger OBs (and by implication, larger OR repertoires) than non-avian dinosaurs of comparable size would have had (electronic supplementary material, figure S2).
The larger estimates of repertoire sizes for dinosaurian superfamilies are coincident with trends of increasing body size in these lineages [34]. Yet this is not simply an association between larger size and more OR genes. The positive correlation indicates that, as these dinosaurian lineages became larger, the relative size of the OB increased, implying increased investment in olfaction as a sensory modality. There is a subsequent shift away from olfaction along the lineage leading to modern birds, coincident with the fundamental change in the OB ratio scaling to body size, probably representing a shift to vision as the dominant sensory modality in the avian lineage [35].
(c). Inferred olfactory receptor repertoires of extinct taxa
Using a variety of methods, we estimated OR repertoire size for all extinct taxa. These suggest a minimum estimate of approximately 500 OR genes in the ancestral dinosaur genome. The reconstructions are dependent on high-quality gene sequences. Given the large number of pseudogenes in the OR repertoire, only highly conserved OR genes will be found. Any OR genes that were subsequently lost and obscured to the point of not being recognizable cannot be recovered from genomic data. Additionally, unique ORs that emerged in specific dinosaurian lineages will not be identified. Therefore, these estimates must represent minima for the complete ancestral OR repertoire. The PGLS models assume that dinosaurs reflect avian OB ratio/OR repertoire scaling, rather than that of the crocodilian or testudine lineages, and therefore may not completely reflect OR repertoire size among theropods. However, the other estimates do not rely on this assumption about OB ratio/OR repertoire correlations, and the minimum estimates we reconstruct across methods are consistent with the 400–600 OR gene range found in many extant vertebrates.
The largest reconstructed repertoires were for Tyrannosauroidea, specifically the large-bodied taxa, such as Tyrannosaurus rex (electronic supplementary material, table S1). The substantially smaller Dilong paradoxus has OBs that are both absolutely and relatively much smaller than Tyrannosaurus rex, consistent with the body size scaling relationship. Among tyrannosaurs, the large taxa had larger OR repertoires than any other non-avian dinosaurs, whereas Dilong paradoxus falls within the inner 95th quantiles of the modern bird distribution, even if the outlier taxa (zebra finch, chicken and budgerigar) are excluded. This overall increase in olfactory capability within Tyrannosauroidea may reflect ecological adaptation, allowing the tracking of prey over large distances, as in modern wolves [36], or to effectively scavenge carrion, as in the modern turkey vulture (electronic supplementary material, table S1).
Some of the smallest theropod repertoire sizes were inferred for the ornithomimosaurs (417–437 OR genes), with a similar repertoire size in the oviraptorid Citipati osmolskae. Omnivorous diets have been inferred for these taxa and the mean reconstructed OR repertoire is significantly smaller than the distribution of reconstructions in carnivorous non-avian dinosaurs (t = 5.403, p < 0.001). Interestingly, the inferred repertoire of the therizinosaur Erlikosaurus andrewsi was larger than most dromaeosaurids and Troodon formosus, despite a transition to herbivory. The expanded gene repertoire potentially signals a transition to complex sociality and/or low visual acuity, as has been inferred for this taxon [26]. OR repertoires for non-avian dinosaurs with inferred herbivorous diets are not significantly smaller than those of carnivorous taxa (t = 1.055, p = 0.306). It should be noted that olfaction also plays a crucial role in ecological adaptations other than diet, such as conspecific communication, which may explain similar repertoire sizes across different dietary niches. This has been observed in eutherian mammals, where OR repertoire size is also a function of pheromone detection and sociality [10]: e.g. the herbivorous and highly social African elephant (Loxodonta africana) possesses the largest known vertebrate OR repertoire (greater than 4000 OR genes) and a large OB [37].
(d). Minimum repertoire estimates using orthology
When comparing putative orthologues shared across extant birds and sauropsids, a minimum of 363 OR genes were estimated for the ancestral sauropsid genome, consistent with previous estimates [38]. As with terrestrial mammals, Class II air-borne odorant binding genes were more numerous than the aquatic odorant binding Class I genes [5,10]. The low number of genes in OR families 1/3/7 and 14, inferred for the ancestral species compared to extant birds suggests expansion of these families in the lineage leading to birds [18]. However, the expansion of family 14 may be the result of convergent expansions of this group in a limited number of extant taxa, as this pattern appears to be driven by the zebra finch, chicken and budgerigar [18]. While it is not yet known what odorants family 14 binds, there is also evidence for extensive gene expansion in this family among various ‘reptile’ lineages and monotreme mammals [5,39]. Families 2/13, 4, 6, 10 and 12 have probably undergone significant contraction in birds. A number of the receptors in these families and their odorant ligands have had ‘odorant descriptors’ characterized in tests of human subjects. Associated odours range from fruity to rancid, and include a compound found in blood (heptanal, OR6A2 [38]). This documents a decrease in the reliance on sensing these odorants among modern birds, but defines a plesiomorphic amniote odorant space. Putative detectable ligands and their odorant descriptors are given in the electronic supplementary material, table S9.
This study complements a growing literature on combining comparative genomics with morphological data to infer genome composition in ancient organisms (e.g. OR repertoires in Neanderthals and Denisovans [40] and Smilodon [41]). In addition to demonstrating a shift in the relationship between body mass and OB ratio along the lineage leading to modern birds, the results presented here demonstrate that variable OR gene repertoires were present in different dinosaurs, potentially highlighting adaptations to different ecological strategies over time. Genetic information from long extinct taxa will forever be lost to us. However by combining comparative genomics and morphology, we show that it is possible to investigate evolutionary trajectories in ancient genomes.
Supplementary Material
Acknowledgements
We thank E.C. Teeling for helpful comments and advice during the course of this study. Unmodified Phylopic.org images for alligator (Scott Hartman), Ceratosaurus (Scott Hartman), Allosaurus (Scott Hartman), Ichthyornis (Matt Matyniuk) and penguin (Noah Schlottman, photo by Reinhard Jahn) made available under a CC BY-NC-SA 3.0 license (https://creativecommons.org/licenses/by-nc-sa/3.0/). Images of Euoplocephalus (Andrew Farke), Oviraptor (Jaime Headdon), Deinonychus (Manabu Sakamoto), downy woodpecker (Gareth Monger), hoatzin (photography by Warren H, vectorization by T. Michael Keesey), falcon (Rebecca Groom) and white-tailed tropicbird (Paul Baker (photo), John E. McCormack, Michael G. Harvey, Brant C. Faircloth, Nicholas G. Crawford, Travis C. Glenn, Robb T. Brumfield and T. Michael Keesey) are made available under a CC BY 3.0 license (http://creativecommons.org/licenses/by/3.0/). Images for Tyrannosaurus rex (Emily Willoughby), ostrich (Matt Martyniuk, vectorized by T. Michael Keesey), owl (John E. McCormack, Michael G. Harvey, Brant C. Faircloth, Nicholas G. Crawford, Travis C. Glenn, Robb T. Brumfield & T. Michael Keesey), zebra finch (photography by Jim Bendon, vectorization by T. Michael Keesey), and great cormorant (L. Shyamal) are made available under a CC BY-SA 3.0 license (https://creativecommons.org/licenses/by-sa/3.0/).
Data accessibility
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.63bd5r6 [42]. All supplemental files are available as part of the electronic supplementary material.
Authors' contributions
This project was conceived, carried out and written by G.M.H. and J.A.F.
Competing interests
The authors declare no competing interests.
Funding
G.M.H. is funded by a Science Foundation Ireland Fellowship 17/IFA/5303.
References
- 1.Gill F, Donsker D. 2016. IOC World Bird List (v. 6.2). ( 10.14344/IOC.ML.6.2) See https://www.worldbirdnames.org (Accessed 2018). [DOI]
- 2.Brusatte SL, Lloyd GT, Wang SC, Norell MA. 2014. Gradual assembly of avian body plan culminated in rapid rates of evolution across the dinosaur-bird transition. Curr. Biol. 24, 1–7. ( 10.1016/j.cub.2014.08.034) [DOI] [PubMed] [Google Scholar]
- 3.Zhang G, et al. 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346, 1311–1320. ( 10.1126/science.1251385) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson AT, Soberon J, Sanchez-Cordero V. 1999. Conservation of ecological niches in evolutionary time . Science 285, 1265–1267. ( 10.1126/science.285.5431.1265) [DOI] [PubMed] [Google Scholar]
- 5.Hayden S, Bekaert M, Crider TA, Mariani S, Murphy WJ, Teeling EC. 2010. Ecological adaptation determined functional mammalian olfactory subgenomes. Genome Res. 20, 1–9. ( 10.1101/gr.099416.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nevitt GA. 2008. Sensory ecology on the high seas: the odor world of the procellariiform seabirds. J. Exp. Biol. 211, 1706–1713. ( 10.1242/jeb.015412) [DOI] [PubMed] [Google Scholar]
- 7.Nevitt GA, Losekoot M, Weimerskirch H. 2008. Evidence for olfactory search in wandering albatross, Diomedea exulans. Proc. Natl Acad. Sci. USA 105, 4576–4581. ( 10.1073/pnas.0709047105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steiger SS, Fidler AE, Valcu M, Kempenaers B. 2008. Avian olfactory receptor gene repertoires: evidence for a well-developed sense of smell in birds? Proc. R. Soc. B 275, 2309–2317. ( 10.1098/rspb.2008.0607) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corfield JR, Price K, Iwaniuk AN, Gutiérrez-Ibáñez C, Birkhead T, Wylie DR. 2015. Diversity in olfactory bulb size in birds reflects allometry, ecology, and phylogeny. Front. Neuroanat. 9, 102 ( 10.3389/fnana.2015.00102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes GM, Bostom ESM, Finarelli JA, Murphy WJ, Higgins DG, Teeling EC. 2018. The birth and death of olfactory receptor gene families in mammalian niche adaptation. Mol. Biol. Evol. 35, 1390–1406. ( 10.1093/molbev/msy028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nei M, Rooney AP. 2006. Concerted and birth-and-death evolution of multigene families. Annu. Rev. Gen. 39, 121–152. ( 10.1146/annurev.genet.39.073003.112240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niimura Y. 2012. Olfactory receptor multigene family in vertebrates: from the viewpoint of evolutionary genetics. Curr. Gen. 13, 103–114. ( 10.2174/138920212799860706) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cobb. 1960. Observations on the comparative anatomy of the avian brain. Perspect. Biol. Med. 3, 383–408. ( 10.1353/pbm.1960.0053) [DOI] [PubMed] [Google Scholar]
- 14.Giles S, Friedman M. 2014. Virtual reconstruction of endocast anatomy in early ray-finned fishes (Osteichthyes, Actinopterygii). J. Paleontol. 88, 636–651. ( 10.1666/13-094) [DOI] [Google Scholar]
- 15.Gittleman JL. 1990. Carnivore olfactory bulb size: allometry, phylogeny and ecology. J. Zool. 225, 253–272. ( 10.1111/j.1469-7998.1991.tb03815.x) [DOI] [Google Scholar]
- 16.Cobb S. 1959. A note on the size of the avian olfactory bulb. Epilepsia 1, 394–402. ( 10.1111/j.1528-1157.1959.tb04276.x) [DOI] [PubMed] [Google Scholar]
- 17.Zelenitsky DK, Therrien F, Kobayashi Y. 2009. Olfactory acuity in theropods: paleobilogical and evolutionary implications. Proc. R. Soc. B 276, 667–673. ( 10.1098/rspb.2008.1075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan I, et al. 2015. Olfactory receptor sub- genomes linked with broad ecological adaptations in Sauropsida. Mol. Biol. Evol. 32, 2832–2843. ( 10.1093/molbev/msv155) [DOI] [PubMed] [Google Scholar]
- 19.Chang BSW, Jonsson K, Kazmi MA, Donoghue MJ, Sakmar TP. 2002. Recreating a functional ancestral archosaur visual pigment. Mol. Biol. Evol. 19, 1483–1489. ( 10.1093/oxfordjournals.molbev.a004211) [DOI] [PubMed] [Google Scholar]
- 20.Chang BSW. 2003. Ancestral gene reconstruction and synthesis of ancient rhodopsins in the laboratory. Integr. Comp. Biol. 43, 500–507. ( 10.1093/icb/43.4.500) [DOI] [PubMed] [Google Scholar]
- 21.Harms MJ, Thornton JW. 2010. Analyzing protein structure and function using ancestral gene reconstruction. Curr. Opin Struct. Biol. 20, 360–366. ( 10.1016/j.sbi.2010.03.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ouangraoua A, Tannier E, Chauve C. 2011. Reconstructing the architecture of the ancestral amniote genome. Bioinformatics 19, 2664–2671. ( 10.1093/bioinformatics/btr461) [DOI] [PubMed] [Google Scholar]
- 23.Romanov MN, et al. 2014. Reconstruction of gross avian genome structure, organization and evolution suggests that the chicken lineage most closely resembles the dinosaur avian ancestor. BMC Genomics 15, 1060 ( 10.1186/1471-2164-15-1060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Connor RE, et al. 2018. Reconstruction of the diapsid ancestral genome permits chromosome evolution tracing in avian and non-avian dinosaurs. Nat Comm. 9, 1883 ( 10.1038/s41467-018-04267-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zelenitsky DK, Therrien F, Ridgely RC, McGee AR, Witmer LM. 2011. Evolution of olfaction in non-avian theropod dinosaurs and birds. Proc. R. Soc. B 278, 3625–3634. ( 10.1098/rspb.2011.0238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lautenschlager S, Rayfield EJ, Altangerel P, Zanno LE, Witmer LM. 2012. The endocranial anatomy of Therizinosauria and its implications for sensory and cognitive function. PLoS ONE 7, e52289 ( 10.1371/journal.pone.0052289) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paulina-Carabajal A, Lee Y-N, Jacobs LL. 2016. Endocranial morphology of the primitive nodosaurid dinosaur Pawpawsaurus campbelli from the Early Cretaceous of North America. PLoS ONE 11, e0150845 ( 10.1371/journal.pone.0150845) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulina-Carabajal A, Filippi L. 2018. Neuroanatomy of the abelisaurid theropod Viavenator: the most complete reconstruction of a cranial endocast and inner ear for a South American representative of the clade. Cretac. Res. 83, 84–94. ( 10.1016/j.cretres.2017.06.013) [DOI] [Google Scholar]
- 29.Lislevand T, Figuerola J, Székely T. 2007. Avian body sizes in relation to fecundity, mating system, display behaviour, and resource sharing. Ecology 88, 1605 ( 10.1890/06-2054) [DOI] [Google Scholar]
- 30.Parr CS, et al. 2014. The encyclopedia of life v2: providing global access to knowledge about life on earth . Biodivers. Data J. 2, e1079 ( 10.3897/BDJ.2.e1079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myers PR, Espinosa R, Parr CS, Jones T, Hammond GS, Dewey TA. 2018. The Animal Diversity Web (online). See https://animaldiversity.org.
- 32.Glusman G, Bahar A, Sharon D, Pilpel Y, White J, Lancet D. 2000. The olfactory receptor gene superfamily: data mining, classification, and nomenclature. Mamm. Genome. 11, 1016–1023. ( 10.1007/s003350010196) [DOI] [PubMed] [Google Scholar]
- 33.Brightly WH. 2014. Olfactory ratio as a potential proxy for behavior in Theropoda. Undergraduate honors theses, p. 103, The College of William and Mary, Williamsburg, VA, USA.
- 34.Benson RBJ, Hunt G, Carrano MT, Campione N. 2018. Cope's rule and the adaptive landscape of dinosaur body size evolution. Paleontology 61, 13–48. ( 10.1111/pala.12329) [DOI] [Google Scholar]
- 35.Healy S, Guildford T. 1990. Olfactory-bulb size and nocturnality in birds. Evolution 44, 339–346. ( 10.1111/j.1558-5646.1990.tb05203.x) [DOI] [PubMed] [Google Scholar]
- 36.Lord K. 2013. A comparison of the sensory development of wolves (Canis lupus) and dogs (Canis lupus familiaris). Ethology 119, 110–120. ( 10.1111/eth.12044) [DOI] [Google Scholar]
- 37.Niimura Y, Matsui A, Touhara K. 2014. Extreme expansion of the olfactory receptor gene repertoire in African elephants and the evolutionary dynamics of orthologous gene groups in 13 placental mammals. Gen. Res. 24, 1485–1496. ( 10.1101/gr.169532.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moran JK, Dietrich DR, Elbert T, Pause BM, Kubler L, Weierstall R. 2015. The scent of blood: a driver of human behavior? PLoS ONE 10, e0137777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandewege MW, Mangum SF, Gabaldón T, Castoe TA, Ray DA, Hoffmann FG. 2016. Contrasting patterns of evolutionary diversification in the olfactory repertoires of reptile and bird genomes. Genome Biol Evol. 8, 470–480. ( 10.1093/gbe/evw078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes GM, Teeling EC, Higgins DG. 2014. Loss of olfactory receptor function in hominin evolution. PLoS ONE 9, e84714 ( 10.1371/journal.pone.0084714) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bird DJ, Murphy WJ, Fox-Rosales L, Hamid I, Eagle RA, Valkenburgh BV. 2018. Olfaction written in bone: cribriform plate size parallels olfactory receptor gene repertoires in Mammalia. Proc. R. Soc. B 285, 20180100 ( 10.1098/rspb.2018.0100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hughes GM, Finarelli JA. 2019. Data from: Olfactory receptor repertoire size in dinosaurs Dryad Digital Repository. ( 10.5061/dryad.63bd5r6) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Hughes GM, Finarelli JA. 2019. Data from: Olfactory receptor repertoire size in dinosaurs Dryad Digital Repository. ( 10.5061/dryad.63bd5r6) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Data available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.63bd5r6 [42]. All supplemental files are available as part of the electronic supplementary material.