The structure of Aspergillus aculeatus β-1,4-galactanase, a glycoside hydrolase belonging to family 53, has been determined in complex with galactobiose at subsites −1 and −2.
Keywords: glycoside hydrolases; specificity; pectin; oligosaccharides; Aspergillus aculeatus; β-1,4-galactanases
Abstract
β-1,4-Galactanases are glycoside hydrolases that are involved in the degradation of pectin and belong to family 53 in the classification of glycoside hydrolases. Previous studies have elucidated the structures of several fungal and two bacterial galactanases, while biochemical studies have indicated differences in the product profiles of different members of the family. Structural studies of ligand complexes have to date been limited to the bacterial members of the family. Here, the first structure of a fungal galactanase in complex with a disaccharide is presented. Galactobiose binds to subsites −1 and −2, thus improving our understanding of ligand binding to galactanases.
1. Introduction
β-1,4-Galactanases are glycoside hydrolases (EC 3.2.1.89) that degrade galactan and arabinogalactan side chains of rhamnogalacturonan I, the most studied and abundant component of pectin found in non-woody plant cell walls (Lau et al., 1985 ▸). The hydrolysis takes place by a double-displacement retaining mechanism involving two carboxylic acid residues, one functioning as a catalytic nucleophile and the other as a general acid/base. Among other applications, galactanases can be used as supplements for biomass degradation (de Lima et al., 2016 ▸). According to the CAZY classification, the enzymes are classified into family GH53 (Lombard et al., 2014 ▸) in clan GH-A (Jenkins et al., 1995 ▸; Davies & Henrissat, 1995 ▸). Structures of β-1,4-galactanases have been reported previously from four fungi: Thermothelomyces thermophila (also known as Myceliophthora thermophila; MtGal; GenBank ID AAE73520.1; PDB entries 1hjs and 1hju; Le Nours et al., 2003 ▸), Humicola insolens (HiGal; GenBank ID AAN99815.1; PDB entry 1hjq; Le Nours et al., 2003 ▸), Aspergillus aculeatus KSM 510 (AaGal; GenBank ID AAA32692.1; PDB entries 1fhl and 1fob; Ryttersgaard et al., 2002 ▸) and the closely related A. nidulans FGSC A4 (EnGal; GenBank ID ABF50874.1; PDB entry 4bf7; Otten et al., 2013 ▸). Additionally, a galactanase from the soil bacterium Bacillus licheniformis has been structurally characterized in complex with galactooligosaccharides (BlGal; GenBank ID AAO31370.1; PDB entries 1r8l, 1ur0, 1ur4, 2ccr, 2gft and 2j74; Ryttersgaard et al., 2004 ▸; Le Nours et al., 2009 ▸), and very recently (during review of this manuscript) the first galactanase structure from a human gut bacterium, Bacteroides thetaiotaomicron, has also been determined (BtGH53; GenBank ID AAO79773.1; PDB entries 6gp5 and 6gpa; Böger et al., 2019 ▸). Two of the structurally characterized fungal enzymes are thermophilic, and the available structures have also been used to design or rationalize thermostability mutants in two fungal enzymes (Larsen et al., 2015 ▸; Torpenholt et al., 2015 ▸).
A number of fungal and bacterial β-1,4-galactanases have also been characterized biochemically. From the studies conducted so far, even though fungal galactanases attack the internal glycosidic linkages in the polysaccharides and are thus true endo enzymes, they eventually release shorter products and can hydrolyse small substrates, both oligosaccharides and small chromogenic substrates. As an example, AaGal and Meripilus giganteus β-1,4-galactanase are able to hydrolyse both 2-nitrophenyl-1,4-β-d-galactopyranoside (with accompanying transglycosylation) and 4-nitrophenyl-1,4-β-d-thiogalactobioside, while BlGal was completely inactive on these substrates (Torpenholt et al., 2011 ▸). Even earlier work showed that BlGal could not degrade galactotriose, while AaGal was able to (Ryttersgaard et al., 2004 ▸). Another bacterial galactanase from Geobacillus stearothermophilus produces mostly galactotetraose from galactan, and was also shown not to degrade galactotriose to smaller oligosaccharides (Tabachnikov & Shoham, 2013 ▸), while the galactanase from Bifidobacterium longum was reported to produce galactotriose by a processive mechanism (Hinz et al., 2005 ▸). However, in a more recent study a bacterial galactanase from Bacteroides thetaiotaomicron showed the ability to degrade potato galactan to galactobiose (77%) and galactose (23%) (van Bueren et al., 2017 ▸; Böger et al., 2019 ▸), thus showing that bacteria possess galactanases with diverse degradation patterns. Structural comparison suggests that the differences in product profile between the fungal and BlGal galactanases and the inability of the latter to degrade galatotriose is owing to the fact that BlGal has a more extended glycone substrate-binding site and thus can bind small oligosaccharides nonproductively (Ryttersgaard et al., 2004 ▸; Torpenholt et al., 2011 ▸). Although crystallographic complexes with oligosaccharides have been obtained with BlGal (Ryttersgaard et al., 2004 ▸; Le Nours et al., 2009 ▸), no ligand complex of a fungal galactanase has been reported in the literature to date.
2. Materials and methods
2.1. Macromolecule production
The cloning and expression of AaGal (GenBank ID AAA32692.1) is described in detail in Dorreich et al. (1995 ▸). Wild-type AaGal was purified from the cell extract (provided by Novozymes A/S) in a similar way to that described in Christgau et al. (1995 ▸). 300 mg of the cell extract was dissolved in 9.5 ml 50 mM Tris pH 6.5, passed through a 0.22 µm Millex-GV filter and purified on a Q Sepharose 16/10 column using a gradient of 1 M sodium chloride in 50 mM Tris pH 6.5. The pooled fractions containing protein were reduced in volume by centrifugation at 4500 rev min−1 through an Amicon Ultra-15 10 000 molecular-weight cutoff filter unit and filtrated through a 0.22 µm Ultrafree-MC filter. The purity was judged by SDS–PAGE on polyacrylamide gels (4–20% from Bio-Rad or 4–12% from NuPAGE) after heat denaturation in the presence of DTT. The gels were stained with Instant Blue from Expedeon. The enzyme was greater than 95% pure as judged from the SDS–PAGE gel.
The purified protein was dialysed into 50 mM citrate pH 4.5 using a 10 000 molecular-weight cutoff Slide-A-Lyzer dialysis cassette. The protein concentration was determined from absorbance measurements at 280 nm using an NanoDrop 1000 spectrophotometer from Fisher Scientific, and the final concentration was determined to be 6.5 mg ml−1. The protein stock was further concentrated prior to crystallization.
2.2. Crystallization
Crystallization was carried out by hanging-drop vapour diffusion at room temperature using a 24-well tray from Hampton Research. During our structural studies of fungal galactanases, ligand soaking of galactobiose, galactotriose and modified carbohydrate analogues was attempted with several enzymes and different crystallization conditions, initially with little success. For AaGal, the original structure was determined at pH 7.5 in the presence of 0.2 M calcium chloride and PEG 400 as the main precipitant (Ryttersgaard et al., 1999 ▸). As no carbohydrate ligand could be observed in crystals grown close to these conditions and soaked with ligands, it was hypothesized that the use of a pH far from the optimum for this enzyme and/or the high concentration of calcium, which is found bound near the enzyme active site in the native structure, could be interfering. Thus, a number of two-dimensional screens were carried out inspired by the original crystallization conditions, exploring PEGs of different molecular weights as precipitants and using lower pH values and concentrations of divalent metal ions than were used to obtain the native structure.
The crystal that was utilized for the successful ligand complex in this study was grown as follows: 1 µl AaGal stock at 7.3 mg ml−1 concentration in 50 mM acetate pH 4.0 was mixed with 3 µl reservoir solution and equilibrated against 1.0 ml reservoir solution consisting of 0.1 M calcium chloride, 0.1 M sodium acetate pH 4.5, 30% PEG 3000 at room temperature. Prior to soaking the crystal in a galactobiose-containing solution, the pH in the drop was measured to be 4.2. The crystal was soaked for 1 min in 0.2 M β-1,4-galactobiose (Megazyme) in 30% PEG 3000 and was cryoprotected in 25% PEG 400 in reservoir solution before data collection.
2.3. Data collection and processing
Data were collected from a cryocooled crystal (100 K) mounted in a cryoloop on beamline I911-2 at MAX-lab, Lund, Sweden. The data set was obtained in 2011, and thus was collected and processed rather conservatively with respect to resolution limit. Processing was carried out with XDS and XSCALE (Kabsch, 2010 ▸). Details of the data-processing statistics are given in Table 1 ▸. Reprocessing using a current version of XDS gives a CC1/2 of over 90% in the outer resolution shell.
Table 1. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | I911-2, MAX-lab |
| Wavelength (Å) | 1.03973 |
| Temperature (K) | 100 |
| Detector | MAR 165 CCD |
| Crystal-to-detector distance (mm) | 150 |
| Rotation range per image (°) | 1.0 |
| Total rotation range (°) | 200 |
| Exposure time per image (s) | 30 |
| Space group | I4122 |
| a, b, c (Å) | 200.2, 200.2, 106.8 |
| α, β, γ (°) | 90, 90, 90 |
| Mosaicity (°) | 0.33 |
| Resolution range (Å) | 50.0–2.69 (2.76–2.69) |
| Total No. of reflections | 475968 |
| No. of unique reflections | 29998 |
| Completeness (%) | 98.5 (94.3) |
| Multiplicity | 15.9 (12.6) |
| 〈I/σ(I)〉 | 23.6 (5.7) |
| R r.i.m. | 0.132 (0.595) |
| Overall B factor from Wilson plot (Å2) | 52.38 |
2.4. Structure solution and refinement
The structure was determined using the known structure of AaGal obtained at pH 7.5 (100 K; PDB entry 1fob) as a search model for molecular replacement. Molecular replacement was carried out using Phaser-MR in PHENIX (Adams et al., 2010 ▸). The rotation-function Z scores for the two molecules were 15.7 and 16.7 and the translation-function Z scores were 50.3 and 88.1, with no clashes, with a final log-likelihood gain of 4366. The structure was refined using REFMAC5 in the CCP4 package (Winn et al., 2011 ▸) together with visual inspection using Coot (Emsley et al., 2010 ▸). The final model contained galactobiose at the active site and a glycosylation at Asn112 modelled by two consecutive N-acetylglucosamines (NAG) and a β-mannose (BMA). Seven calcium ions were modelled as well as some other ligands consisting of acetate and several PEG fragments of different lengths from the crystallization/cryoprotecting liquor. The glycosylation as well as the galactobiose bound at the active site were validated in Privateer (Agirre et al., 2015 ▸) and exhibited an acceptable level of confidence. The galactobiose displayed a real-space correlation coefficient (RSCC) value of greater than 0.90, while the NAG constituents of the glycosylation exhibited values of at least 0.89. However, the BMA constituents only gave values of 0.75 for chain A and 0.61 for chain B, which is most likely owing to disorder. The refinement statistics are summarized in Table 2 ▸.
Table 2. Structure solution and refinement.
Values in parentheses are for the outer shell.
| Resolution range (Å) | 50.00–2.69 (2.76–2.69) |
| Completeness (%) | 98.5 (94.4) |
| No. of reflections, working set | 26963 (2013) |
| No. of reflections, test set | 1517 (81) |
| Final R cryst | 0.145 (0.219) |
| Final R free | 0.211 (0.289) |
| Cruickshank DPI | 0.4008 |
| No. of non-H atoms | |
| Protein | 5341 |
| Ions | 7 |
| Other ligands | 146 |
| Water | 222 |
| Total | 5716 |
| R.m.s. deviations | |
| Bonds (Å) | 0.0142 |
| Angles (°) | 1.7663 |
| Average B factors (Å2) | |
| Protein, chain A | 38.41 |
| Protein, chain B | 41.06 |
| Galactobiose, chain A | 44.05 |
| Galactobiose, chain B | 42.95 |
| Glycosylation, chain A | 56.97 |
| Glycosylation, chain B | 58.03 |
| Ions | 64.93 |
| Other ligands | 71.24 |
| Water | 35.41 |
| Ramachandran plot† | |
| Most favoured (%) | 96.37 |
| Allowed (%) | 3.63 |
The Ramachandran plot was generated using dynarama in Coot (Emsley et al., 2010 ▸).
3. Results and discussion
The structure of AaGal in complex with galactobiose was determined to a maximum resolution of 2.7 Å (Fig. 1 ▸). As a member of the GH-A clan, the overall fold of the enzyme has a (β/α)8-barrel architecture. The two catalytic glutamic acid residues are located at the end of β-strands 4 (acid/base, Glu136) and 7 (nucleophile, Glu246), and the glycosylation is positioned at Asn112. In a previously published AaGal structure (PDB entry 1fob) a Ca2+ ion is present in the active site. This is not observed in the current structure as galactobiose is able to compete with this particular position of the ion under the new crystallization conditions. However, several calcium ions are present in the structure. The space group was I4122, with two molecules in the asymmetric unit. The previously reported structures of AaGal (PDB entry 1fob at 100 K and PDB entry 1fhl at 293 K) belonged to space group I222 with one molecule in the asymmetric unit (Ryttersgaard et al., 2002 ▸). Both chain A and chain B superimpose well on PDB entry 1fob, with root-mean-square deviations (r.m.s.d.s) of 0.6 Å for all atoms and 0.4 Å for Cα atoms. Superimposition of chain A on chain B gives r.m.s.d.s of 0.47 Å for all atoms and 0.19 Å for Cα atoms, which indicate that the two molecules in the asymmetric unit are close to identical.
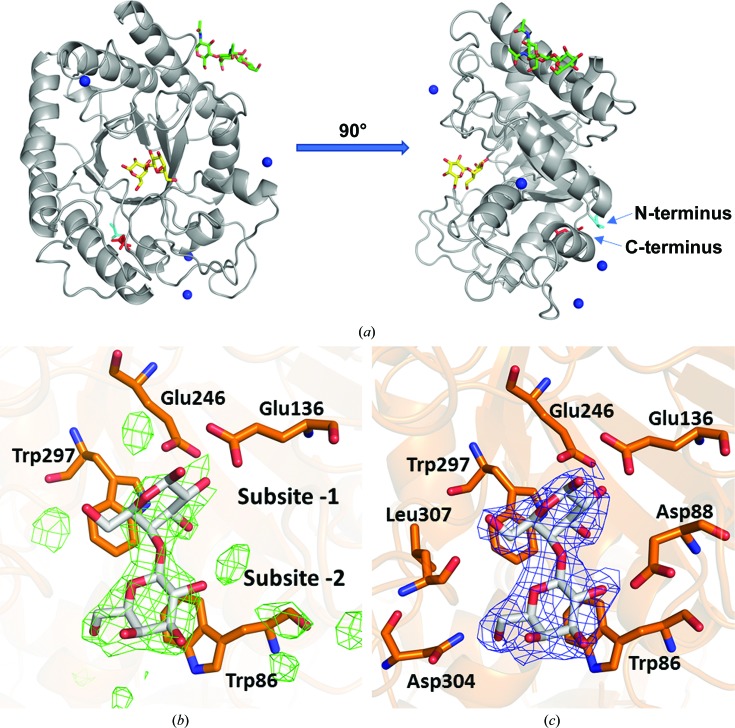
Figure 1.
Galactobiose binding site in chain A of AaGal. (a) Overview of the enzyme with galactobiose. The galactobiose and glycosylation are shown in stick representation (with yellow and green C atoms, respectively). Ca2+ ions are depicted as blue spheres and the polypeptide chain is shown as a grey ribbon. The N-terminus and C-terminus are labelled and are coloured cyan and red, respectively. (b) The difference map before modelling of galactobiose (F obs − F calc map contoured at the 3.0σ level). (c) Electron density for galactobiose (weighted 2F obs − F calc map contoured at the 1.0σ level).
Galactobiose binding identified two subsites, −1 and −2, defined using the standard nomenclature (Davies et al., 1997 ▸) with minus subsites towards the reducing end and plus subsites towards the nonreducing end of the substrate. Hydrolysis occurs between subsites −1 and +1.
3.1. Subsite −1
This subsite, which is occupied for the first time in a GH53 crystal structure, is located near the catalytic active-site residues Glu136 and Glu246. O∊1 and O∊2 of both Glu136 and Glu246 are within hydrogen-bonding distance of the O1 and O2 hydroxyl groups, respectively, of the galactose unit. Potential hydrogen bonds are also observed between O2 of galactose and Nδ2 of Asn135, and between O6 of galactose and the backbone O atom of Leu307, the hydroxyl group of Tyr218 and a water molecule. O1 and O3 in the galactose unit are hydrogen-bonded to water molecules. Trp297 serves as an aromatic platform at the −1 subsite. The modelled galactose occupying the −1 subsite is slightly skewed compared with the regular chair conformation in both chain A and chain B. This is observed both by visual inspection in Coot and as seen in the coordinates given by Privateer. This is likely to be owing to the enzyme forcing the substrate into a higher energy conformation or simply owing to the limitations of modelling at 2.7 Å resolution. However, it is important to note that additional distortion would be needed in a complex with a substrate spanning the −1/+1 subsites, since in the current product complex the catalytic nucleophile does not reach the scissile bond.
3.2. Subsite −2
This subsite is located between Trp86 and Gly308, where Trp86 serves as an aromatic platform to stack with the galactose unit. The O6 of galactose is hydrogen-bonded to Nδ2 of Asn304, while O2 is hydrogen-bonded to Oδ2 of Asp88.
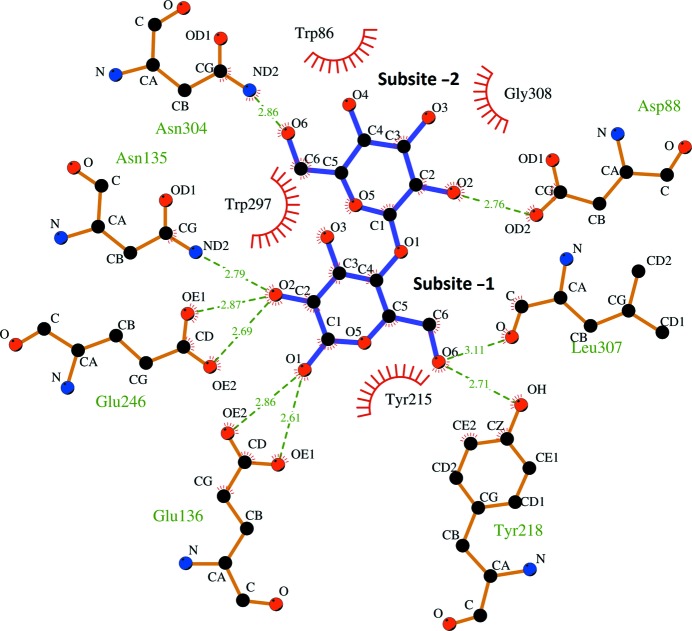
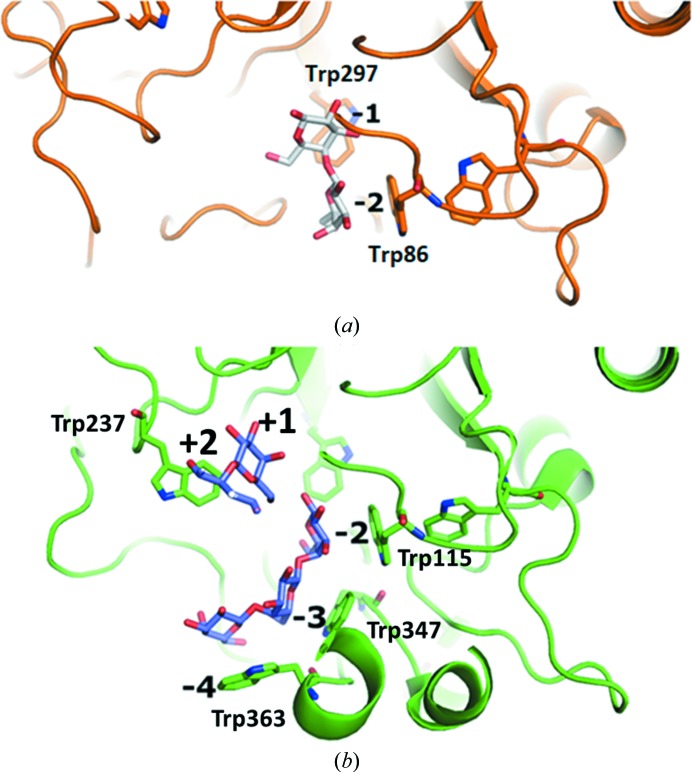
The structure of AaGal in complex with galactobiose at subsites −1 and −2 is the first structure of a fungal galactanase in complex with a product. The ability of AaGal to bind galactobiose at subsites −1 and −2 (Fig. 2 ▸) fits together well with the observed degradation (Ryttersgaard et al., 2004 ▸) and transglycosidation (Torpenholt et al., 2011 ▸) patterns of AaGal. AaGal has been shown to degrade galactotriose and galactotetraose completely to galactose and galactobiose and to produce galactose, galactobiose and 2-nitrophenol from the hydrolysis of 2-nitrophenyl-1,4-β-d-galactopyranoside at high concentrations, where galactobiose is thought to be a product of transglycosidation (Torpenholt et al., 2011 ▸). In the structure AaGal binds galactobiose nonproductively in subsites −1 and −2, presumably as a result of the high concentrations used for soaking, but the fact that galactose is observed at the −1 subsite, which is rarely occupied in product complexes of glycoside hydrolases, indicates a relatively high affinity at this subsite which makes transglycosidation with the small chromogenic substrate possible. In contrast, the previously determined structure (PDB entry 2ccr; Ryttersgaard et al., 2004 ▸) of BlGal, a bacterial galactanase, in complex with galactobiose and galactotriose showed no binding at subsite −1, which fits well with the inability of BlGal to degrade (Ryttersgaard et al., 2004 ▸) or transglycosidate (Torpenholt et al., 2011 ▸) small substrates. The amino-acid residues Asn135, Trp297 and Tyr215 constructing subsite −1, Trp86 and Asp88 constructing subsite −2 and the two catalytic glutamates Glu136 and Glu246 are conserved in AaGal and BlGal. Asn304, Leu307 and Gly308 are observed in AaGal and constitute a turn. Leu307 and Asn304 are hydrogen bonded to the O6 hydroxyl group of galactose in subsites −1 and −2, respectively. This turn is not observed in BlGal, as the corresponding residues in BlGal are part of an extra-long loop 8 containing subsites −3 and −4. BlGal has an additional lysine (Lys120) that interacts with the O3 hydroxyl group of galactose at subsite −2 but lacks an appropriate residue corresponding to Tyr218 in AaGal for interaction with the O6 hydroxyl group of galactose in subsite −1 (Fig. 2 ▸). Alignment of the available galactanase sequences has shown that Lys120 is conserved in most bacterial galactanases and that Asn135, Trp297, Trp86, Asp88, Leu307, Gly308 and Asn304 are conserved in fungal galactanases. Tyr218 is conserved in many fungal galactanases and is a tryptophan in BlGal.
Figure 2.
Detailed view of the interactions between AaGal and galactobiose (chain A). This figure was made using LigPlot+ (Laskowski & Swindells, 2011 ▸).
Some disorder is observed in the structure, mainly at calcium-bridging carboxylate side chains, which could indicate that numerous possibilities are available for rearranging the water molecules around the bridges. Trp163 also seems to be disordered and is most likely to adopt several conformations, but owing to the relatively low resolution it has not been possible to model it satisfactorily.
Supplementary Material
PDB reference: β-1,4-galactanase in complex with galactobiose, 6q3r
Figure 3.
Comparison of the binding grooves of (a) AaGal (PDB entry 6q3r) and (b) BlGal (PDB entry 2ccr), with tryptophan residues highlighted.
Acknowledgments
We thank Dorthe Boelskifte for help with crystallization. LLL’s group is part of ISBUC (Integrative Structural Biology at the University of Copenhagen; https://isbuc.ku.dk/).
Funding Statement
This work was funded by Strategiske Forskningsråd grant 2106-07-0030.
References
- Adams, P. D., Afonine, P. V., Bunkóczi, G., Chen, V. B., Davis, I. W., Echols, N., Headd, J. J., Hung, L.-W., Kapral, G. J., Grosse-Kunstleve, R. W., McCoy, A. J., Moriarty, N. W., Oeffner, R., Read, R. J., Richardson, D. C., Richardson, J. S., Terwilliger, T. C. & Zwart, P. H. (2010). Acta Cryst. D66, 213–221. [DOI] [PMC free article] [PubMed]
- Agirre, J., Iglesias-Fernández, J., Rovira, C., Davies, G. J., Wilson, K. S. & Cowtan, K. D. (2015). Nature Struct. Mol. Biol. 22, 833–834. [DOI] [PubMed]
- Böger, M., Hekelaar, J., van Leeuwen, S. S., Dijkhuizen, L. & Lammerts van Bueren, A. (2019). J. Struct. Biol. 205, 1–10. [DOI] [PubMed]
- Christgau, S., Sandal, T., Kofod, L. V. & Dalbøge, H. (1995). Curr. Genet. 27, 135–141. [DOI] [PubMed]
- Davies, G. & Henrissat, B. (1995). Structure, 3, 853–859. [DOI] [PubMed]
- Davies, G. J., Wilson, K. S. & Henrissat, B. (1997). Biochem. J. 321, 557–559. [DOI] [PMC free article] [PubMed]
- Dorreich, K., Dalbøge, H., Mikkelsen, J. M., Mischler, M. & Christensen, F. M. (1995). US Patent 5474922.
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Hinz, S. W. A., Pastink, M. I., van den Broek, L. A. M., Vincken, J.-P. & Voragen, A. G. J. (2005). Appl. Environ. Microbiol. 71, 5501–5510. [DOI] [PMC free article] [PubMed]
- Jenkins, J., Lo Leggio, L., Harris, G. & Pickersgill, R. (1995). FEBS Lett. 362, 281–285. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Lammerts van Bueren, A., Mulder, M., Leeuwen, S. V. & Dijkhuizen, L. (2017). Sci. Rep. 7, 40478. [DOI] [PMC free article] [PubMed]
- Larsen, D. M., Nyffenegger, C., Swiniarska, M. M., Thygesen, A., Strube, M. L., Meyer, A. S. & Mikkelsen, J. D. (2015). Appl. Microbiol. Biotechnol. 99, 4245–4253. [DOI] [PubMed]
- Laskowski, R. A. & Swindells, M. B. (2011). J. Chem. Inf. Model. 51, 2778–2786. [DOI] [PubMed]
- Lau, J. M., McNeil, M., Darvill, A. G. & Albersheim, P. (1985). Carbohydr. Res. 137, 111–125.
- Le Nours, J., De Maria, L., Welner, D., Jørgensen, C. T., Christensen, L. L. H., Borchert, T. V., Larsen, S. & Lo Leggio, L. (2009). Proteins, 75, 977–989. [DOI] [PubMed]
- Le Nours, J., Ryttersgaard, C., Lo Leggio, L., Østergaard, P. R., Borchert, T. V., Christensen, L. L. H. & Larsen, S. (2003). Protein Sci. 12, 1195–1204. [DOI] [PMC free article] [PubMed]
- Lima, E. A. de, Machado, C. B., Zanphorlin, L. M., Ward, R. J., Sato, H. H. & Ruller, R. (2016). Appl. Biochem. Biotechnol. 179, 415–426. [DOI] [PubMed]
- Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M. & Henrissat, B. (2014). Nucleic Acids Res. 42, D490–D495. [DOI] [PMC free article] [PubMed]
- Otten, H., Michalak, M., Mikkelsen, J. D. & Larsen, S. (2013). Acta Cryst. F69, 850–854. [DOI] [PMC free article] [PubMed]
- Ryttersgaard, C., Le Nours, J., Lo Leggio, L., Jørgensen, C. T., Christensen, L. L. H., Bjørnvad, M. & Larsen, S. (2004). J. Mol. Biol. 341, 107–117. [DOI] [PubMed]
- Ryttersgaard, C., Lo Leggio, L., Coutinho, P. M., Henrissat, B. & Larsen, S. (2002). Biochemistry, 41, 15135–15143. [DOI] [PubMed]
- Ryttersgaard, C., Poulsen, J.-C. N., Christgau, S., Sandal, T., Dalbøge, H. & Larsen, S. (1999). Acta Cryst. D55, 929–930. [DOI] [PubMed]
- Tabachnikov, O. & Shoham, Y. (2013). FEBS J. 280, 950–964. [DOI] [PubMed]
- Torpenholt, S., De Maria, L., Olsson, M. H., Christensen, L. H., Skjøt, M., Westh, P., Jensen, J. H. & Lo Leggio, L. (2015). Comput. Struct. Biotechnol. J. 13, 256–264. [DOI] [PMC free article] [PubMed]
- Torpenholt, S., Le Nours, J., Christensen, U., Jahn, M., Withers, S., Østergaard, P. R., Borchert, T. V., Poulsen, J.-C. & Lo Leggio, L. (2011). Carbohydr. Res. 346, 2028–2033. [DOI] [PubMed]
- Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., Keegan, R. M., Krissinel, E. B., Leslie, A. G. W., McCoy, A., McNicholas, S. J., Murshudov, G. N., Pannu, N. S., Potterton, E. A., Powell, H. R., Read, R. J., Vagin, A. & Wilson, K. S. (2011). Acta Cryst. D67, 235–242. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: β-1,4-galactanase in complex with galactobiose, 6q3r