Abstract
The goal of this work was to develop a label free, comprehensive and reproducible high resolution LC-MS-based untargeted lipidomic workflow using a single instrument, which could be applied to biomarker discovery in both basic and clinical studies. For this, we have i) optimized lipid extraction and elution to enhance coverage of polar and non-polar lipids as well as resolution of their isomers, ii) ensure MS signal reproducibility and linearity, and iii) developed a bioinformatic pipeline to correct remaining biases. Workflow validation is reported for 48 replicates of a single human plasma sample: 1,124 reproducible LC-MS signals were extracted (median signal intensity RSD=10%), 50% of which are redundant due to adducts, dimers, in-source fragmentation, contaminations, or positive and negative ion duplicates. From the resulting 578 unique compounds, 428 lipids were identified by MS/MS, including acyl chain composition, of which 394 had RSD < 30% inside their linear intensity range, thereby enabling robust semi-quantitation. MS signal intensity spanned 4 orders of magnitude, covering 16 lipid subclasses. Finally, the power of our workflow is illustrated by a proof-of-concept study in which 100 samples from healthy human subjects were analyzed and the dataset investigated using three different statistical testing strategies in order to compare their capacity in identifying the impact of sex and age on circulating lipids.
Keywords: Lipidomics, Workflow, Mass spectrometry, Lipids, Untargeted, Lipid isomers, Coverage, Reproducibility, Annotation, WGCNA
INTRODUCTION
Lipidomics is a subset of metabolomics and an emerging discipline that aims at profiling and characterizing all lipids in a biological system. Lipids play crucial biological roles, as part of cell membrane structure, acting as signaling molecules, nerve conduction, etc. Hence, lipidomics offers great promise to extend our potential for the discovery of early disease biomarkers or new mechanisms that could be targets for therapy, as evidenced by studies in diabetes, cardiovascular and neurological diseases.1,2 Mass spectrometry-(MS)-based methods are most commonly used for lipidomics since they offer several advantages such as greater sensitivity and coverage, particularly when coupled to liquid chromatography (LC).3–6 However, the application of lipidomics remains challenging particularly due to the diversity of lipid classes in term of structures and polarity as well as the wide dynamic range of lipid concentrations in biological matrices, from pM to mM7 and the limited availability of commercial standards. Currently, the LIPID MAPS classification system8 subdivides lipids in 8 categories: fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, sterols, prenols, saccharolipids and polyketides. Adding to this diversity in structure and chemical properties, the nature and position (sn-1 vs. sn-2) of the acyl side chains on the head group as well as the localization of the double bond on these acyl side chains generate a multitude of isomers, which could have important different biological properties,9 while increasing the complexity of the analysis.
To tackle the challenges of lipidomics, both targeted and untargeted approaches are used. The targeted approach monitors selected known lipid species or classes of interest usually by multiple reaction monitoring (MRM).10,11 Its advantages are selectivity, specificity, as well as absolute and precise quantification using standards. Using this approach, 588 lipid species in human plasma have been quantitated using several methods and LC-MS platforms by the LIPID MAPS Consortium.12 The targeted approach is, however, limited on, a priori, selected lipids resulting in a low potential for new biomarker discovery. In contrast, an untargeted approach, which measures by survey scan all chemical compounds that are detectable in a single sample offers a greater potential as an unbiased discovery platform.
Most untargeted lipidomic workflows (reviewed in3,5,13,13) use Folch,14 Bligh-Dyer15 and, more recently, methyl tert-butyl ether (MTBE)16 for lipid extraction, followed by elution on a reverse phase C18 column and running time often >30 min for LC, plus electrospray ionization in positive and negative modes using high resolution for MS analysis. Some limitations and challenges exist, however, irrespective of workflows. These include: (i) the dynamic range of available MS instruments (~5-fold), which is less than the ~8-fold range12 for lipid concentrations in biological samples, (ii) limited lipid coverage, (iii) ion suppression resulting from co-elution of lipid species of high vs. low abundance, and (iv) variability in data processing.17 Furthermore, it is often difficult to discriminate the actual number of unique lipid species vs. the huge number of extracted MS signals that can be found in a given biological matrix using some untargeted lipidomic workflows. Lipids identification can also be problematic; annotation based solely on using accurate mass only with existing database can lead to numerous potential hits that cannot be distinguished even with the highest resolution MS instruments and is not equivalent to identification with MS/MS analysis. Recent studies have provided valuable insights into workflow harmonization through the comparison of results between (i) different laboratory-based methods (31 in total)18 and (ii) LC-MS instruments (9 in total)19 for the analysis of human plasma samples. Using MTBE-based extraction, the latter study reported between 200 and 300 unique lipids detected with relative standard deviation (RSD) < 30% for a 15-min gradient on a single instrument. Nevertheless, enhancing coverage of lipids measured on a single instrument, while meeting criteria for linearity, precision and reproducibility remains an important objective.
We herein present a comprehensive and reproducible label-free untargeted lipidomic workflow using a single high resolution LC-QTOF, which was developed to further optimize recovery, coverage and chromatographic resolution of both polar and non-polar lipids, lipid isomers, while minimizing false discoveries. Taking advantage of lipid isomers resolution under our optimized conditions, we emphasize the identification of acyl side chains though manual interpretation of MS/MS spectra.
MATERIALS AND METHODS
Materials
LC-MS reagent methanol, acetonitrile, isopropanol, ethyl acetate and HPLC grade MTBE were purchased from J.T. Baker (USA), HPLC grade hexane from Laboratoire Mat (Quebec, QC, Canada), HPLC grade chloroform and formic acid Fisher Scientific (Ottawa, ON, Canada) and ammonium formate from Sigma-Aldrich (St. Louis, MO, USA). Ultra-pure water was produced using a Milli-Q system (Millipore, Billerica, MA, USA). Synthetic lipid standards were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL, USA) and are listed in Supplemental Table 1. Lipids were dissolved in either hexane/isopropanol (3:4, v/v) or chloroform/methanol (2:1, v/v) at concentrations ranging from 5 mM to 5μM and stored at −80°C.
Human plasma and serum samples
Method optimisation and validation:
Protocol was approved by the Montreal Heart Institute Research Ethics Committee and written informed consent was obtained. Non-fasting blood samples were collected in EDTA tubes, and immediately put on ice for 30 min, prior to centrifugation at 2,000 g for 15 min. Plasma was immediately put on ice, divided in 125 μL aliquots prior to storage at −80°C until further analysis. To assess linearity and determine the amount of sample to be injected onto the LC-QTOF, loading experiments were conducted using 5 different volumes of the same plasma sample (n=3), corresponding to (in plasma equivalent): 0.13, 0.20, 0.26, 0.33, 0.40 μL in positive mode and 0.33, 0.46, 0.56, 0.66 and 0.76 μL in negative mode. To assess robustness and reproducibility, we have tested 48 replicates of a single human plasma sample (30 year-old healthy female), which were processed through the entire workflow, from extraction (which was split on two consecutive days), to data processing. Finally, paired plasma and serum samples from three individuals were also processed simultaneously for comparison of signals between these two matrices.
Proof-of-concept study:
The usefulness of our lipidomic workflow is illustrated using samples from a human cohort. Serum samples (100 μL) from 100 non-fasted healthy individuals North American or European background (53 females: median age: 43 years, interquartile range: 28–53; and 47 males: median age: 38 years, interquartile range: 26.5–48.5) were obtained from the NIDDK IBD Genetics Consortium Biobank (http://ibdgc.uchicago.edu/data/repository.html) and that were collected to match inflammatory bowel disease (IBD) cases (not studied herein). All subjects provided informed consent. Recruitment protocols and consent forms were approved by local Institutional Review Boards at all participating institutions. All serum samples and data in this study were denominalized.
Lipidomic workflow using LC-QTOF
This section reports the final validated protocols that were selected for our untargeted label-free lipidomic workflow using high resolution LC-QTOF and which were used for its validation and application. For the lipid extraction and chromatographic elution steps, the optimized protocols were also compared to selected published ones. For the MS data processing, the bioinformatic pipeline was optimized through manual curation. Details about the experimental procedures used for method comparisons as well as about the various steps of the bioinformatic pipeline are in the Supplementary file.
Sample preparation:
Extraction of lipids from plasma or serum samples spiked with 6 lipid standards (2.5 nmol of monoacylglycerosphosphocholine (LPC) 13:0, diacylglycerophosphocholine (PC) 14:0/14:0 and 19:0/19:0, phosphatidylserine (PS) 12:0/12:0, diacylglycerophosphopethanolamine (PE) 17:0/17:0, and diacylglycerophosphoglycerol (PG) 15:0/15:0)) was achieved using a modification of the MTBE-based protocol of Matyash et al.,16 as follows: Step 1: Plasma samples (100 μL) were added to 15 mL glass tubes containing lipid standards that had first been dried under nitrogen and resolubilized in 750 μL methanol followed by 20 μL 1M formic acid. Tubes were vortexed 10 sec prior to addition of (i) 2.5 mL MTBE and (ii) deionized water (625 μL), followed each time by mixing with a multi-pulse vortexer (Glas-Col, Terre-Haute, IN, USA) for 5 and 3 min, respectively. Samples were centrifuged at 1000 g for 5 min and the upper phase was collected. Step 2: The lower phase was subjected to three additional extractions each including 3-min vortex and 5-min centrifugation steps with: (i) 2 mL of the upper phase of a prepared solution of MTBE/methanol/water (10:3:2.5, v/v/v), (ii) 2 mL ethyl acetate containing saturated NaCl and 10 μL HCl 10 M and (iii) 2 mL ethyl acetate. Step 3: The four upper organic phases were combined and dried overnight in a vacuum centrifuge (Christ RVC 2–25, Osterode, Germany). Lipids were resolubilized by sequential addition of 200 and 100 μL methanol/chloroform (2:1, v/v); following each addition, the sample was subjected to vortex (8 sec), sonication (5 min), vortex (15 sec) and centrifugation (30 sec) prior to transfer to a glass vial (Agilent Technologies Inc., Santa Clara, CA, USA). Then, the combined volume for each sample was centrifuged 3 minutes before being aliquoted in 6 glass vials and stored at −80°C until analysis.
LC-MS analyses:
Frozen samples were thawed on ice, sonicated 1 min and vortexed 30 sec before injection onto the LC-MS. Samples were injected on a 1290 Infinity HPLC coupled with a 6530 (for workflow validation and lipidome characterization) or 6550 (workflow application) QTOF equipped with a dual ESI source (Agilent Technologies Inc) and analyzed in positive and negative mode. Injected sample volumes, which were optimized in loading experiments to minimize MS signal saturation while maximizing lipid coverage, were: 1 and 0.7 μL in the positive mode for workflow validation and application respectively, and 2 μL in the negative mode in both cases (corresponding to 0.33, 0.23, and 0.66 μL of plasma-equivalent, respectively). Lipids were eluted on a Zorbax Eclipse plus C18, 2.1 × 100 mm, 1.8μm (Agilent Technologies Inc., Santa Clara, USA), heated at 40°C with a constant flow rate at 0.45 mL/min, using a gradient of mobile phase A (0.2% formic acid and 10 mM ammonium formate in water), and B (0.2% formic acid and 5 mM ammonium formate in methanol/acetonitrile/MTBE, 55:35:10 (v/v/v)). The solvent program was: 0–2 min, 50% B; 2–11 min, 50–85% B; 11–33 min, 85–94% B; 33–35 min, 94–98% B, 35–38 min; 98–99.5% B; 38–82 min, 98–99.5% B; 82–83 min, 99.5–50% B, plus an equilibration of 8 min. MS conditions were: gas source temperature: 325°C; drying gas rate: 8 L/min; nebulizer: 35 psi; spray voltage: +/− 3500 V; cone voltage: 150V. Mass correction was applied during data acquisition using 2 reference compounds (Agilent Technologies Inc.): C5H4N4 and C18H18O6N3P3F24. Mass spectra were acquired from mass-to-charge ratio (m/z) 50 to 1700 and MS scans were collected during 1 sec. Samples were analyzed in randomized order and MS data were acquired sequentially in the positive and negative mode. Quality control (QC) was assessed by: (1) Injecting 3 control samples (mix of 6 internals standards) before starting the project to assess RSD; (2) Injecting “in–house” plasma pool QC sample at the beginning, the end and at every 20 runs; (3) Injecting blanks every 20 runs; (4) Monitoring the 6 internals standards spiked in samples for signal intensity, mass accuracy and retention time (RT).
MS data processing:
All LC-MS results pertaining to a given experiment were analyzed using the software package of Mass Hunter Qualitative Analysis (version B.06 for 6530-QTOF, and version B.07 for 6550-QTOF, Agilent, Santa Clara, USA) for peak picking. In addition, we have developed a bioinformatic script, which is encoded in Perl language and includes several steps in R language such as an algorithm to optimize MS peak alignment between chromatographic runs. The data processing steps can be summarized as follows (cf. Supplemental file for more details): Step 1: MS features, defined by mass-to-charge ratio (m/z), signal intensity and RT, were extracted from the raw MS signals for each data run using the peak picking Mass Hunter Molecular Feature Extractor (MFE); Step 2: MS features between chromatographic runs were aligned by selecting features that were present in all samples with no isobars (defined by mass ± 20 ppm) in a RT window of ± 2 min, spread over the gradient as references for RT correction prior to MS features alignment; Step 3: Recursive peak extraction on MS features was performed using Mass Hunter Find by Formula in order to minimize missing data; Step 4: The refined set of features from all runs was re-aligned using endogenous reference compounds as described in step 2; Step 5: Duplicate features filtering; Step 6: Features coming from multiples adducts ([M+Na]+, [M+K]+, [M+NH4]+, [M+NaCOOH]+ in positive mode and [M+FA]− and [M+NaCOOH]− in negative mode) were filtered by retaining in the final dataset only the highest MS intensity signal from each cluster, since errors can be created by pooling intensities, which are not in the linear range20; Step 7: Abundance values for all MS features from both negative and positive ionization were merged; Step 8: Filter of presence; Step 9: Normalization using cyclic loess21 algorithm (R library Limma22 from bioconductor); Step 10: Imputation of missing values using k-nearest neighbour on scaled data (R library impute23 from bioconductor, k=5); Step 11: Batch effect correction using Combat.24 The resulting dataset, which lists features detected in a given experiment, with their mass, corrected signal intensity and RT, is ready for further data analysis.
Lipid identification with acyl side chains for plasma lipidome characterization:
Data-dependent MS/MS scans were collected during 500 ms on each m/z selected with a RT window of 4 minutes. The resulting MS/MS files were aligned with the previously acquired MS files from the same study. Then, the MS/MS scans corresponding to selected features were interpreted manually in silico similar to Godzien et al.25 Briefly, identification of lipid structure, including fatty acyl side chains, was performed as follows. First, the m/z of selected features listed in the dataset resulting from data processing was searched in METLIN or LIPID MAP databases, considering that the highest intensity ion observed for the following lipid (sub)class lipids are: (i) in positive ionization, PC, PE, sphingomyelin (SM): as [M+H]+; fatty Acids (FA), phosphatidylinositol (PI): [M+NH4]+; steryl esters (CE), glycerolipids: [M+NH4]+ or [M+Na]+; and (ii) in negative ionization: FA, PE, PI: [M-H]−; PC, SM, Ceramide (Cer): [M+HCOO]−. Second, from the MS/MS spectra, the polar head group was identified by their signature fragment ion: (i) in positive ionization: PC, LPC, SM: m/z 184.073; PE, LPE: from the loss of 141.019; PS: from the loss of 185.009; CE: m/z 369.352; (ii) in negative ionization: PI: m/z 241.011. Finally, the acyl chains were identified by their representative fragment ion: (i) in positive ionization: sphingolipids: RCN+ (e.g. 264.269 for d18:1) and for glycerolipids: from the loss of RCOO; and (ii) in negative ionization: glycerophospholipids: RCOO− (e.g. 281.247 for 18:1 acyl chain). If only one acyl chain fragment ion was observed, the identity of the other acyl chain was deduced by accurate mass search in METLIN or LIPID MAP databases. A putative identification of the sn-1 and sn-2 position of fatty acyl side chains of all lipid classes, except lysophospholipids, was also attempted by considering the relative intensity of fragments from the acyl chains reported using lipid standards. For glycerophospholipids, the peak intensity for the sn-2 fragment is stronger in negative ionization mode,26,27 except for those containing as side chains a saturated plus a highly polyunsaturated fatty acids28 and for PS.29 Similarly, for triacylglycerols, the sn-2 fragment gives the lowest signal intensity.30 In some cases, lipid structure identification could be ascertained using the retention time behavior,31,32 which predicts that there is a linear relationship between the RT (x axis) and mass (y axis) of all lipids within a given class and which depends on the carbon chain length and degree of unsaturation of the two acyl side chains. Since acyl side chain positions were not validated with synthetic standards, glycerophospholipids and glycerolipids were annotated along with the identified structure for the acyl side chains using the underscore convention, and reported with their highest intensity MS/MS fragment.
Lipid annotation and identification for the proof-of-concept study:
For annotation of lipids in the workflow application step, the MS dataset obtained from the analysis of samples from the 100 healthy subjects from the NIDDK IBD Genetics Consortium was aligned manually with the one obtained for the 48 replicate samples for which we had characterized the lipidome in the previously described method validation step. From the generated list of lipids annotated though manual peak alignment; we selected 25 of them as being representative of lipid subclasses distribution in the whole workflow application dataset for further validation of their identity by MS/MS.
Statistical analysis
Workflow validation:
The output text file containing the processed dataset, was imported into Mass Professional Pro (MPP: version 12.6.1; Agilent Technologies Inc.) software to calculate individual RSD and average intensity values for each feature.
Proof-of-concept study:
The dataset was investigated using the following three different statistical testing strategies to compare their capacity in identifying the impact of sex and age on the circulating lipidome. Strategies 1 and 2 are commonly used, while strategy 3 has only been recently applied to metabolomics data. (a) Strategy 1: Independent testing of each feature using regression analysis. (b) Strategy 2: Biologically informed grouping of MS features based on subclass of lipids and their fatty acyl groups, followed by within-group principal component analysis. Each class was then represented by its first two principal components, and then tested for association with the sex and age variables. (c) Strategy 3: Unsupervised and completely data-driven grouping of correlated MS features into modules, using weighted correlation network analysis (WGCNA) R tool. Each module is then represented as its first two principal components and tested for association with the variables sex and age. WGCNA33,34 has been created for and mostly applied to the analysis of gene expression data but, more recently, also to metabolomics data.33,35–38 Significance thresholds and multiple testing: A suggestive significance threshold of P-value < 0.05 was chosen instead of a very stringent one, given the correlated nature of MS features and in order to optimize power and maximize the potential of discovery for this limited-scale study (n=100). Multiple testing was accounted for independently for each strategy and each phenotype, by evaluating false discovery rate (FDR), which is relevant in a context of discovery and exploratory analyses. For strategy I, we report q-values as an estimate of the FDR. For strategy II and III, computing q-values could have been misleading, because there are fewer tests, non-trivial correlation structure and high enrichment of lower P-values. Marginal P-value threshold of 0.05 and R2 > 0.2 for one of the principal component were used to declare association between a group or module and a phenotype. A clear enrichment is observed using these criteria, which indicates low FDR. As an example, if we consider association to age, for strategy II there are 20 groups (each with 2 tests) one would expect about 2 groups to be false positives in average if all these tests were independent and under the null. We observe 10 groups to be significant, which is much more than expected under the null.
RESULTS AND DISCUSSION
Overview
We have developed a label free untargeted LC-MS-based lipidomic workflow that combines the following characteristics: (i) untargeted MS data acquisition to enable its use in biomarker discovery, (ii) optimized extraction recovery across multiple classes of polar and non-polar lipids (for e.g. glycerophospholipids vs. glycerolipids), (iii) chromatographic resolution within lipid isomers characterized by fatty acyl side chains of different chain length (for e.g. PC(18:2_20:4) and PC(16:0_22:6)), or position of double bonds (for e.g. PC(18:0_20:4n-3) and PC(18:0_20:4n-6)), (iv) data processing method that incorporated scripts to optimize MS data alignment, filtering and correction, and (v) lipid identification, including acyl side chains, through MS/MS. The performance characteristics of this workflow as well as characterization and reproducibility of all extracted LC-MS signal and referred thereafter as features are presented. Thereafter, the usefulness and power of the workflow in identifying changes in circulating lipid isomers are illustrated through the comparison of sex and age in a cohort of 100 healthy human subjects.
Workflow optimisation
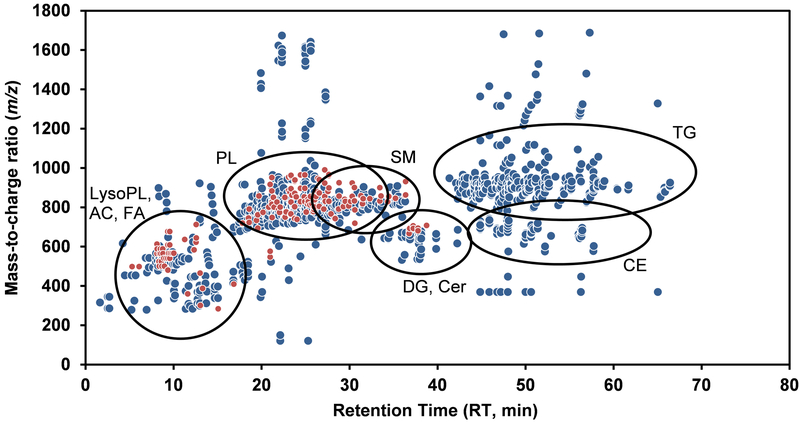
A two-dimensional map (m/z vs. retention time (RT)), presented in Figure 1, depicts the 1089 features detected throughout the LC-MS analysis performed in positive mode (blue) and negative mode (red) with our optimized workflow. In brief, the developed LC-MS method enables detection of a wide range of both polar and non-polar lipid subclasses, 16 in total (cf. Section on “Characterization of the Human Lipidome” and Table 1 for more details). While the elution pattern of these lipids resembles those reported by others under similar conditions,31,39 we have optimized conditions for the lipid extraction and chromatographic elution steps to enhance recovery, coverage and chromatographic resolution of MS features as described in details in the Supplementary file. In addition, to enable robust MS data processing and thereby minimize false discovery, we have developed a bioinformatic pipeline, which complements the Agilent softwares, and includes several scripts that covers MS data extraction, alignment, filtration and correction. Despite using an 83-min LC-MS gradient, we also found that one challenging and crucial step was MS peak alignment; this is often confounded by the presence of multiple isomers with similar elution time (Supplemental Figure S-4). Hence, a RT correction algorithm was developed, which enables alignment using multiple endogenous compounds as reference (cf. Supplementary file for details). Finally, following data normalization, we included an algorithm to account for batch effect, since we observed some inter-day variations in sample preparation (Supplemental Figure S-5).
Figure 1. Representative two-dimensional map (m/z vs. retention time (RT)) of extracted MS signals or features obtained in a human plasma sample processed through our workflow.
This figure shows the 1089 features (defined by mass-to-charge ratio (m/z), signal intensity and RT; depicted as circles) acquired under our LC-MS conditions in positive (blue) and negative (red) ionization mode. The main (sub)classes of lipids detected as well as their elution time period are highlighted. Details about the identified lipids are provided in Table 1 and in the Supplemental Excel file. AC: Acylcarnitine, CE: steryl esters, Cer: ceramides, DG: diacylglycerols, FA: fatty acyls, LysoPL: lysophospholipids, PL: glycerophospholipids, SM: sphingomyelins and TG: triacylglycerols.
Table 1.
Number of lipids identified for robust semi-quantitation in human plasma sample according to the various (sub)classes.
| Category | (Sub)class | identified lipids # | semi-quantifiable lipids # |
|---|---|---|---|
| Fatty acyls (FA) | Fatty acids and conjugates Fatty acylcarnitines |
7 2 |
6 2 |
| Glycerophospholipids (GP) | Monoacylglycerophosphocholines (LPC)
Monoacylglycerophosphoethanolamines
(LPE) Diacylglycerophosphocholines (PC) Diacylglycerophosphoethanolamines (PE) Diacylglycerophosphoinositols (PI) Diacylglycerophosphoserines (PS) |
41 8 150 (+1)* 12 15 1 |
32 8 145 12 14 1 |
| Sphingolipids (SP) | Sphingomyelins (SM) Ceramides (Cer) Neutral glycosphingolipids |
38 (+3)* 9 7 |
40 6 7 |
| Sterols Lipids (ST) | Steryls esters (CE) Cholesterol & derivatives |
13 2 |
6 2 |
| Glycerolipids (GP) | Diacylglycerols (DG) Triacylglycerols (TG) |
6 116 |
5 107 |
| Prenol Lipids (PR) | Ubiquinones | 1 | 1 |
| TOTAL | 432 | 394 |
Data processing of the 1,624 MS features acquired over the 48 LC-MS analysis of replicates of human plasma samples resulted in 1,124 MS features following filtering and removal of adducts, of which 428 were identified by MS/MS and 4 (indicated by *) with RT behavior. This table reports the number of lipids identified by category and (sub)classes. The complete list of identified lipids is provided in the Supplemental Excel file.
Finally, for acyl side chain identification, although we initially used SIMLipid440 and LipidBlast41 with some success to identify some unknown lipids through matching of peaks resulting from MS/MS fragmentation with a database entry, we have chosen to manually interpret each MS/MS spectrum. This was important to ensure optimal identification of lipids, especially their acyl chains given the presence of (i) multiple lipid isomers which produced multiple matches, and (ii) non-specific ion fragments (for e.g. m/z 184,073 for PC and SM). Furthermore, we were able to take advantage of the property of some lipids to provide information in both ionization modes. For example, while PCs have better signal intensity in positive mode, their MS/MS fragmentation provides information on their acyl chain composition only in the negative mode. Finally, we were also able to putatively assign acyl chain position, although this was found to be more difficult in complex matrices such as plasma or serum than with standards due to the low intensity of some lipids and the presence of numerous co-eluting lipids.
Workflow validation
Based on results from loading experiments (cf. Supplemental file and Supplemental Table S-3), a volume of 1 and 2 μL was selected for validation in the positive and negative ionization mode (corresponding to 0.33 and 0.66 μL of plasma-equivalent). Injection of volume > 2 μL did not result in any signal saturation in the negative ionization mode, albeit we noticed some carry over with time. Approximately 70% of the lipid features measured have a linear correlation (vs. injected volume) R2 > 0.98, and 98.5% have R2 > 0.96; those with lower R2 (0.98–0.96) are predominantly of low MS signal intensity. A linear dynamic range of almost 4 orders of magnitude was obtained. Using the 48 human plasma sample replicates, a total of 1,624 MS signals or features, defined by a mass-to-charge ratio (m/z) and RT, were detected over the 48 samples, of which 1,124 features (895 positive ions and 229 negative ions) remained after data filtering for presence in 80% in all samples and elimination of features coming from selected adducts (cf. Materials and Methods) retaining only the feature with the highest intensity signal from each adduct’s cluster. The median intra- and inter-assay coefficients of variations (RSD) of raw data for each of the 1,124 features detected over the 48 samples were, respectively, 14.4 % and 15.7%. Following data processing, these RSDs decreased to 9.5% and 9.7%, respectively. Median RSD for RT values assessed for 497 reference features (cf. Supplemental Files for details on reference features), representative of the dataset, was 0.2%. Overall, these results demonstrate the robustness and excellent reproducibility of our untargeted lipidomic workflow, thereby providing good confidence for the application of this label-free approach for semi-quantitative analysis of lipid signals in plasma under various conditions.
Characterization of the human circulating lipidome
Prior to the application of our untargeted lipidomic workflow to a specific discovery-based study, we have ascertained the quality of the final dataset obtained from the analysis of the 48 human plasma sample replicates by characterizing the lipidome. Specifically, from the 1,124 MS features that remained after robust data processing, we aimed to (i) determine the proportion of unique MS compounds, and (ii) identify these compounds, which include characterization of the acyl side chains of the various lipids.
Number of features vs. unique lipids detected:
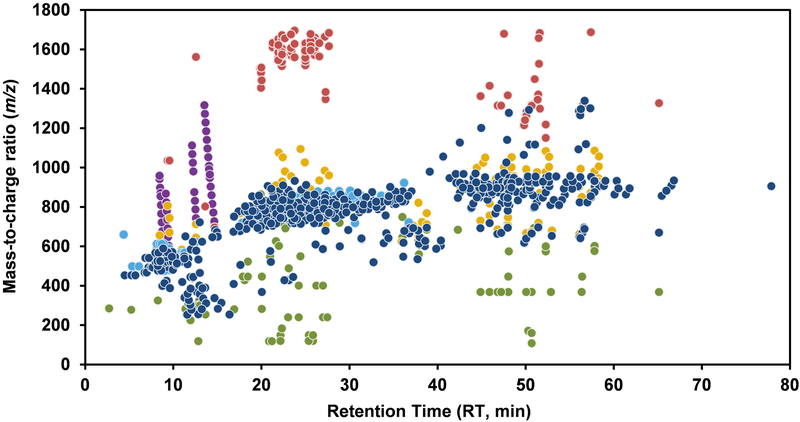
Given that all MS features detected do not correspond to unique lipid species, we aimed to identify and eliminate redundant ones. From the 1,124 MS features aligned and filtered through data processing, >50% were from the following sources or processes as illustrated in Figure 2: (A) Contamination: We found 46 contaminating ions from polymers (PEG: repeating mass unit of 44.026); (B) Duplicate ions: Through manual inspection of the extracted ion chromatogram, we found duplicates of the same feature based on several criteria such as similar peak shape and identical RT values. These duplicates ions were attributed to the following processes: (i) Dimerization (102 features): this occurs during the ionization process with ESI;42 it is concentration-42,43 and instrument-42,44 dependant and can be reduced by tuning the cone voltage.42,43 Dimers can be homodimers (for e.g. 2 PC) or heterodimers (for e.g. steryl ester plus cholesterol). (ii) In-source fragmentation (95 features): this is concentration- and lipid-dependant and can also be reduced by adjusting the cone voltage45. (iii) Adduct formation (151 features): while 287 adducts were filtered during data processing, there were 151 features that remained and were attributed mostly to unknown adducts or their combination resulting by an added mass of 37.065 (39 features) and 59.047 (36 features) in positive mode and mass 203.962 in negative mode (43 features). (iv) Redundancy between positive and negative ionization signals (150 features): some lipid species such as fatty acids, glycerophospholipids and sphingolipids, are detected in both ionization modes. Theses replicates have a mass difference of 2.02 (H2) for PE, 17.026 (NH3) for FA and PI and 43.989 (COO) for PC, SM and Cer and present similar chromatographic profiles. Removing all duplicates and contaminant MS signals from the initial 1,124 MS features resulted in 580 unique compounds. It is noteworthy that dimer formation and in-source fragmentation are processes that are concentration-dependant and, hence, injecting a larger sample volume will increase the number of features detected but will also result in a greater proportion of redundant features vs. unique compounds.
Figure 2. Two-dimensional maps depicting the different sources of MS signals.
This figure shows the 1,124 features (shown as filled circles of different colours) detected in 80% of 48 replicates of human samples using our LC-MS conditions. The figure highlights in blue 580 features identified as unique lipids and 544 other features classified as follows: contaminants (46 features, purple), dimers (102 features, red), in-source fragmentation (95 features, green), adducts not filtered out by selecting known adducts with our data processing pipeline, (151 features, yellow) and duplicates in positive and negative mode (150 features, turquoise).
Identification of human plasma lipids including their acyl side chains:
The aforementioned 580 unique compounds were processed for annotation using METLIN and LIPID MAPS databases and subsequent identification by MS/MS. Among them, 134 features were classified as unknown since they had no valid correspondence at +/− 5ppm in databases when taking into consideration the proposed adduct for the lipid class, for example a [NH4]+ adduct for PC is unlikely. Targeted MS/MS was achieved for all remaining 446 features. When a given feature was annotated to a lipid, which ionizes in positive and negative mode, MS/MS was achieved in both modes to complement the interpretation process. Finally, 428 different lipids were identified, namely 9 fatty acyls, 227 glycerophospholipids, 54 sphingolipids, 15 sterols, 122 glycerolipids and 1 prenol lipid (Table 1). A complete list of identified lipids is provided in the Supplementary Excel file. It is noteworthy that we report only one PS. This subclass of glycerophospholipids is less abundant than PC, PE and PI12 and probably resulted in too low signal intensity for their robust detection using our injection volume, which was selected to limit MS signal saturation of abundant lipids (cf. section “Worfklow Validation”).
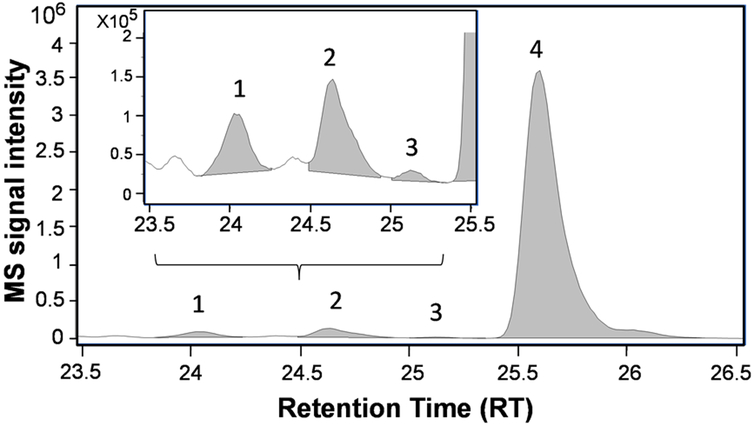
The structure of acyl side chains for all lipids was identified from MS/MS spectra.26,27 This structural information is very important since classes of fatty acids have different biological roles, for example n-3 and n-6 polyunsaturated fatty acids, such as docosahexaenoic and arachidonic acid, respectively, have anti- and pro-inflammatory properties.46 The identification of acyl side chains was facilitated by our long chromatographic gradient, which separated many lipid isomers that differed in their acyl side chain composition. We were also able to assign n-6 vs. n-3 C20:4 or C22:5 fatty acyl side chains on 12 PC. This is illustrated for PC38:4 in Figure 3, which depicts the extraction chromatogram (EIC) of m/z 810.597 and highlight the elution and peak shape of the 4 identified isomers. MS/MS analysis on features 810.597 at RT 25.11 (peak #3) and RT 25.58 (peak #4) identified two PC(18:0_20:4) with a similar MS/MS signal intensity pattern for the two fragments corresponding to their fatty acyl side chain: higher at m/z 303.23 for C20:4, most likely on sn-2 position, and lower at m/z 283.26 for C18:0. This suggests that these two PC are not positional isomers. Therefore, based on the following considerations, we reasoned that the peak #3 and 4 contains a 20:4 fatty acyl side chain of the n-3 and n-6 series, respectively: (i) total plasma levels of C20:4n-3 (eicosatetraenoic acid) are ~120-fold less abundant than C20:4n-6 (arachidonic acid) in healthy human plasma47 and (ii) under our chromatographic conditions (RP), lipids containing fatty acyl side chain of n-3 series elute prior to their n-6 isomer counterparts. Even if we were not able to identify all the isomers defined by their double bond location characteristics due to the limitation of our MS/MS technology vs. MS3 for example, one clear advantage of our long chromatographic run is that it enabled separation of some of these isomers. It is noteworthy, however, that definitive identification of all these lipid isomers require synthetic standards, which to the best of our knowledge are not currently commercially available.
Figure 3. Extracted ion chromatogram of m/z 810.597 (PC38:4 + H+) depicting the resolution and peak shape of four PC38:4 isomers.
The structure of the fatty acyl side chains was identified by MS/MS: Peak#1: PC(20:2_18:2), Peak#2: PC(18:1_20:3), Peak#3 & 4: PC(18:0_20:4) (See text for more details).
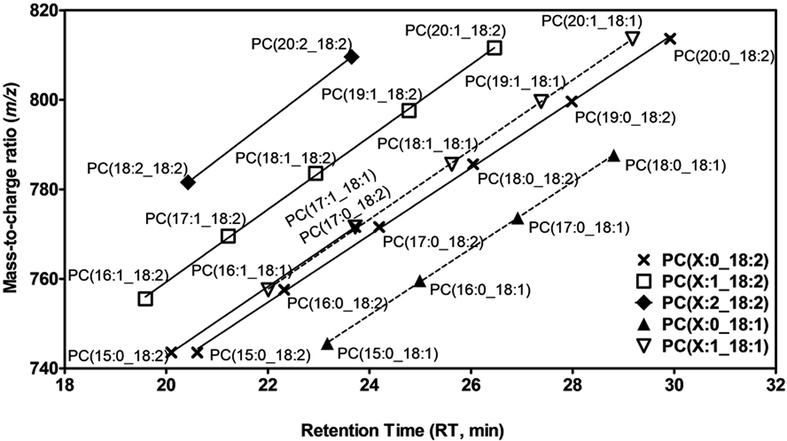
To further ascertain identification of some lipids, we used the RT behavior approach,31,32 which was particularly useful for low abundance MS/MS fragments. In practice, this approach involves positioning RT (x axis) and mass (y axis) of lipids from the same class on a graph: lipids elute according to the carbon chain length and the degree of unsaturation of the two acyls in sn-1 and sn-2 position. This is illustrated in Figure 4 for some PC. This approach enabled the identification of 3 SM and 1 PC plasmalogen (PCO-) (data not shown).
Figure 4. RT behavior for some glycerophosphatidylcholines with different fatty acyl side chains.
The linear relationship between retention time (RT: x axis) and mass (m/z: y axis) is shown for different PC containing C18:1 and C18:2.
Robust semi-quantitative analysis of lipids in human plasma:
The Supplemental Excel file lists all 432 lipids identified, 428 by MS/MS and 4 using RT behavior, along with their respective RSD, signal intensity as well as correlation factor value calculated as described in the workflow validation section above. Among the 432 identified lipids, 360 lipids have an intensity signal with a RSD < 20%, which is the upper limit tolerated by the FDA48 and 403 lipids have RSD < 30%, which is accepted by many laboratories19,49 for untargeted analysis. Among these lipids, 7 CE and 2 TG showed signal saturation, but the remaining 394 are meeting criteria for robust semi-quantitative analysis: namely, 8 fatty acyls, 212 glycerophospholipids, 53 sphingolipids, 8 sterols, 112 glycerolipids and 1 prenol lipid (Table 1). The use of replicates of a single vs. a pool of human plasma samples may have limited the number and diversity of lipids measured. Nevertheless, compared to Quehenberger et al.,12 who quantified 588 lipids with multiple platforms using a targeted approach, we cover (i) more lipid (sub)classes with numerous isomeric forms such as glycerophospholipids (212 vs. 160) and glycerolipids (112 vs. 73), but (ii) less low abundant ones such as fatty acyls, which include acylcarnitines and eicosanoids (8 vs. 107), or prenols (1 vs. 8), since we are limited by the signal intensity dynamic range of our single instrument. Using a similar instrument, Cajka et al.19 reported only 118 glycerophospholipids with RSD < 30%, but this could be explained by the short 15 min–gradient as their focus was more on reproducibility than coverage.
Proof of concept: Impact of sex and age on the circulating lipidome
This section reports data from our proof-of-concept study conducted in a cohort of healthy males (n=47, median age: 38 years, interquantile range: 26.5–48.5) and females (n=53, median age: 43 years, interquantile range: 28–53) from the NIDDK IBD Genetics Consortium Biobank in order to illustrate the robustness and power of the developed untargeted lipidomic workflow in identifying the impact of sex and age on circulating lipid subclasses, but more specifically on their fatty acyl side chains.
MS lipid feature annotation:
Following data processing, the final data set had 854 features. Because of the similarity in plasma and serum lipid profiles (Supplemental Figure S-6), we took advantage of our prior identification of 428 lipids by MS/MS in plasma (cf. section “Identification of plasma lipidome” above) and manually aligned the datasets generated from the workflow validation and those of the workflow application and this enabled the annotation of 295 unique lipids in the latter. Using MS/MS analysis, we have ascertained the identity of 25 of these lipids, selected for representing subclasses in the whole dataset (8 PC, 6 SM, 4 TG, 3 PE, 2 Cer, 1 diacylglycerol (DG), 1 glucose/galactose (Glc/Gal)-Cer), except for a misidentification of the fatty acyl side chain composition for 1 SM. Because statistical testing is achieved on the entire dataset and not only on identified lipids, we also manually looked for signal redundancy for those 295 lipids in the entire data set consisting of 854 features (cf. section above) and identified 485 MS features. Collectively, these results demonstrate the potential of building an in-house database for rapid lipid identification in an untargeted lipidomic discovery study instead of conducting MS/MS analysis multiple times on different datasets from the same or similar matrix, which is more time consuming.
Impact of sex and age on the circulating lipidome:
This section compares results obtained using three different statistical testing strategies on our lipidomic dataset.
The first two strategies are commonly used in metabolomics, namely individual testing of features (here using regression analysis) followed by evaluation of FDR (Strategy 1) as well as biologically informed grouping (Strategy 2). The third strategy, namely data-driven grouping of MS features using WGCNA (Strategy 3), was created for and mostly applied to the analysis of gene expression data but, only more recently, applied to metabolomics.33,35–38 It is noteworthy that since 55 of the 854 MS features were not attributed to any module during WGCNA analysis, the 3 different strategies were compared using the same dataset consisting of 799 features.
Individual testing of features (Strategy 1):
Figure 5 shows all 799 MS features of the dataset as a Volcano plot, in which the impact of sex or age is reported on lipid feature subclasses (Figure 5A & C) or fatty acyl side chains (Figure 5B & D), both indicated by differently colored symbols. Figure 5A & B compare male vs. female data adjusted for age. Using a threshold of P-value < 0.05 (corresponding to an estimated FDR of 8.5%), 219 of the 799 MS features were significantly different between males and females. Among these, higher values were found for 40 glycerolipids, 10 ceramides, 7 PC and 4 PC plasmalogens (PCO-) in males vs. 18 SM, 18 PC, 5 PE, 2 PI, 15 LPC and 3 FFA in females. As for the fatty acyl side chains, among the 219 lipid features discriminating males from females, higher levels were found for lipids containing (number of entities in brackets): C18:2 (15), C20:4 (12) and C22:4 (4) in males vs. C20:4 (3), C20:5 (6) and C22:6 (26) higher in females. Figure 5C and D depict the impact of a person’s age on the abundance of all lipid features, expressed as a 1-year difference and adjusted for sex. Using a threshold of P-value < 0.05 (indicated by the dotted line; corresponding to a FDR of 17%) 160 of the 799 features were significantly affected by age. Among these, abundance with age was higher for 4 Cer, 5 SM, 8 FFA, 6 CE and free cholesterol, but lower for 14 PCO-, 3 PE, 2 PI and 6 LPE. For fatty acyl side chains, abundances were higher with age for C22:5 (2), C20:5 (2) and C22:4, but lower for C18:2 (44) and C20:4 (4).
Figure 5. Volcano plots depicting the impact of sex (A & B) and age (C & D) on circulating lipids (A & C) and their fatty acyl side chains (B & D) from 100 healthy individuals using independent testing (strategy 1).
All panels depict the 799 features obtained following MS data processing from which 295 lipids were annotated using manual peak alignment with previously acquired dataset for which we had characterized the lipidome using MS/MS, including their fatty acyl side chains, as shown by the different colours. The dotted line indicates the P-value <0.05 threshold, which corresponds to FDR values of 8.5% (impact of sex) and 17% (impact of age, expressed on a one-year difference).
Biologically informed grouping of features (Strategy 2):
Table 2 reports results from supervised correlation analyses between identified subclasses and fatty acyl groups with sex and age. Significant associations were found for the following lipid subclasses for (i) sex: di- and triacylglycerols, ceramides, PE, PI and Glc/Gal-Cer, and for their fatty acyl side chains: C18:2, C20:5, C22:4 and C22:6 (Table 2), and (ii) age: diacylglycerols, SM, PCO-, PE, PI, LPE and for their fatty acyl side chains C18:2 and C22:5 (Table 2).
Table 2.
Impact of sex and age on the circulating lipidome of 100 healthy individuals as revealed by supervised correlation analysis between identified groups of lipid subclasses and fatty acyl side chains (strategy 2).
| P-value/correlation | |||||||
|---|---|---|---|---|---|---|---|
| PC1 | PC2 | PC1 | PC2 | ||||
| Category | Subclass | Features # | Unique lipids # | Sex | Age | ||
| FA | FFA | 13 | 12 | 1E-02/−0.27 | 6E-01/0.04 | 1E-02/0.25 | 1E-01/0.15 |
| GP | LPC | 73 | 37 | 2E-01/−0.14 | 8E-02/0.18 | 8E-01/0.07 | 8E-01/0.02 |
| LPCO- | 3 | 3 | 2E-01/−0.13 | 9E-01/0.00 | 9E-01/0.02 | 1E-01/−0.16 | |
| LPE | 22 | 8 | 6E-01/0.07 | 5E-01/−0.08 | 1E-01/−0.16 | 4E-02/0.21 | |
| PC | 129 | 75 | 6E-02/−0.18 | 5E-01/−0.08 | 1E-01/−0.15 | 1E-01/0.15 | |
| PCO- | 35 | 27 | 5E-01/0.09 | 3E-01/−0.09 | 1E-02/−0.26 | 4E-01/−0.07 | |
| PE | 14 | 8 | 4E-02/−0.21 | 2E-2/0.25 | 4E-01/−0.08 | 2E-03/−0.30 | |
| PI | 8 | 8 | 6E-02/−0.18 | 6E-01/0.03 | 2E-01/−0.13 | 3E-06/0.28 | |
| SP | SM | 55 | 29 | 1E-01/−0.18 | 1E-01/0.14 | 3E-01/0.10 | 4E-02/0.21 |
| Cer | 25 | 9 | 2E-02/0.23 | 5E-01/−0.08 | 1E-01/0.16 | 2E-01/0.13 | |
| Glc-Cer | 6 | 6 | 8E-01/−0.02 | 8E-4/−0.34 | 4E-01/−0.09 | 1E00/−0.16 | |
| ST | CE | 25 | 7 | 1E-01/−0.15 | 3E-01/−0.13 | 9E-01/0.02 | 6E-3/0.28 |
| GL | DG | 5 | 4 | 1E-02/0.25 | 2E-01/0.16 | 3E-01/0.10 | 1E-2/0.26 |
| TG | 72 | 62 | 3E-04/0.36 | 4E-01/−0.09 | 9E-01/−0.01 | 7E-01/−0.04 | |
| Fatty acyls | Features # | Unique lipids # | Sex | Age | |||
| C18:2 | 92 | 53 | 9E-01/−0.02 | 2E-02/0.25 | 3E-06/−0.45 | 9E-01/−0.02 | |
| C20:4 | 75 | 45 | 8E-01/−0.02 | 2E-01/0.13 | 8E-01/−0.02 | 5E-01/−0.07 | |
| C20:5 | 15 | 9 | 1E-01/−0.18 | 1E-03/0.33 | 9E-02/−0.02 | 4E-01/−0.09 | |
| C22:4 | 10 | 8 | 5E-02/0.19 | 5E-02/−0.20 | 2E-01/0.14 | 9E-01/−0.01 | |
| C22:5 | 11 | 9 | 6E-01/0.04 | 2E-01/−0.13 | 2E-02/0.24 | 2E-01/0.12 | |
| C22:6 | 44 | 28 | 7E-04/−0.34 | 4E-02/−0.22 | 4E-01/−0.07 | 1E-01/0.15 | |
This table reports P-value and correlation coefficient of identified subclasses from the 485 features (corresponding to 295 unique lipids) including their fatty acyl side chains, for sex and age effect on two principal components: PC1, PC2. Associations with sex or age which have a P-value < 0.05 are highlighted in bold. For abbreviations of lipid categories and (sub)classes, please refer to Table 1.
Data-driven grouping of MS features using WGCNA (Strategy 3):
Table 3 reports the 18 most significant modules (P-value < 0.05) for one of the principal components from the 22 modules created by WGCNA analysis (Strategy 3). These modules contained between 12 and 124 MS features. Using WGCNA, we identified 10 modules that were strongly correlated with sex containing predominantly the same lipid subclasses as those found with strategy 1 and 2, namely di- and triacylglycerols (modules 3 and 11); SM (modules 5, 14 and 22), PCO- (module 17), PE (module 18), Cer (module 6), FFA (module 21) and the C22:6 fatty acyl side chain (module 15). Modules that correlated the most with age contained also di- and triacylglycerols (module 11), SM (module 5), PCO- (module 17), PE (module 18), PC and PI (modules 8 & 12) (Table 3).
Table 3.
Impact of sex and age on the circulating lipidome of 100 healthy individuals as revealed by WGCNA (strategy 3).
| P-value/correlation | |||||||
|---|---|---|---|---|---|---|---|
| PC1 | PC2 | PC1 | PC2 | ||||
| Modules 22 total |
Features # (% identified) |
Predominant Category (%) |
Predominant (Sub)class or FA |
Sex | Age | ||
| M1 | 124 (35%) | GP (23%) | LPC (21%) | 1E-02/0.26 | 3E-01/0.09 | 6E-01/0.05 | 7E-01/0.04 |
| M2 | 70 (20%) | GP (13%) | PC (10%) | 1E-01/0.17 | 2E-01/0.12 | 3E-02/−0.22 | 9E-01/0.01 |
| M3 | 58 (80%) | GL (50%) | TG (43%) | 7E-04/0.34 | 7E-01/0.02 | 6E-01/0.06 | 2E-02/0.23 |
| M4 | 54 (55%) | GP (43%) | C18:2 & C20:4 (39%) | 9E-02/0.19 | 4E-01/0.09 | 4E-03/−0.29 | 3E-01/−0.11 |
| M5 | 49 (63%) | SL (47%) | SM (41%) | 1E-01/−0.17 | 9E-03/−0.27 | 7E-01/−0.03 | 4E-05/0.39 |
| M6 | 46 (59%) | SL (54%) | Cer (50%) | 3E-02/0.21 | 2E-01/0.15 | 7E-02/0.18 | 2E-01/−0.13 |
| M8 | 36 (72%) | GP (67%) | PC (64%) | 7E-01/−0.05 | 2E-01/0.16 | 5E-01/0.07 | 2E-03/−0.31 |
| M9 | 35 (83%) | GP (83%) | LPC & LPE (83%) | 9E-01/0.01 | 4E-02/0.21 | 4E-01/0.08 | 7E-01/−0.04 |
| M10 | 30 (83%) | GP (74%) | C18:2 (67%) | 5E-01/−0.03 | 8E-02/0.18 | 2E-04/−0.37 | 9E-01/0.02 |
| M11 | 30 (70%) | GL (67%) | C18:2 & C20:4 (47%) | 5E-03/0.29 | 2E-01/−0.13 | 6E-01/−0.05 | 8E-03/0.27 |
| M12 | 28 (34%) | GP (61%) | PC & PI (57%) | 2E-01/−0.12 | 1E00/0.02 | 7E-01/−0.03 | 7E-03/0.28 |
| M14 | 27 (55%) | SL (55%) | SM (55%) | 9E-01/−0.04 | 2E-03/−0.32 | 2E-2/0.24 | 2E-01/0.11 |
| M15 | 26 (92%) | GP (86%) | C22:6 (92%) | 8E-03/−0.33 | 8E-02/0.18 | 4E-01/−0.09 | 5E-01/0.06 |
| M16 | 21 (0%) | Unknown | Unknown | 6E-1/0.07 | 3E-02/0.22 | 4E-02/−0.21 | 7E-01/−0.03 |
| M17 | 19 (84%) | GP (79%) | PCO-(79%) | 2E-01/0.14 | 6E-04/0.32 | 6E-02/−0.19 | 1E-02/0.24 |
| M18 | 17 (88%) | GP (88%) | PE (88%) | 7E-02/−0.17 | 9E-03/0.27 | 2E-01/−0.12 | 1E-03/−0.31 |
| M21 | 14 (57%) | FA (57%) | FFA (57%) | 3E-02/−0.23 | 7E-01/0.05 | 1E-02/0.25 | 9E-01/−0.01 |
| M22 | 12 (100%) | SL (100%) | SM (75%) | 8E-01/−0.02 | 3E-02/0.23 | 6E-01/−0.06 | 4E-01/−0.08 |
This table reports P-value and correlation coefficient of the 18 significant modules (P-value <0.05) from the 22 generated by WGCNA. All 18 modules have P-value <0.05 (highlighted in bold) for at least one of the variable tested (sex and age) for one of the two principal components: PC1, PC2. For each module, we report the predominant lipid category, subclass or fatty acyl side chain that was identified.
Taken altogether, results from all 3 testing strategies converge to a similar conclusion regarding classes of lipids or their fatty acyl side chains that were most affected by sex and age, although there are some subtle differences. It should, however, be kept in mind that in this proof-of concept study, data analyses did not take into account factors such as body mass index and medications, which could influence the sex and age comparison50. Nevertheless, our results corroborate the fact that lipids of various classes containing a fatty acyl side chain of the n-3 series such as C22:6 and C20:5 are significantly more abundant in females vs. those of the n-6 series such as C18:2, C20:4 and C22:4 that are more abundant in males. To the best of our knowledge, such a detailed analysis of the impact of sex on the fatty acyl side chain composition of lipids has not been previously reported. Second, they also identify changes in lipid subclasses with sex, which are consistent with those previously reported by Weir et al.51 (cohort of 1076 Mexican Americans) and Sales et al.50 (cohort of 71 healthy Caucasians), except for diacylglycerols, for which higher levels were not reported in males. Interestingly, although 18 individual SM were more abundant in females using strategy 1, when all grouped as a class, it was not correlated to sex (R2=−0.18, P-value=0.1; strategy 2). There was, however, a better correlation, both negative and positive, for sex with modules containing SM (modules 5 & 14: R2= −0.27 & −0.32; P-value=0.009 & 0.0016, module 22: R2=0.23, P-value=0.026). Hence, these results suggest that some, but not all, SM are associated with sex. In this regard, Mielke et al.52 have emphasized the importance of measuring SM individually, along with their carbon-chain length instead of measuring total SM as a class, to distinguish their cellular functions since SM metabolism is highly compartmentalized.53
Based on the aforementioned results obtained with the 3 different testing strategies, we conclude that all of them provide relevant and complementary information about the impact of sex and age on lipidomic data acquired using an untargeted-discovery based workflow. However, compared to strategy 2, strategy 1 (testing of individual features with a cut-off that is not too stringent) and strategy 3 (WGCNA) appear to yield results that are more representative of the dataset, more data-driven and more suitable for untargeted-based lipidomic approach since they do not require prior annotation of MS lipid features. The constraint behind strategy 1 is, however, the burden of multiple testing due to the high number of features, thus reducing power and/or increasing the risk of false positive. Strategy 2 (testing associations between pre-defined lipid subgroups and a selected variable) is attractive for its biological relevance, but needs accurate annotation of features and is likely to miss important information on the differential impact of a given condition (i.e. sex or age in this study) within a given lipid subgroup, as it was illustrated in this study for the impact of sex on SM.
One of the unique advantages of using WGCNA for untargeted lipidomic data analysis is that lipid features are grouped in modules based on correlation prior to testing of their association with the selected variable(s), thereby decreasing the burden of dimensionality (multiple testings) and allowing easier control over type-one error rates. It is noteworthy that similarly to DiLeo et al.,35 who applied WGCNA to the analysis of tomato fruit metabolome, we found that for a given unique lipid, duplicate features such as precursors and product ions tend to be in the same module. In this study, we found that most modules of correlated lipid features are not necessarily part of the same subclass. For example, in this study, module 15 contained lipids from various subclasses with a C22:6 fatty acyl side chain; this module was strongly associated with sex. For a data-driven study, the identified modules provide a framework for prioritizing the identification of lipid features that are most associated with the selected variables.
The ultimate challenge remains, however, the biological interpretation of these data. In this regard, modules can be hypothesis-generating. For example, in this study we found that some modules containing SM correlated more strongly with sex than others. While the biological basis for this result remains to be investigated, this could suggest that sex impacts only a subgroup of SM. Data interpretation could become, however, more difficult when different classes/types of lipids are present within the same module. One way to approach this would be to do a pathway enrichment analysis on the module of interest as it was done by Sue et al.37 and Heiland et al.38 for metabolomic data, or by Keck et al.54 for proteomic data. For this approach, the database that is mostly used is the Kyoto Encyclopedia of Genes and Genomics (KEGG55). The current coverage of KEGG database for lipid metabolites and metabolism is, however, limited and not sufficient for the analysis of comprehensive lipidomic data. In this regard, we tried to find some pathways associated with specific lipid-containing modules using MetaboAnalyst 3.0,56 but without success. Furthermore, while the LIPID MAPS website offers extensive information on lipid metabolites, it contains only a few metabolic pathways and it is not currently designed for pathway mining of lipidomic dataset. Some of these issues may, however, be solved using the latest version of RECON3D57, which includes lipid pathways. Clearly, much remains to be developed in terms of bioinformatics for mining of comprehensive lipidomic datasets as well as their biological interpretation.
Future perspectives
Through the application of our lipidomic workflow to the plasma and serum samples, we encountered some constraints and limitations, which represent perspectives for future development of this workflow. For example, while our long chromatographic run (~83 min) was found to be essential for optimal resolution of lipid isomers, addition of other more recent technologies such as ion mobility58 or SFC59,60 would be expected to decrease the runtime while keeping optimal resolution of separation of lipids isomers. Similarly, in this study, the position of double bonds of the lipid fatty acyl side chains was only suggested using MS/MS on lipids with saturated fatty acyl side chain on the sn-1 position. Future development of our lipidomic workflow could take advantage of other technologies such as ozone-induced dissociation61 or MS3 scanning for this identification. In addition, to enhance throughput, future perspective will include the development of a bioinformatic script for automatizing the manual peak alignment of identified lipids between projects. Finally, the lipid coverage of our developed workflow (lipids from ~100 pg/mL to μg/mL12), is limited by instrument dynamic range as well as the presence of high concentration lipids. Hence, additional LC-MS analyses would be required to include lipid (sub)categories present at lower concentration such as isoprostanes and prostaglandins using separate samples that have been processed through pre-fractionation steps.62,63
CONCLUSIONS
We herein present a comprehensive and reproducible label-free untargeted workflow for unbiased LC-MS-based lipidomic analysis in hundreds of samples on a single instrument. This workflow includes modifications of commonly used lipid extraction and elution protocols as well as a newly developed data processing subroutine and thereby extends the current coverage of polar and non-polar lipids as well as lipid isomer separation that can be reproducibly measured inside a linear intensity range. We herein provide a list of 432 different circulating lipids identified, including acyl side chain, of which 394 had RSD < 30% with no signal intensity saturation, which enable their robust semi-quantitative analysis required for the application of an untargeted workflow. The usefulness, power and robustness of the developed workflow were demonstrated through the analysis of 100 serum samples from healthy individuals and the comparison of three different testing strategies in their capacity to identify the impact of sex and age on the circulating lipidome. While all strategies provided relevant information, the use of unsupervised and data-driven approaches (i.e. strategy 1 and 3) was found to be most suitable for untargeted lipidomics since it does not require prior annotation of MS features, of which strategy 3 (i.e. WGCNA) identifies also correlated lipids, which may be biologically informative. Nevertheless, irrespective of the testing approach, this analysis revealed sex and age differences in lipids of various classes, particularly those containing polyunsaturated fatty acids of the n-3 (C22:6 and C20:5) and n-6 (C18:2, C20:4 and C22:4) series as acyl side chains. While additional work is needed to identify the remaining 134 unidentified features, one would expect that the advent of new technologies such as ion mobility, SFC and MSn will further extent the number of lipid features detected on a single instrument in a shorter time frame and facilitate the lipid identification process over time.
Supplementary Material
Figure S-1: Comparison of lipid recovery using the modified vs. original MTBE-based extraction protocols.
Figure S-2: Comparison of lipid recovery using the modified vs. original MTBE-based extraction protocols in plasma.
Figure S-3: RP-HPLC-ESI-MS total ion chromatogram of human plasma samples using 4 different mixtures of solvents as elution buffer.
Figure S-4: Example of possible misalignment.
Figure S-5: Principal component analysis of the lipidomic dataset obtained for the 48 human plasma sample replicates processed without (A) and with (B) batch correction using Combat algorithm.
Figure S-6: Volcano plot representing the lipidomic dataset obtained from plasma vs. serum samples collected from same subject and processed in parallel through our lipidomic workflow.
Table S-1: List of lipid standards.
Table S-2: Capacity factor k, and selectivity α of 2 isomers of m/z=784.584 for the four buffers of elution B.
Table S-3: Number of features and lipids with saturated intensity for different loading volume of plasma.
Excel file with Identified Lipids.
ACKNOWLEDGMENTS
We are also thankful to Ms. Julie Thompson-Legault for her help with Ethics study approval and sample handling logistics as well as Ms. Lucie Lefebvre and Sara Hamad for editorial and secretarial assistance, and Dr. Alain Lesimple for helpful advices in setting up the lipidomic workflow.
GRANT SUPPORT
This study was supported by the Canadian Institutes of Health Research (CIHR Grants # 9575 & 102168 to C.D.R.), the Montreal Heart Institute Foundation and the “Fondation Grand Défi Pierre Lavoie” and benefited from infrastructure supported by the Canadian Foundation for Innovation (to J-C. T, J.D.R & C.D.R). The iGenoMed Consortium would also like to acknowledge the financial support of the NIDDK-IBD Genetics Consortium as well as Génome Québec, Genome Canada, the government of Canada, the Ministère de l’enseignement supérieur, de la recherche, de la science et de la technologie du Québec, the Canadian Institutes of Health Research (with contributions from the Institute of Infection and Immunity, the Institute of Genetics, and the Institute of Nutrition, Metabolism and Diabetes), Genome BC, and Crohn’s Colitis Canada as well as Agilent Technologies.
ABBREVIATIONS
- AC
acylcarnitine
- ACN
acetonitrile
- CE
steryl ester
- Cer
ceramide
- DG
diacylglycerol
- ESI
electrospray source ionization
- EIC
extracted ion chromatogram
- FDR
false discovery rate
- Glc-Cer
glucosyl ceramide
- IPA
isopropyl alcohol
- LPC
monoacylglycerosphosphocholine
- LPE
monoacylglycerosphosphoethanolamine
- MRM
multiple reaction monitoring
- MTBE
methyl-tert-butyl ether
- m/z
mass-to-charge ratio
- PA
diacylglycerophosphatidic acid
- PC
diacylglycerophosphocholine
- PE
diacylglycerophosphopethanolamine
- PG
diacylglycerophosphoglycerol
- PI
diacylglycerophosphoinositol
- PL
phospholipid
- QC
quality control
- RP
reverse phase
- RSD
relative standard deviation
- RT
retention time
- TIC
Total ion chromatogram
Footnotes
DISCLOSURE
Dr Tardif holds the Canada Research Chair in translational and personalized medicine and the Université de Montréal Pfizer-endowed research chair in atherosclerosis. Dr Rioux holds a Canada Research Chair in Genetics and Genomic Medicine.
Reference List
- 1.Zhao YY; Miao H; Cheng XL; Wei F Lipidomics: Novel insight into the biochemical mechanism of lipid metabolism and dysregulation-associated disease. Chem. Biol. Interact 2015, 240, 220–238. [DOI] [PubMed] [Google Scholar]
- 2.Yang L; Li M; Shan Y; Shen S; Bai Y; Liu H Recent advances in lipidomics for disease research. J. Sep. Sci 2016, 39 (1), 38–50. [DOI] [PubMed] [Google Scholar]
- 3.Yang K; Han X Lipidomics: Techniques, Applications, and Outcomes Related to Biomedical Sciences. Trends Biochem. Sci 2016, 41 (11), 954–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li M; Yang L; Bai Y; Liu H Analytical methods in lipidomics and their applications. Anal. Chem 2014, 86 (1), 161–175. [DOI] [PubMed] [Google Scholar]
- 5.Cajka T; Fiehn O Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. Trends Analyt. Chem 2014, 61, 192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird SS; Marur VR; Sniatynski MJ; Greenberg HK; Kristal BS Serum lipidomics profiling using LC-MS and high-energy collisional dissociation fragmentation: focus on triglyceride detection and characterization. Anal. Chem 2011, 83 (17), 6648–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rustam YH; Reid GE Analytical Challenges and Recent Advances in Mass Spectrometry Based Lipidomics. Anal. Chem 2017, 90 (1), 374–397. [DOI] [PubMed] [Google Scholar]
- 8.Fahy E; Subramaniam S; Murphy RC; Nishijima M; Raetz CR; Shimizu T; Spener F; van MG; Wakelam MJ; Dennis EA Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res 2009, 50 Suppl, S9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emken E Human studies using isotope labeled fatty acids: answered and unanswered questions. J. Oleo. Sci 2013, 62 (5), 245–255. [DOI] [PubMed] [Google Scholar]
- 10.Li J; Hu C; Zhao X; Dai W; Chen S; Lu X; Xu G Large-scaled human serum sphingolipid profiling by using reversed-phase liquid chromatography coupled with dynamic multiple reaction monitoring of mass spectrometry: method development and application in hepatocellular carcinoma. J. Chromatogr. A 2013, 1320, 103–110. [DOI] [PubMed] [Google Scholar]
- 11.Kwak HS; Han JY; Ahn HK; Kim MH; Ryu HM; Kim MY; Chung HJ; Cho DH; Shin CY; Velazquez-Armenta EY; Nava-Ocampo AA Blood levels of phosphatidylethanol in pregnant women reporting positive alcohol ingestion, measured by an improved LC-MS/MS analytical method. Clin. Toxicol. (Phila) 2012, 50 (10), 886–891. [DOI] [PubMed] [Google Scholar]
- 12.Quehenberger O; Armando AM; Brown AH; Milne SB; Myers DS; Merrill AH; Bandyopadhyay S; Jones KN; Kelly S; Shaner RL; Sullards CM; Wang E; Murphy RC; Barkley RM; Leiker TJ; Raetz CR; Guan Z; Laird GM; Six DA; Russell DW; McDonald JG; Subramaniam S; Fahy E; Dennis EA Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res 2010, 51 (11), 3299–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu T; Zhang JL Mass-spectrometry-based lipidomics. J. Sep. Sci 2017, 41 (1), 351–372. [DOI] [PubMed] [Google Scholar]
- 14.FOLCH J; LEES M; SLOANE STANLEY GH A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem 1957, 226 (1), 497–509. [PubMed] [Google Scholar]
- 15.BLIGH EG; DYER WJ A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol 1959, 37 (8), 911–917. [DOI] [PubMed] [Google Scholar]
- 16.Matyash V; Liebisch G; Kurzchalia TV; Shevchenko A; Schwudke D Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res 2008, 49 (5), 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurdeniz G; Kristensen M; Skov T; Dragsted LO The Effect of LC-MS Data Preprocessing Methods on the Selection of Plasma Biomarkers in Fed vs. Fasted Rats. Metabolites 2012, 2 (1), 77–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowden JA; Heckert A; Ulmer CZ; Jones CM; Koelmel JP; Abdullah L; Ahonen L; Alnouti Y; Armando A; Asara JM; Bamba T; Barr JR; Bergquist J; Borchers CH; Brandsma J; Breitkopf SB; Cajka T; Cazenave-Gassiot A; Checa A; Cinel MA; Colas RA; Cremers S; Dennis EA; Evans JE; Fauland A; Fiehn O; Gardner MS; Garrett TJ; Gotlinger KH; Han J; Huang Y; Neo AH; Hyotylainen T; Izumi Y; Jiang H; Jiang H; Jiang J; Kachman M; Kiyonami R; Klavins K; Klose C; Kofeler HC; Kolmert J; Koal T; Koster G; Kuklenyik Z; Kurland IJ; Leadley M; Lin K; Maddipati KR; McDougall D; Meikle PJ; Mellett NA; Monnin C; Moseley MA; Nandakumar R; Oresic M; Patterson RE; Peake D; Pierce JS; Post M; Postle AD; Pugh R; Qui Y; Quehenberger O; Ramrup P; Rees J; Rembiesa B; Reynaud D; Roth MR; Sales S; Schuhmann K; Schwartzman ML; Serhan CN; Shevchenko A; Somerville SE; John-Williams LS; Surma MA; Takeda H; Thakare R; Thompson JW; Torta F; Triebl A; Trotzmuller M; Ubhayasekera SJK; Vuckovic D; Weir JM; Welti R; Wenk MR; Wheelock CE; Yao L; Yuan M; Zhao XH; Zhou S Harmonizing Lipidomics: NIST Interlaboratory Comparison Exercise for Lipidomics using Standard Reference Material 1950 Metabolites in Frozen Human Plasma. J. Lipid Res 2017, 58 (12), 2275–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cajka T; Smilowitz JT; Fiehn O Validating Quantitative Untargeted Lipidomics Across Nine Liquid Chromatography-High-Resolution Mass Spectrometry Platforms. Anal. Chem 2017, 89 (22), 12360–12368. [DOI] [PubMed] [Google Scholar]
- 20.Layre E; Sweet L; Hong S; Madigan CA; Desjardins D; Young DC; Cheng TY; Annand JW; Kim K; Shamputa IC; McConnell MJ; Debono CA; Behar SM; Minnaard AJ; Murray M; Barry CE III; Matsunaga I; Moody DB A comparative lipidomics platform for chemotaxonomic analysis of Mycobacterium tuberculosis. Chem. Biol 2011, 18 (12), 1537–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ballman KV; Grill DE; Oberg AL; Therneau TM Faster cyclic loess: normalizing RNA arrays via linear models. Bioinformatics 2004, 20 (16), 2778–2786. [DOI] [PubMed] [Google Scholar]
- 22.Ritchie ME; Phipson B; Wu D; Hu Y; Law CW; Shi W; Smyth GK limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43 (7), e47 DOI: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hastie T; Tibshirani R; Narasimhan B; Chu G impute: Imputation for microarray data. R package version 1.51.0 2017. https://www.bioconductor.org/packages/devel/bioc/html/impute.html
- 24.Johnson WE; Li C; Rabinovic A Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8 (1), 118–127. [DOI] [PubMed] [Google Scholar]
- 25.Godzien J; Ciborowski M; Martinez-Alcazar MP; Samczuk P; Kretowski A; Barbas C Rapid and Reliable Identification of Phospholipids for Untargeted Metabolomics with LC-ESI-QTOF-MS/MS. J. Proteome. Res 2015, 14 (8), 3204–3216. [DOI] [PubMed] [Google Scholar]
- 26.Hou W; Zhou H; Bou KM; Seebun D; Bennett SA; Figeys D Lyso-form fragment ions facilitate the determination of stereospecificity of diacyl glycerophospholipids. Rapid Commun. Mass Spectrom 2011, 25 (1), 205–217. [DOI] [PubMed] [Google Scholar]
- 27.Ham BM; Jacob JT; Keese MM; Cole RB Identification, quantification and comparison of major non-polar lipids in normal and dry eye tear lipidomes by electrospray tandem mass spectrometry. J. Mass Spectrom 2004, 39 (11), 1321–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berdeaux O; Juaneda P; Martine L; Cabaret S; Bretillon L; Acar N Identification and quantification of phosphatidylcholines containing very-long-chain polyunsaturated fatty acid in bovine and human retina using liquid chromatography/tandem mass spectrometry. J. Chromatogr. A 2010, 1217 (49), 7738–7748. [DOI] [PubMed] [Google Scholar]
- 29.Hsu FF; Turk J Studies on phosphatidylserine by tandem quadrupole and multiple stage quadrupole ion-trap mass spectrometry with electrospray ionization: structural characterization and the fragmentation processes. J. Am. Soc. Mass Spectrom 2005, 16 (9), 1510–1522. [DOI] [PubMed] [Google Scholar]
- 30.Lisa M; Holcapek M Characterization of triacylglycerol enantiomers using chiral HPLC/APCI-MS and synthesis of enantiomeric triacylglycerols. Anal. Chem 2013, 85 (3), 1852–1859. [DOI] [PubMed] [Google Scholar]
- 31.Sandra K; Pereira AS; Vanhoenacker G; David F; Sandra P Comprehensive blood plasma lipidomics by liquid chromatography/quadrupole time-of-flight mass spectrometry. J. Chromatogr. A 2010, 1217 (25), 4087–4099. [DOI] [PubMed] [Google Scholar]
- 32.Ovcacikova M; Lisa M; Cifkova E; Holcapek M Retention behavior of lipids in reversed-phase ultrahigh-performance liquid chromatography-electrospray ionization mass spectrometry. J. Chromatogr. A 2016, 1450, 76–85. [DOI] [PubMed] [Google Scholar]
- 33.Pei G; Chen L; Zhang W WGCNA Application to Proteomic and Metabolomic Data Analysis. Methods Enzymol. 2017, 585, 135–158. [DOI] [PubMed] [Google Scholar]
- 34.Zhang B; Horvath S A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol 2005, 4, Article17. DOI: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 35.DiLeo MV; Strahan GD; den BM; Hoekenga OA Weighted correlation network analysis (WGCNA) applied to the tomato fruit metabolome. PLoS. One 2011, 6 (10), e26683 DOI: 10.1371/journal.pone.0026683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sui X; Niu X; Shi M; Pei G; Li J; Chen L; Wang J; Zhang W Metabolomic analysis reveals mechanism of antioxidant butylated hydroxyanisole on lipid accumulation in Crypthecodinium cohnii. J. Agric. Food Chem 2014, 62 (51), 12477–12484. [DOI] [PubMed] [Google Scholar]
- 37.Su Y; Wang J; Shi M; Niu X; Yu X; Gao L; Zhang X; Chen L; Zhang W Metabolomic and network analysis of astaxanthin-producing Haematococcus pluvialis under various stress conditions. Bioresour. Technol 2014, 170, 522–529. [DOI] [PubMed] [Google Scholar]
- 38.Heiland DH; Worner J; Haaker JG; Delev D; Pompe N; Mercas B; Franco P; Gabelein A; Heynckes S; Pfeifer D; Weber S; Mader I; Schnell O The integrative metabolomic-transcriptomic landscape of glioblastome multiforme. Oncotarget 2017, (8), 49178–49190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao X; Zhang Q; Meng D; Isaac G; Zhao R; Fillmore TL; Chu RK; Zhou J; Tang K; Hu Z; Moore RJ; Smith RD; Katze MG; Metz TO A reversed-phase capillary ultra-performance liquid chromatography-mass spectrometry (UPLC-MS) method for comprehensive top-down/bottom-up lipid profiling. Anal. Bioanal. Chem 2012, 402 (9), 2923–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meitei N; Biswas A; Apte A; Madden S; Sartain M Automatic lipid characterization using SimLipid® from normal phase and reverse phase liquid chromatography-MS, MS/MS data acquired in variable ion modes 2015. http://www.premierbiosoft.com
- 41.Cajka T; Fiehn O LC-MS-Based Lipidomics and Automated Identification of Lipids Using the LipidBlast In-Silico MS/MS Library. Methods Mol. Biol 2017, 1609, 149–170. [DOI] [PubMed] [Google Scholar]
- 42.James PF; Perugini MA; O’Hair RA Sources of artefacts in the electrospray ionization mass spectra of saturated diacylglycerophosphocholines: from condensed phase hydrolysis reactions through to gas phase intercluster reactions. J. Am. Soc. Mass Spectrom 2006, 17 (3), 384–394. [DOI] [PubMed] [Google Scholar]
- 43.Ding J; Anderegg RJ Specific and nonspecific dimer formation in the electrospray ionization mass spectrometry of oligonucleotides. J. Am. Soc. Mass Spectrom 1995, 6 (3), 159–164. [DOI] [PubMed] [Google Scholar]
- 44.Pan H A non-covalent dimer formed in electrospray ionisation mass spectrometry behaving as a precursor for fragmentations. Rapid Commun. Mass Spectrom 2008, 22 (22), 3555–3560. [DOI] [PubMed] [Google Scholar]
- 45.Alonso MC, Tracing polar benzene DB - and naphthalenesulfonates in untreated industrial effluents and water treatment works by ion-pair chromatography-fluorescence and electrospray-mass spectrometry. Anal. Chim. Acta 1999, 400, 211–231. [Google Scholar]
- 46.Calder PC Omega-3 fatty acids and inflammatory processes: from molecules to man. Biochem. Soc. Trans 2017, 45 (5), 1105–1115. [DOI] [PubMed] [Google Scholar]
- 47.Siguel EN; Chee KM; Gong JX; Schaefer EJ Criteria for essential fatty acid deficiency in plasma as assessed by capillary column gas-liquid chromatography. Clin. Chem 1987, 33 (10), 1869–1873. [PubMed] [Google Scholar]
- 48.FDA. Guidance for Industry, Bioanalytical Method Validation 2018. https://www.fda.gov/downloads/drugs/guidances/ucm070107.Pdf
- 49.Naz S; Vallejo M; Garcia A; Barbas C Method validation strategies involved in non-targeted metabolomics. J. Chromatogr. A 2014, 1353, 99–105. [DOI] [PubMed] [Google Scholar]
- 50.Sales S; Graessler J; Ciucci S; Al-Atrib R; Vihervaara T; Schuhmann K; Kauhanen D; Sysi-Aho M; Bornstein SR; Bickle M; Cannistraci CV; Ekroos K; Shevchenko A Gender, Contraceptives and Individual Metabolic Predisposition Shape a Healthy Plasma Lipidome. Sci. Rep 2016, 6, 27710 DOI: 10.1038/srep27710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weir JM; Wong G; Barlow CK; Greeve MA; Kowalczyk A; Almasy L; Comuzzie AG; Mahaney MC; Jowett JB; Shaw J; Curran JE; Blangero J; Meikle PJ Plasma lipid profiling in a large population-based cohort. J. Lipid Res 2013, 54 (10), 2898–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mielke MM; Bandaru VV; Han D; An Y; Resnick SM; Ferrucci L; Haughey NJ Factors affecting longitudinal trajectories of plasma sphingomyelins: the Baltimore Longitudinal Study of Aging. Aging Cell 2015, 14 (1), 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hannun YA; Obeid LM Many ceramides. J. Biol. Chem 2011, 286 (32), 27855–27862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keck M; Androsova G; Gualtieri F; Walker A; von Ruden EL; Russmann V; Deeg CA; Hauck SM; Krause R; Potschka H A systems level analysis of epileptogenesis-associated proteome alterations. Neurobiol. Dis 2017, 105, 164–178. [DOI] [PubMed] [Google Scholar]
- 55.Kanehisa M; Furumichi M; Tanabe M; Sato Y; Morishima K KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45 (D1), D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xia J; Wishart DS Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis . Curr. Protoc. Bioinformatics 2016, 55 (1), 14.10.1–14.10.91. [DOI] [PubMed] [Google Scholar]
- 57.Brunk E; Sahoo S; Zielinski DC; Altunkaya A; Drager A; Mih N; Gatto F; Nilsson A; Preciat Gonzalez GA; Aurich MK; Prlic A; Sastry A; Danielsdottir AD; Heinken A; Noronha A; Rose PW; Burley SK; Fleming RMT; Nielsen J; Thiele I; Palsson BO Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat. Biotechnol 2018, 36 (3), 272–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bowman AP; Abzalimov RR; Shvartsburg AA Broad Separation of Isomeric Lipids by High-Resolution Differential Ion Mobility Spectrometry with Tandem Mass Spectrometry. J. Am. Soc. Mass Spectrom 2017, 28 (8), 1552–1561. [DOI] [PubMed] [Google Scholar]
- 59.Sen A; Knappy C; Lewis MR; Plumb RS; Wilson ID; Nicholson JK; Smith NW Analysis of polar urinary metabolites for metabolic phenotyping using supercritical fluid chromatography and mass spectrometry. J. Chromatogr. A 2016, 1449, 141–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takeda H; Koike T; Izumi Y; Yamada T; Yoshida M; Shiomi M; Fukusaki E; Bamba T Lipidomic analysis of plasma lipoprotein fractions in myocardial infarction-prone rabbits. J. Biosci. Bioeng 2015, 120 (4), 476–482. [DOI] [PubMed] [Google Scholar]
- 61.Brown SH; Mitchell TW; Blanksby SJ Analysis of unsaturated lipids by ozone-induced dissociation. Biochim. Biophys. Acta 2011, 1811 (11), 807–817. [DOI] [PubMed] [Google Scholar]
- 62.Slatter DA; Aldrovandi M; O’Connor A; Allen SM; Brasher CJ; Murphy RC; Mecklemann S; Ravi S; Darley-Usmar V; O’Donnell VB Mapping the Human Platelet Lipidome Reveals Cytosolic Phospholipase A2 as a Regulator of Mitochondrial Bioenergetics during Activation. Cell Metab 2016, 23 (5), 930–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adkins Y; Belda BJ; Pedersen TL; Fedor DM; Mackey BE; Newman JW; Kelley DS Dietary Docosahexaenoic Acid and trans-10, cis-12-Conjugated Linoleic Acid Differentially Alter Oxylipin Profiles in Mouse Periuterine Adipose Tissue. Lipids 2017, 52 (5), 399–413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S-1: Comparison of lipid recovery using the modified vs. original MTBE-based extraction protocols.
Figure S-2: Comparison of lipid recovery using the modified vs. original MTBE-based extraction protocols in plasma.
Figure S-3: RP-HPLC-ESI-MS total ion chromatogram of human plasma samples using 4 different mixtures of solvents as elution buffer.
Figure S-4: Example of possible misalignment.
Figure S-5: Principal component analysis of the lipidomic dataset obtained for the 48 human plasma sample replicates processed without (A) and with (B) batch correction using Combat algorithm.
Figure S-6: Volcano plot representing the lipidomic dataset obtained from plasma vs. serum samples collected from same subject and processed in parallel through our lipidomic workflow.
Table S-1: List of lipid standards.
Table S-2: Capacity factor k, and selectivity α of 2 isomers of m/z=784.584 for the four buffers of elution B.
Table S-3: Number of features and lipids with saturated intensity for different loading volume of plasma.
Excel file with Identified Lipids.