Abstract
Approximately 5–10% of melanoma cases occur in a familial context. CDKN2A/CDK4 were the first high- penetrance melanoma genes identified. The aims of this study were to evaluate CDKN2A/CDK4 variants in Greek familial melanoma patients and to correlate the mutational status with specific clinico-epidemiological characteristics. A cross-sectional study was conducted by genotyping CDKN2A/CDK4 variants and selected MC1R polymorphisms in 52 melanoma-prone families. Descriptive statistics were calculated and comparisons were made using the X2 test, Fisher’s exact test and Student’s t-test for statistical analysis, as appropriate. CDKN2A variants were detected in 46.2% of melanoma-prone families, while a CDK4 variant was found in only one family. This study confirmed that, in the Greek population, the age at melanoma diagnosis was lower in patients carrying a variant in CDKN2A compared with wild-type patients. No statistically significant associations were found between CDKN2A mutational status and MC1R polymorphisms.
Keywords: familial melanoma, CDKN2A, CDK4, MC1R, Greece
Melanoma is one of the most aggressive types of skin cancer because of its tendency to metastasize (1). The incidence of melanoma is increasing rapidly in Caucasian populations (2), with more than 230,000 new cases and 55,000 deaths estimated worldwide in 2012 (Globocan, 2012, World Health Organization (WHO); http://globocan.iarc.fr). The aetiology of melanoma is complex and is driven by the interaction of environmental, phenotypic, and genetic factors. The main environmental risk factor for melanoma is excessive ultraviolet (UV) radiation exposure, either from sunlight or from indoor tanning beds (3). Phenotypic characteristics, such as red or blond hair, blue or green eyes, fair skin with low tanning ability, eye colour, hair colour, freckles, multiple melanocytic naevi, and the presence of clinically atypical naevi, are associated with an increased risk of developing melanoma (4). A personal history of melanoma increases the risk of developing a second melanoma by 5–8% (5, 6), while a family history has been associated with a 1.74 relative risk for melanoma (7), supporting the role of genetic risk factors. A recent study of familial cancer risk in twins from Nordic countries estimated that cutaneous malignant melanoma (CMM) had the highest heritability (58%; 95% confidence interval (CI): 43–73%) among all common cancers (8).
Approximately 5–10% of melanoma cases occur in a familial context (2). Although the underlying genetic basis in the majority of melanoma-prone families is unknown, certain highly penetrant genes, such as the cyclin-dependent kinase inhibitor 2A (CDKN2A) gene (9, 10) and cyclin-dependent kinase 4 (CDK4), have been implicated (11–13). CDKN2A is responsible for melanoma susceptibility in approximately 10% of 2-case melanoma families and 30–40% of families with 3 or more cases of melanoma up to third-degree relatives (14), whereas its incidence among sporadic melanomas is very low (less than 3%) (15). It encodes 2 distinct proteins: p16INK4A (p16) and p14ARF (p14), both of which function in cell cycle regulation (15). CDKN2A mutation carriers are also at increased risk of the development of pancreatic cancer compared with the general population (14, 16, 17).
Variants in the CDK4 gene have been observed in only a small number of melanoma families (11–13). All variants occur in codon 24, with families either carrying a p.R24C substitution (13) or a p.R24H substitution (12). Families carrying variants in CDKN2A and CDK4 exhibit similar characteristics, such as cases of early-onset cutaneous melanoma and multiple primary melanomas (11). Other high-penetrance genes that have been associated with familial melanoma in recent years include BRCA-1 associated protein 1 (BAP-1), telomerase reverse transcriptase (TERT) gene, protection of telomeres 1 (POT1), adrenocortical dysplasia protein homolog (ACD) and telomeric repeat-binding factor 2-interacting protein (TERF2IP). Furthermore, common variants in melanocortin 1 receptor (MC1R) and a single variant (p.E318K) in microphthalmia-associated transcription factor (MITF) confer a moderately increased risk of melanoma (2, 18–20).
The aims of this study were to report the incidence of CDKN2A and CDK4 variants in a series of Greek familial melanoma families and to examine their association with epidemiological and clinical factors, as well as MC1R polymorphisms.
METHODS
Description of familial melanoma cases
The study group consisted of patients with a diagnosis of histologically confirmed cutaneous invasive melanoma in subjects with a confirmed history of a first-, second- or third-degree relative also affected by histologically confirmed invasive melanoma. Patients were recruited consecutively at A. Sygros Hospital, a large referral centre for melanoma and skin cancer in Athens, Greece, and collaborating centres from 2000 to 2016.
Patients recruited for the study were first evaluated for familial occurrence of melanoma using a questionnaire to interview probands about their relatives. Melanoma families were identified if at least 2 melanoma-affected members existed within the family. All patients were informed about the aims and limits of the study and provided written informed consent prior to participation. The study protocol was approved by the Scientific and Ethics Committee of A. Sygros Hospital. The variables included in the analyses were: age of onset, number of primary melanomas, number of melanoma cases within the family and the presence of internal cancers in first- and second-degree relatives of the patients with melanoma. The different types of cancers reported within melanoma-prone families are depicted in Fig. S11. The pedigree of each patient, as well as a family and personal history of cancer, was obtained from a dermatologist upon melanoma diagnosis or during the patient follow-up. Non-melanoma skin cancers were excluded from the analysis.
Phenotypic characteristics were also recorded. Eye and skin colour were evaluated by direct inspection and were subsequently classified into different categories based on a standardized colour system (blue, green, light-brown, dark-brown for eye colour; white, light-brown and dark for skin colour of the inner part of the upper arms). Hair colour at the age of 18 years was assessed on a 5-category scale based on a colour sample chart and separated into 5 categories (blonde, red, light-brown, dark-brown, black). Skin phototype was determined by an interview based on Fitzpatrick’s classification into 4 categories, i.e. (i) always burns, never tans, (ii) always burns, tans lightly, (iii) seldom burns, tans well, and (iv) never burns, tans deeply. In addition, all subjects underwent a complete skin examination by experienced dermatologists for the presence of common and atypical melanocytic naevi.
CDKN2A/CDK4 variant analysis
Patients donated 3 ml peripheral blood for variant analysis. Genomic DNA was obtained from peripheral lymphocytes of the familial melanoma patients using the QIAamp DNA blood mini kit (Qiagen, Hilden, Germany). DNA concentration was quantified in samples prior to genotyping by using Quant-iT dsDNA HS Assay kit (ThermoFischer Scientific, Massachusetts, USA). The CDKN2A locus (exon 1α, 1β, 2) and CDK4exon 2 were amplified by PCR followed by Sanger sequencing (21, 22). In particular, CDKN2A exon 1α, 1β, 2 and the part of the intron between exon 2 and 3 and CDK4 exon 2 were amplified for variant analysis. Exon 1α, 1β, 2 and CDK4 exon 2 were amplified using primers, as described previously (21, 22). The primers for intron amplifications were: P16IV2–7453F (5’ GTT GTA AAA CGA CGG CCA GTA CCA GGG AGGTGT GGG AGA G 3’), and P16IV2–7647R (5’CAC AGG AAA CAG CTATGA CCT GGTTCT TTC AAT CGG GGATG 3’). M13 forward were added to the 5’ of the 7453F primer and M13 reverse sequence were added to 7647 R primer for sequencing purposes. In addition, specific polymorphisms for MC1R were selected and genotyped based on previous work (23). MC1R status was categorized as 0=wild-type (WT) and 1 = at least one of rs11547464, rs1805005, rs1805007, rs1805009, rs2228479, rs1805006 MC1R variants.
Statistical analysis
The collected data were: age of onset, phenotypic characteristics, number of primary melanomas, number of melanoma cases within the family, and the presence of internal cancers in first- and second-degree relatives of the melanoma patients. Data were analysed with Stata 13 software (StataCorp. 2013. Stata Statistical Software: Release 13. College Station, TX: StataCorp LP). Differences in age at onset between CDKN2A+ vs CDKN2A− status were assessed using a Student’s t-test, whilst differences in categorical data (sex, multiple primary melanomas and phenotypic characteristics) were assessed using either a 2-sided χ2 test or a Fisher’s exact test (the latter when any of expected cell counts was lower than 5). CDK4 variants were discovered in only one family; therefore no further statistical analysis was performed. A total of 11 independent statistical tests were performed and a Bonferroni-derived p-value threshold of 0.05/11 = 0.005 was applied to call a result statistically significant.
RESULTS
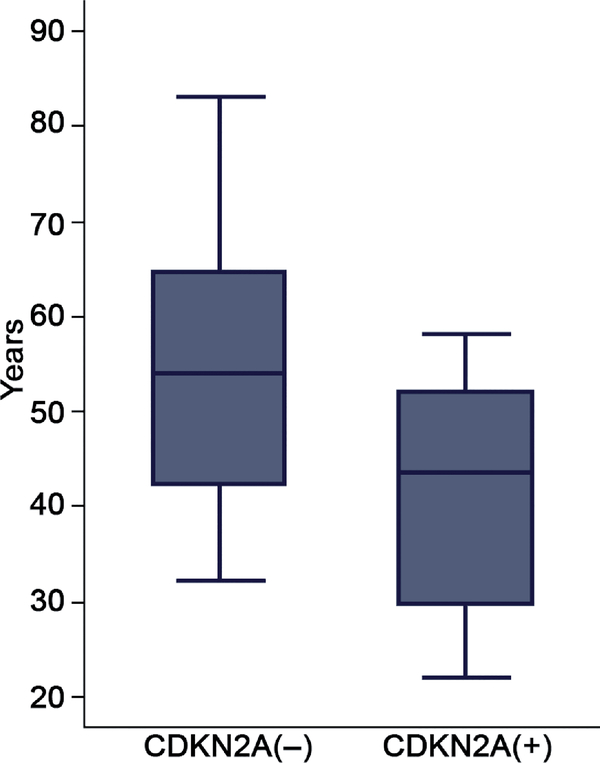
The total number of familial melanoma patients was 66, belonging to 52 families. Since related patients share similar genetic background, only one affected patient (the index case) from each family was included in the analysis. Of these 52 patients, 17 were men and 35 women, with a male-to-female ratio of 1:2. Mean age was 44.6 years for men and 49.4 years for women. Table I presents the distribution of sex, age, history of multiple primary melanomas and phenotypic characteristics among CDKN2A+ and CDKN2A− patients. Age at diagnosis was statistically significantly lower in patients harbouring a CDKN2A variant compared with wild-type patients (41.1 vs 53.6 years, p = 0.001) (Fig. 1). The presence of multiple primary melanomas was not statistically significantly different among CDKN2A+ compared with CDKN2A− patients. There was a trend towards higher number of naevi and more multiple primary melanomas in the CDKN2A carriers but due to the small sample size, this did not reach statistical significance. In addition, there were no statistically significant differences in the phenotypic characteristics (eye, hair and skin colour, tanning, phototype and naevi) between the 2 groups. CDKN2A variants were identified in 36.1% of families with 2 affected members and in 80% of families with at least 3 melanoma-affected members (Table SI1).
Table I.
Demographic characteristics, history of multiple primary melanomas and phenotypic characteristics according to CDKN2A status
| CDKN2A+ | CDKN2A− | Total | ||
|---|---|---|---|---|
| (n = 24) | (n = 28) | (n = 52) | ||
| Sex, n (%) | ||||
| Male | 11 (45.8) | 6 (21.4) | 17 (32.7) | 0.061 |
| Female | 13 (54.2) | 22 (78.6) | 35 (67.3) | |
| Age, mean ± SD | 41.1 ± 11.5 | 53.6 ± 14.2 | 47.8 ± 14.4 | 0.001 |
| Multiple primary melanomas,n (%) | ||||
| Yes | 6 (25.0) | 1 (3.6) | 7 (13.5) | 0.040* |
| No | 18 (75.0) | 27 (96.4) | 45 (86.5) | |
| Phenotypic characteristics, n (%) | ||||
| Eyes (missing values: 3) | ||||
| Blue | 1 (4.6) | 7 (25.9) | 8 (16.3) | 0.109* |
| Green | 9 (40.9) | 5 (18.5) | 14 (28.6) | |
| Light-brown | 3 (13.6) | 6 (22.2) | 9 (18.4) | |
| Dark-brown | 9 (40.9) | 9 (33.3) | 18 (36.7) | |
| Hair (missing values: 2) | ||||
| Blonde | 4 (18.2) | 2 (7.1) | 6 (12.0) | 0.134* |
| Red | 1 (4.6) | 0 (0) | 1 (2.0) | |
| Light-brown | 10 (45.5) | 9 (32.1) | 19 (38.0) | |
| Dark-brown | 7 (31.8) | 13 (46.4) | 20 (40.0) | |
| Black | 0 (0) | 4 (13.8) | 4 (8.0) | |
| Skin colour (missing values : 2) | ||||
| White | 17 (77.3) | 17 (60.7) | 34 (68.0) | 0.213 |
| Brown (light, dark) | 5 (22.7) | 11 (39.3) | 16 (32.0) | |
| Phototype (missing values: 2) | ||||
| Type IIa | 14 (63.6) | 14 (50.0) | 28 (56.0) | |
| Type IIIb | 6 (27.3) | 6 (21.4) | 12 (24.0) | |
| Type IVc | 2 (9.1) | 8 (28.6) | 10 (20.0) | 0.253* |
| Number of naevi (missing values: 6) | ||||
| <30 | 8 (47.1) | 20 (74.1) | 28 (63.6) | 0.070 |
| ≥30 | 9 (52.9) | 7 (25.9) | 16 (36.4) | |
Always burn, minimal tan.
Burn then tan well.
No burn, tan well.
Fisher’s exact test. SD: standard deviation.
Fig. 1.
Difference in distribution at the age of melanoma onset according to CDKN2A status.
CDKN2A and CDK4 variant analysis
Overall, 8 CDKN2A variants (G101R, W110X, 23fsX25, R24P, A148T, c.41–43del, G101E and R87W) were identified (Table II). Five variants were missense found in 18 families (34.6%); 2 were frameshift in 3 families (5.8%) and 1 was nonsense in 4 families (7.7%). In total, 19.7% of CDKN2A variants were in exon 1α, whereas 25.8% were in exon 2. Among the 52 melanoma-prone families, 24 families (46.2%) were found to have 1 variant of CDKN2A and 2 of these families carried 2 different variants of this gene simultaneously. R24P and A148T were the most abundant variants, each of which was found in 8 families respectively, followed by W110X present in 4 families, respectively. CDK4 variant (Arg24His) was observed in only one family.
Table II.
CDKN2A mutations detected in Greek melanoma-prone families
| Variants | Presence (n) in families | Frequency (%) in the 52 families | Effect of mutation | Biological significance (ClinVar) | Allele frequencies in European (non-Finnish) (GnomAD) | |
|---|---|---|---|---|---|---|
| CDKN2A gene | ||||||
| exon 1a | R24P (c.71G>C) | 8 | 15.38 | Missense | Pathogenic | 0.00003834 |
| exon 2 | A148T (c. 442G>A) | 8 | 15.38 | Missense | Uncertain significance | 0.03281 |
| exon 2 | W110X (c.330G>C) | 4 | 7.69 | Nonsense | Not recorded | 0.000 |
| exon 1a | G23fsX25 (c.68delG) | 1 | 1.92 | Frameshift | Not recorded | Not available |
| exon 2 | G101R (c.301G>C) | 1 | 1.92 | Missense | Uncertain significance | 0.000009541 |
| exon 1a | c.41_43delins CCG TGG CTG GCC ACG GCC AC) | 2 | 3.84 | Frameshift | Not recorded | Not available |
| exon 2 | G101E (c.302G>A) | 1 | 1.92 | Missense | Uncertain significance | 0.000009541 |
| exon 2 | R87W (c.259C>T) | 1 | 1.92 | Missense | Likely pathogenic | 0.000 |
| CDK4 gene | ||||||
| exon 2 | R24H (c.71G>A) | 1 | 1.92 | Missense | Likely pathogenic | Not available |
MC1R status among melanoma-prone families
Forty-two out of 52 familial melanoma patients had been genotyped for 6 polymorphisms of MC1R (rs11547464, rs1805007, rs1805009, rs1805006, rs1805005, and rs2228479) in a previous study by our team (23). Table III presents the MC1R status in correlation with CDKN2A variants among familial melanoma families. A MC1R mutation was found in 15 of 18 (83.3%) CDKN2A patients and there was no association of MC1R status with CDKN2A status after Bonferroni correction (p = 0.083). Distribution of major MC1R variants R (rs11547464, rs1805007, rs1805009, rs1805006) and minor MC1R variants r (rs1805005, rs2228479) polymorphisms did not differ statistically significantly among CDKN2A mutant and wild-type families (Table SII1), although CDKN2A mutation carriers had a trend of an increased frequency of R polymorphisms compared with CDKN2A wild types (p = 0.164).
Table III.
MC1R status in association with CDKN2A status in familial melanoma families
|
CDKN2A status |
||||
|---|---|---|---|---|
| MC1R status | CDKN2A+ (n =18) n (%) | CDKN2A− (n = 24) n (%) | Total | p-value |
| Wild-type | 3 (16.7) | 10 (41.7) | 13 (31.0) | 0.083 |
| R or r* | 15 (83.3) | 14 (58.3) | 29 (69.1) | |
rs11547464, rs1805005, rs1805007, rs1805009, rs2228479, rs18050067.
DISCUSSION
This study presents the epidemiological, clinical and genetic characterization of the largest reported series of Greek melanoma-prone families so far. There is a high diversity in the prevalence of CDKN2A variants among families across different geographical areas (14). In the study by GenoMel the highest frequency of variants was observed in Europe (57%) and the lowest in Australia (20%) (14). In areas with high melanoma incidence a combined effect of moderate or low-penetrance susceptibility genes and higher levels of sun exposure may influence the occurrence of familial cases (14). The high percentage (46.2%) of CDKN2A variants detected in our families is consistent with previous findings in European populations, albeit higher than expected for a low incidence population, underscoring the importance of genetic risk factors in the low incidence Greek population. Studies have shown that awareness of CDKN2A variant status among melanoma patients and relatives increases their compliance in sun protection and rigorous mole screening (24). With respect to early diagnosis of internal cancers in patients with CDKN2A variants, there are some encouraging investigational screening protocols for the early detection of pancreatic cancer in high-risk groups (25); however, there is no conclusive evidence to suggest that current methods of pancreatic screening offer any survival benefit (26). Thus, the overall impact of a positive genetic testing for CDKN2A/CDK4 in the early detection and improved outcome of internal cancers and, in particular, of pancreatic cancer remains undetermined.
Our current study did not reveal any new variants or mutations of CDKN2A. A148T and R24P variant were the most frequent variants in the Greek familial melanoma patients. A148T has been cited as a common polymorphism in the general population predominantly of European ancestry in many studies (27–30), while, in some geographical areas, it has also been strongly correlated with familial melanoma (29, 31). In a previous study (32) we identified this polymorphism in 22/304 of sporadic cases (7.2%) and in 3/9 of familial cases (33.3%). R24P has also been reported in other European populations, mainly British (14, 33), but shows a higher frequency in our Greek familial cases. In our familial cases we identified 2 distinct p.G101 mutations, p.G101R and p.G101E, whereas all other studies report a W protein change at the same position (1, 15, 34). Overall, these changes pinpoint that this position could be a hotspot for mutagenesis.
The age of onset was lower in CDKN2A+ patients compared with CDKN2A wild-type, as expected. Specifically, the mean age at melanoma diagnosis was shifted a decade earlier in carriers of CDKN2A variants, highlighting the need for intense vigilance in these individuals from a young age. Presence of MC1R variations was more frequent among CDKN2A+ patients, but not at a statistically significant level. It has been shown recently that MC1R variants increase the penetrance of CDKN2A variants, especially with respect to multiple MC1R variants and to the presence of R alleles (35, 36). It is unclear whether other common variants that have been associated with melanoma, particularly other pigmentation genes, such as SLC45A2, TYR or TYRP-1, may also exhibit a similar modifying effect CDKN2A penetrance similar to MC1R.
Our study has certain limitations. Although it presents the most extensive and best characterized series of Greek melanoma probands published to date, the number of patients and corresponding families is relatively small compared with other published studies on familial melanoma. In addition, we did not genotype the rest of the known high-risk genes involved in familial melanoma in order to investigate the full spectrum of melanoma-related variants in our series. Nevertheless, a subset (49 cases) of these probands has undergone germline whole exome sequencing and no variants in the other high-risk loci were discovered (37). Finally, we did not include in our study other high-risk individuals, such as patients with early onset of disease, as the goal of this study was to present a comprehensive analysis of familial melanoma cases. A more detailed genetic characterization of our expanding melanoma-prone patient series will be our goal for the near future.
In conclusion, our study delineates the important role of CDKN2A variants in Greek familial melanoma families, as almost half of our studied population carried a disease-causing variant in this gene. Intense vigilance and early intervention are important, since age of diagnosis is low, especially among CDKN2A mutants, while medical advice on healthy lifestyle and prompt screening for other cancers, such as pancreatic cancer, is essential.
Supplementary Material
SIGNIFICANCE.
Melanoma is the most aggressive skin cancer. Although the majority of affected individuals are sporadic cases, a minor group is at high risk because of its family history. Cases among the same family often share the same mutations in melanoma-associated genes. Our study represents the largest familial melanoma series in Greece and delineates the need for screening members of affected families for particular genetic alterations. Carriers should undergo frequent clinical follow-up, since they are at high risk for developing melanoma at an early age.
ACKNOWLEDEGMENTS
The authors acknowledge the generous assistance and support of the patients in this study.
Funding sources: This research was financed by Greek national funds through the Operational Program “Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF)-Research Funding Program: Aristeia I-1094. Part of the work has been also funded by the Institute of Dermatologic Research and Education (Athens, Greece).
Footnotes
The authors have no conflicts of interest to declare.
REFERENCES
- 1.Puig S, Potrony M, Cuellar F, Puig-Butille JA, Carrera C, Aguilera P, et al. Characterization of individuals at high risk of developing melanoma in Latin America: bases for genetic counseling in melanoma. Genet Med 2016; 18: 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Potrony M, Puig-Butille JA, Aguilera P, Badenas C, Tell-Marti G, Carrera C, et al. Prevalence of MITF p.E318K in Patients with melanoma independent of the presence of CDKN2A causative mutations. JAMA Dermatol 2016; 152: 405–412. [DOI] [PubMed] [Google Scholar]
- 3.Whiteman DC, Green AC. Melanoma and sun exposure: where are we now? Int J Dermatol 1999; 38: 481–489. [DOI] [PubMed] [Google Scholar]
- 4.Belbasis L, Stefanaki I, Stratigos AJ, Evangelou E. Non-genetic risk factors for cutaneous melanoma and keratinocyte skin cancers: an umbrella review of meta-analyses. J Dermatol Sci 2016; 84: 330–339. [DOI] [PubMed] [Google Scholar]
- 5.Levi F, Randimbison L, Te VC, La Vecchia C. High constant incidence rates of second cutaneous melanomas. Int J Cancer 2005; 117: 877–879. [DOI] [PubMed] [Google Scholar]
- 6.Ferrone CR, Ben Porat L, Panageas KS, Pasquini P, Zanetti R, Masini C, Boyle P, Melchi CF. Clinicopathological features of and risk factors for multiple primary melanomas. JAMA 2005; 294: 1647–1654. [DOI] [PubMed] [Google Scholar]
- 7.Gandini S, Sera F, Cattaruzza MS,Pasquini P, Zanetti R, Masini C et al. Meta-analysis of risk factors for cutaneous melanoma: III. Family history, actinic damage and phenotypic factors. Eur J Cancer 2005; 41: 2040–2059. [DOI] [PubMed] [Google Scholar]
- 8.Mucci LA, Hjelmborg JB, Harris JR, Czene K, Havelick DJ, Scheike T, et al. Familial risk and heritability of cancer among twins in nordic countries. JAMA 2016; 315: 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hussussian CJ, Struewing JP, Goldstein AM, Higgins PA, Ally DS, Sheahan MD, et al. Germline p16 mutations in familial melanoma. Nat Genet 1994; 8: 15–21. [DOI] [PubMed] [Google Scholar]
- 10.Kamb A, Shattuck-Eidens D, Eeles R, Liu Q, Gruis NA, Ding W, et al. Analysis of the p16 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat Genet 1994; 8: 23–26. [DOI] [PubMed] [Google Scholar]
- 11.Puntervoll HE, Yang XR, Vetti HH, Bachmann IM, Avril MF, Benfodda M, et al. Melanoma prone families with CDK4 germline mutation: phenotypic profile and associations with MC1R variants. J Med Genet 2013; 50: 264–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soufir N, Avril MF, Chompret A, Demenais F, Bombled J, Spatz A, et al. Prevalence of p16 and CDK4 germline mutations in 48 melanoma-prone families in France. The French Familial Melanoma Study Group. Hum Mol Genet 1998; 7: 209–216. [DOI] [PubMed] [Google Scholar]
- 13.Zuo L, Weger J, Yang Q, Goldstein AM, Tucker MA, Walker GJ, et al. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat Genet 1996; 12: 97–99. [DOI] [PubMed] [Google Scholar]
- 14.Goldstein AM, Chan M, Harland M, Hayward NK, Demenais F, Bishop DT, et al. Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. J Med Genet 2007; 44: 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harland M, Cust AE, Badenas C, Chang YM, Holland EA, Aguilera P, et al. Prevalence and predictors of germline CDKN2A mutations for melanoma cases from Australia, Spain and the United Kingdom. Hered Cancer Clin Pract 2014; 12: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Snoo FA, Bishop DT, Bergman W, van Leeuwen I, van der Drift C, van Nieuwpoort FA, et al. Increased risk of cancer other than melanoma in CDKN2A founder mutation (p16- Leiden)-positive melanoma families. Clin Cancer Res 2008; 14: 7151–7157. [DOI] [PubMed] [Google Scholar]
- 17.Lynch HT, Fusaro RM, Lynch JF, Brand R. Pancreatic cancer and the FAMMM syndrome. Fam Cancer 2008; 7: 103–112. [DOI] [PubMed] [Google Scholar]
- 18.Bertolotto C, Lesueur F, Giuliano S, Strub T, de Lichy M, Bille K, et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature 2011; 480: 94–98. [DOI] [PubMed] [Google Scholar]
- 19.Scherer D, Kumar R. Genetics of pigmentation in skin cancer - a review. Mutat Res 2010; 705: 141–153. [DOI] [PubMed] [Google Scholar]
- 20.Yokoyama S, Woods SL, Boyle GM, Aoude LG, MacGregor S, Zismann V, et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature 2011; 480: 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niendorf KB, Goggins W, Yang G, Tsai KY, Shennan M, Bell DW, et al. MELPREDICT: a logistic regression model to estimate CDKN2A carrier probability. J Med Genet 2006; 43: 501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hogg D, Liu L, Lassam N. Genetic testing in familial melanoma: epidemiologic/genetic assessment of risks and role of CDKN2A analysis. Methods Mol Med 2001; 61: 109–122. [DOI] [PubMed] [Google Scholar]
- 23.Kypreou KP, Stefanaki I, Antonopoulou K, Karagianni F, Ntritsos G, Zaras A, et al. prediction of melanoma risk in a southern European population based on a weighted genetic risk score. J Invest Dermatol 2016; 136: 690–695. [DOI] [PubMed] [Google Scholar]
- 24.Aspinwall LG, Taber JM, Leaf SL, Kohlmann W, Leachman SA. Genetic testing for hereditary melanoma and pancreatic cancer: a longitudinal study of psychological outcome. Psychooncology 2013; 22: 276–289. [DOI] [PubMed] [Google Scholar]
- 25.Lynch HT, Brand RE, Hogg D, Deters CA, Fusaro RM, Lynch JF, et al. Phenotypic variation in eight extended CDKN2A germline mutation familial atypical multiple mole melanoma- pancreatic carcinoma-prone families: the familial atypical mole melanoma-pancreatic carcinoma syndrome. Cancer 2002; 94: 84–96. [DOI] [PubMed] [Google Scholar]
- 26.Maheu C, Vodermaier A, Rothenmund H, Gallinger S, Ardiles P, Semotiuk K, et al. Pancreatic cancer risk counselling and screening: impact on perceived risk and psychological functioning. Fam Cancer 2010; 9: 617–624. [DOI] [PubMed] [Google Scholar]
- 27.Bakos RM, Besch R, Zoratto GG, Godinho JM, Mazzotti NG, Ruzicka T, et al. The CDKN2A p.A148T variant is associated with cutaneous melanoma in Southern Brazil. Exp Dermatol 2011; 20: 890–893. [DOI] [PubMed] [Google Scholar]
- 28.Goldstein AM, Stacey SN, Olafsson JH, Jonsson GF, Helgason A, Sulem P, et al. CDKN2A mutations and melanoma risk in the Icelandic population. J Med Genet 2008; 45: 284–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koulermou G, Shammas C, Vassiliou A, Kyriakides TC, Costi C, Neocleous V, et al. CDKN2A and MC1R variants found in Cypriot patients diagnosed with cutaneous melanoma. J Genet 2017; 96: 155–160. [DOI] [PubMed] [Google Scholar]
- 30.Pjanova D, Engele L, Randerson-Moor JA, Harland M, Bishop DT, Newton Bishop JA, et al. CDKN2A and CDK4 variants in Latvian melanoma patients: analysis of a clinic-based population. Melanoma Res 2007; 17: 185–191. [DOI] [PubMed] [Google Scholar]
- 31.Cossu A, Casula M, Cesaraccio R, Lissia A, Colombino M, Sini MC, et al. Epidemiology and genetic susceptibility of malignant melanoma in North Sardinia, Italy. Eur J Cancer Prev 2017; 26: 263–267. [DOI] [PubMed] [Google Scholar]
- 32.Nikolaou V, Kang X, Stratigos A, Gogas H, Latorre MC, Gabree M, et al. Comprehensive mutational analysis of CDKN2A and CDK4 in Greek patients with cutaneous melanoma. Br J Dermatol 2011; 165: 1219–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldstein AM, Chan M, Harland M, Gillanders EM, Hayward NK, Avril MF, et al. High-risk melanoma susceptibility genes and pancreatic cancer, neural system tumors, and uveal melanoma across GenoMEL. Cancer Res 2006; 66: 9818–9828. [DOI] [PubMed] [Google Scholar]
- 34.Di Lorenzo S, Fanale D, Corradino B, Calo V, Rinaldi G, Bazan V, et al. Absence of germline CDKN2A mutation in Sicilian patients with familial malignant melanoma: Could it be a population-specific genetic signature? Cancer Biol Ther 2016; 17: 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams PF, Olsen CM, Hayward NK, Whiteman DC. Melanocortin 1 receptor and risk of cutaneous melanoma: a metaanalysis and estimates of population burden. Int J Cancer 2011; 129: 1730–1740. [DOI] [PubMed] [Google Scholar]
- 36.Fargnoli MC, Gandini S, Peris K, Maisonneuve P, Raimondi S. MC1R variants increase melanoma risk in families with CDKN2A mutations: a meta-analysis. Eur J Cancer 2010; 46: 1413–1420. [DOI] [PubMed] [Google Scholar]
- 37.Artomov M, Stratigos AJ, Kim I, Kumar R, Lauss M, Reddy BY, et al. Rare variant, gene-based association study of hereditary melanoma using whole-exome sequencing. J Natl Cancer Inst 2017; 109: djx083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.