Abstract
Exosomes are nano-sized membranous vesicles of endosomal origin that carry nucleic acids, lipids and proteins. The cargo of exosomes is cell origin specific and the release of these exosomes and uptake by an acceptor cell is seen as a vital element of cell-cell communication. Here, we sought to investigate the diagnostic and prognostic value of the expression of glypican 3 (GPC3) on primary gastro-esophageal adenocarcinoma (GEA) tissue (tGPC3) and corresponding serum exosomes (eGPC3). Circulating exosomes were extracted from serum samples of 49 patients with GEA and 56 controls. Extracted exosomes were subjected to flow cytometry for the expression of eGPC3 and GPC3 expression on primary GEA tissue samples was determined by immunohistochemistry and correlated to clinicopathological parameters. We found decreased eGPC3 levels in GEA patients compared to healthy controls (p < 0.0001) and high tGPC3 expression. This was significantly associated with poor overall survival (high vs. low eGPC3: 87.40 vs. 60.93 months, p = 0.041, high vs. low tGPC3: 58.03 vs. 84.70 months, p = 0.044). Cox regressional analysis confirmed tGPC3 as an independent prognostic biomarker for GEA (p = 0.02) and tGPC3 expression was validated in two independent cohorts. Our findings demonstrate that eGPC3 and tGPC3 can be used as potential diagnostic and prognostic biomarkers for GEA.
Keywords: exosomes; gastro-esophageal adenocarcinoma; glypican 3; prognostic biomarker, diagnostic biomarker
1. Introduction
Gastro-esophageal adenocarcinomas (GEA) are one of the most common causes of cancer-related mortality [1], despite an overall decline in incidence [2]. True non-cardia carcinomas and cancers of the esophagogastric junction develop asymptomatically and at time of diagnosis, most patients present with loco-regional or metastatic disease, limiting the possibility for curative interventions [3]. GEA is a heterogeneous disease with distinct clinical subtypes and the complex molecular mechanisms underlying the initiation and progression of these cancers is not very well understood. Thus, there is an urgent need to better characterize the molecular mechanism underlying this disease and develop effective screening systems to facilitate earlier diagnosis and treatment [3].
Liquid biopsies are a non-invasive tool to provide an easily accessible surrogate for tumor tissue and represent a promising approach to detect malignancies at early stage and monitor the response to treatment [4,5]. In gastric cancer patients, a large number of novel blood-based biomarkers have been reported for their potential role in clinical practice [6]. However, none of these have been established in the clinic for early detection or disease monitoring. The most commonly used biomarkers for GEA are serum carbohydrate antigen 19-9 (CA-19-9), carbohydrate antigen 72-4 (CA-72-4) and carcinoembryonic antigen (CEA), although it has been shown that they vary in their sensitivity and specificity and strongly depend on tumor burden [7].
Exosomes comprise a subgroup of extracellular vesicles, defined by their endosomal origin and size ranging from 20–150 nm [8]. As exosomes are spherical, lipid-bilayered vesicles, generated through a complex regulated process within multivesicular bodies, they may resemble the molecular landscape of the donor tissue and its pathologic characteristics [9]. Several studies have highlighted the crucial role that exosomes play in the pathobiology of GEA [10,11,12]. These vesicles carry nucleic acids, lipids and proteins which can act as signal molecules for intercellular communication [13], thereby promoting the activation of the PI3K-AKT and MAPK pathway, which significantly impacts cancer cell proliferation and invasion [10]. These characteristics of exosomes may provide unique opportunities for the detection of malignancies and identification of diagnostic and prognostic biomarkers [14].
Glypicans belong to a group of heparan-sulfate proteoglycans, a large family of plasma membrane associated glycoproteins, which are involved in the crosstalk between cancer cells and their microenvironment [15]. Exosomes express glypicans on their cellular surface [16] and recently glypican 1+ (GPC1) exosomes have been identified for the detection of pancreatic cancer independent of staging [17] in multiple studies [18,19], emphasizing the great potential of exosome-based biomarkers for gastrointestinal cancers. Glypican 3 (GPC3), another member of the glypican protein family, has also been shown to act as a regulator of different cellular functions including proliferation and differentiation [20]. However, there is conflicting data in the literature regarding the role of GPC3 in gastric and esophageal cancer. Several studies have reported, that GPC3 is an oncofetal protein, which is associated with the alpha-fetoprotein-producing (AFP) hepatoid phenotype of gastric adenocarcinoma and gastric adenocarcinoma of enteroblastic differentiation [21,22,23]. Intriguingly, the common gastric adenocarcinomas express the oncofetal proteins GPC3 and AFP [24]. In addition, 25% of esophageal adenocarcinomas (2/8) and 26.5% of esophageal squamous cell carcinoma (SCC; (5/19) showed increased expression of GPC3 [25] in contrast to previous studies that reported lower frequencies of positive staining for GPC3 in esophageal SCC. Moreover, Zhu et al. have reported that expression of glypican 3 is markedly decreased in gastric cancer but not in esophageal cancer [26]. This data supports the finding by Han et al. that GPC3 is a potential suppressor of metastasis as loss of GPC3 was associated with increased lymph node metastasis and poor overall survival (OS) [27].
Here in this study, we sought to examine the expression of GPC3 in primary GEA tissue and matched blood for the presence of GPC3 positive exosomes (eGPC3+). Given that the expression of glypicans is altered during tumorigenesis and glycoproteins have been suggested as a promising target of biomarkers in cancer [28], we want to determine whether GPC3 may serve as a non-invasive diagnostic and prognostic tool for GEA.
2. Materials and Methods
The study was in accordance with the reporting recommendations for tumor marker prognostic studies (REMARK) guidelines [29].
2.1. Patient Characteristics and Data Collection
The Ethics committee of the Technical University of Dresden approved the study (EK 96032017). The experimental cohort consisted of 49 patients with GEA who underwent surgical resection between 2007 and 2013 at the Department of General, Thoracic and Vascular Surgery, University Hospital Carl Gustav Carus, Technical University Dresden, Dresden, Germany. Blood serum samples collected from patients without evidence of neoplastic disease (n = 25) and healthy donors (n = 31) were included as the control cohort. Two independent patient cohorts were used as validation cohorts as previously described [30,31]. The publicly available database Human Protein Atlas [32] includes a human pathology atlas section with a metadata study of the TCGA transcriptome data of 354 patients. GPC3 was one of the 171 genes associated with an unfavorable diagnosis [30]. The expression cutoff was 3.16 FPKM. The second database analyzed for GPC3 mRNA levels of primary GEA tumors was the Kaplan Meier Plotter (n = 876 [31]). To generate the Kaplan Meier graphs, we used the default settings for affyID 209200, which is GPC3.
2.2. Blood Collection and Isolation of Serum Exosomes
Venous blood samples were taken immediately before surgical resection using serum separator tubes. The tubes were centrifuged at 2500 × g for 10 min and the serum was aliquoted and stored at −80 °C. Serum exosomes were isolated using the Total Exosome Isolation Reagent (InvitrogenTM, Thermo Fisher Scientific®, Waltham, MA, USA, Cat-Nr.: 4478360) according to the manufacturer’s instructions. Briefly, 40 µl isolation reagent was added to 200 µl of serum and vortexed. After incubation at 4 °C for 30 min, the samples were centrifuged at 10,000 × g for 10 min at room temperature (RT). The exosome pellet was retrieved after removal of the supernatant and resuspended in 100 µl 1x phosphate buffered saline (PBS). The presence of exosomes has been confirmed and validated with standard and fluorescent nanoparticle tracking analysis (NTA) (ZetaView®, Particle Metrix GmbH, Meerbusch, Germany), Western blotting with exosomal markers and transmission electron microscopy (TEM) (see sections below).
2.3. Nanoparticle Tracking Analysis (NTA) and Fluorescent Nanoparticle Tracking Analysis (fNTA)
Nanoparticle tracking analysis (NTA) was conducted using the ZetaView® to determine the concentration and size of the extracted exosomes according to the manufacturer’s instructions. Additionally, the fluorescent mode of the ZetaView® was employed to measure exosomal markers (CD9, CD63 and TSG101) of the extracted particles. For this analysis, 1 × 1010 exosomes in 100 µl 1x PBS were bound to 0.3 µm polystyrene latex beads (Sigma-Aldrich®, Merck, Darmstadt, Germany) and incubated for 15 min. Subsequently, this suspension was diluted to a total of 1 ml with 1x PBS and incubated on a horizontal shaker at 600 rpm for 60 min. The reaction was stopped by adding 100 mM glycine and 1% BSA in 1x PBS followed by centrifugation at 12,000 rpm for 3 min at RT. The exosome bound bead pellet was diluted in 40 µl 1x PBS. The final suspension was divided in two equal volumes. One aliquot was used for the detection of exosomal markers (CD9 monoclonal rabbit, dilution 1:20, Abcam, Cambridge, UK, Cat. Nr.: ab92726; CD63 polyclonal rabbit, dilution 1:20, Santa Cruz, Dallas, TX, USA, Cat. Nr. sc-15363; and TSG101, monoclonal mouse, dilution: 1:100, Abcam, Cat. Nr.: ab83). The second aliquot was used as a control for unspecific binding (secondary antibody alone). After 1 h of incubation, the reaction was stopped by adding a blocking solution of 100 mM glycine and 1% BSA in 1x PBS. Subsequently, the suspension was centrifuged at 12,000 rpm for 3 min at RT and the exosomes-bound beads were recovered as pellet. Alexa-488 secondary antibody (Life Technologies, Carlsbad, CA, USA; anti-rabbit: A11034 or anti-mouse: A11029) was added and incubated for 60 min at RT. After three washing steps in 1 x PBS/2% BSA, the suspension was diluted in 1 ml 1x PBS. The fluorescent exosome-bound beads (positive and negative control) were analyzed using the ZetaView® (Particle Metrix GmbH, Meerbusch, Germany) with the ZetaView® 8.03.04.01 Software.
2.4. Western Blot (Immunoblotting)
Immunoblot analyses were performed as previously described with minor modifications [33]. Briefly, exosomal total protein was harvested using RIPA buffer including 1x Halt™ Protease- and Phosphatase-Inhibitor-Cocktail (ThermoFisher Scientific, Waltham, MA, USA). Using Bradford quantification, 25 µg of protein was denaturized and separated by a 4–12% gradient Bis-Tris polyacrylamide gel electrophoresis (InvitrogenTM, ThermoFisher Scientific). The proteins were then transferred onto a nitrocellulose blotting membrane (AmershamTM ProtranTM, Amershan, UK) using a wet blotting approach. After blocking for 1 hour at RT, the blots were incubated with following primary antibodies: anti-CD9 (ERP2949) (Abcam), anti-CD63 (H-193) (Santa Cruz), anti-CD81 (NBP2-20564) (Novus Biologicals, Centennial, CO, USA), anti-TSG101 (4A10) (Abcam), anti-GPC-3 (AF2119, R&D Systems) and anti-calreticulin (Cell Signaling, Danvers, MA, USA; Cat. Nr. 2891) overnight at 4 °C on an orbital shaker. The next day, the blots were washed with 1 x PBS/0.05% Tween for at least 30 min, followed by the incubation with horseradish peroxidase (HRP) coupled secondary antibodies (Cell Signaling, anti-mouse Cat. Nr. 7076, anti-rabbit Cat. Nr. 7074) for 1 hour at RT. The blots were washed again and developed with Immobilon Western chemiluminescent HRP substrate using a G:Box XT4 imager (Syngene, Cambridge, UK).
2.5. Transmission Electron Microscopy (TEM)
Fixed specimens at an optimal concentration were placed onto 400-mesh carbon/formvar coated grids and allowed to absorb to the formvar for a minimum of 1 min. After rinsing in 1x PBS and distilled water the grids were allowed to dry and stained for contrast using uranyl acetate. The samples were viewed with a Tecnai Bio Twin transmission electron microscope (TEI) and images were taken with an AMT CCD Camera (Advanced Microscopy Techniques, Woburn, MA, USA).
2.6. Flow Cytometry Analysis (FC)
Exosomes were attached to 4 mm aldehyde/sulphate latex beads (InvitrogenTM) by mixing 100 µl of exosomes solution with 10 µl volume of beads for 30 min at 4 °C. This suspension was diluted to a total of 1 ml with PBS and incubated for 30 min rotating at RT. The reaction was stopped adding 100 mM glycine and 2% BSA in PBS and further rotating for 30 min at RT. Exosome-bound beads were washed once in 5% Milk in PBS and centrifuged for 1 min at 14,800× g, blocked with 10% BSA / 1x PBS by rotating for 30 min at RT, washed a second time in 2% BSA / 1x PBS and centrifuged for 1 min at 14,800× g, and incubated with anti-GPC3 (R&D Systems, Minneapolis, MN, USA; AF2119, 15 µl of antibody in 15 µl PBS) for 30 min at 4 °C. Beads were centrifuged for 1 min at 14,800× g, the supernatant was discarded and beads were washed in 2% BSA and centrifuged for 1 min at 14,800× g. Alexa-488 secondary antibodies (Life Technologies, 3 µl of antibody in 20 ml of 2% BSA) were incubated for 30 min at 4 °C. Incubation of the secondary antibody alone was used as control to identify the population of beads with GPC3-bound exosomes. The percentage of positive beads was calculated to the total number of beads analyzed per sample (20,000 events) and therein referred to as the percentage of beads with GPC3+ exosomes.
2.7. Immunohistochemistry (IHC)
Immunohistochemical staining of GPC3 on formalin-fixed, paraffin-embedded whole tissue sections was performed as described previously [34]. All slides were stained in the Autostainer Lab Vision 408S using the anti-GPC3 antibody (dilution 1:25 in 1x PBS, MSK067-05; Zytomed, Berlin, Germany). The endogenous peroxidase was blocked with peroxidase block (CellMarque, Merck) and BrightVision + Poly-AP-anti mouse/rabbit IgG biotin-free antibody kit (VWRKDPVB110AP; ImmunoLogic, Palo Alto, CA, USA) was used. Two independent researchers blindly scored the expression of GPC3 and consensus was reached for each slide. The staining intensity of GPC3 was classified as absent: 0, weak: 1, medium: 2 and strong: 3. The scoring for GPC3 expression was based on several publications assessing GPC levels in a broad spectrum of tumor types [35,36,37,38].
2.8. CA19-9, CA72-4 and CEA and Immunoassays
Protein levels were measured in patients with GEA, individuals with non-malignant disease and healthy donors using a Human Sandwich-Chemiluminescence-Immunoessay for the cancer antigen CA19-9 (Diasorin, Saluggia, Italy; 314171), CA72-4 (Cobas, Roche, Basel, Switzerland; ms_11776258122V12.0) and the CEA (Diasorin, 314311) according to the manufacturer’s directions.
2.9. Statistical Analyses
All statistical analyses have been carried out with the IBM SPSS Statistics software version 25 (SPSS, Chicago, IL, USA), GraphPad Prism version 6 (GraphPad Software, San Diego, CA, USA) and MedCalc® (MedCalc Software bvba, Ostend, Belgium, Version 18.11.6). Categorical data were expressed as absolute and relative frequencies and compared using Chi-square-test or Fisher´s exact test. Continuous data were reported as arithmetic mean with the corresponding standard deviation. Analysis of variance (ANOVA) tests or student´s t tests were performed to calculate differences of multiple serum factors in human serum samples as wells as the area under the curve (AUC) through receiver operating characteristics (ROC) curve analysis. Differences between ROC curves were calculated MedCalc® software. Overall survival was calculated from the date of surgery for GEA to the date of death or the date of last follow-up information. The Kaplan-Meier method was used to construct survival curves that were compared using the log-rank test. A Cox proportional hazards regression analysis was used to assess independent predictors of overall survival. A p-value ≤ 0.05 was considered statistically significant.
3. Results
3.1. Study Design and Study Population
To conduct the study, we used a cohort of 49 patients with GEA who underwent surgical resection at the Department of General, Thoracic and Vascular Surgery of the University Hospital Carl Gustav Carus Dresden, Technical University Dresden, Dresden, Germany. Clinicopathologic characteristics of the GEA patients are summarized in Table 1.
Table 1.
Clinicopathologic characteristics of included patients.
| Characteristics | Total Number of Cases | eGPC3 | tGPC3 | ||||
|---|---|---|---|---|---|---|---|
| eGPC3 low | eGPC3 high | P-Value | tGPC3 low | tGPC3 high | P-Value | ||
| Age | |||||||
| < median = 63 years | 24 | 15 | 9 | 0.156 | 11 | 13 | 0.778 |
| ≥ median = 63 years | 25 | 10 | 15 | 13 | 12 | ||
| Gender | |||||||
| Male | 34 | 16 | 18 | 0.538 | 18 | 16 | 0.538 |
| Female | 15 | 9 | 6 | 6 | 9 | ||
| Classification of Lauren | |||||||
| Intestinal | 23 | 11 | 12 | 0.342 | 9 | 14 | 0.560 |
| Diffuse | 14 | 8 | 6 | 8 | 6 | ||
| Mixed | 5 | 1 | 4 | 2 | 3 | ||
| Missing | 7 | 5 | 2 | 5 | 2 | ||
| UICC stage | |||||||
| I and II | 37 | 18 | 19 | 0.742 | 20 | 17 | 0.321 |
| III and IV | 12 | 7 | 5 | 4 | 8 | ||
| Tumor size | |||||||
| T1 and T2 | 34 | 16 | 18 | 0.538 | 17 | 17 | 1.000 |
| T3 and T4 | 15 | 9 | 6 | 7 | 8 | ||
| Lymph node status | |||||||
| N0 and N1 | 39 | 9 | 15 | 0.089 | 20 | 19 | 0.572 |
| N2 and N3 | 10 | 16 | 9 | 4 | 6 | ||
| Metastasis status | |||||||
| M0 | 43 | 20 | 23 | 0.189 | 23 | 20 | 0.189 |
| M1 | 6 | 5 | 1 | 1 | 5 | ||
| Resection margin status | |||||||
| R0 | 43 | 20 | 23 | 0.189 | 22 | 21 | 0.667 |
| R1 and R2 | 6 | 5 | 1 | 2 | 4 | ||
| Tumor differentiation | |||||||
| G1 and G2 | 17 | 9 | 8 | 0.751 | 4 | 13 | 0.050 |
| G3 and G4 | 22 | 10 | 12 | 13 | 9 | ||
| CEA | |||||||
| < Median | 21 | 9 | 12 | 0.538 | 12 | 9 | 0.215 |
| > Median | 20 | 11 | 9 | 7 | 13 | ||
| CA 19-9 | |||||||
| < Median | 21 | 11 | 10 | 0.758 | 8 | 13 | 0.354 |
| > Median | 20 | 9 | 11 | 11 | 9 | ||
| CA 72-4 | |||||||
| < Median | 24 | 12 | 12 | 0.732 | 11 | 13 | 0.292 |
| > Median | 12 | 5 | 7 | 3 | 9 | ||
| Adjuvant therapy | |||||||
| Yes | 22 | 13 | 8 | 0.244 | 11 | 16 | 0.244 |
| No | 27 | 11 | 16 | 13 | 8 | ||
| Neoadjuvant therapyes | |||||||
| Yes | 27 | 16 | 11 | 0.256 | 9 | 13 | 0.393 |
| No | 22 | 9 | 13 | 15 | 12 | ||
Abbreviations: Glypican 3 (GPC3), tissue expression of GPC3 (tGPC3) and corresponding serum exosomes (eGPC3), carcinoembryonic antigen (CEA), carbohydrate antigen (CA), union of international cancer control (UICC). Significant p values are highlighted using bold font.
The pathological analysis revealed that the majority of patients were male (34/49), the intestinal, diffuse or mixed type of GEA was 47%, 29% and 10% of the cases according to the Lauren classification. Most of the patients presented with GEA of UICC stage I and II (37/49), tumor size of T1 and T2 (34/49) and a lymph node status of N0 and N1 (39/49). Six out of 49 patients had synchronous, metastatic disease and the median follow-up duration was 36.4 (0.1–93.3) months.
3.2. Confirmation of the Presence of Exosomes
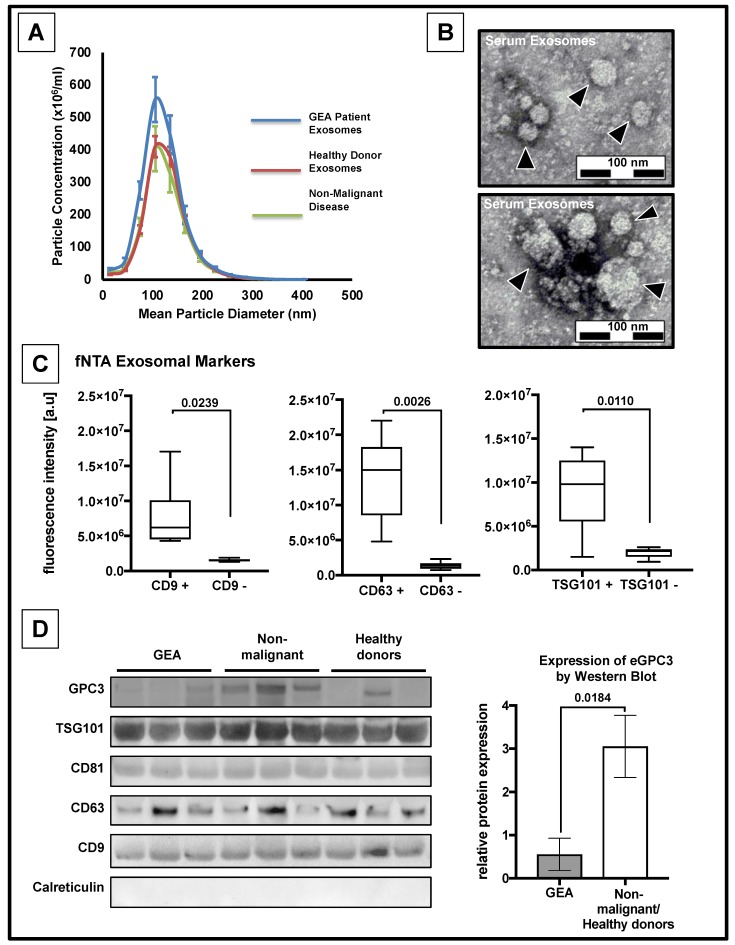
Initially, we sought to determine whether it is possible to measure exosomes from GEA patients. Therefore, serum and tissue samples from patients with GEA, serum samples from patients with non-malignant diseases (n = 25) and healthy donors (n = 31) were collected and subjected to exosome extraction. We performed nanoparticle tracking analysis (NTA) of selected specimens of 44 patients with GEA, 25 patients with non-malignant disease and 29 healthy donors. Moreover, we confirmed medium size of the isolated particles of about 105 nm in diameter in nine representative specimens of each subgroup of patients and healthy donors (Figure 1A), matching the size of exosomes ranging from 20 to 150 nm [8]. We observed an increased concentration of exosomes in patients with GEA in comparison to patients with non-malignant disease or healthy donors (p < 0.05, respectively) (Supplementary Figure S1A). In contrast, the median size of exosomes was significantly increased in healthy donors and patients with a non-malignant disease in comparison to patients with GEA (p < 0.05, respectively) (Supplementary Figure S1B). By using Transmission Electron Microscope (TEM), we were able to visualize the presence of vesicular structures with a bilayered lipid membrane and within the size range of exosomes (Figure 1B). To validate that these vesicles are exosomes, we analyzed the expression of exosomal marker proteins using immunoblotting and fluorescent nanoparticle tracking analysis (fNTA) (Figure 1C,D). We detected the presence of exosome-associated markers TSG101, CD9 and CD63 with both approaches, whilst CD81 was only detected by immunoblotting. To assess the expression of GPC3 on serum derived exosomes (eGPC3) in these samples, we performed immunoblot analysis of GPC3. eGPC3 was enriched in the samples from healthy donors and patients with a non-malignant disease compared to the samples from patients with GEA (p < 0.05, Figure 1D). To eliminate cross-contamination of proteins of endosomal origin, we stained for calreticulin and found no detectable protein expression for calreticulin suggesting that the levels of eGPC3 are indeed from the exosomes. Taken together, we were able to visualize exosomes in the serum of patients with GEA according to the recommendations for the characterization of exosomes [39,40].
Figure 1.
Decreased expression levels of eGPC3 in patients with gastro-esophageal adenocarcinomas (GEA). (A) Nanoparticle tracking analysis (NTA) diagram of nanoparticle concentration and average particle size from serum samples of patients with GEA (n = 9; blue line), healthy donors (n = 9; red line) and patients with non-malignant disease (n = 9; green line). (B) Transmission electron microscopy (TEM) images of serum derived vesicles (black arrowheads). (C) Quantification of exosome-associated proteins CD9, CD63 and TSG101 determined by fluorescent NTA (p < 0.05, two-paired student´s t-test). (D) Protein expression analyses of exosome-derived GPC3, TSG101, CD81, CD63, and CD9. Calreticulin was used as a marker for cytoplasm-derived contamination (bottom panel). Representative quantification of GPC3 levels of serum-derived exosomes from patients with GEA compared to serum-derived exosomes from healthy donors and patients with non-malignant disease (right panel, p < 0.05 two-paired student´s t-test).
3.3. eGPC3 Outperforms Current Serum Biomarkers of GEA And Negatively Correlates with Overall Survival
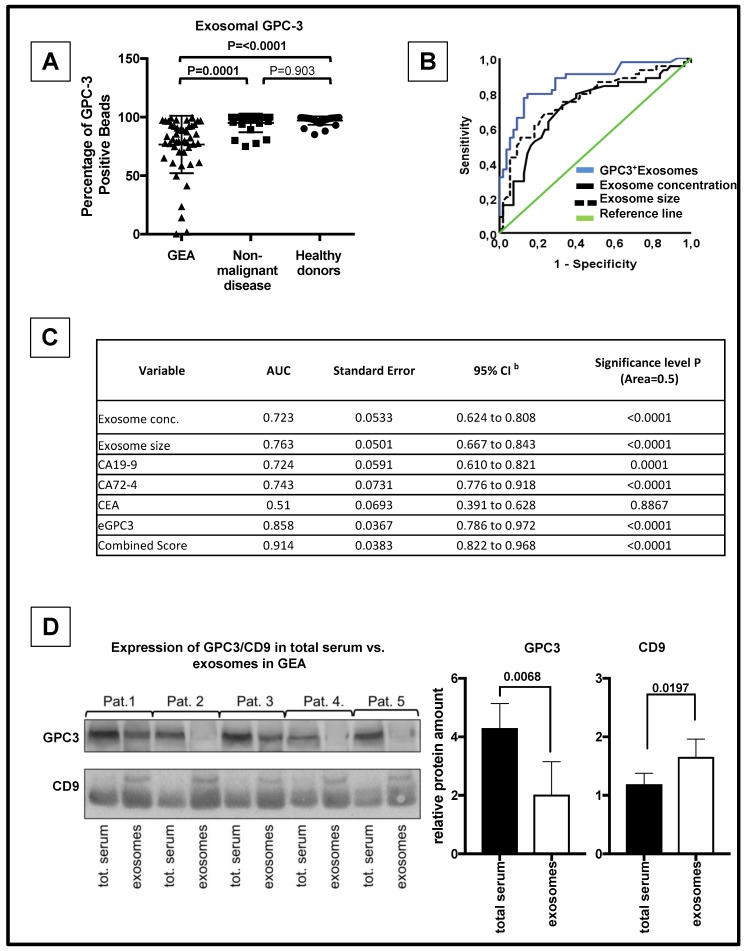
To determine whether eGPC3 expression can be used as a diagnostic marker of GEA, we conducted flow cytometry analysis of GPC3 on exosome-bound latex beads in healthy donors (n =3 1), patients with a non-malignant disease (n = 25) and patients with GEA from our Dresden cohort (n = 49). The count of GPC3 positive exosome-bound latex beads was significantly lower in GEA patients compared to healthy donors or patients with non-malignant disease (eGPC3: 97.04% in healthy donors, 95.03% in patients with non-malignant disease, 76.56% in GEA patients, ANOVA, p < 0.0001) (Figure 2A, Supplementary Figure S1C) confirming the results of the immunoblot analysis of exosome GPC3 shown in Figure 1D. ROC curve analysis was performed and resulted in an AUC of 0.85 with a sensitivity of 85.7% and a specificity of 75.5% for patients with GEA vs. control (healthy donors and patients with a non-malignant disease) (Figure 2B,C). In contrast, protein expression of routine biomarkers including CEA, CA 72-4 and CA 19-9 did not show any significant difference between our three different sample cohorts (Supplementary Figure S2A–C). A pairwise comparison of ROC curves revealed that the AUC of CEA and CA 19-9 was significantly inferior to the AUC of eGPC3 (p < 0.05, respectively) (Figure 2C, Supplementary Figure S2D,E, and Supplementary Figure S3A). In contrast, pairwise comparison of ROC curves between eGPC3 and CA 72-4 failed to be significant (p = 0.09) (Figure 2C, Supplementary Figure S2F and Supplementary Figure S3A). Intriguingly, by conducting a ROC curve analysis using a combined score of eGPC3, CA 72-4 and CA 19-9 ((eGPC3* CA 72-4)/CA 19-9), our data analysis yielded an AUC of 0.914 with a sensitivity of 80.6% and a specificity of 97.0% (Figure 2C). This combined score outperformed significantly the AUC of CA 72-4 and CA 19-9 (p < 0.05) but was not significantly increased when compared to the AUC of eGPC3 (p = 0.84) (Supplementary Figure S3A).
Figure 2.
Decreased serum levels of eGPC3 in GEA patients. (A) Dot plot analysis of the percentage of GPC3 positive exosome-bound latex beads in serum from patients with GEA, patients with non-malignant disease and healthy donors. (B) Receiver operating characteristics (ROC) curve analysis of eGPC3 (blue), exosome concentration (black), and exosome size (dashed line) for patients with GEA vs. patients with non-malignant disease and healthy donors. (C) ROC curve analysis showing the area under the curve (AUC) for eGPC3, exosome concentration, exosome size, serum CA 19-9, serum CA 72-4, serum CEA and a combined score (eGPC3* CA 72-4)/CA 19-9) for patients with GEA vs. patients with non-malignant disease and healthy donors. (D) Protein expression analysis of GPC3 and CD9 in total serum and corresponding exosomes from five patients with GEA (left panel). Quantification of GPC3 and CD9 protein expression is shown on the left panel (right panel; p < 0.05).
To further elucidate the decreased expression of eGPC3 in patients with gastric cancer, we sought to compare the expression of GPC3 and CD9 in total serum and corresponding isolated exosomes from five patients with GEA. Protein expression analysis showed increased GPC3 levels in the total serum fraction compared to the exosomal fraction (p < 0.05) (Figure 2D). In contrast, we found the exosomal marker CD9 was significantly elevated in the exosomal fraction than in the total serum fraction (p < 0.05) (Figure 2D). These data might suggest that eGPC3 in patients with GEA can be cleaved by different proteases in the bloodstream and is therefore mainly present in a soluble form.
3.4. eGPC3 Negatively Correlates with tGPC3 and Is Associated with Dismal Overall Survival
We next sought to determine whether the tissue expression of GPC3 (tGPC3) in our GEA patient cohort demonstrated any correlation with the eGPC3 levels. tGPC3 expression was examined at the protein level in paraffin embedded healthy appearing gastric tissue adjacent to the tumor and primary GEA tumors by immunohistochemistry. Expression of GPC3 was found in 79.6% of all cases (39/49) and absent in 20.4% (10/49) compared to healthy tissue, which only showed positive staining in 8% of cases (Supplementary Figure S3B, representative figures, right upper panel). Next, we dichotomized the positive GPC3 expression according to the staining intensity (Low: staining intensity 0–1 vs. high: staining intensity 2–3) and found 28.6% of all cases with low staining intensity and 51% of cases with strong staining intensity (Supplementary Figure S3B, representative images). Spearman correlation analysis showed a strong inverse correlation between expression of tGPC3 and GPC3 on exosome-bound latex beads in patients with GEA (correlation coefficient: −0.304, p = 0.034).
GPC3 has been described as a biomarker in several other cancer types. Therefore, we sought to identify clinical and histopathological prognostic markers in our cohort including the expression of exosomal and tissue GPC3. We correlated the expression of tGPC3 and eGPC3 to the clinicopathological data that was available. A summary of the results is shown in Table 1. We observed a significant correlation between tumor grade and high expression of tGPC3 (p = 0.050) whereas none of the other clinical or histopathological parameters showed a significant association. We further analyzed if grade 1 and 2 tumors showed a differential expression of tGPC3 and eGPC3 versus grade 3 and 4 tumors. However, this analysis revealed that there is no tumor grade-specific difference regarding tGPC3 or eGPC3 (Supplementary Figure S3C,D). Moreover, the Chi-Square test revealed that there is no significant correlation between tumor grading and tissue or exosome expression of GPC3 (Supplementary Figure S3E,F).
GPC3 has been reported to be a prognostic marker in several other cancer types. Therefore, we sought to identify clinical and histopathological prognostic marker in our cohort including the expression of exosomal and tissue GPC3. Univariate analysis revealed that lymph node status (p = 0.014), tumor size (p = 0.003), M stage (p = 0.003), L stage (p = 0.003), V stage (p = 0.001) and Resection status (p = 0.009) were associated significantly with overall survival (OS) (Table 2).
Table 2.
Univariate analysis (log-rank test) of prognostic parameters in GEA for median overall survival.
| Characteristics | 95%-Confidence-Interval | |||||
|---|---|---|---|---|---|---|
| No. of Cases | Mean Survival (Months) | Standard Error | Lower Limit | Upper Limit | P-Value | |
| Adjuvant Chemotherapy | ||||||
| Not received | 27 | 62.6 | 4.3 | 54.2 | 71.0 | 0.386 |
| Received | 22 | 69.6 | 8.4 | 53.1 | 86.1 | |
| Neoadjuvant Chemotherapy | ||||||
| Not received | 22 | 78.1 | 7.3 | 63.8 | 92.4 | 0.746 |
| Received | 27 | 68.8 | 6.5 | 56.1 | 81.4 | |
| Lymph node status | ||||||
| Negative | 24 | 87.9 | 4.0 | 80.0 | 95.8 | 0.014 |
| Positive | 25 | 56.2 | 7.4 | 41.6 | 70.7 | |
| Tumor size | ||||||
| T1 and T2 | 34 | 83.1 | 4.8 | 73.7 | 92.4 | 0.003 |
| T3 and T4 | 15 | 38.7 | 7.1 | 24.7 | 52.6 | |
| M stage | ||||||
| M0 | 43 | 76.2 | 5.3 | 65.8 | 86.6 | 0.003 |
| M1 | 6 | 9.7 | 0.000 | 9.7 | 9.7 | |
| L stage | ||||||
| L0 | 27 | 87.2 | 4.4 | 78.6 | 95.7 | 0.003 |
| L1 | 20 | 52.5 | 8.9 | 35.0 | 70.0 | |
| V stage | ||||||
| V0 | 39 | 81.6 | 4.8 | 72.3 | 91.0 | 0.001 |
| V1 | 8 | 26.4 | 7.1 | 12.5 | 40.2 | |
| Tumor differentiation | ||||||
| G1 and G2 | 17 | 70.4 | 6.2 | 58.3 | 82.6 | 0.432 |
| G3 and G4 | 22 | 71.1 | 8.0 | 55.4 | 86.8 | |
| UICC stage | ||||||
| I and II | 37 | 76.4 | 5.7 | 65.2 | 87.5 | 0.392 |
| III and IV | 12 | 46.7 | 8.1 | 30.7 | 62.6 | |
| Resection status | ||||||
| R0 | 43 | 77.8 | 5.2 | 67.50 | 88.0 | 0.009 |
| R1 and R2 | 6 | 24.0 | 8.6 | 7.1 | 40.9 | |
| eGPC3 | ||||||
| Low | 25 | 60.9 | 7.4 | 46.4 | 75.4 | 0.041 |
| High | 24 | 87.4 | 4.5 | 78.6 | 96.2 | |
| tGPC3 | ||||||
| Low | 24 | 84.7 | 4.9 | 75.0 | 94.4 | 0.044 |
| High | 25 | 58.0 | 8.2 | 41.9 | 74.2 | |
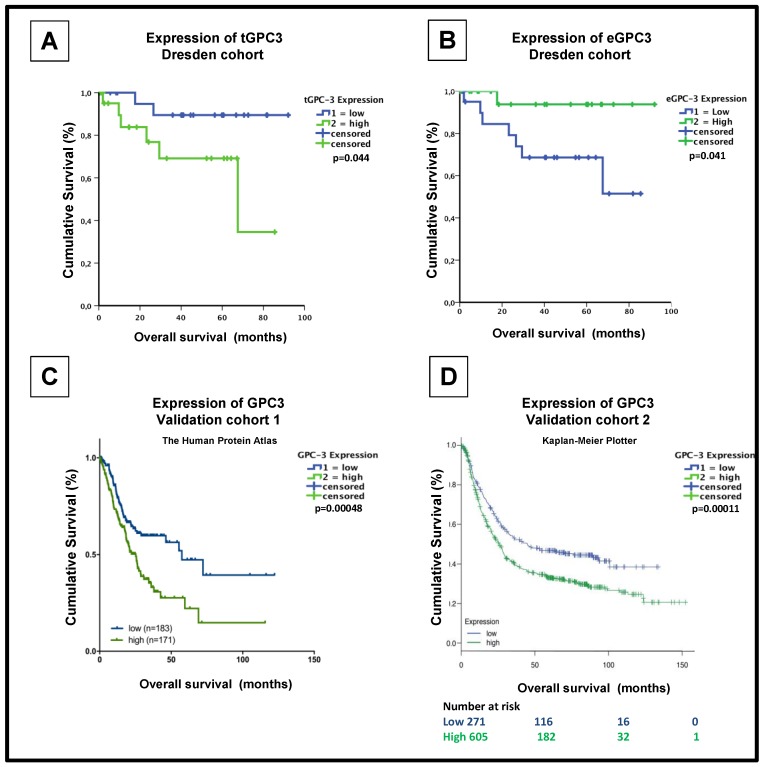
When we compared the differential expression of tGPC3, we found that it was associated with a dismal prognosis in patients with GEA (low expression of tGPC3: median OS: 84.7 months, high expression of tGPC3: median OS: 58.0 months, p = 0.044.) (Figure 3A, Table 2). Interestingly, univariate analysis showed that high expression of GPC3 on exosome-bound latex beads was associated with improved OS (low expression of eGPC3: median OS: 60.926 months, high expression of eGPC3: median OS: 87.403 months, p = 0.041) (Figure 3B, Table 2) Further univariate analysis revealed that there was no significant correlation between aberrant expression of CEA, CA 72-4 or CA 19-9 and overall survival (p > 0.05, respectively, Supplementary Figure S4A–C).
Figure 3.
Expression of GPC3 in GEA tumor tissues correlates to poor overall survival. (A) Kaplan-Meier curve (months) from Dresden GEA cohort. High expression of GPC3 protein levels (green line) on GEA tissue conferring a poorer prognosis compared to patients with low GPC3 expression (blue line). (B) Kaplan-Meier curve (months) depicting overall survival in patients with high (green line) and low (blue line) percentage of expression of exosome-bound latex beads stained against GPC3. (C) Kaplan-Meier curve (months) from the Human Protein Atlas conferring overall survival in patients with high (green line) and low (blue line) expression of GPC3 in primary GEA cancer (n = 354; source: v18.proteinatlas.org/ENSG00000147257-GPC3/pathology) [30]. (D) Kaplan-Meier curve (months) depicting overall survival in patients with high (green line) and low (blue line) mRNA expression of GPC3 in gastric cancer (n = 876; source: [31])
In multivariate analysis, high expression of tGPC3 remained formal statistical significance as a prognostic marker for poor OS in patients with GEA (p = 0.026; Table 3). However, multivariate analysis failed to reach the formal statistical significance between expression of GPC3 on exosome-bound latex beads and OS (p = 0.607; Table 3). To confirm the prognostic impact of tGPC3 from our GEA cohort, we used data from two independent publicly available databases. A cohort of 354 patients with GEA from the Human Protein Atlas [32] showed increased expression of tGPC3, which correlated with a poor OS (low expression of tGPC3: 5-year survival: 47%, high expression of tGPC3: 5-year survival: 22%, p < 0.0001; Figure 3C) [30]. The second validation cohort of 876 patients with GEA [31] also revealed that patients with high GPC3 mRNA levels had significantly lower OS (low expression of tGPC3: median overall survival: 45.1 months, high expression of tGPC3: median overall survival: 26 months, p = 0.00011; Figure 3D) [31].
Table 3.
COX proportional hazard models for multivariate analyses of eGPC3 and tGPC3 for overall survival.
| Characteristics | 95%-Confidence-Interval | ||||
|---|---|---|---|---|---|
| Category | P-Value | Hazard Ratio | Lower | Upper | |
| eGPC3 | High vs. low | 0.361 | 0.306 | 0.024 | 3.890 |
| tGPC3 | High vs. low | 0.026 | 30.334 | 1.497 | 614.752 |
| N | N0 vs. N+ | 0.781 | 0.635 | 0.026 | 15.587 |
| M | M0 vs. M1 | 0.992 | 0.000 | 0.000 | 0.000 |
| L | L0 vs. L1 | 0.016 | 68.320 | 2.204 | 2118.091 |
| V | V0 vs. V1 | 0.473 | 0.462 | 0.056 | 3.816 |
| R | R0 vs. R+ | 0.158 | 13.888 | 0.359 | 537.946 |
4. Discussion
In this study, we evaluated the expression of GPC3 on serum-derived exosomes and corresponding primary tumor tissue in patients with GEA. We demonstrate that increased expression of GPC3 in GEA tissue is associated with decreased overall survival in the Dresden cohort and two independent cohorts including a total of 1279 patients.
These data contradict two previously published reports that have proposed GPC3 acts as a tumor suppressor gene. The expression of GPC3 was markedly decreased in gastric but not in esophageal cancer [26], suggesting an active downregulation of GPC3 in gastric tumor cells. However, Zhu et al. only examined the RNA expression of GPC3 with no further investigation of whether mRNA is functionally expressed on the protein level [26]. The influence of post-translational modification may explain the discrepancy between our results and the findings by Zhu et al. In addition, Zhu et al. have investigated the expression of GPC3 in esophageal squamous and adenocarcinomas, whilst our cohort only consisted of gastro-esophageal adenocarcinomas. The difference in these histological subtypes may explain the discrepancy between Zhu et al. and our results. Additionally, Han et al. reported that GPC3 is absent in invasive gastric tumors and lymph node metastases implying GPC3 is a potent metastasis suppressor in GEA [27]. Their cohort included a heterogeneous group of patients with gastric adenocarcinoma including patients with signet ring cell carcinoma. The expression of GPC3 was much lower in signet ring cell carcinomas in comparison to other adenocarcinomas. As this histological subtype is known to be more aggressive and associated with a poor prognosis [41], it may have biased the results and led to a different conclusion with regards to our study. A recent study by Yamazawa and colleagues has investigated a panel of stem cell and oncofetal markers including GPC3 in 386 patients with gastric cancer [42]. In concordance with our current data, the authors show that positive staining for these panel markers including GPC3 was associated with poor prognosis and was an independent risk factor for disease-free survival. Intriguingly, our evaluation between the expression of GPC3 and histopathological data revealed high expression of tissue-derived GPC3 correlated with a lower tumor grade. This data is in contradiction to previous studies that show a positive correlation between the expression of GPC3 and tumor grading in other solid malignancies such as hepatocellular or urothelial carcinomas [43,44]. Our opposing findings could be explained by intratumoral sampling variability as it has been shown for hepatocellular carcinomas [45]. The variability of tumor grading in a heterogenous tumor bulk can lead to a biased interpretation of the results. To further assess whether the expression of GPC3 can be employed as a differential diagnostic marker for tumor grading, additional studies would be necessary with a more exclusive focus on the tissue expression of GPC3 in gastric cancer.
Our study shows that GPC3 is significantly decreased on serum-derived exosomes from GEA patients in comparison to healthy donors or patients with a non-malignant disease, when we analyzed the occurrence of GPC3 on circulating extracellular vesicles. These data concur with previous studies that provide evidence for a discrepant expression of proteins between tumor cells and corresponding serum exosomes. For example, CD47 is highly expressed in breast cancer tissue samples [46]. However, when Kibria et al. measured the expression of CD47 on circulating exosomes, this surface marker was unexpectedly detected in healthy donors and only minimal CD47 expression was observed in circulating exosomes from breast cancer patients [47]. Likewise, while the epithelial cell adhesion molecule (EpCAM) is strongly expressed in breast cancer tissue [48], it seems to be decreased on serum-derived exosomes of breast cancer patients [49]. Thus far, in our study, we have not further elucidated the inverse expression of GPC3 in tumor tissue and corresponding serum exosomes in GEA. However, we show that GPC3 is more abundant in total serum than in the exosomal fraction. Based on these findings, it could be hypothesized that GPC3 can be cleaved by different proteases in the bloodstream. This mechanism has been observed by Rupp et al. who have provided data that the expression of EpCAM on serum-derived exosomes is decreased since it is cleaved from exosomes via metalloproteinases in the circulation [49]. In this context, our findings would not be contradictory to studies that have described an overexpression of GPC3 in the serum of patients with hepatocellular carcinoma [50,51], assuming that the soluble form of GPC3 was measured. An alternative theory might be that the significant inverse correlation of tGPC3 and eGPC3 in patients with GEA is based on the active suppression of the presence of GPC3 in the systemic circulation by tumor cells. This suppression would prevent the degradation of these exosomes in the blood in order to deliver their tumorigenic load to distant organs for the formation of pre-metastatic niches. Moreover, the presence of GPC3 on serum exosomes in patients with GEA could be suppressed through a modified loading process of protein cargo including GPC3 into multivesicular bodies and exosomes. Villarroya-Beltri et al. described the modification of the loading process of exosomes in cancer cells [52]. This alternate loading of cancer cell derived exosomes may in turn contribute to our observation of the accumulation of GPC3 in GEA cancer cells. In this context, the low exosomal GPC3 levels could be linked to a failed release resulting in an intracellular accumulation, as we have observed in our cohort and as it has been described in the independent validation cohorts.
However, despite the lack of more profound information about the functional role of GPC3-positive exosomes in GEA, we show a potential role as a minimal-invasive prognostic biomarker that can be utilized for molecular risk stratification and personalized postoperative therapy guidance. Moreover, we show that low expression of GPC3 on serum exosomes is superior to standard serum markers such as CEA or CA 19-9 in order to discriminate between patients with GEA and the control group. Additionally, we can show that a combined panel of serum biomarkers including exosomal GPC3 can increase the diagnostic power for the non-invasive discrimination of patient with GEA vs. healthy donors of patients with a non-malignant disease. Therefore, though in isolation, GPC3 may not have clinical utility, it might be an interesting candidate for a multi-parametric exosomal biomarker panel to facilitate the diagnosis of GEA. For clinical integration of eGPC3 and tGPC3 as biomarkers for GEA, however, further validation of the above given findings in prospective clinical trials is necessary.
Acknowledgments
We would like to thank Thomas Kurth, Head of the Electron Microscopy and Histology Facility, Centre for Regenerative Therapies Dresden (CRTD) for technical help regarding the capture of electron microscope images.
Supplementary Materials
The supplemental data is available online at https://www.mdpi.com/2077-0383/8/5/696/s1, Figure S1: Comparison of mean exosome concentration and mean diameter in GEA patients, patients with non-malignant disease and healthy donors and representative histograms of flow cytometry analysis. Figure S2: Diagnostic significance of the expression of current laboratory markers for GEA in venous blood. Figure S3: Comparison of the AUC of ROC curves and impact of tumor grading on eGPC3 and tGPC3 expression. Figure S4: Expression of current laboratory markers for GEA in venous blood in association to overall survival outcomes.
Author Contributions
Conceptualization: M.R., J.W., N.N.R. and C.K.; Methodology: M.R., D.A., H.S., V.R., H.G., N.Z., N.N.R. and C.K.; Software: M.R. and C.K.; Validation: M.P., K.L., J.W., N.N.R. and C.K.; Data analysis: M.R., F.B. and C.K.; Investigation: M.R., D.A., H.S., O.T., A.C., T.T., V.R., N.Z., H.G. and K.L.; Resources: M.P., D.A., T.T., H.G., J.W. and C.K.; Data curation: M.R., H.G., O.T., A.C., K.L., F.B. and C.K.; Writing—original draft preparation: M.R.; Writing—review and editing: D.A., H.S., V.R., N.Z., K.L., J.W., N.N.R., F.B. and C.K.; Visualization: M.R. and C.K.; Supervision: J.W., N.N.R. and C.K.; Project administration: J.W., N.N.R. and C.K.; Funding acquisition: J.W. and C.K.
Funding
This work was supported by the Roland-Ernst-Stiftung.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; the collection, analyses, or interpretation of data; the writing of the manuscript, or in the decision to publish these results.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Steliarova-Foucher E., Lortet-Tieulent J., Rosso S., Coebergh J.W.W., Comber H., Forman D., Bray F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur. J. Cancer. 2013;49:1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 3.Van Cutsem E., Sagaert X., Topal B., Haustermans K., Prenen H. Gastric cancer. Lancet. 2016;388:2654–2664. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 4.Liu C., Yang Y., Wu Y. Recent Advances in Exosomal Protein Detection Via Liquid Biopsy Biosensors for Cancer Screening, Diagnosis, and Prognosis. AAPS J. 2018;20:41. doi: 10.1208/s12248-018-0201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siravegna G., Marsoni S., Siena S., Bardelli A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017;14:531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 6.Uchoa Guimaraes C.T., Ferreira Martins N.N., Cristina da Silva Oliveira K., Almeida C.M., Pinheiro T.M., Gigek C.O., Roberto de Araujo Cavallero S., Assumpcao P.P., Cardoso Smith M.A., Burbano R.R., et al. Liquid biopsy provides new insights into gastric cancer. Oncotarget. 2018;9:15144–15156. doi: 10.18632/oncotarget.24540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim D.H., Oh S.J., Oh C.A., Choi M.G., Noh J.H., Sohn T.S., Bae J.M., Kim S. The relationships between perioperative CEA, CA 19-9, and CA 72-4 and recurrence in gastric cancer patients after curative radical gastrectomy. J. Surg. Oncol. 2011;104:585–591. doi: 10.1002/jso.21919. [DOI] [PubMed] [Google Scholar]
- 8.Cocucci E., Meldolesi J. Ectosomes and exosomes: Shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25:364–372. doi: 10.1016/j.tcb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Kourembanas S. Exosomes: Vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu. Rev. Physiol. 2015;77:13–27. doi: 10.1146/annurev-physiol-021014-071641. [DOI] [PubMed] [Google Scholar]
- 10.Qu J.L., Qu X.J., Zhao M.F., Teng Y.E., Zhang Y., Hou K.Z., Jiang Y.H., Yang X.H., Liu Y.P. Gastric cancer exosomes promote tumour cell proliferation through PI3K/Akt and MAPK/ERK activation. Dig. Liver Dis. 2009;41:875–880. doi: 10.1016/j.dld.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Ohshima K., Inoue K., Fujiwara A., Hatakeyama K., Kanto K., Watanabe Y., Muramatsu K., Fukuda Y., Ogura S., Yamaguchi K., et al. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS ONE. 2010;5:e13247. doi: 10.1371/journal.pone.0013247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C., Liu D.R., Li G.G., Wang H.H., Li X.W., Zhang W., Wu Y.L., Chen L. CD97 promotes gastric cancer cell proliferation and invasion through exosome-mediated MAPK signaling pathway. World J. Gastroenterol. 2015;21:6215–6228. doi: 10.3748/wjg.v21.i20.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pitt J.M., Kroemer G., Zitvogel L. Extracellular vesicles: Masters of intercellular communication and potential clinical interventions. J. Clin. Investig. 2016;126:1139–1143. doi: 10.1172/JCI87316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberson C.D., Atay S., Gercel-Taylor C., Taylor D.D. Tumor-derived exosomes as mediators of disease and potential diagnostic biomarkers. Cancer Biomark. 2010;8:281–291. doi: 10.3233/CBM-2011-0211. [DOI] [PubMed] [Google Scholar]
- 15.Pinho S.S., Reis C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer. 2015;15:540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 16.Christianson H.C., Svensson K.J., van Kuppevelt T.H., Li J.P., Belting M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl. Acad. Sci. USA. 2013;110:17380–17385. doi: 10.1073/pnas.1304266110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melo S.A., Luecke L.B., Kahlert C., Fernandez A.F., Gammon S.T., Kaye J., LeBleu V.S., Mittendorf E.A., Weitz J., Rahbari N., et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang K.S., Im H., Hong S., Pergolini I., Del Castillo A.F., Wang R., Clardy S., Huang C.H., Pille C., Ferrone S., et al. Multiparametric plasma EV profiling facilitates diagnosis of pancreatic malignancy. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis J.M., Vyas A.D., Qiu Y., Messer K.S., White R., Heller M.J. Integrated Analysis of Exosomal Protein Biomarkers on Alternating Current Electrokinetic Chips Enables Rapid Detection of Pancreatic Cancer in Patient Blood. ACS Nano. 2018;12:3311–3320. doi: 10.1021/acsnano.7b08199. [DOI] [PubMed] [Google Scholar]
- 20.Sarrazin S., Lamanna W.C., Esko J.D. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ushiku T., Uozaki H., Shinozaki A., Ota S., Matsuzaka K., Nomura S., Kaminishi M., Aburatani H., Kodama T., Fukayama M. Glypican 3-expressing gastric carcinoma: Distinct subgroup unifying hepatoid, clear-cell, and alpha-fetoprotein-producing gastric carcinomas. Cancer Sci. 2009;100:626–632. doi: 10.1111/j.1349-7006.2009.01108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikeda H., Sato Y., Yoneda N., Harada K., Sasaki M., Kitamura S., Sudo Y., Ooi A., Nakanuma Y. alpha-Fetoprotein-producing gastric carcinoma and combined hepatocellular and cholangiocarcinoma show similar morphology but different histogenesis with respect to SALL4 expression. Hum. Pathol. 2012;43:1955–1963. doi: 10.1016/j.humpath.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 23.Hishinuma M., Ohashi K.I., Yamauchi N., Kashima T., Uozaki H., Ota S., Kodama T., Aburatani H., Fukayama M. Hepatocellular oncofetal protein, glypican 3 is a sensitive marker for alpha-fetoprotein-producing gastric carcinoma. Histopathology. 2006;49:479–486. doi: 10.1111/j.1365-2559.2006.02522.x. [DOI] [PubMed] [Google Scholar]
- 24.Kinjo T., Taniguchi H., Kushima R., Sekine S., Oda I., Saka M., Gotoda T., Kinjo F., Fujita J., Shimoda T. Histologic and immunohistochemical analyses of alpha-fetoprotein--producing cancer of the stomach. Am. J. Surg. Pathol. 2012;36:56–65. doi: 10.1097/PAS.0b013e31823aafec. [DOI] [PubMed] [Google Scholar]
- 25.Mounajjed T., Zhang L., Wu T.T. Glypican-3 expression in gastrointestinal and pancreatic epithelial neoplasms. Hum. Pathol. 2013;44:542–550. doi: 10.1016/j.humpath.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 26.Zhu Z., Friess H., Kleeff J., Wang L., Wirtz M., Zimmermann A., Korc M., Buchler M.W. Glypican-3 expression is markedly decreased in human gastric cancer but not in esophageal cancer. Am. J. Surg. 2002;184:78–83. doi: 10.1016/S0002-9610(02)00884-X. [DOI] [PubMed] [Google Scholar]
- 27.Han S., Ma X., Zhao Y., Zhao H., Batista A., Zhou S., Zhou X., Yang Y., Wang T., Bi J., et al. Identification of Glypican-3 as a potential metastasis suppressor gene in gastric cancer. Oncotarget. 2016;7:44406. doi: 10.18632/oncotarget.9763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costa J. Glycoconjugates from extracellular vesicles: Structures, functions and emerging potential as cancer biomarkers. Biochim. Biophys. Acta Rev. Cancer. 2017;1868:157–166. doi: 10.1016/j.bbcan.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 29.McShane L.M., Altman D.G., Sauerbrei W., Taube S.E., Gion M., Clark G.M., Statistics Subcommittee of the N.C.I.E.W.G.o.C.D. Reporting recommendations for tumor marker prognostic studies (REMARK) J. Natl. Cancer Inst. 2005;97:1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 30.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 31.Szasz A.M., Lanczky A., Nagy A., Forster S., Hark K., Green J.E., Boussioutas A., Busuttil R., Szabo A., Gyorffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The Human Protein Atlas Project. Knut & Alice Wallenberg Foundation; Stockholm, Sweden: 2017. Version: 18.1. [Google Scholar]
- 33.Kahlert C., Melo S.A., Protopopov A., Tang J., Seth S., Koch M., Zhang J., Weitz J., Chin L., Futreal A., et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem. 2014;289:3869–3875. doi: 10.1074/jbc.C113.532267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gailey M.P., Bellizzi A.M. Immunohistochemistry for the novel markers glypican 3, PAX8, and p40 (DeltaNp63) in squamous cell and urothelial carcinoma. Am. J. Clin. Pathol. 2013;140:872–880. doi: 10.1309/AJCP4NSKW5TLGTDS. [DOI] [PubMed] [Google Scholar]
- 35.Moek K.L., Fehrmann R.S.N., van der Vegt B., de Vries E.G.E., de Groot D.J.A. Glypican 3 Overexpression across a Broad Spectrum of Tumor Types Discovered with Functional Genomic mRNA Profiling of a Large Cancer Database. Am. J. Pathol. 2018;188:1973–1981. doi: 10.1016/j.ajpath.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 36.Chen G., Chen Y.C., Reis B., Belousov A., Jukofsky L., Rossin C., Muehlig A., Xu C., Essioux L., Ohtomo T., et al. Combining expression of GPC3 in tumors and CD16 on NK cells from peripheral blood to identify patients responding to codrituzumab. Oncotarget. 2018;9:10436–10444. doi: 10.18632/oncotarget.23830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirabayashi K., Kurokawa S., Maruno A., Yamada M., Kawaguchi Y., Nakagohri T., Mine T., Sugiyama T., Tajiri T., Nakamura N. Sex differences in immunohistochemical expression and capillary density in pancreatic solid pseudopapillary neoplasm. Ann. Diagn. Pathol. 2015;19:45–49. doi: 10.1016/j.anndiagpath.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 38.Tretiakova M., Zynger D.L., Luan C., Andeen N.K., Finn L.S., Kocherginsky M., Teh B.T., Yang X.J. Glypican 3 overexpression in primary and metastatic Wilms tumors. Virchows Arch. 2015;466:67–76. doi: 10.1007/s00428-014-1669-4. [DOI] [PubMed] [Google Scholar]
- 39.Witwer K.W., Soekmadji C., Hill A.F., Wauben M.H., Buzas E.I., Di Vizio D., Falcon-Perez J.M., Gardiner C., Hochberg F., Kurochkin I.V., et al. Updating the MISEV minimal requirements for extracellular vesicle studies: Building bridges to reproducibility. J. Extracell. Vesicles. 2017;6:1396823. doi: 10.1080/20013078.2017.1396823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lotvall J., Hill A.F., Hochberg F., Buzas E.I., Di Vizio D., Gardiner C., Gho Y.S., Kurochkin I.V., Mathivanan S., Quesenberry P., et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pernot S., Voron T., Perkins G., Lagorce-Pages C., Berger A., Taieb J. Signet-ring cell carcinoma of the stomach: Impact on prognosis and specific therapeutic challenge. World J. Gastroenterol. 2015;21:11428–11438. doi: 10.3748/wjg.v21.i40.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamazawa S., Ushiku T., Shinozaki-Ushiku A., Hayashi A., Iwasaki A., Abe H., Tagashira A., Yamashita H., Seto Y., Aburatani H., et al. Gastric Cancer With Primitive Enterocyte Phenotype: An Aggressive Subgroup of Intestinal-type Adenocarcinoma. Am. J. Surg. Pathol. 2017;41:989–997. doi: 10.1097/PAS.0000000000000869. [DOI] [PubMed] [Google Scholar]
- 43.Aydin O., Yildiz L., Baris S., Dundar C., Karagoz F. Expression of Glypican 3 in low and high grade urothelial carcinomas. Diagn. Pathol. 2015;10:34. doi: 10.1186/s13000-015-0266-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J., Zhang M., Ma H., Song X., He L., Ye X., Li X. Overexpression of glypican-3 is a predictor of poor prognosis in hepatocellular carcinoma: An updated meta-analysis. Medicine. 2018;97:e11130. doi: 10.1097/MD.0000000000011130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Senes G., Fanni D., Cois A., Uccheddu A., Faa G. Intratumoral sampling variability in hepatocellular carcinoma: A case report. World J. Gastroenterol. 2007;13:4019–4021. doi: 10.3748/wjg.v13.i29.4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willingham S.B., Volkmer J.P., Gentles A.J., Sahoo D., Dalerba P., Mitra S.S., Wang J., Contreras-Trujillo H., Martin R., Cohen J.D., et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc. Natl. Acad. Sci. USA. 2012;109:6662–6667. doi: 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kibria G., Ramos E.K., Lee K.E., Bedoyan S., Huang S., Samaeekia R., Athman J.J., Harding C.V., Lotvall J., Harris L., et al. A rapid, automated surface protein profiling of single circulating exosomes in human blood. Sci. Rep. 2016;6:36502. doi: 10.1038/srep36502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soysal S.D., Muenst S., Barbie T., Fleming T., Gao F., Spizzo G., Oertli D., Viehl C.T., Obermann E.C., Gillanders W.E. EpCAM expression varies significantly and is differentially associated with prognosis in the luminal B HER2(+), basal-like, and HER2 intrinsic subtypes of breast cancer. Br. J. Cancer. 2013;108:1480–1487. doi: 10.1038/bjc.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rupp A.K., Rupp C., Keller S., Brase J.C., Ehehalt R., Fogel M., Moldenhauer G., Marme F., Sultmann H., Altevogt P. Loss of EpCAM expression in breast cancer derived serum exosomes: Role of proteolytic cleavage. Gynecol. Oncol. 2011;122:437–446. doi: 10.1016/j.ygyno.2011.04.035. [DOI] [PubMed] [Google Scholar]
- 50.Zhu Z.W., Friess H., Wang L., Abou-Shady M., Zimmermann A., Lander A.D., Korc M., Kleeff J., Buchler M.W. Enhanced glypican-3 expression differentiates the majority of hepatocellular carcinomas from benign hepatic disorders. Gut. 2001;48:558–564. doi: 10.1136/gut.48.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu J.W., Zuo X.L., Wang S. Diagnosis accuracy of serum Glypican-3 level in patients with hepatocellular carcinoma and liver cirrhosis: A meta-analysis. Eur. Rev. Med. Pharmacol. Sci. 2015;19:3655–3673. [PubMed] [Google Scholar]
- 52.Villarroya-Beltri C., Baixauli F., Gutierrez-Vazquez C., Sanchez-Madrid F., Mittelbrunn M. Sorting it out: Regulation of exosome loading. Semin. Cancer Biol. 2014;28:3–13. doi: 10.1016/j.semcancer.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.