Abstract
There are large knowledge gaps regarding how to control stem cells growth and differentiation. The limitations of currently available technologies, such as growth factors and/or gene therapies has led to the search of alternatives. We explore here how a cell’s epigenome influences determination of cell type, and potential applications in tissue engineering. A prevalent epigenetic modification is the acetylation of DNA core histone proteins. Acetylation levels heavily influence gene transcription. Histone deacetylase (HDAC) enzymes can remove these acetyl groups, leading to the formation of a condensed and more transcriptionally silenced chromatin. Histone deacetylase inhibitors (HDACis) can inhibit these enzymes, resulting in the increased acetylation of histones, thereby affecting gene expression. There is strong evidence to suggest that HDACis can be utilised in stem cell therapies and tissue engineering, potentially providing novel tools to control stem cell fate. This review introduces the structure/function of HDAC enzymes and their links to different tissue types (specifically bone, cardiac, neural tissues), including the history, current status and future perspectives of using HDACis for stem cell research and tissue engineering, with particular attention paid to how different HDAC isoforms may be integral to this field.
Subject terms: Stem-cell biotechnology, Stem-cell research
Introduction
There is a largely unmet clinical need for the repair and regeneration of human tissues and organs. Due to the limitations of conventional clinical therapies, tissue engineering, a multidisciplinary area of research, has gained prominence, as it is likely to offer novel solutions to healthcare problems. Some early success has been achieved in transplanting relatively simple tissues or organs such as the trachea1 and the bladder.2 These early accomplishments demonstrate the massive potential of utilising tissue engineering approaches to restore tissue and organ function, although much further research is needed, especially for more complicated organs and tissues.
Stem cells are an exciting cell source, with the potential to be differentiated into the specific lineages required for the repair and restoration of functional tissues. Increased reporting of the limitations of current methods of using stem cells in tissue engineering has galvanised research to investigate new methods of controlling stem cell fate. However, effectively controlling stem cell proliferation and differentiation, with minimum side effects, is very challenging.
It has become apparent in recent years that ‘epigenetics’ plays a massive role in cell fate. ‘Epigenetics’ refers to the post-genetic modifications made to DNA sequences and related proteins, where chemical functional groups such as methyl or acetyl are transferred onto the DNA or such related protein (e.g., the histone scaffolds, which DNA is wrapped around). Epigenetic modifications make no changes to the actual base genomic code, and epigenetic changes are potentially heritable.3,4 Many diseases, perhaps most notably cancers, have epigenetics as a key mediator.5–7
More specifically, research has discovered that modifications to the histone, such as those by histone deacetylation enzymes (HDACs), affect chromatin structure, and thus stem cell properties such as potency and differentiation.3,8,9 As such, the epigenetic make-up of a cell may be crucial for cell-based therapies and tissue engineering. For example, inhibition of HDACs has been utilised to improve in vitro expansion methods of human hematopoietic stem cells,10 to improve bone differentiation of mesenchymal stem cells (MSCs),11 to enhance the efficiency of induced pluripotent stem cell (iPSC) generation,8 and to increase the efficiency of cellular therapies.12
This review will explore background knowledge of epigenetics, the structure, and function of HDACs and their inhibitors, highlight the current uses of and potential of these compounds in cell-based therapies and tissue engineering and suggest where this field may find itself in the future.
Epigenetics: post-genetic modifications of DNA sequence
DNA, the set of ‘rules’ which control the cell in its most basic form, exists as the famous double helix, is packed into tight but flexible assemblies.13 This flexibility allows constant structural alteration so that DNA can be utilised in transcription, replication, and repair.14 The 147 base units of DNA wrap around histone proteins, forming nucleosomes. These can be organised further, with the ever-changing histone epigenome affecting how compact or how relaxed these structures are.3
Acetylation is one of the most abundant post-translational modifications, and genome-wide analysis of cells has shown that acetylation is as frequent as phosphorylation.9,15 The lysine residues on the histone proteins which DNA wraps around are especially important sites of acetylation. The defined combination of these histone modifications has been termed the ‘histone code’.16 The histone code can be “read” by proteins, such as bromodomains, but can also be erased and re-written,17 predominantly by two classifications of cellular enzymes; histone deacetylases (HDACs) and histone acetyl transferases (HATs). If bromodomains are an example of acetylation ‘readers’, HATs are the ‘writers’, and HDACs are the ‘erasers’.15 HATs mediate the transfer of acetyl groups onto the lysine residues (histone acetylation), which results in more open and more transcriptionally active chromatin structures, as the interactions between the nucleosomes are altered, and the histone tails are released from the linker DNA. HDAC enzymes remove those acetyl groups (histone deacetylation), repressing transcription due to the formation of a more condensed and transcriptionally silenced chromatin.3 HDAC inhibitors (HDACis) inhibit the action of HDAC enzymes, resulting in increased acetylation levels in the cell. This may affect the chromatin compaction, linked to the potency, or differentiation potential of stem cells.9 Despite the misleading name, containing the word ‘histone’, there are a wide range of non-histone targets of these enzyme.18
The structure and activity of HDACs
There are 18 human enzymes known to have deacetylation activity, conventionally numbered 1–18 (e.g., ‘HDAC8’). Based on molecular phylogenetic analysis of primary structures, their location in the cell and their homology to yeast enzymes RPD3 and HDA1, these 18 HDACs are grouped into four classes,19 which can be further subdivided into two categories— classical (classes I, II and IV) (see Tables 1 and 2 for a summary) and sirtuins (class III). The activity of the classical HDACs depends on zinc ions (Zn2+), whereas the sirtuins utilise nicotinamide adenine dinucleotide (NAD+), a phosphate linked dinucleotide coenzyme. As they have entirely different mechanisms of action, sirtuins fall outside the scope of this review; only the classical HDACs are reviewed.20
Table 1.
Summary of Class I zinc-dependent HDAC isoforms
| HDAC | Cellular location | Tissue distribution | Known biological functions |
|---|---|---|---|
| HDAC1 | Nucleus193 | Ubiquitous194 |
Cellular proliferation,195,196 and cell cycle regulation and haematopoiesis197,198 DNA damage response,199 cardiac development115,119 Oligodendrocyte, glial cell, synapse and neuronal cell development and function,146,148,154,155,158,200 regulating Schwann cells156,201 Osteogenic and skeletal muscle development,99,202 cartilage formation43 Modulating T cell response,203 epidermal formation,204 adipogenesis205 |
| HDAC2 | Nucleus193 | Ubiquitous194 |
Cell cycle regulation,198 DNA damage response,199 cardiac development115,117 Oligodendrocyte, glial cell, synapse and neuronal cell development and function,146,148,154,155,158,160,206 regulating Schwann cells151,201 Skeletal muscle development,202 regulating cartilage structure43 |
| HDAC3 | Nucleus and cytoplasm193 | Ubiquitous194 |
Cell cycle regulation,207,208 Liver function,38,209 cardiac function,120,121 osteogenic development,99 bone mass and bone formation,67–70,99 including osteoclast suppression,82 chondroprogenitor differentiation to cartilage68 Brain function162 and proliferation/differentiation of neural cells145,208 hematopoietic stem cell growth10 |
| HDAC8 | Nucleus and cytoplasm193 | Ubiquitous194 | Craniofacial, skull and bone formation in embryonic development210 |
Table 2.
Summary of Class II and IV zinc-dependent HDAC isoforms
| Class | HDAC | Cellular location | Tissue distribution | Known biological functions |
|---|---|---|---|---|
| IIa | HDAC4 | Nucleus and cytoplasm193 | Brain, heart and skeletal muscle,211,212 prehypertrophic chondrocytes,78 retina,213 neurons214–216 |
Myofibroblast development,40 chondrocyte hypertrophy and endochondral ossification,78 muscular differentiation,212 retinal neuronal function,213 regulation of neuronal activity, cell death and survival214,215 |
| HDAC5 | Nucleus and cytoplasm193 |
Heart, skeletal muscle and brain,211,217 neurons216 |
Differentiation of neural stem cells,145 and neuronal activity,216 myocardial and endothelial functions,217 memory function218 |
|
| HDAC7 | Nucleus and cytoplasm193 | Thymus,219 heart, muscle and lung211 |
In embryonic endothelial cells of developing heart, blood vessels, mesenchyme and myocardial layers of heart and in lung tissue,118 role in developing thymocytes,219,220 |
|
| HDAC9 | Nucleus and cytoplasm193 | Heart, skeletal muscle and brain211,222 |
Redundant role in heart development,123 controls genes affected by motor innervation in muscles167 |
|
| IIb | HDAC6 | Mainly Cytoplasm193 | Muscle,223 brain,166 heart,224 liver,225 kidneys,226 and teste227 | Neuroprotection and neurodegeneration,166 muscular differentiation,223 arterial modelling,224 tubulin acetylation, bone mass regulation and immune response modulation,227 involved in cellular response to stress,228,229 and macro-autophagy,230 platelet activation231 |
| HDAC10 | Nucleus and cytoplasm193 | Liver, spleen, kidney,232 skin233 |
Expressed in the developing brain with neural oligodendrocyte cells,168 melanin production in the skin,233 promotes autophagy-mediated cell survival in neuroblastoma cells234 |
|
| IV | HDAC11 | Nucleus and cytoplasm193 | Brain,168 heart, skeletal muscle, kidney,235 T cells236 | Influences immune activation versus immune tolerance236 |
HDACis and specificity
HDACis are typically small-molecular compounds which can bind to and block the action of HDAC enzymes. Some have been isolated from natural sources, such as Trichostatin A (TSA), or designed and synthesised in a laboratory, such as MS-275.21
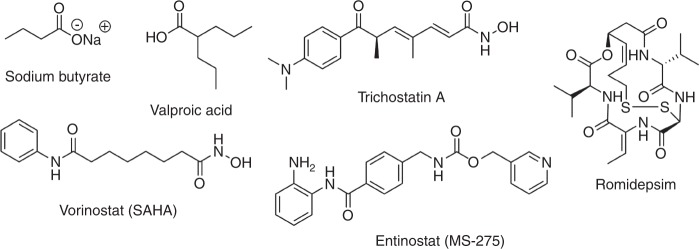
When HDACis bind to, or block, the active site (e.g., the zinc ion) of HDAC enzymes, they act to block the deacetylation action of the zinc ion. To date, research has largely focused on non-specific HDACi compounds (pan-HDACis); broad spectrum HDACis which target many of the HDAC isoforms.22 It is widely accepted that most first generation HDACis, such as Vorinostat (suberanilohydroxamic acid, SAHA) and Romidepsin23,24 are relatively isoform unselective.25 However, the findings can be conflicting, for example, Bradner and co-workers (2010) found some of these HDACis to be selective for certain HDAC isoforms. Discrepancies can arise due to a lack of knowledge of HDAC structure and problems with screening techniques.19 The structure of several commercially available HDACis can be seen in Fig. 1, showing Sodium butyrate, Valproic acid (VPA), Trichostatin A (TSA), Romidepsin, Entinostat (MS-275) and Vorinostat (also known as Zolinza® or suberoylanilide hydroxamic acid, SAHA).
Fig. 1.
Structures of HDACis commonly found in the literature. Clockwise from top left–Sodium butyrate, Valproic acid, Trichostatin A, Romidepsin, Entinostat (MS-275) and Vorinostat (SAHA)
Whilst some pan-HDACis have proven effective drugs,23,24 research is beginning to focus on isoform-selective inhibitors. The structural differences between HDAC enzymes mean that HDACis can be designed to be selective for specific enzymes. It has been argued by many that targeting specific HDACs is key to the development of future HDAC therapeutics.22,25,26 Table 3 summarises the HDAC class/isoform specificity of some commonly used HDACis.
Table 3.
Summary of the HDAC class/isoform specificity of commonly used HDACis
| HDACi | HDACs targeted | Notes and references |
|---|---|---|
| Sodium butyrate | Class I and II | Early pan-HDACi237 |
| Valproic acid | Class I and II | Early pan-HDACi237 |
| Trichostatin A | HDAC1, HDAC2, HDAC3, HDAC5, HDAC6 and HDAC7 (or “Class I and Class II”) | Conflicting reports: with a low efficacy to HDAC8 in some studies,238 or as specific to HDAC1, HDAC3 and HDAC8,239 or a high level of targeting many Class I and Class II HDACs19 |
| Romidepsin | Class I | FDA anticancer agent used to treat various lymphomas237 |
| Entinostat (MS-275) | HDAC1, HDAC2 and HDAC3 | Widely reported as an isoform-specific HDACi237,238 |
| Vorinostat (SAHA) | HDAC1, HDAC2, HDAC3, HDAC6 and possibly HDAC7 | Specificity has conflicting reports–some report as Class I, II and IV, others just HDAC1, HDAC2, HDAC3 and HDAC6.19 Multiple reports of just targetting HDAC7 also reported30 |
Some HDACis, (perhaps most extensively Vorinostat), have been explored in clinical trials as cancer treatments. High levels of deacetylation activity have been linked to tumour pathology and utilising HDACis to reduce deacetylation activity which allows cancer suppressing genes to work.27 The anti-cancer action of HDACis is also due to many other effects of the compounds on cancer cells, including induction of senescence, activation of apoptosis and mitotic cell death, growth arrest and anti-angiogenesis effects.28
Moving on from their applications in oncology, recent publications have shown the huge potential of using HDACis in other fields, such as tissue engineering. The effects of HDACis can be selective for cancerous cells, even at very low doses, and so the concentrations optimised to affect tumour cells typically do not affect normal cells, and their effect on stem/progenitor/normal somatic cells is often unexplored.24,29,30
The potential of using HDACis in tissue engineering
HDACis affect many cellular properties, such as the cell cycle, proliferation rates, gene expression, differentiation potential, accumulation of reactive oxygen species and changes in cell death pathways. This is dependent on the cell type and state, choice of HDACi, and application factors (e.g., exposure time and concentration of inhibitor).28
Work has begun attempting to utilise HDACis for the regeneration of different tissues such as bone,11 cardiac,31 neural/nervous,32 adipose,33 dental,34,35 liver,36–39 skin,40 pancreas,41 muscle42 and cartilage.43 Also worth noting from a translational tissue engineering perspective are the reports of HDACis demonstrating pain relief,44–48 anti-microbial,49 immune-modulatory50–53 and anti-inflammatory properties.7,53–55
Relative to other tissues, a reasonable volume of research has been undertaken into the link between different HDAC enzymes and bone, cardiac and neural/nervous tissues, and further, tissue engineering. Thus, this review article will focus on these three key areas, looking at the history, current status of in vitro and in vivo research, and finally with a nod to future perspectives.
HDACs and HDACis in bone tissue formation and regeneration
There is still an unmet major clinical need to regenerate bone tissue for fracture repair and the restoration of bone loss due to injury, congenital disorders and degenerative diseases. Researchers currently typically utilise chemicals such as dexamethasone, ascorbic acid and phosphate sources,56,57 as well as osteogenic growth factors such as bone morphogenetic proteins to stimulate osteogenic differentiation of stem cells toward bone forming cells.58 These osteoblast-like cells can produce collagen matrix and modulate matrix mineralisation and maturation.59 However, the current technologies either have limited efficacy, or adverse side effects, resulting in a search for additional or alternative approaches enhancing bone tissue engineering.
History
Osteoblasts and osteoclasts are the two main cell types responsible for bone formation (osteoblasts), resorption (osteoclasts) and remodelling (both). Research into the effect of HDACis on bone first began in the 1990s, when sodium butyrate (NaB) was found to promote osteoblast bone formation.60
Since then, historically, a large number of studies involving HDACis have been to investigate their effect on bone densities, due to initial concerns about the side-effects of HDACis utilised in cancer therapies on bone tissue and density.61–64 Some research indicated that SAHA inhibited the growth of bone-related cell populations, such as human bone marrow stem cells, whilst also having the potential to increase osteogenic differentiation in vitro. However, a study found that SAHA did not affect bone mineral density, and could be a promising tool for both oncological and tissue engineering applications.65 However, in different animal species, HDACis may affect bone density in different ways.62
HDAC isoforms and their links with bone
It has been suggested that several HDAC isoforms are linked to osteoblast activity, in particular, HDAC3 is thought to be closely linked to bone tissue formation.66–69 HDAC3 interacts with Runx2, suppressing osteocalcin production and regulating progenitors to differentiate into osteoblasts.67,70,71 During osteoblast differentiation, HDAC3 activity is reduced, resulting in acetylation of histones 3 and 4. This balance of acetylation can be altered in cells by over-expressing or under-expressing Nuclear Factor of Activated T Cells (NFAT) signalling.72 Further linking HDAC3 to bone formation, H3, as well as H4, acetylation can promote osteocalcin production, which is essential for bone formation.73 In vivo HDAC3 knockout reduced osteoblast levels and increased fatty deposits in bone marrow, seriously compromising skeletal health in mice.68 Another study found HDAC3 to be essential for bone maintenance during ageing when knocked down the mouse bone mass was reduced.69 Similarly, HDAC1 is downregulated during osteoblast differentiation, making it another potentially interesting bone linked HDAC enzyme to be targeted with specific inhibitors to induce osteoblast differentiation.74 Another class 1 HDAC, HDAC8 is known to suppress osteogenic-related genes expression, and inhibition of this with VPA promoted osteogenic expression of rat bone marrow stromal cells.75
Jointly HDAC4 and HDAC5 have been found to play a role in osteoblast differentiation.76,77 HDAC4 also specifically controls chondrocyte hypertrophy, which is involved in endochondral ossification during bone formation.78 Moreover, HDAC6 is linked to the differentiation of MSCs into osteogenic lineages.79
Moving on from osteoblasts, both class I and class II HDAC enzymes are required for osteoclast differentiation,80 and Romidepsin, which preferentially inhibits HDAC1 and HDAC2, inhibits osteoclastogenesis.81 HDAC3 and HDAC7 appear to have opposite effects: suppression of HDAC7 accelerates osteoclast differentiation, while suppression of HDAC3 inhibits osteoclast differentiation.82 Similarly, other researchers have suggested that HDAC5 and HDAC6 activity will reduce osteoclast differentiation.83
The Wnt signalling pathway is an extensively studied pathway with a key role in the promotion of osteogenic differentiation of MSCs. The Wnt pathway cross-talks with pathways such as the BMP, Notch and Hedgehog pathways, and causes upregulation of osteogenic mediators Runx2, Dl5x and Osterix, as well as suppressing PPARγ. It is likely that increased knowledge of how HDAC enzymes interact with the Wnt pathway will result in improvements in bone tissue engineering protocols.84
Current status of in vitro research for HDACis in bone tissue engineering
Although the specificity of different HDACis is possibly questionable,19 it is clear that there are obvious differences in effects between different HDACis on bone-related cells. Reports on the effects of HDACis on bone cells can be conflicting. One study showed that high levels of HDAC acetylation were found to block vitamin D stimulation of osteocalcin production, and thus osteoblast differentiation.85 Lee HW and colleagues (2006) showed that NaB and TSA increased osteogenic characteristics in rat cell lines, as well as the upregulation of osteoblast marker genes by NaB.74 NaB can also promote osteogenic differentiation of periodontal ligament fibroblasts, and has the added benefit of modulating inflammatory reactions.55 Similarly, TSA, MS-275 and VPA have been shown to upregulate pre-osteoblast’s osteogenic gene expression.86 However, other reports suggest that TSA may not induce osteoblasts maturation.60,66 Another study found that HDAC2-specific MS-275 stimulates bone regeneration both in vitro and in vivo.87,88 Thus, the effects of HDACis on cells are still very complex and require further study.
As previously highlighted, several publications demonstrate that HDACis, such as Trichostatin A (TSA) and NaB, inhibit osteoclast differentiation.60,89,90 In 2007, T Yi and J Baek reported that TSA could actually cause the death of osteoclasts,91 and Kim et al. (2012) suggested that HDAC2-specific inhibitor (MS-275) can reduce bone absorption.88 These inhibitory effects on osteoclasts could be utilised to prevent bone loss in inflammatory disease.92 Although, contradictorily, it has also been shown that TSA promoted the expression of RANKL, a ligand integral to osteoclast formation, function and survival.93 Another HDACi, Largazole was found to increase the in vitro osteogenic potential of C2C12 cells, a murine mesenchymal progenitor cell line, the authors believe this to be due to an increased expression of Runx2 and BMPs.94
MSCs can also be derived from dental pulp and have shown great potential for tissue engineering particularly for mineralised tissues such as bone.56 VPA has been shown to improve the generation of mineral matrix in dental pulp MSCs, as well as promoting the expression of some osteogenic bone markers such as osteopontin (OPN) and bone sialoprotein (BSP), albeit with a decrease in osteocalcin (OC) expression. It was also demonstrated that shRNA silencing of HDAC2 lead to increased expression of OPN and BSP, but also the downregulation of OC.95 TSA has also been shown to promote proliferation and differentiation odontoblasts in DPSCs,96 and VPA and TSA have been demonstrated to increase mineralisation of murine dental pulp-derived cell line (MDPC-23) cells.35 A recent review of this area has suggested how bone-related markers and HDACs in hPDLs and DPSCs change during odonto/osteogenic differentiation.97
It is clear that the choice of HDACi, delivery system and the length of time that cells are exposed to the HDACis may be vital for utilising HDACis in bone tissue engineering.98 Several in vitro studies demonstrate the importance of controlling the length of time cells that are exposed to the HDACis, leading to researchers often developing pre-treatment strategies to effect differentiation in in vitro protocols.99–101
Current status of in vivo research for HDACis in bone tissue engineering
In the literature, it is not uncommon for in vivo and in vitro studies to contradict each other. Building on previous work by the group,101 one of the key first studies utilising HDACis in bone tissue engineering in 2006 reported that the pan-HDACis TSA and NaB increased osteogenic differentiation of MSCs in vitro and ex vivo, however, these results could not be recreated in in vivo conditions.11
In 2010, Hung HM et al. observed an increased osteoblast differentiation when the α-calcium scaffolds with HDACis TSA and NaB were placed in rat bone defect models.98 Elsewhere in both an in vitro and in vivo study, HDACis NaB and VPA have been used in combination with reduced oxygen environments and adipose-derived stem cells to achieve the regeneration of bone tissue.100 Backing up their in vitro findings, Lee et al. (2011) found that when collagen scaffolds soaked in Largazole solution were implanted in mouse models, there was improved bone formation.94
HDACs and HDACis in heart tissue formation and regeneration
There is a great demand to be able to develop cardiac tissue engineering and regeneration therapies, as heart disease is a major killer worldwide.102 It has been suggested that HDAC proteins are key in cardiac tissue development and repair,103 as well as the prevention of degenerative cardiac diseases.104 For example, HDAC activity has been linked to ischaemic injury, which kills 1 million Americans a year.105 HDACis have potential as drugs for treatments of cardiac diseases,106–110 as well as in improving cardiac tissue engineering,31,104,111 although there is a complicated epigenetic landscape to be understood with each indicated disease associated with the heart.112
History
Cardiac tissue engineering is challenged with selecting a suitable cell type and obtaining and maintaining enough viable cardiomyocytes from stem cell populations. Currently, chemicals such as 5-azacytidine, or other methods such as the coculture of MSCs with mature cardiac muscle cells have been utilised, in an attempt to control stem cell differentiation.102,113,114 However, these methods have yielded inconsistent results. Some studies show that there may be a link between HDAC enzymes and cardiac differentiation, therefore a potential to utilise HDACis for cardiac tissue engineering and regeneration.31,104,111 Also, such processes are very complicated, and research is far from being able to create functional cardiac tissue.
HDAC isoforms and their links with cardiac tissue
HDAC1 knockout mice die early in embryogenesis at embryonic day 9.5 due to cardiac defects. HDAC2 deletion leads to survival to the perinatal stage (shortly after birth), but a subsequent death due to a variety of cardiac defects. Cardiac tissue-specific deletion of HDAC1 or HDAC2 alone resulted in no effect on the heart, but deletion of both together resulted in cardiac defects.115 Some reports suggested that activity of the class IIa HDACs (HDAC4, 5, 7 and 9) suppress cardiac hypertrophy (heart wall thickening)116 whereas class I enzymes (HDAC1 and 2) promote hypotrophy.117 Knocking out HDAC7 leads to vascular fatalities in mice.118
During coculture with cardiomyocytes, expression of HDAC1 decreases in bone marrow MSCs. When HDAC1 was knocked down in these cells, the expression of cardiomyocyte related genes was then significantly increased, further indicating the key role of HDAC1 in the differentiation down this lineage.119 In both mice and humans HDAC3 is upregulated during endothelial differentiation, and HDAC7 was upregulated during smooth muscle differentiation.103 HDAC3 is a regulator of cardiac myocyte proliferation during cardiac development in mice120 and, also in mice, is essential to the maintenance of cardiac function and cell metabolism.121 HDAC4, and HDAC5 in particular are known to be highly expressed in the heart tissue.122 Individually knocking out HDAC5 and HDAC9 caused mice to become sensitive to cardiac stress signals, and knocking down both resulted in severe defects, suggesting overlapping functions in cardiac tissue development.123
To date, the vast majority of reports studying HDAC isoforms and heart tissue have been carried out in mouse models; however, there are obvious differences between the mouse and the human.124 Different methods of assessing the role of HDAC knockdown may also affect results.125 Thus, the findings of these studies may not represent the true role of HDACis on human heart disease and development.
Current status of in vitro research for HDACis in cardiac tissue engineering
TSA is the most studied HDACi in this field; along with SAHA, it has been found to have a strong effect on cardiogenic induction of rat MSCs when compared with the conventional 5-azacytidine protocol.111,126 Interestingly, pre-treatment of rat MSCs with 5-azacytidine enhances the effect of TSA on MSC carcinogenesis, and improvements were also demonstrated when adding TSA treatment to the coculture of MSCs and neonatal cardiomyocytes protocol.111 These researchers suggest that the combination of HDACis with existing methods could be the future of this research.
TSA has been utilised to stimulate the differentiation of human adipose-derived stem cells into cardiomyocytes.127 TSA has also been shown to induce other cardiac cell type differentiation (e.g., endothelial and smooth muscle cells) from cardiac side population stem cells.128 In 2010, Kaichi et al. reported that TSA could reduce inconsistencies and greatly improve the quality of mouse cardiomyocytes differentiated from mouse iPSC lines.129 Perhaps even more interestingly, a short term treatment of human or mouse embryonic stem cells (ESCs) with TSA can dramatically promote the differentiation of these ESCs into cardiomyocytes, and further increase the cardiac function of the resulting cardiomyocytes.130–132 Furthermore, the use of TSA with mouse embryonic carcinoma stem cells promoted cardiac development; however, the upregulation of HDAC4 specifically decreased cardiomyogenesis.133
Aside from TSA, Chow and co-workers (2013) used VPA to promote the maturation of human ESC-derived ventricular cardiomyocytes. Interestingly, they also suggest that this may be context dependent and that the developmental stage of the cells being treated could influence the outcome.124
Current status of in vivo research for HDACis in cardiac tissue engineering
There has been limited in vivo validation of the potential of HDACis in cardiac repair. Lee et al. (2007) proved that VPA and tributyrin had a positive effect on the remodelling of damage rat ventricles.12 Further work has shown TSA can improve ventricular function recovery in a defect model, which was found to be c-kit signalling dependent.134
HDACs and HDACis in the brain, and nervous tissue formation and regeneration
The typical mammalian nervous system of the body is made up of several distinctly different tissue types: the brain and the spinal cord make up the central nervous system (CNS), separate from the peripheral nervous system (PNS), which exists to relay information to and from the CNS.135 Researchers are struggling to develop effective strategies to heal injuries, or generate tissue from both the CNS and the PNS. Investigations have shed some light onto key HDAC enzymes in these tissues and cell populations, often drawing on the fact HDACis (particularly VPA) have historically actually been used for the treatment of various neurological diseases.136,137
History
HDACis have been used in the treatment of neurodegenerative diseases since the 1970s, primarily used to prevent neuron damage.138 Although VPA earned FDA approval in 1987 for its use as an anticonvulsant and mood stabiliser,21 it was not until 2001 that histone deacetylation inhibition was identified as the mode of action of VPA, resulting in its reclassification as a HDACi.139,140
Tissue engineering of these very complicated tissues is a relatively undeveloped area (especially when compared to the previously discussed bone and heart tissues), and differentiation protocols are still being refined and developed. For nervous tissue engineering, current research mainly focuses on the use of nerve guide conduits in combination with different growth factors.141 Hu et al. (2012) reported that there might be potential for epigenetic control to be exploited in nervous tissue engineering.142 However, the CNS has a much lower regenerative capacity than the PNS, and current therapies focus mainly on preventing neurodegeneration, rather than actual full regeneration. Regeneration with stem cells and biodegradable and bio-absorbable materials such as polyethene, collagen, gelatin, and chitosan have been explored in the brain and spinal cord, but there is a vast volume of work to be done.143
HDAC isoforms and their links with neural and nervous tissue
HDAC enzymes play a key role in neural cell formation in mouse ESCs.144 Some HDACs (e.g., HDAC1, HDAC3, HDAC5 and HDAC7) are highly expressed in neural stem cells but are downregulated as neuronal cells differentiate, and therefore are thought to play a role in neuronal differentiation.145,146
More specifically, HDAC1 induces the differentiation of retinal progenitor cells and motor neurons in zebrafish development.147 HDAC1 and HDAC2 are linked to neural stem and progenitor cells and synaptic formation, but only HDAC1 is expressed in fully differentiated glia.146,148 However, HDAC2 is upregulated in neuronal differentiation, while HDAC1, HDAC3, HDAC5 and HDAC7 are downregulated.149–153 In mice, HDAC1 and HDAC2 activity are required for oligodendrocyte differentiation, as well as Schwann cell development.154,155 They are also regulators of myelination in the peripheral nervous system.156 However, other researchers report that, in general, HDAC activity inhibits the differentiation of oligodendrocytes to neurons.157 HDAC2 activity may be related to the differentiation of neural progenitors,151,153 and can inhibit astrocyte differentiation.154 Knockout of both HDAC1 and HDAC2 in mice neuronal precursor cells leads to cell abnormalities, disorganisation and postnatal mouse fatality.158 Individually knocking down HDAC1 or HDAC2 also affects neuronal development of mice.159,160
HDAC3 is key to neuronal survival, control of apoptosis, and is linked to neural disease,161 and memory function.162 HDAC3, together with HDAC1 and HDAC2, has been linked to cortical cell differentiation.154 HDAC8 is not found to be expressed in normal cells from the central or peripheral nervous system, but is linked to related cancers, notably childhood neuroblastoma, so could be a promising target in this field.26,163
Caution should be exercised when exploring HDACis in this field, as it has been demonstrated that targeting class I HDACs with inhibitors in human oligodendrocyte progenitor cells leads to a reduction in progenitor recruitment via proliferation arrest, and inhibition of differentiation into oligodendrocytes. Consequently, Conway and co-workers emphasis that it is also important to realise that there are subtle differences between species in regulation of HDAC activity.157
Of the class II HDACs, HDAC4 is key, to neuronal activity, and highly expressed in the brain, in neurodegenerative disorders, and HDAC4 activity is thought to provide the neurons protection from cell death by preventing abortive cell cycle progression.164 HDAC5, HDAC6 and HDAC7 are all linked to neural diseases, such as Huntingdon’s disease, protein aggregated diseases and to neural mitochondrial-related diseases respectively.163 HDAC5 mediated pathways are linked to early-stage neural cell fate.165 HDAC6 has a complicated role in these systems with both a neuroprotective and neurodegenerative role.166 HDAC9 is highly expressed in the brain and is proposed to be important in the formation of nerve synapses.167
Interestingly, the only class IV HDAC, HDAC11, is barely expressed in precursors to neural cells, but largely features in mature neurons, and therefore is thought to be key to postnatal development of neurons.168
Current status of in vitro research for HDACis in neural and nervous tissue engineering
Neural stem cells were first isolated and reported in the early 90s,169,170 and subsequently several related other distinct cell populations have since been discovered.171,172 Due to the importance of different HDAC isoforms in neuronal and nervous tissue development, the use of specific HDACis could be an especially promising approach for improving the current approaches in tissue engineering. However, so far, only limited studies into the effects of HDACis on relevant neuronal cells have been undertaken.
As Valproic acid (VPA) obtained FDA approval for treating neurological diseases in the 1980s, there has been a relatively large volume of work on the effects of VPA on cells of these systems.173 VPA can induce neuronal differentiation of adult rat hippocampal neural progenitor cells whilst inhibiting glial cell differentiation; favouring neuronal differentiation.32 VPA can alter cell function and microglial phenotype of fully developed human glial cells, although it does not result in apoptosis as shown in rodent cells.174 In a different study, VPA was shown to enhance rat cerebral cortex neural progenitor cell neural differentiation both in vitro and in vivo.175 Moreover, VPA has been shown to control oligodendrocyte progenitor differentiation.150 VPA induces proneural factors in rat cells, specifically it has been shown that early stage progenitor cells are more sensitive to the effects of HDAC inhibition, which is thought to be linked to H4 acetylation.176 Juliandi and co-workers (2012) reported that VPA treatment enhanced the differentiation of mouse-derived ESCs into neuronal lineage layers.177 However, VPA has also been demonstrated to inhibit proliferation, cell “stemness” and the ability to form neurospheres of mice adult subventricular zone cells, a separate neural cell population.178 Finally, it has been demonstrated that VPA increases axonal growth of neurons in vitro.179
Similar to VPA, TSA has also been utilised to induce mouse neural cells (in minimal supplement media) to express some of the properties of fully functioning neurons.180 In a separate study, Siebzehnrubl et al. (2007) found that the HDACis MS-275, M344 and SAHA induced the neuronal differentiation of mouse forebrain precursor cells.181 Also, HDACi treatment has been combined with extrinsic transcription factors to promote mouse neural cells to differentiate into dopaminergic neurons.182 When treated with HDACis, mouse cells showed increased plasticity, reversed lineage commitment and upregulation of 13 genes associated with neural stem cell states, e.g., Sox2.183 Interestingly, HDAC1 and HDAC2 inhibition can be detrimental to oligodendrocyte differentiation.157,183 However, in comparison, HES5 (a notch regulator) was not upregulated in human cells. Thus, once again in this field, it is important to note that not all animal cell models may be directly translatable into human models.157
Current status of in vivo research for HDACis in nervous tissue engineering
Unsurprisingly, given the limited number of in vitro studies, there is only a handful of in vivo studies specifically related to tissue engineering and/or regeneration. An in vivo model of the aforementioned in vitro study demonstrated that VPA regulates differentiation and proliferation of rat cerebral cortex neural progenitor cells in the cerebral cortex of developing rat embryos.175 VPA has also been proven to reduce cell apoptosis and increase locomotion recovery in a rat spinal cord injury model.184 VPA has also been shown to increase the differentiation of NSCs into neurons, not glial cells, along with improved limb function, in mouse spinal cord injury models.185
A brief discussion of the role of HDACs and HDACis in skeletal muscle
Skeletal muscle tissue formation is also regulated by signalling pathways such as Wnt, BMP and the Class II HDACs (HDAC4, HDAC5, HDAC7 and HDAC9) are highly expressed in skeletal muscle. HDACs play a key role in the epigenetic regulation of embryonic myogenesis and adult muscle regeneration.186 For example Class II HDACs are known to directly bind myocyte enhancer factor-2 (MEF2) and consequently repress expression of MEF2-dependent genes.187,188 HDAC enzymes and their signalling pathways play a key role in regulating myocyte differentiation and remodelling of muscle tissue.187,189 HDAC4, in particular, is necessary for maintaining skeletal muscle homoeostasis and function. And it has been identified as a potential target for treatment of skeletal muscle disorders such as cancer cachexia, amyotrophic lateral sclerosis (ALS) or muscular dystrophy.190
Summary
In these three key tissue areas, it has been demonstrated HDACis could be very useful tools to improve tissue engineering strategies. Since bone tissue engineering has found early success with studies combining MSCs with HDACis,11,100,101 there is potential to translate this research into clinical settings. Similarly, for cardiac tissue engineering, the many studies involving TSA is a strong beginning for this field,108 and when isoform-specific strategies begin, HDAC4 in particular has proven a potentially interesting target for cardiac tissue engineering.122 Regarding neural tissue engineering, most of the studies have explored the use of VPA, which is considered a pan-HDACi. Therefore, it is believed that selective isoform targeting may lead to improvements in this field.163,191
Presently, researchers aim to combine suitable cells (e.g., MSCs) with scaffolds, chemical cues (e.g., growth factors) and environmental stimuli (e.g., mechanical stimulation) to general functional tissue, such as new nerves, bone and other muscular-skeletal tissues in dentistry and/or orthopaedics. This can be carried out in vitro before being implanted/transplanted in vivo, where the naturally occurring cells can populate the scaffold and chemical cues aid the growth of the targeted tissues. Therefore, HDACis could be utilised as the chemical cues to improve the efficacy of current tissue engineering techniques.
Future perspectives
There has been some early success stories utilising HDACis in tissue engineering, but looking to the future, the rise of isoform-specific HDACis may lead this research to new heights. There is still plenty to be said for the classical HDACis; however, the connection between tissues and specific HDAC isoforms should lead to more examples of targeted tissue engineering scenarios.
The current limitations of the technologies must be considered:
a lack of understanding of the pathways HDAC enzymes themselves are involved in, and the effects of these enzymes on cellular properties and behaviour;
a fundamental lack of knowledge of the function of individual HDAC enzymes, in different cell/tissue types;
the mechanism of action of many HDACis, i.e., how pan-HDACi’s function, what HDAC isoforms each actually inhibits, and therefore their true mechanisms of action;
full optimisation of the desired concentration and treatment period of each HDACi on individual cells, to give the best outcome; and
the need for extensive in vivo testing in clinically relevant animal models, with particular attention to the side effects, before the translation of any HDACis for clinical therapies in this area.
Conclusion
In conclusion, the field of HDACis in tissue engineering is still in its infancy compared with cancer research where at least 4 HDACis have achieved clinical approval, with many others in late-stage clinical trials.192 As demonstrated herein, easily synthesised small-molecular compounds in low concentrations could massively improve the efficacy of stem cell therapies and tissue engineering, potentially without the risks associated with genome altering therapies. As discussed at length in this review, there is a potential paradigm shift in this field with the rise of isoform-specific HDACis. As more regulatory barriers are cut, due to increased clinical approval of HDACis, the potential to translate these compounds in other filed such as in tissue engineering has been increased.
Acknowledgements
This research was funded by the EPSRC Centre for Doctoral Training in Tissue Engineering and Regenerative Medicine–Innovation in Medical and Biological Engineering, multidisciplinary collaboration of Faculties at the University of Leeds. Grant number EP/L014823/1. X.Y. and L.L. were partially sponsored by the EU [FP7/2007-2013] for ‘SkelGEN’ consortium (n° 318553) and the British Council. Emeritus Professor Ronald Grigg at Leeds School of Chemistry for his supervision to L.L. and proofreading of the manuscript.
Competing interests
The authors declare no competing interests.
References
- 1.Orlando G, et al. Regenerative medicine and organ transplantation: past, present, and future. Transplantation. 2011;91:1310–1317. doi: 10.1097/TP.0b013e318219ebb5. [DOI] [PubMed] [Google Scholar]
- 2.Atala A, Bauer SB, Soker S, Yoo JJ, Retik AB. Tissue-engineered autologous bladders for patients needing cystoplasty. Lancet. 2006;367:1241–1246. doi: 10.1016/S0140-6736(06)68438-9. [DOI] [PubMed] [Google Scholar]
- 3.Tollervey JR, Lunyak VV. Epigenetics: judge, jury and executioner of stem cell fate. Epigenetics. 2012;7:823–840. doi: 10.4161/epi.21141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 5.Dinarello CA, Fossati G, Mascagni P. Histone deacetylase inhibitors for treating a spectrum of diseases not related to cancer. Mol. Med. 2011;17:333–352. doi: 10.2119/molmed.2011.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ropero S, Esteller M. The role of histone deacetylases (HDACs) in human cancer. Mol. Oncol. 2007;1:19–25. doi: 10.1016/j.molonc.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shakespear MR, Halili Ma, Irvine KM, Fairlie DP, Sweet MJ. Histone deacetylases as regulators of inflammation and immunity. Trends Immunol. 2011;32:335–343. doi: 10.1016/j.it.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kretsovali A, Hadjimichael C, Charmpilas N. Histone deacetylase inhibitors in cell pluripotency, differentiation, and reprogramming. Stem Cells Int. 2012;2012:10. doi: 10.1155/2012/184154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elizalde C, et al. Histone deacetylase 3 modulates the expansion of human hematopoietic stem cells. Stem Cells Dev. 2012;21:2581–2591. doi: 10.1089/scd.2011.0698. [DOI] [PubMed] [Google Scholar]
- 11.Boer J, Licht R, Bongers M. Inhibition of histone acetylation as a tool in bone tissue engineering. Tissue Eng. 2006;12:2927–2937. doi: 10.1089/ten.2006.12.2927. [DOI] [PubMed] [Google Scholar]
- 12.Lee T, Lin M, Chang N. Inhibition of histone deacetylase on ventricular remodeling in infarcted rats. Am. J. Physiol. 2007;293:968–977. doi: 10.1152/ajpheart.00891.2006. [DOI] [PubMed] [Google Scholar]
- 13.Watson J, Crick F. A structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 14.Hondele M, Ladurner AG. The chaperone-histone partnership: for the greater good of histone traffic and chromatin plasticity. Curr. Opin. Struct. Biol. 2011;21:698–708. doi: 10.1016/j.sbi.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Filippakopoulos P, Knapp S. The bromodomain interaction module. FEBS Lett. 2012;586:2692–2704. doi: 10.1016/j.febslet.2012.04.045. [DOI] [PubMed] [Google Scholar]
- 16.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 17.Dhalluin C, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 18.Ocker M. Deacetylase inhibitors-focus on non-histone targets and effects. World J. Biol. Chem. 2010;1:55–61. doi: 10.4331/wjbc.v1.i5.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradner JE, et al. Chemical phylogenetics of histone deacetylases. Nat. Chem. Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uciechowska U, Sippl W, Jung M. NAA+-dependent histone deacetylases (Sirtuins) as novel therapeutic targets. Med. Res. Rev. 2009;30:861–889. doi: 10.1002/med.20178. [DOI] [PubMed] [Google Scholar]
- 21.Hahnen E, et al. Histone deacetylase inhibitors: possible implications for neurodegenerative disorders. Expert Opin. Investig. Drugs. 2008;17:169–184. doi: 10.1517/13543784.17.2.169. [DOI] [PubMed] [Google Scholar]
- 22.Balasubramanian S, Verner E, Buggy JJ. Isoform-specific histone deacetylase inhibitors: the next step? Cancer Lett. 2009;280:211–221. doi: 10.1016/j.canlet.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Bertino EM, Otterson GA. Romidepsin: a novel histone deacetylase inhibitor for cancer. Expert Opin. Investig. Drugs. 2011;20:1151–1158. doi: 10.1517/13543784.2011.594437. [DOI] [PubMed] [Google Scholar]
- 24.Richardson P, et al. Phase I trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) in patients with advanced multiple myeloma. Leuk. Lymphoma. 2008;49:502–507. doi: 10.1080/10428190701817258. [DOI] [PubMed] [Google Scholar]
- 25.Jones P. Development of second generation epigenetic agents. Med. Chem. Commun. 2012;3:135. [Google Scholar]
- 26.Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: what are the cancer relevant targets? Cancer Lett. 2009;277:8–21. doi: 10.1016/j.canlet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 27.Bertrand P. Inside HDAC with HDAC inhibitors. Eur. J. Med. Chem. 2010;45:2095–2116. doi: 10.1016/j.ejmech.2010.02.030. [DOI] [PubMed] [Google Scholar]
- 28.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 29.Nebbioso A, et al. Tumor-selective action of HDAC inhibitors involves TRAIL induction in acute myeloid leukemia cells. Nat. Med. 2005;11:77–84. doi: 10.1038/nm1161. [DOI] [PubMed] [Google Scholar]
- 30.Dokmanovic M, et al. Histone deacetylase inhibitors selectively suppress expression of HDAC7. Mol. Cancer Ther. 2007;6:2525–2534. doi: 10.1158/1535-7163.MCT-07-0251. [DOI] [PubMed] [Google Scholar]
- 31.Ohtani K, Dimmeler S. Epigenetic regulation of cardiovascular differentiation. Cardiovasc. Res. 2011;90:404–412. doi: 10.1093/cvr/cvr019. [DOI] [PubMed] [Google Scholar]
- 32.Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc. Natl Acad. Sci. USA. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Catalioto R-M, Maggi CA, Giuliani S. Chemically distinct HDAC inhibitors prevent adipose conversion of subcutaneous human white preadipocytes at an early stage of the differentiation program. Exp. Cell Res. 2009;315:3267–3280. doi: 10.1016/j.yexcr.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 34.Duncan HF, Smith AJ, Fleming GJP, Cooper PR. HDACi: cellular effects, opportunities for restorative dentistry. J. Dent. Res. 2011;90:1377–1388. doi: 10.1177/0022034511406919. [DOI] [PubMed] [Google Scholar]
- 35.Duncan HF, Smith AJ, Fleming GJP, Cooper PR. Histone deacetylase inhibitors induced differentiation and accelerated mineralization of pulp-derived cells. J. Endod. 2012;38:339–345. doi: 10.1016/j.joen.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 36.Niki T, et al. A histone deacetylase inhibitor, trichostatin A, suppresses myofibroblastic differentiation of rat hepatic stellate cells in primary culture. Hepatology. 1999;29:858–867. doi: 10.1002/hep.510290328. [DOI] [PubMed] [Google Scholar]
- 37.Snykers S, De Kock J, Rogiers V, Vanhaecke T. In vitro differentiation of embryonic and adult stem cells into hepatocytes: state of the art. Stem Cells. 2009;27:577–605. doi: 10.1634/stemcells.2008-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knutson SK, et al. Liver-specific deletion of histone deacetylase 3 disrupts metabolic transcriptional networks. EMBO J. 2008;27:1017–1028. doi: 10.1038/emboj.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurinna S, Barton MC. Cascades of transcription regulation during liver regeneration. Int. J. Biochem. Cell Biol. 2011;43:189–197. doi: 10.1016/j.biocel.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glenisson W, Castronovo V, Waltregny D. Histone deacetylase 4 is required for TGFβ1 -induced myofibroblastic differentiation. Biochim. Biophys. Acta. 2007;1773:1572–1582. doi: 10.1016/j.bbamcr.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 41.Avrahami D, Kaestner KH. Epigenetic regulation of pancreas development and function. Semin. Cell Dev. Biol. 2012;23:693–700. doi: 10.1016/j.semcdb.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim M-S, et al. Protein kinase D1 stimulates MEF2 activity in skeletal muscle and enhances muscle performance. Mol. Cell. Biol. 2008;28:3600–3609. doi: 10.1128/MCB.00189-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hong S, Derfoul A, Pereira-Mouries L, Hall DJ. A novel domain in histone deacetylase 1 and 2 mediates repression of cartilage-specific genes in human chondrocytes. FASEB J. 2009;23:3539–3552. doi: 10.1096/fj.09-133215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, et al. Expression of acetylated histone 3 in the spinal cord and the effect of morphine on inflammatory pain. Neural Regen. Res. 2012;7:517–522. doi: 10.3969/j.issn.1673-5374.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naguib M, Bie B, Ting AH. Fundamental concepts of epigenetics for consideration in anesthesiology. Curr. Opin. Anaesthesiol. 2012;25:434–443. doi: 10.1097/ACO.0b013e3283556211. [DOI] [PubMed] [Google Scholar]
- 46.Denk F, McMahon SB. Chronic pain: emerging evidence for the involvement of epigenetics. Neuron. 2012;73:435–444. doi: 10.1016/j.neuron.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiechio S, et al. Epigenetic modulation of mGlu2 receptors by histone deacetylase inhibitors in the treatment of inflammatory pain. Mol. Pharmacol. 2009;75:1014–1020. doi: 10.1124/mol.108.054346. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Z, Cai Y-Q, Zou F, Bie B, Pan ZZ. Epigenetic suppression of GAD65 expression mediates persistent pain. Nat. Med. 2011;17:1448–1455. doi: 10.1038/nm.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steinmann J, Halldórsson S, Agerberth B, Gudmundsson GH. Phenylbutyrate induces antimicrobial peptide expression. Antimicrob. Agents Chemother. 2009;53:5127–5133. doi: 10.1128/AAC.00818-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schildberg FA, Hagmann CA, Böhnert V, Tolba RH. Improved transplantation outcome by epigenetic changes. Transpl. Immunol. 2010;23:104–110. doi: 10.1016/j.trim.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 51.Kinugasa F, et al. Effect of a new immunosuppressant histon deacetylase (HDAC) inhibitor FR276457 in a rat cardiac transplant model. Biol. Pharm. Bull. 2008;31:1723–1726. doi: 10.1248/bpb.31.1723. [DOI] [PubMed] [Google Scholar]
- 52.Reddy P, et al. Histone deacetylase inhibitor suberoylanilide hydroxamic acid reduces acute graft-versus-host disease and preserves graft-versus-leukemia effect. Proc. Natl Acad. Sci. USA. 2004;101:3921–3926. doi: 10.1073/pnas.0400380101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi S, Reddy P. HDAC inhibition and graft versus host disease. Mol. Med. 2011;17:404–416. doi: 10.2119/molmed.2011.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhavsar P, Ahmad T, Adcock IM. The role of histone deacetylases in asthma and allergic diseases. J. Allergy Clin. Immunol. 2008;121:580–584. doi: 10.1016/j.jaci.2007.12.1156. [DOI] [PubMed] [Google Scholar]
- 55.Kim T-I, Han J-E, Jung H-M, Oh J-H, Woo KM. Analysis of histone deacetylase inhibitor-induced responses in human periodontal ligament fibroblasts. Biotechnol. Lett. 2013;35:129–133. doi: 10.1007/s10529-012-0992-6. [DOI] [PubMed] [Google Scholar]
- 56.El-Gendy R, Yang X. Osteogenic differentiation of human dental pulp stromal cells on 45S5 Bioglass® based scaffolds in vitro and in vivo. Tissue Eng. Part A. 2012;19:707–715. doi: 10.1089/ten.tea.2012.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saha S, et al. Osteochondral tissue engineering in vivo: a comparative study using layered silk fibroin scaffolds from mulberry and nonmulberry silkworms. PLoS ONE. 2013;8:e80004. doi: 10.1371/journal.pone.0080004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang X, et al. Induction of human osteoprogenitor chemotaxis, proliferation, differentiation, and bone formation by osteoblast stimulating factor-1/pleiotrophin: osteoconductive biomimetic scaffolds for tissue engineering. J. Bone Miner. Res. 2003;18:47–57. doi: 10.1359/jbmr.2003.18.1.47. [DOI] [PubMed] [Google Scholar]
- 59.Vater C, Kasten P, Stiehler M. Culture media for the differentiation of mesenchymal stromal cells. Acta Biomater. 2011;7:463–477. doi: 10.1016/j.actbio.2010.07.037. [DOI] [PubMed] [Google Scholar]
- 60.Iwami K, Moriyama T. Effects of short chain fatty acid, sodium butyrate, on osteoblastic cells and osteoclastic cells. Int. J. Biochem. 1993;25:1631–1635. doi: 10.1016/0020-711x(93)90522-g. [DOI] [PubMed] [Google Scholar]
- 61.Nissen-Meyer LSH, et al. Levetiracetam, phenytoin, and valproate act differently on rat bone mass, structure, and metabolism. Epilepsia. 2007;48:1850–1860. doi: 10.1111/j.1528-1167.2007.01176.x. [DOI] [PubMed] [Google Scholar]
- 62.Senn SM, et al. Adverse effects of valproate on bone: defining a model to investigate the pathophysiology. Epilepsia. 2010;51:984–993. doi: 10.1111/j.1528-1167.2009.02516.x. [DOI] [PubMed] [Google Scholar]
- 63.McGee-Lawrence ME, et al. Suberoylanilide hydroxamic acid (SAHA; vorinostat) causes bone loss by inhibiting immature osteoblasts. Bone. 2011;48:1117–1126. doi: 10.1016/j.bone.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pratap J, et al. The histone deacetylase inhibitor, vorinostat, reduces tumor growth at the metastatic bone site and associated osteolysis, but promotes normal bone loss. Mol. Cancer Ther. 2010;9:3210–3220. doi: 10.1158/1535-7163.MCT-10-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu S, et al. Effect of the HDAC inhibitor vorinostat on the osteogenic differentiation of mesenchymal stem cells in vitro and bone formation in vivo. Acta Pharmacol. Sin. 2013;34:699–709. doi: 10.1038/aps.2012.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schroeder TM, Westendorf JJ. Histone deacetylase inhibitors promote osteoblast maturation. J. Bone Miner. Res. 2005;20:2254–2263. doi: 10.1359/JBMR.050813. [DOI] [PubMed] [Google Scholar]
- 67.Schroeder TM, Kahler RA, Li X, Westendorf JJ. Histone deacetylase 3 interacts with runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. J. Biol. Chem. 2004;279:41998–42007. doi: 10.1074/jbc.M403702200. [DOI] [PubMed] [Google Scholar]
- 68.Razidlo DF, et al. Histone deacetylase 3 depletion in osteo/chondroprogenitor cells decreases bone density and increases marrow fat. PLoS ONE. 2010;5:e11492. doi: 10.1371/journal.pone.0011492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McGee-Lawrence ME, et al. Histone deacetylase 3 is required for maintenance of bone mass during aging. Bone. 2013;52:296–307. doi: 10.1016/j.bone.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hesse E, et al. Zfp521 controls bone mass by HDAC3-dependent attenuation of Runx2 activity. J. Cell Biol. 2010;191:1271–1283. doi: 10.1083/jcb.201009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lamour V, Detry C, Sanchez C. Runx2-and histone deacetylase 3-mediated repression is relieved in differentiating human osteoblast cells to allow high bone sialoprotein expression. J. Biol. Chem. 2007;282:36240–36249. doi: 10.1074/jbc.M705833200. [DOI] [PubMed] [Google Scholar]
- 72.Choo M, Yeo H, Zayzafoon M. NFATc1 mediates HDAC-dependent transcriptional repression of osteocalcin expression during osteoblast differentiation. Bone. 2009;45:579–589. doi: 10.1016/j.bone.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shen J, et al. Histone acetylation in vivo at the osteocalcin locus is functionally linked to vitamin D-dependent, bone tissue-specific transcription. J. Biol. Chem. 2002;277:20284–20292. doi: 10.1074/jbc.M112440200. [DOI] [PubMed] [Google Scholar]
- 74.Lee HW, et al. Histone deacetylase 1-mediated histone modification regulates osteoblast differentiation. Mol. Endocrinol. 2006;20:2432–2443. doi: 10.1210/me.2006-0061. [DOI] [PubMed] [Google Scholar]
- 75.Fu Y, et al. Histone deacetylase 8 suppresses osteogenic differentiation of bone marrow stromal cells by inhibiting histone H3K9 acetylation and RUNX2 activity. Int. J. Biochem. Cell Biol. 2014;54:68–77. doi: 10.1016/j.biocel.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 76.Kang JS, Alliston T, Delston R, Derynck R. Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. EMBO J. 2005;24:2543–2555. doi: 10.1038/sj.emboj.7600729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jeon E-J, et al. Bone morphogenetic protein-2 stimulates Runx2 acetylation. J. Biol. Chem. 2006;281:16502–16511. doi: 10.1074/jbc.M512494200. [DOI] [PubMed] [Google Scholar]
- 78.Vega RB, et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 79.Huang S, et al. Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression. Stem Cells Dev. 2012;21:2531–2540. doi: 10.1089/scd.2012.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cantley MD, et al. Inhibitors of histone deacetylases in class I and class II suppress human osteoclasts in vitro. J. Cell. Physiol. 2011;226:3233–3241. doi: 10.1002/jcp.22684. [DOI] [PubMed] [Google Scholar]
- 81.Nakamura T, et al. Inhibition of histone deacetylase suppresses osteoclastogenesis and bone destruction by inducing IFN-beta production. J. Immunol. 2005;175:5809–5816. doi: 10.4049/jimmunol.175.9.5809. [DOI] [PubMed] [Google Scholar]
- 82.Pham L, et al. HDAC3 and HDAC7 have opposite effects on osteoclast differentiation. J. Biol. Chem. 2011;286:12056–12065. doi: 10.1074/jbc.M110.216853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim JH, et al. RANKL induces NFATc1 acetylation and stability via histone acetyltransferases during osteoclast differentiation. Biochem. J. 2011;436:253–262. doi: 10.1042/BJ20110062. [DOI] [PubMed] [Google Scholar]
- 84.Desiderio V, Tirino V, Papaccio G, Paino F. Bone defects: molecular and cellular therapeutic targets. Int. J. Biochem. Cell Biol. 2014;51:75–78. doi: 10.1016/j.biocel.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 85.Montecino M, et al. Chromatin hyperacetylation abrogates vitamin D-mediated transcriptional upregulation of the tissue-specific osteocalcin gene in vivo. Biochemistry. 1999;38:1338–1345. doi: 10.1021/bi982171a. [DOI] [PubMed] [Google Scholar]
- 86.Schroeder TM, Nair AK, Staggs R, Lamblin A-F, Westendorf JJ. Gene profile analysis of osteoblast genes differentially regulated by histone deacetylase inhibitors. BMC Genom. 2007;8:362. doi: 10.1186/1471-2164-8-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim H-N, et al. Histone deacetylase inhibitor MS-275 stimulates bone formation in part by enhancing Dhx36-mediated TNAP transcription. J. bone Miner. Res. 2011;26:2161–2173. doi: 10.1002/jbmr.426. [DOI] [PubMed] [Google Scholar]
- 88.Kim H-N, et al. MS-275, a benzamide histone deacetylase inhibitor, prevents osteoclastogenesis by down-regulating c-Fos expression and suppresses bone loss in mice. Eur. J. Pharmacol. 2012;691:69–76. doi: 10.1016/j.ejphar.2012.07.034. [DOI] [PubMed] [Google Scholar]
- 89.Rahman MM, et al. Two histone deacetylase inhibitors, trichostatin A and sodium butyrate, suppress differentiation into osteoclasts but not into macrophages. Blood. 2003;101:3451–3459. doi: 10.1182/blood-2002-08-2622. [DOI] [PubMed] [Google Scholar]
- 90.Takada Y, Gillenwater A, Ichikawa H, Aggarwal BB. Suberoylanilide hydroxamic acid potentiates apoptosis, inhibits invasion, and abolishes osteoclastogenesis by suppressing nuclear factor-kappaB activation. J. Biol. Chem. 2006;281:5612–5622. doi: 10.1074/jbc.M507213200. [DOI] [PubMed] [Google Scholar]
- 91.Yi T, Baek J. Trichostatin A-mediated upregulation of p21 osteoclast apoptosis. Exp. Mol. Med. 2007;39:213–221. doi: 10.1038/emm.2007.24. [DOI] [PubMed] [Google Scholar]
- 92.Cantley MD, Bartold PM, Fairlie DP, Rainsford KD, Haynes DR. Histone deacetylase inhibitors as suppressors of bone destruction in inflammatory diseases. J. Pharm. Pharmacol. 2012;64:763–774. doi: 10.1111/j.2042-7158.2011.01421.x. [DOI] [PubMed] [Google Scholar]
- 93.Fan X, et al. Regulation of RANKL promoter activity is associated with histone remodeling in murine bone stromal cells. J. Cell. Biochem. 2004;93:807–818. doi: 10.1002/jcb.20217. [DOI] [PubMed] [Google Scholar]
- 94.Lee S-U, et al. In vitro and in vivo osteogenic activity of largazole. ACS Med. Chem. Lett. 2011;2:248–251. doi: 10.1021/ml1002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Paino F, et al. Histone deacetylase inhibition with valproic acid downregulates osteocalcin gene expression in human dental pulp stem cells and osteoblasts: evidence for HDAC2 involvement. Stem Cells. 2014;32:279–289. doi: 10.1002/stem.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jin H, Park J, Choi H, Choung P. HDAC inhibitor trichostatin a promotes proliferation and odontoblast differentiation of human dental pulp stem cells. Tissue Eng. Part A. 2013;19:613–624. doi: 10.1089/ten.TEA.2012.0163. [DOI] [PubMed] [Google Scholar]
- 97.Huynh NCN, Everts V, Ampornaramveth RS. Histone deacetylases and their roles in mineralized tissue regeneration. Bone Rep. 2017;7:33–40. doi: 10.1016/j.bonr.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jung H-M, et al. Modulation of the resorption and osteoconductivity of alpha-calcium sulfate by histone deacetylase inhibitors. Biomaterials. 2010;31:29–37. doi: 10.1016/j.biomaterials.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 99.Maroni P, et al. Chemical and genetic blockade of HDACs enhances osteogenic differentiation of human adipose tissue-derived stem cells by oppositely affecting osteogenic and adipogenic transcription factors. Biochem. Biophys. Res. Commun. 2012;428:271–277. doi: 10.1016/j.bbrc.2012.10.044. [DOI] [PubMed] [Google Scholar]
- 100.Xu Y, et al. Inhibition of histone deacetylase activity in reduced oxygen environment enhances the osteogenesis of mouse adipose-derived stromal cells. Tissue Eng. Part A. 2009;15:3697–3707. doi: 10.1089/ten.tea.2009.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cho HH, et al. Induction of osteogenic differentiation of human mesenchymal stem cells by histone deacetylase inhibitors. J. Cell. Biochem. 2005;96:533–542. doi: 10.1002/jcb.20544. [DOI] [PubMed] [Google Scholar]
- 102.Nunes SS, Song H, Chiang CK, Radisic M. Stem cell-based cardiac tissue engineering. J. Cardiovasc. Transl. Res. 2011;4:592–602. doi: 10.1007/s12265-011-9307-x. [DOI] [PubMed] [Google Scholar]
- 103.Zhou B, Margariti A, Zeng L, Xu Q. Role of histone deacetylases in vascular cell homeostasis and arteriosclerosis. Cardiovasc. Res. 2011;90:413–420. doi: 10.1093/cvr/cvr003. [DOI] [PubMed] [Google Scholar]
- 104.McKinsey Ta. Therapeutic potential for HDAC inhibitors in the heart. Annu. Rev. Pharmacol. Toxicol. 2012;52:303–319. doi: 10.1146/annurev-pharmtox-010611-134712. [DOI] [PubMed] [Google Scholar]
- 105.Granger A, et al. Histone deacetylase inhibition reduces myocardial ischemia-reperfusion injury in mice. FASEB J. 2008;22:3549–3560. doi: 10.1096/fj.08-108548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McKinsey TA, Olson EN. Cardiac histone acetylation -therapeutic opportunities abound. Trends Genet. 2004;20:206–213. doi: 10.1016/j.tig.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 107.Hewitson Ruth, Dargan James, Collis David, Green Aneta, Moorjani Narain, Ohri Sunil, Townsend Paul A. Heart failure: The pivotal role of histone deacetylases. The International Journal of Biochemistry & Cell Biology. 2013;45(2):448–453. doi: 10.1016/j.biocel.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 108.McKinsey TA. Isoform-selective HDAC inhibitors: closing in on translational medicine for the heart. J. Mol. Cell. Cardiol. 2011;51:491–496. doi: 10.1016/j.yjmcc.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 109.Lehmann Lorenz H., Worst Barbara C., Stanmore David A., Backs Johannes. Histone deacetylase signaling in cardioprotection. Cellular and Molecular Life Sciences. 2013;71(9):1673–1690. doi: 10.1007/s00018-013-1516-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cardinale JP, et al. HDAC inhibition attenuates inflammatory, hypertrophic, and hypertensive responses in spontaneously hypertensive rats. Hypertension. 2010;56:437–444. doi: 10.1161/HYPERTENSIONAHA.110.154567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang G, et al. Trichostatin a promotes cardiomyocyte differentiation of rat mesenchymal stem cells after 5-azacytidine induction or during coculture with neonatal cardiomyocytes via a mechanism independent of histone deacetylase inhibition. Cell Transplant. 2012;21:985–996. doi: 10.3727/096368911X593145. [DOI] [PubMed] [Google Scholar]
- 112.Papait R, et al. Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proc. Natl Acad. Sci. USA. 2013;110:20164–20169. doi: 10.1073/pnas.1315155110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vunjak-Novakovic G, et al. Challenges in cardiac tissue engineering. Tissue Eng. Part B. Rev. 2010;16:169–187. doi: 10.1089/ten.teb.2009.0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Teng M, Zhao X, Huang Y. Regenerating cardiac cells: insights from the bench and the clinic. Cell Tissue Res. 2012;350:189–197. doi: 10.1007/s00441-012-1484-7. [DOI] [PubMed] [Google Scholar]
- 115.Montgomery RL, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang C, McKinsey T, Chang S, Antos C. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Trivedi CM, et al. HDAC2 regulates the cardiac hypertrophic response by modulating Gsk3 beta activity. Nat. Med. 2007;13:324–331. doi: 10.1038/nm1552. [DOI] [PubMed] [Google Scholar]
- 118.Chang S, et al. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell. 2006;126:321–334. doi: 10.1016/j.cell.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 119.Lu D-F, et al. Knockdown of the HDAC1 promotes the directed differentiation of bone mesenchymal stem cells into cardiomyocytes. PLoS ONE. 2014;9:e92179. doi: 10.1371/journal.pone.0092179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Trivedi CM, Lu MM, Wang Q, Epstein JA. Transgenic overexpression of Hdac3 in the heart produces increased postnatal cardiac myocyte proliferation but does not induce hypertrophy. J. Biol. Chem. 2008;283:26484–26489. doi: 10.1074/jbc.M803686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Montgomery RL, et al. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J. Clin. Invest. 2008;118:3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen HP, et al. HDAC inhibition promotes cardiogenesis and the survival of embryonic stem cells through proteasome-dependent pathway. J. Cell. Biochem. 2011;112:3246–3255. doi: 10.1002/jcb.23251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chang S, McKinsey T. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell. Biol. 2004;24:8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chow MZY, et al. Epigenetic regulation of the electrophysiological phenotype of human embryonic stem cell-derived ventricular cardiomyocytes: insights for driven maturation and hypertrophic growth. Stem Cells Dev. 2013;22:2678–2690. doi: 10.1089/scd.2013.0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Reichert N, Choukrallah M-A, Matthias P. Multiple roles of class I HDACs in proliferation, differentiation, and development. Cell. Mol. Life Sci. 2012;69:2173–2187. doi: 10.1007/s00018-012-0921-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Feng C, et al. Suberoylanilide hydroxamic acid promotes cardiomyocyte differentiation of rat mesenchymal stem cells. Exp. Cell Res. 2009;315:3044–3051. doi: 10.1016/j.yexcr.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 127.Choi YS, et al. Differentiation of human adipose-derived stem cells into beating cardiomyocytes. J. Cell. Mol. Med. 2010;14:878–889. doi: 10.1111/j.1582-4934.2010.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oyama T, et al. Cardiac side population cells have a potential to migrate and differentiate into cardiomyocytes in vitro and in vivo. J. Cell Biol. 2007;176:329–341. doi: 10.1083/jcb.200603014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kaichi S, et al. Cell line-dependent differentiation of induced pluripotent stem cells into cardiomyocytes in mice. Cardiovasc. Res. 2010;88:314–323. doi: 10.1093/cvr/cvq189. [DOI] [PubMed] [Google Scholar]
- 130.Kawamura T, et al. Acetylation of GATA-4 is involved in the differentiation of embryonic stem cells into cardiac myocytes. J. Biol. Chem. 2005;280:19682–19688. doi: 10.1074/jbc.M412428200. [DOI] [PubMed] [Google Scholar]
- 131.Kaichi S, et al. Cyclin-dependent kinase 9 forms a complex with GATA4 and is involved in the differentiation of mouse ES cells into cardiomyocytes. J. Cell. Physiol. 2011;226:248–254. doi: 10.1002/jcp.22336. [DOI] [PubMed] [Google Scholar]
- 132.Otsuji TG, Kurose Y, Suemori H, Tada M, Nakatsuji N. Dynamic link between histone H3 acetylation and an increase in the functional characteristics of human ESC/iPSC-derived cardiomyocytes. PLoS ONE. 2012;7:e45010. doi: 10.1371/journal.pone.0045010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Karamboulas C, et al. HDAC activity regulates entry of mesoderm cells into the cardiac muscle lineage. J. Cell Sci. 2006;119:4305–4314. doi: 10.1242/jcs.03185. [DOI] [PubMed] [Google Scholar]
- 134.Zhang L, et al. Inhibition of histone deacetylase-induced myocardial repair is mediated by c-kit in infarcted hearts. J. Biol. Chem. 2012;287:39338–39348. doi: 10.1074/jbc.M112.379115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lee S, Wolfe S. Peripheral nerve injury and repair. J. Am. Acad. Orthop. Surg. 2000;8:243–252. doi: 10.5435/00124635-200007000-00005. [DOI] [PubMed] [Google Scholar]
- 136.Chen S, Wu H, Klebe D, Hong Y, Zhang J. Valproic acid: a new candidate of therapeutic application for the acute central nervous system injuries. Neurochem. Res. 2014;39:1621–1633. doi: 10.1007/s11064-014-1241-2. [DOI] [PubMed] [Google Scholar]
- 137.Hao Y, et al. Mood stabilizer valproate promotes ERK pathway-dependent cortical neuronal growth and neurogenesis. J. Neurosci. 2004;24:6590–6599. doi: 10.1523/JNEUROSCI.5747-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Brochier C, et al. Specific acetylation of p53 by HDAC inhibition prevents DNA damage-induced apoptosis in neurons. J. Neurosci. 2013;33:8621–8632. doi: 10.1523/JNEUROSCI.5214-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Phiel CJ, et al. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J. Biol. Chem. 2001;276:36734–36741. doi: 10.1074/jbc.M101287200. [DOI] [PubMed] [Google Scholar]
- 140.Göttlicher M, et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bell JHA, Haycock JW. Next generation nerve guides: materials, fabrication, growth factors, and cell delivery. Tissue Eng. Part B Rev. 2012;18:116–128. doi: 10.1089/ten.TEB.2011.0498. [DOI] [PubMed] [Google Scholar]
- 142.Hu X-L, Wang Y, Shen Q. Epigenetic control on cell fate choice in neural stem cells. Protein Cell. 2012;3:278–290. doi: 10.1007/s13238-012-2916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Forraz N, Wright KE, Jurga M, Mcguckin CP. Experimental therapies for repair of the central nervous system : stem cells and tissue engineering. J. Tissue Eng. Regen. Med. 2013;7:523–536. doi: 10.1002/term.552. [DOI] [PubMed] [Google Scholar]
- 144.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 145.Sun G, Yu RT, Evans RM, Shi Y. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc. Natl Acad. Sci. USA. 2007;104:15282–15287. doi: 10.1073/pnas.0704089104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.MacDonald JL, Roskams AJ. Histone deacetylases 1 and 2 are expressed at distinct stages of neuro-glial development. Dev. Dyn. 2008;237:2256–2267. doi: 10.1002/dvdy.21626. [DOI] [PubMed] [Google Scholar]
- 147.Yamaguchi M, et al. Histone deacetylase 1 regulates retinal neurogenesis in zebrafish by suppressing Wnt and Notch signaling pathways. Development. 2005;132:3027–3043. doi: 10.1242/dev.01881. [DOI] [PubMed] [Google Scholar]
- 148.Akhtar MW, et al. Histone deacetylases 1 and 2 form a developmental switch that controls excitatory synapse maturation and function. J. Neurosci. 2009;29:8288–8297. doi: 10.1523/JNEUROSCI.0097-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Romm E, Nielsen JA, Kim JG, Hudson LD. Myt1 family recruits histone deacetylase to regulate neural transcription. J. Neurochem. 2005;93:1444–1453. doi: 10.1111/j.1471-4159.2005.03131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J. Cell Biol. 2005;169:577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bai S, et al. DNA methyltransferase 3b regulates nerve growth factor-induced differentiation of PC12 cells by recruiting histone deacetylase 2. Mol. Cell. Biol. 2005;25:751–766. doi: 10.1128/MCB.25.2.751-766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 152.Ballas N, et al. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- 153.Roopra A, et al. Transcriptional repression by neuron-restrictive silencer factor is mediated via the Sin3-histone deacetylase complex. Mol. Cell. Biol. 2000;20:2147–2157. doi: 10.1128/mcb.20.6.2147-2157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Humphrey GW, et al. Complementary roles for histone deacetylases 1, 2, and 3 in differentiation of pluripotent stem cells. Differentiation. 2008;76:348–356. doi: 10.1111/j.1432-0436.2007.00232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ye F, et al. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat. Neurosci. 2009;12:829–838. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chen Y, et al. HDAC-mediated deacetylation of NF-κB is critical for Schwann cell myelination. Nat. Neurosci. 2011;14:437–441. doi: 10.1038/nn.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Conway GD, O’Bara Ma, Vedia BH, Pol SU, Sim FJ. Histone deacetylase activity is required for human oligodendrocyte progenitor differentiation. Glia. 2012;60:1944–1953. doi: 10.1002/glia.22410. [DOI] [PubMed] [Google Scholar]
- 158.Montgomery RL, Hsieh J, Barbosa AC, Richardson JA, Olson EN. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc. Natl Acad. Sci. USA. 2009;106:7876–7881. doi: 10.1073/pnas.0902750106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zupkovitz G, et al. Negative and positive regulation of gene expression by mouse histone deacetylase 1. Mol. Cell. Biol. 2006;26:7913–7928. doi: 10.1128/MCB.01220-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jawerka M, et al. The specific role of histone deacetylase 2 in adult neurogenesis. Neuron Glia Biol. 2010;6:93–107. doi: 10.1017/S1740925X10000049. [DOI] [PubMed] [Google Scholar]
- 161.Panteleeva I, et al. HDAC-3 participates in the repression of e2f-dependent gene transcription in primary differentiated neurons. Ann. N. Y. Acad. Sci. 2004;1030:656–660. doi: 10.1196/annals.1329.076. [DOI] [PubMed] [Google Scholar]
- 162.McQuown SC, et al. HDAC3 is a critical negative regulator of long-term memory formation. J. Neurosci. 2011;31:764–774. doi: 10.1523/JNEUROSCI.5052-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Morrison BE, Majdzadeh N, D’Mello SR. Histone deacetylases: focus on the nervous system. Cell. Mol. Life Sci. 2007;64:2258–2269. doi: 10.1007/s00018-007-7035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Majdzadeh N, et al. HDAC4 inhibits cell cycle progression and protects neurons from cell death. Dev. Neurobiol. 2008;68:1076–1092. doi: 10.1002/dneu.20637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Schneider JW, et al. Small-molecule activation of neuronal cell fate. Nat. Chem. Biol. 2008;4:408–410. doi: 10.1038/nchembio.95. [DOI] [PubMed] [Google Scholar]
- 166.d’Ydewalle C, Bogaert E, Van Den Bosch L. HDAC6 at the intersection of neuroprotection and neurodegeneration. Traffic. 2012;13:771–779. doi: 10.1111/j.1600-0854.2012.01347.x. [DOI] [PubMed] [Google Scholar]
- 167.Méjat A, et al. Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nat. Neurosci. 2005;8:313–321. doi: 10.1038/nn1408. [DOI] [PubMed] [Google Scholar]
- 168.Liu H, Hu Q, Kaufman A, D’Ercolem J, Ye P. Developmental expression of histone deacetylase 11 in the murine brain. J. Neurosci. Res. 2008;86:537–543. doi: 10.1002/jnr.21521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lois C, Alvarez-Buylla A. Proliferating subventricular zone cells in the adult mammalian forebrain can differentiate into neurons and glia. Proc. Natl Acad. Sci. USA. 1993;90:2074–2077. doi: 10.1073/pnas.90.5.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Reynolds B, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 171.Alvarez-Buylla A, Temple S. Stem cells in the developing and adult nervous system. J. Neurobiol. 1998;36:105–110. [PubMed] [Google Scholar]
- 172.McKay R. Stem cells in the central nervous system. Science. 1997;276:66–71. doi: 10.1126/science.276.5309.66. [DOI] [PubMed] [Google Scholar]
- 173.Gurvich N. Histone deacetylase is a target of valproic acid-mediated cellular differentiation. Cancer Res. 2004;64:1079–1086. doi: 10.1158/0008-5472.can-03-0799. [DOI] [PubMed] [Google Scholar]
- 174.Gibbons HM, et al. Valproic acid induces microglial dysfunction, not apoptosis, in human glial cultures. Neurobiol. Dis. 2011;41:96–103. doi: 10.1016/j.nbd.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 175.Jung G-A, et al. Valproic acid induces differentiation and inhibition of proliferation in neural progenitor cells via the beta-catenin-Ras-ERK-p21Cip/WAF1 pathway. BMC Cell Biol. 2008;9:66. doi: 10.1186/1471-2121-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yu IT, et al. Valproic acid promotes neuronal differentiation by induction of proneural factors in association with H4 acetylation. Neuropharmacology. 2009;56:473–480. doi: 10.1016/j.neuropharm.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 177.Juliandi B, et al. Induction of superficial cortical layer neurons from mouse embryonic stem cells by valproic acid. Neurosci. Res. 2012;72:23–31. doi: 10.1016/j.neures.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 178.Zhou Q, Dalgard CL, Wynder C, Doughty ML. Valproic acid inhibits neurosphere formation by adult subventricular cells by a lithium-sensitive mechanism. Neurosci. Lett. 2011;500:202–206. doi: 10.1016/j.neulet.2011.06.037. [DOI] [PubMed] [Google Scholar]
- 179.Lv L, Han X, Sun Y, Wang X, Dong Q. Valproic acid improves locomotion in vivo after SCI and axonal growth of neurons in vitro. Exp. Neurol. 2012;233:783–790. doi: 10.1016/j.expneurol.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Balasubramaniyan V, et al. Effects of histone deacetylation inhibition on neuronal differentiation of embryonic mouse neural stem cells. Neuroscience. 2006;143:939–951. doi: 10.1016/j.neuroscience.2006.08.082. [DOI] [PubMed] [Google Scholar]
- 181.Siebzehnrubl FA, et al. Histone deacetylase inhibitors increase neuronal differentiation in adult forebrain precursor cells. Exp. Brain Res. 2007;176:672–678. doi: 10.1007/s00221-006-0831-x. [DOI] [PubMed] [Google Scholar]
- 182.Rössler R, Boddeke E, Copray S. Differentiation of non-mesencephalic neural stem cells towards dopaminergic neurons. Neuroscience. 2010;170:417–428. doi: 10.1016/j.neuroscience.2010.07.023. [DOI] [PubMed] [Google Scholar]
- 183.Lyssiotis CA, et al. Inhibition of histone deacetylase activity induces developmental plasticity in oligodendrocyte precursor cells. Proc. Natl Acad. Sci. USA. 2007;104:14982–14987. doi: 10.1073/pnas.0707044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lv L, et al. Valproic acid improves outcome after rodent spinal cord injury: potential roles of histone deacetylase inhibition. Brain Res. 2011;1396:60–68. doi: 10.1016/j.brainres.2011.03.040. [DOI] [PubMed] [Google Scholar]
- 185.Abematsu M, et al. Neurons derived from transplanted neural stem cells restore disrupted neuronal circuitry in a mouse model of spinal cord injury. J. Clin. Invest. 2010;120:3255–3266. doi: 10.1172/JCI42957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Jin W, Peng J, Jiang S. The epigenetic regulation of embryonic myogenesis and adult muscle regeneration by histone methylation modification. Biochem. Biophys. Rep. 2016;6:209–219. doi: 10.1016/j.bbrep.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- 188.McKinsey TA, Zhang CL, Olson EN. Control of muscle development by dueling HATs and HDACs. Curr. Opin. Genet. Dev. 2001;11:497–504. doi: 10.1016/s0959-437x(00)00224-0. [DOI] [PubMed] [Google Scholar]
- 189.Mckinsey TA, Zhang C, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Moresi V, Marroncelli N, Coletti D, Adamo S. Regulation of skeletal muscle development and homeostasis by gene imprinting, histone acetylation and micro RNA. Biochim. Biophys. Acta. 2015;1849:309–316. doi: 10.1016/j.bbagrm.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 191.Fischer A, Sananbenesi F, Mungenast A, Tsai L-H. Targeting the correct HDAC(s) to treat cognitive disorders. Trends Pharmacol. Sci. 2010;31:605–617. doi: 10.1016/j.tips.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 192.Suraweera A, O’Byrne KJ, Richard DJ. Combination therapy with histone deacetylase inhibitors (HDACi) for the treatment of cancer: achieving the full therapeutic potential of HDACi. Front. Oncol. 2018;8:1–15. doi: 10.3389/fonc.2018.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.De Ruijter AJM, van Gennip AH, Caron HN, Kemp S, van Kuilenburg ABP. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yang W-M, Yao Y-L, Sun J-M, Davie JR, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J. Biol. Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 195.Lagger G, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zupkovitz G, et al. The cyclin-dependent kinase inhibitor p21 is a crucial target for histone deacetylase 1 as a regulator of cellular proliferation. Mol. Cell. Biol. 2010;30:1171–1181. doi: 10.1128/MCB.01500-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wilting RH, et al. Overlapping functions of Hdac1 and Hdac2 in cell cycle regulation and haematopoiesis. EMBO J. 2010;29:2586–2597. doi: 10.1038/emboj.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yamaguchi T, et al. Histone deacetylases 1 and 2 act in concert to promote the G1-to-S progression. Genes Dev. 2010;24:455–469. doi: 10.1101/gad.552310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Miller KM, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kim D, et al. Deregulation of HDAC1 by p25/Cdk5 in neurotoxicity. Neuron. 2008;60:803–817. doi: 10.1016/j.neuron.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Jacob C, et al. HDAC1 and HDAC2 control the transcriptional program of myelination and the survival of Schwann cells. Nat. Neurosci. 2011;14:429–436. doi: 10.1038/nn.2762. [DOI] [PubMed] [Google Scholar]
- 202.Moresi V, et al. Histone deacetylases 1 and 2 regulate autophagy flux and skeletal muscle homeostasis in mice. Proc. Natl Acad. Sci. USA. 2012;109:1649–1654. doi: 10.1073/pnas.1121159109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Grausenburger R, et al. Conditional deletion of histone deacetylase 1 in T cells leads to enhanced airway inflammation and increased Th2 cytokine production. J. Immunol. 2010;185:3489–3497. doi: 10.4049/jimmunol.0903610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.LeBoeuf Matthew, Terrell Anne, Trivedi Sohum, Sinha Satrajit, Epstein Jonathan A., Olson Eric N., Morrisey Edward E., Millar Sarah E. Hdac1 and Hdac2 Act Redundantly to Control p63 and p53 Functions in Epidermal Progenitor Cells. Developmental Cell. 2010;19(6):807–818. doi: 10.1016/j.devcel.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Haberland M, Carrer M, Mokalled MH, Montgomery RL, Olson EN. Redundant control of adipogenesis by histone deacetylases 1 and 2. J. Biol. Chem. 2010;285:14663–14670. doi: 10.1074/jbc.M109.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Guan J-S, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Bhaskara S, et al. Deletion of Histone Deacetylase 3 reveals critical roles in S-phase progression and DNA damage control. Mol. Cell. 2009;30:61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Jiang Y, Hsieh J. HDAC3 controls gap 2/mitosis progression in adult neural stem/progenitor cells by regulating CDK1 levels. Proc. Natl Acad. Sci. USA. 2014;111:13541–13546. doi: 10.1073/pnas.1411939111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1320. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Haberland M, Mokalled MH, Montgomery L, Olson EN. Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev. 2009;23:1625–1630. doi: 10.1101/gad.1809209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Verdin E, Dequiedt F, Kasler HG. Class II histone deacetylases: versatile regulators. Trends Genet. 2003;19:286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 212.Zhao X, et al. The modular nature of histone deacetylase HDAC4 confers phosphorylation-dependent intracellular trafficking. J. Biol. Chem. 2001;276:35042–35048. doi: 10.1074/jbc.M105086200. [DOI] [PubMed] [Google Scholar]
- 213.Chen B, Cepko CL. HDAC4 regulates neuronal survival in normal and diseased retinas. Science. 2009;323:256–259. doi: 10.1126/science.1166226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Bolger T, Yao T-P. Intracellular trafficking of histone deacetylase 4 regulates neuronal cell death. J. Neurosci. 2005;25:9544–9553. doi: 10.1523/JNEUROSCI.1826-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Bolger T, Zhao X, Cohen TJ, Tsai C-C, Yao T-P. The neurodegenerative disease protein ataxin-1 antagonizes the neuronal survival function of myocyte enhancer factor-2. J. Biol. Chem. 2007;282:29186–29192. doi: 10.1074/jbc.M704182200. [DOI] [PubMed] [Google Scholar]
- 216.Chawla S, Vanhoutte P, Arnold FJL, Huang CL-H, Bading H. Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J. Neurochem. 2003;85:151–159. doi: 10.1046/j.1471-4159.2003.01648.x. [DOI] [PubMed] [Google Scholar]
- 217.Mckinsey TA, Zhang C, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Lett. Nat. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Agis-Balboa RC, Pavelka Z, Kerimoglu C, Fischer A. Loss of HDAC5 impairs memory function: implications for Alzheimer’s disease. J. Alzheimers Dis. 2013;33:35–44. doi: 10.3233/JAD-2012-121009. [DOI] [PubMed] [Google Scholar]
- 219.Dequiedt F, et al. HDAC7, a thymus-specific class II histone deacetylase, regulates Nur77 transcription and TCR-mediated apoptosis. Immunity. 2003;18:687–698. doi: 10.1016/s1074-7613(03)00109-2. [DOI] [PubMed] [Google Scholar]
- 220.Ji H, et al. A comprehensive methylome map of lineage commitment from hematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Shakespear MR, et al. Histone deacetylase 7 promotes Toll-like receptor 4-dependent proinflammatory gene expression in macrophages. J. Biol. Chem. 2013;288:25362–25374. doi: 10.1074/jbc.M113.496281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Zhou X, Marks PA, Rifkind RA, Richon VM. Cloning and characterization of a histone deacetylase, HDAC9. Proc. Natl Acad. Sci. USA. 2001;98:10572–10577. doi: 10.1073/pnas.191375098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Balasubramanian A, et al. Fam65b is important for formation of the HDAC6-dysferlin protein complex during myogenic cell differentiation. FASEB J. 2014;28:2955–2969. doi: 10.1096/fj.13-246470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zhang D, et al. Activation of histone deacetylase-6 induces contractile dysfunction through derailment of α-tubulin proteostasis in experimental and human atrial fibrillation. Circulation. 2014;129:346–358. doi: 10.1161/CIRCULATIONAHA.113.005300. [DOI] [PubMed] [Google Scholar]
- 225.Gradilone SA, et al. HDAC6 is overexpressed in cystic cholangiocytes and its inhibition reduces cystogenesis. Am. J. Pathol. 2014;184:600–608. doi: 10.1016/j.ajpath.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Liu W, et al. HDAC6 regulates epidermal growth factor receptor (EGFR) endocytic trafficking and degradation in renal epithelial cells. PLoS ONE. 2012;7:e49418. doi: 10.1371/journal.pone.0049418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zhang Y, et al. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol. Cell. Biol. 2008;28:1688–1701. doi: 10.1128/MCB.01154-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Boyault C, et al. HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes Dev. 2007;21:2172–2181. doi: 10.1101/gad.436407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Kwon S, Zhang Y, Matthias P. The deacetylase HDAC6 is an essential component of stress granules and plays a critical role in the cellular response to stress Inauguraldissertation. Genes Dev. 2007;21:3381–3394. doi: 10.1101/gad.461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Lee J-Y, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Sadoul K, et al. HDAC6 controls the kinetics of platelet activation. Platelets Thrombopoiesis. 2015;120:4215–4219. doi: 10.1182/blood-2012-05-428011. [DOI] [PubMed] [Google Scholar]
- 232.Kao H-Y, Lee C-H, Komarov A, Han CC, Evans RM. Isolation and characterization of mammalian HDAC10, a novel histone deacetylase. J. Biol. Chem. 2002;277:187–193. doi: 10.1074/jbc.M108931200. [DOI] [PubMed] [Google Scholar]
- 233.Lai I-L, et al. Histone deacetylase 10 relieves repression on the melanogenic program by maintaining the deacetylation status of repressors. J. Biol. Chem. 2010;285:7187–7196. doi: 10.1074/jbc.M109.061861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Oehme I, et al. Histone deacetylase 10 promotes autophagy-mediated cell survival. Proc. Natl Acad. Sci. USA. 2013;110:E2592–E2601. doi: 10.1073/pnas.1300113110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Gao L, Cueto Ma, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J. Biol. Chem. 2002;277:25748–25755. doi: 10.1074/jbc.M111871200. [DOI] [PubMed] [Google Scholar]
- 236.Villagra A, et al. The histone deacetylase HDAC11 regulates the expression of interleukin 10 and immune tolerance. Nat. Immunol. 2008;10:92–100. doi: 10.1038/ni.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Ververis K, Hiong A, Karagiannis TC, Licciardi PV. Histone deacetylase inhibitors (HDACIS): Multitargeted anticancer agents. Biol. Targets Ther. 2013;7:47–60. doi: 10.2147/BTT.S29965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Khan N, et al. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem. J. 2008;409:581–589. doi: 10.1042/BJ20070779. [DOI] [PubMed] [Google Scholar]
- 239.Hu Erding, et al. Identification of novel isoform-selective inhibitors within Class I histone deacetylases. J. Pharmacol. Exp. Ther. 2003;307:720–728. doi: 10.1124/jpet.103.055541. [DOI] [PubMed] [Google Scholar]