Abstract
Aims/hypothesis
Mitochondria are important regulators of the metabolic phenotype in type 2 diabetes. A key factor in mitochondrial physiology is the H+-ATP synthase. The expression and activity of its physiological inhibitor, ATPase inhibitory factor 1 (IF1), controls tissue homeostasis, metabolic reprogramming and signalling. We aimed to characterise the putative role of IF1 in mediating skeletal muscle metabolism in obesity and diabetes.
Methods
We examined the ‘mitochondrial signature’ of obesity and type 2 diabetes in a cohort of 100 metabolically characterised human skeletal muscle biopsy samples. The expression and activity of H+-ATP synthase, IF1 and key mitochondrial proteins were characterised, including their association with BMI, fasting plasma insulin, fasting plasma glucose and HOMA-IR. IF1 was also overexpressed in primary cultures of human myotubes derived from the same biopsies to unveil the possible role played by the pathological inhibition of the H+-ATP synthase in skeletal muscle.
Results
The results indicate that type 2 diabetes and obesity act via different mechanisms to impair H+-ATP synthase activity in human skeletal muscle (76% reduction in its catalytic subunit vs 280% increase in IF1 expression, respectively) and unveil a new pathway by which IF1 influences lipid metabolism. Mechanistically, IF1 altered cellular levels of α-ketoglutarate and L-carnitine metabolism in the myotubes of obese (84% of control) and diabetic (76% of control) individuals, leading to limited β-oxidation of fatty acids (60% of control) and their cytosolic accumulation (164% of control). These events led to enhanced release of TNF-α (10 ± 2 pg/ml, 27 ± 5 pg/ml and 35 ± 4 pg/ml in control, obese and type 2 diabetic participants, respectively), which probably contributes to an insulin resistant phenotype.
Conclusions/interpretation
Overall, our data highlight IF1 as a novel regulator of lipid metabolism and metabolic disorders, and a possible target for therapeutic intervention.
Keywords: Energy metabolism, H+-ATP synthase, Inhibitory factor 1 (IF1), Mitochondria, Obesity, Skeletal muscle, Type 2 diabetes
Introduction
Interest in a pathophysiological role for mitochondria in obesity and type 2 diabetes has increased in response to growing evidence linking metabolic diseases with mitochondrial dysfunction [1, 2], especially in the case of skeletal muscle [3]. Genetic and metabolic factors, oxidative stress, ageing, high-fat diets and sedentary lifestyles may affect mitochondrial function, contributing to the development of insulin resistance [2]. However, molecular mechanisms underlying skeletal muscle mitochondria dysfunction in the setting of obesity and diabetes remain poorly understood.
Mitochondria are key regulators of cellular metabolism, influencing bioenergetics and cell death, and mediating intracellular signalling [4, 5]. Glucose and lipid metabolism are largely dependent on mitochondria. Normoxic cells oxidise pyruvate to CO2, generating the proton electrochemical gradient used for ATP synthesis in oxidative phosphorylation (OXPHOS) [4]. The cellular availability of ATP, NADH and Krebs cycle intermediates coordinates the flux of glucose consumption and lipid metabolism in the short term [4]. A key transducer in these processes is H+-ATP synthase, an enzyme localised to the inner mitochondrial membrane [6] that has emerging roles in coordinating the activation of apoptosis [7], the shape and structure of cristae [8], and reactive oxygen species (ROS)-mediated crosstalk between mitochondria and the nucleus [9, 10].
As a consequence of the multiplicity of H+-ATP synthase functions, alterations in its expression, phosphorylation and activity may contribute to a pathophysiological aetiology common for many chronic diseases, including cancer [4] and type 2 diabetes [11–13]. Modulation of the expression/activity of the H+-ATP synthase physiological inhibitor, ATPase inhibitory factor 1 (IF1), is another strategy employed by cells to control energy metabolism and activate nuclear programmes for survival and proliferation [14, 15]. This protein of the mitochondrial matrix also plays a prominent role in metabolic reprogramming exhibited by human cancer cells [16], as well as by induced pluripotent stem cells and stem cells [17, 18]. However, the basic cell biology of IF1 in obesity and type 2 diabetes remains surprisingly unknown.
Therefore, we aimed to characterise the putative role of IF1 in skeletal muscle, a tissue highly dependent on mitochondrial activity, in the setting of these metabolic disorders. Our hypothesis is that IF1, by inhibiting the H+-ATP synthase, could reroute energy and lipid metabolism, contributing to an insulin resistant phenotype and to the onset and progression of type 2 diabetes.
Methods
Human samples
Muscle biopsies were obtained from 100 participants (32 obese diabetic participants [O-T2D], ten non-obese diabetic participants [nO-T2D], 24 obese nondiabetic participants [O-nT2D] and 34 non-obese non-diabetic participants [nO-nT2D]; Table 1) as previously described [19, 20]. Participants were recruited over a number of years for multiple studies with institutional and ethical approval to use the samples and data collected for additional evaluations. The experimental protocol was approved by the Human Research Protection Programs of the VA San Diego Medical Center and University of California (San Diego, CA, USA). Informed written consent was obtained from all participants. Muscle satellite cells were prepared [19,20], transfected on day 4 after fusion using reported plasmids [9] and experiments were performed at day 6. Where indicated, cells were treated with carnitine (1 mmol/l), etomoxir (100 μmol/l), oligomycin (5 μmol/l), dibutyryl cAMP (dbcAMP; 100 μmol/l) or H89 (10 μmol/l).
Table 1.
Participant characteristics
| Number | Age | Sex (F/M) | BMI (kg/m2) | FG (mmol/l) | FI (pmol/l) | HOMA-IR | |
|---|---|---|---|---|---|---|---|
| O-T2D | 32 | 54 ± 8 | 9/23 | 36.0 ± 6.1 | 8.79 ± 1.78 | 104 ± 15 | 2.00 ± 1.14 |
| nO-T2D | 10 | 56 ± 12 | 4/8 | 26.4 ± 1.1 | 9.31 ± 4.20 | 54 ± 4 | 2.95 ± 1.10 |
| O-nT2D | 24 | 54 ± 8 | 3/21 | 34.2 ± 3.5 | 5.70 ± 0.77 | 42 ± 8 | 1.15 ± 0.69 |
| nO-nT2D | 34 | 51 ± 9 | 7/29 | 25.1 ± 2.0 | 5.01 ± 1.86 | 38 ± 9 | 1.20 ± 0.98 |
Data are presented as mean ± SD
Obese, BMI >30 kg/m2; non-obese, BMI <27 kg/m2
Details for all procedures are provided and the buffers used are specified in the electronic supplementary material (ESM) Methods.
Determination of the activity of OXPHOS complexes
Permeabilised skeletal muscle tissue (electron transport chain [ETC] complexes) or isolated mitochondria (H+-ATP synthase) were used for the spectrophotometric determination of activities [7, 21]. See ESM Methods: Determination of the activity of OXPHOS complexes and Mitochondria isolation and H+-ATP synthase activity.
Glucose uptake assay and lactate determination
Uptake of the non-metabolised analogue 2-deoxyglucose (0.01 mmol/l) was measured in the presence or absence of 100 nmol/l insulin [20]. The rate of lactate production was determined in human primary culture of skeletal muscle cells (hSMC) media [9] using a YSI 2300 STAT Plus analyser (YSI Life Science, Yellow Springs, OH, USA). See ESM Methods: Glucose uptake assay and Lactate determination.
Determination of NEFA oxidation, glutathione, α-KG, NEFA, NAD/NADH and myokine secretion
Fatty acid metabolism was assayed by β-oxidation of the long chain fatty acid palmitate [22] (see ESM Methods: NEFA oxidation). Muscle glutathione [7] and a-ketoglutarate (α-KG) concentrations were spectrophotometrically determined. The NEFA assay kit (Sigma-Aldrich, St Louis, MO, USA), NAD/NADH quantification kit (Sigma-Aldrich) and the RayBio C-Series Human Cytokine Antibody Array C3 (RayBiotech, Norcross, GA, USA) were employed. A Specific Human TNF-α Quantikine HS ELISA (R&D Systems, Minneapolis, MN, USA) was used to quantify TNF-α levels in myotube media. See ESM Methods: Determination of myokine secretion and Determination of glutathione, α-KG, NEFA and NAD/NADH.
Proteomics, immunoblot and immunoprecipitation analysis
Proteins were digested in trypsin, labelled, fractionated and processed by LC-MS2/MS3 as previously described [23]. Data were processed using the Proteome Discoverer 2.1 (Thermo Fisher Scientific, Waltham, MA, USA). See ESM Methods: Proteomics for further details. Proteomic raw data and annotated spectra have been uploaded to the online repository ProteomeXchange through MassIVE (accession numbers: PXD006629 and MSV000081125, respectively).
Tissue and cell lysates were fractionated by SDS-PAGE or incubated with IF1 antibody bound to EZ View Red Protein G Affinity Gel at 4°C [24]. See ESM Methods: Immunoprecipitation assays.
Details of antibodies are specified in ESM Methods: Western blot antibodies and methodology.
Detection and quantification of bands were accomplished using the LiCor Odyssey CxL system and Licor Image Studio analysis software (v.3.1.4).
Statistical analysis
Statistical analyses were performed using a two-tailed Student’s t test. ANOVA and the post hoc Tukey’s test were used for multiple comparisons, employing the SPSS 17.0 and Graphpad Prism 7 software packages. Bonferroni correction was applied to avoid multiple comparison errors. Regression lines were calculated using Excel and Graphpad Prism 7 software. The results shown are means ± SEM or SD. A p < 0.05 was considered to be statistically significant.
Results
Obesity and type 2 diabetes induce dysfunction in skeletal muscle mitochondria
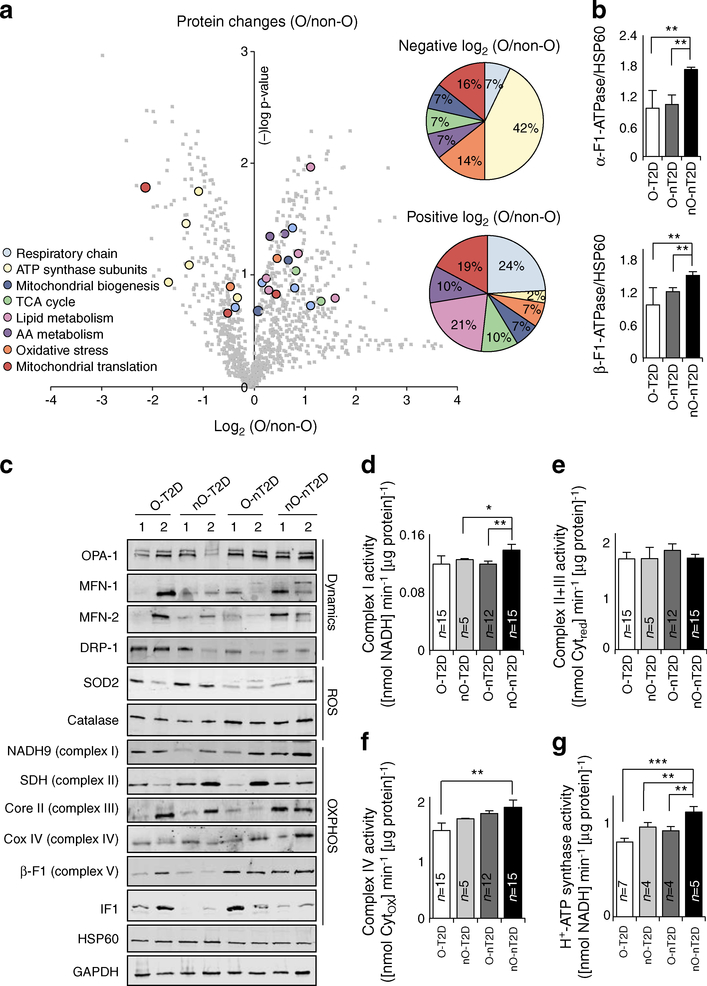
To assess the role of obesity in regulating human muscle mitochondria, we studied the proteomic profile of mitochondrial enriched samples from hSMCs derived from O-nT2D and nO-nT2D participants (Fig. 1a and Table 2). Among the mitochondrial proteins whose expression was highly altered during obesity, 15 were repressed and 32 elevated compared with samples from non-obese participants (Fig. 1a and Table 2). Of obesity-induced downregulated proteins, 42% are represented by subunits of H+-ATP synthase. Other up- or downregulated proteins revealed impairments in the ETC, mitochondrial translation machinery, biogenesis, Krebs cycle, lipid and amino acid metabolism, and oxidative stress pathways (Fig. 1a and Table 2). Immunoblotting of hSMCs confirmed the results from proteomic profiling of the downregulation in obesity of catalytic subunits α- and β-F1-ATPase of H+-ATP synthase (Fig. 1b).
Fig. 1.
Skeletal muscle ‘mitochondrial signatures’ in pathology. (a) Proteomic analysis of mitochondria-enriched hSMCs. Lateral pie charts represent the percentage of mitochondrial downregulated (negative log2[O/non-O]) and upregulated (positive log2[O/non-O]) proteins. A (−)log p value of >1.3 was considered to be statistically significant. (b) Immunoblotting of hSMCs showing catalytic subunits α- and β-F1-ATPase of H+-ATP synthase. (c) Representative blots of mitochondrial proteins related to dynamics, ROS signalling and OXPHOS, and quantification of ETC complex activities (d–f) in 100 skeletal muscle biopsies. (g) H+-ATP synthase activity in isolated mitochondria from hSMCs. AA, amino acid; Core II, ubiquinol-cytochrome c reductase core protein II; COX IV, mitochondrially encoded cytochrome c oxidase IV; MFN-1, mitofusin 1; MFN-2, mitofusin 2; SDH, succinate dehydrogenase complex; TCA, tricarboxylic acid. Histograms show the mean ± SEM of the number of individuals analysed. *p < 0.05, **p < 0.01, ***p < 0.001 by ANOVA multiple comparisons. Non-O, non-obese; O, obese. Also see ESM Figs 1–3 and Tables 1, 2
Table 2.
The ‘mitochondrial signature’ of obesity. Identification of 47 proteins of different mitochondrial pathways that are significantly down- or upregulated in skeletal muscle biopsies from obese (O) compared with non-obese (non-O) healthy participants
| Protein no. | Description | log2(O/non-O) |
|---|---|---|
| Respiratory chain (ETC) | ||
| O96000 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 OS=Homo sapiens GN=NDUFB10 PE=1 SV=3 |
1.10 |
| Q00325 | Phosphate carrier protein, mitochondrial OS=Homo sapiens GN=SLC25A3 PE=1 SV=2 |
0.90 |
| Q9H8M1 | Coenzyme Q-binding protein COQ10 homologue B, mitochondrial OS=Homo sapiens GN=COQ10B PE=2 SV=1 |
0.88 |
| P49821 | NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial OS=Homo sapiens GN=NDUFV1 PE=1 SV=4 |
0.84 |
| O43181 | NADH dehydrogenase [ubiquinone] iron-sulphur protein 4, mitochondrial OS=Homo sapiens GN=NDUFS4 PE=1 SV=1 |
0.72 |
| P31040 | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial OS=Homo sapiens GN=SDHA PE=1 SV=2 |
0.48 |
| P28331 | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial OS=Homo sapiens GN=NDUFS1 PE=1 SV=3 |
0.16 |
| P56556 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6 OS=Homo sapiens GN=NDUFA6 PE=1 SV=3 |
−0.94 |
| ATP synthase subunits | ||
| O75947 | ATP synthase subunit d, mitochondrial OS=Homo sapiens GN=ATP5H PE=1 SV=3 |
0.03 |
| P24539 | ATP synthase F(0) complex subunit B1, mitochondrial OS=Homo sapiens GN=ATP5F1 PE=1 SV=2 |
0.03 |
| P48047 | ATP synthase subunit O, mitochondrial OS=Homo sapiens GN=ATP5O PE=1 SV=1 |
−0.20 |
| F8W0P7 | ATP synthase subunit beta, mitochondrial (Fragment) OS=Homo sapiens GN=ATP5B PE=1 SV=2 |
−0.31 |
| P25705 | ATP synthase subunit alpha, mitochondrial OS=Homo sapiens GN=ATP5A1 PE=1 SV=1 |
−0.50 |
| P06576 | ATP synthase subunit beta, mitochondrial OS=Homo sapiens GN=ATP5B PE=1 SV=3 |
−1.09 |
| O75964 | ATP synthase subunit g, mitochondrial OS=Homo sapiens GN=ATP5L PE=1 SV=3 |
−1.68 |
| ROS | ||
| Q9Y2Q3 | Glutathione S-transferase kappa 1 OS=Homo sapiens GN=GSTK1 PE=1 SV=3 |
1.17 |
| P30044 | Peroxiredoxin-5, mitochondrial OS=Homo sapiens GN=PRDX5 PE=1 SV=4 |
0.45 |
| A0A0C4DFU2 | Superoxide dismutase OS=Homo sapiens GN=SOD2 PE=1 SV=1 |
−0.66 |
| Q9Y6N5 | Sulphide:quinone oxidoreductase, mitochondrial OS=Homo sapiens GN=SQRDL PE=1 SV=1 |
−1.34 |
| Biogenesis | ||
| Q9UJZ1 | Stomatin-like protein 2, mitochondrial OS=Homo sapiens GN=STOML2 PE=1 SV=1 |
0.67 |
| E5KLJ5 | Dynamin-like 120 kDa protein, mitochondrial OS=Homo sapiens GN=OPA-1 PE=1 SV=1 |
0.06 |
| Q04837 | Single-stranded DNA-binding protein, mitochondrial OS=Homo sapiens GN=SSBP1 PE=1 SV=1 |
−0.34 |
| TCA cycle | ||
| P07954 | Fumarate hydratase, mitochondrial OS=Homo sapiens GN=FH PE=1 SV=3 |
1.13 |
| P11498 | Pyruvate carboxylase, mitochondrial OS=Homo sapiens GN=PC PE=1 SV=2 |
0.95 |
| A0A087WZN1 | Isocitrate dehydrogenase [NAD] subunit, mitochondrial OS=Homo sapiens GN=IDH3B PE=1 SV=1 |
0.82 |
| E9PCR7 | 2-oxoglutarate dehydrogenase, mitochondrial OS=Homo sapiens GN=OGDH PE=1 SV=1 |
−0.54 |
| Lipid metabolism | ||
| P49327 | Fatty acid synthase OS=Homo sapiens GN=FASN PE=1 SV=3 |
1.15 |
| Q9NWU1 | 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial OS=Homo sapiens GN=OXSM PE=1 SV=1 |
1.01 |
| A0A0A0MSE2 | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial OS=Homo sapiens GN=HADH PE=1 SV=1 |
0.80 |
| Q9H845 | Acyl-CoA dehydrogenase family member 9, mitochondrial OS=Homo sapiens GN=ACAD9 PE=1 SV=1 |
0.40 |
| Q9Y305 | Acyl-coenzyme A thioesterase 9, mitochondrial OS=Homo sapiens GN=ACOT9 PE=1 SV=2 |
0.24 |
| E7EX59 | Propionyl-CoA carboxylase beta chain, mitochondrial OS=Homo sapiens GN=PCCB PE=1 SV=1 |
0.17 |
| Amino acid metabolism | ||
| Q02252 | Methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial OS=Homo sapiens GN=ALDH6A1 PE=1 SV=2 |
0.60 |
| P35914 | Hydroxymethylglutaryl-CoA lyase, mitochondrial OS=Homo sapiens GN=HMGCL PE=1 SV=2 |
0.33 |
| Q92947 | Glutaryl-CoA dehydrogenase, mitochondrial OS=Homo sapiens GN=GCDH PE=1 SV=1 |
0.31 |
| P30837 | Aldehyde dehydrogenase X, mitochondrial OS=Homo sapiens GN=ALDH1B1 PE=1 SV=3 |
−0.90 |
| Mitochondrial translation | ||
| Q8IYB8 | ATP-dependent RNA helicase SUPV3L1, mitochondrial OS=Homo sapiens GN=SUPV3L1 PE=1 SV=1 |
0.78 |
| Q8TCS8 | Polyribonucleotide nucleotidyltransferase 1, mitochondrial OS=Homo sapiens GN=PNPT1 PE=1 SV=2 |
0.53 |
| P43897 | Elongation factor Ts, mitochondrial OS=Homo sapiens GN=TSFM PE=1 SV=2 |
0.36 |
| J3QLS3 | 28S ribosomal protein S7, mitochondrial OS=Homo sapiens GN=MRPS7 PE=1 SV=1 |
0.35 |
| Q9H9J2 | 39S ribosomal protein L44, mitochondrial OS=Homo sapiens GN=MRPL44 PE=1 SV=1 |
0.28 |
| Q9P015 | 39S ribosomal protein L15, mitochondrial OS=Homo sapiens GN=MRPL15 PE=1 SV=1 |
−1.06 |
| P82912 | 28S ribosomal protein S11, mitochondrial OS=Homo sapiens GN=MRPS11 PE=1 SV=2 |
−2.14 |
| Others | ||
| Q9H2U2 | Inorganic pyrophosphatase 2, mitochondrial OS=Homo sapiens GN=PPA2 PE=1 SV=2 |
0.74 |
| Q9HB07 | UPF0160 protein MYG1, mitochondrial OS=Homo sapiens GN=C12orf10 PE=1 SV=2 |
0.44 |
| Q16822 | Phosphoenolpyruvate carboxykinase [GTP], mitochondrial OS=Homo sapiens GN=PCK2 PE=1 SV=3 |
−1.19 |
| Q5JRX3 | Presequence protease, mitochondrial OS=Homo sapiens GN=PITRM1 PE=1 SV=3 |
−1.23 |
OS, organism name; GN, gene name; PE, protein existence; SV, sequence version; TCA, tricarboxylic acid
These data were confirmed by determining the expression of mitochondrial proteins in skeletal muscle biopsies from O-T2D, nO-T2D, O-nT2D and nO-nT2D individuals (representative blots are presented in Fig. 1c; full quantification is shown in ESM Fig. 1). Skeletal muscle immunoblot data correlated with hSMC protein abundance assessed by proteomics, suggesting that hSMCs are valuable tools for the study of human pathologies. β-F1-ATPase was highly repressed (76%) in both O-T2D and nO-T2D compared with nO-nT2D participants. However, its inhibitor IF1 was overexpressed (280%) in obese participants regardless of the presence of diabetes (Fig. 1c and ESM Fig. 1), suggesting that the inhibition of OXPHOS is a common feature of obesity and diabetes. In addition, we found diabetes-associated changes in the expression of the mitochondrial redox sensor superoxide dismutase 2 (SOD2) and in the fusion/fission regulators mitochondrial dynamin-like GTPase (OPA-1) and dynamin-related protein (DRP-1) (Fig. 1c and ESM Fig. 1). The DRP-1/OPA-1 ratio was significantly correlated with fasting plasma glucose (FG) levels (r2 = 0.191, p = 0.049; ESM Fig. 2), supporting the idea that type 2 diabetes modifies the balance between the machinery involved in the mitochondrial dynamic network in skeletal muscle [25]. See ESM Results for more details.
In addition to their expression levels, even slight changes in OXPHOS complex activities can contribute to the skeletal muscle bioenergetic impairment observed during metabolic pathologies [2]. While neither obesity nor diabetes induced changes in the activity of complexes II+III, complex I activity was significantly reduced in O-nT2D (80%) and nO-T2D (85%) compared with healthy individuals (Fig. 1d, e and ESM Fig. 3). Complex I activity was inversely correlated with BMI (r2 = 0.338, p = 0.042; ESM Fig. 3). While independent of BMI, complex IV activity was impaired in O-T2D individuals (75%) and inversely correlated with fasting plasma insulin (FI) (r2 = 0.272, p = 0.051) and HOMA-IR (r2 = 0.203, p = 0.050; Fig. 1f and ESM Fig. 3). Both obesity and diabetes influenced the activity of H+-ATP synthase, reducing it to 82% and 67% relative to nO-nT2D participants, respectively (Fig. 1g).
Taken together, these data suggest that obesity and diabetes contribute in different ways to impairments in the skeletal muscle mitochondrial network.
Increasing IF1/β-F1-ATPase ratio is a hallmark of obesity and type 2 diabetes
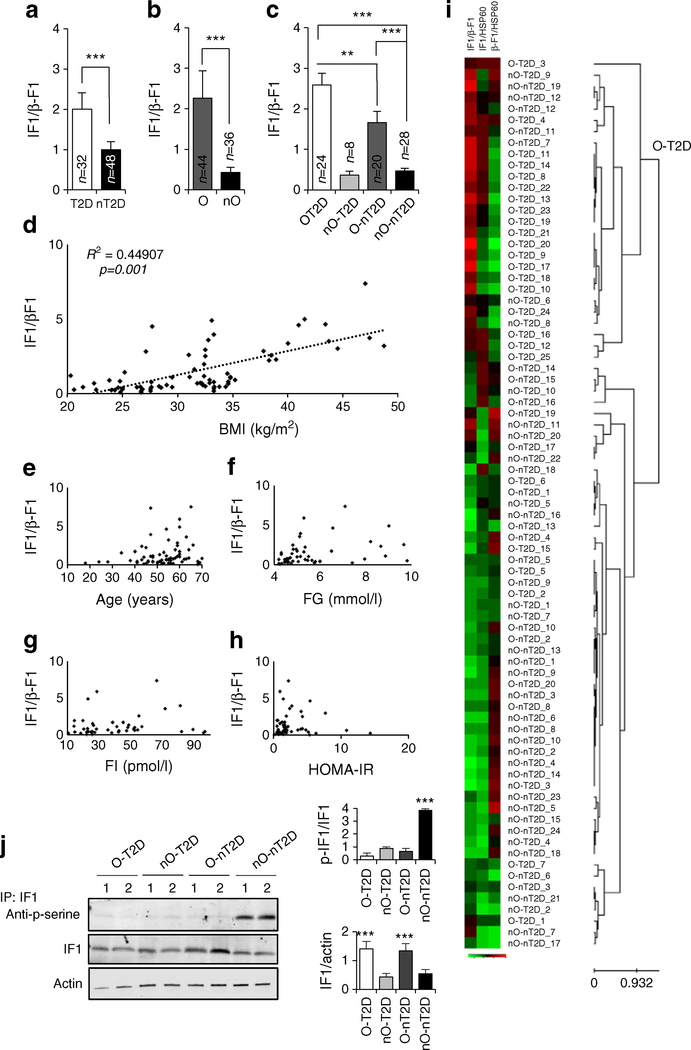
Despite metabolic pathologies acting distinctly on the expression of IF1 and β-F1-ATPase proteins, the ratio of IF1/β-F1-ATPase, which serves as an indicator of impaired H+-ATP synthase activity, was significantly increased in both diabetes (190%) and obesity (380%) compared with nO-nT2D individuals (Fig. 2a–c). However, no changes in the IF1/β-F1-ATPase ratio were observed in the nO-T2D group (Fig. 2c), suggesting that obesity is the primary modulator of the expression of these proteins. Accordingly, the IF1/β-F1-ATPase ratio positively correlated with BMI (r2 = 0.448, p = 0.011; Fig. 2d) but not with age (Fig. 2e), FG (Fig. 2f), FI (Fig. 2g) or HOMA-IR (Fig. 2h).
Fig. 2.
IF1/β-F1-ATPase ratio in skeletal muscle from participants with obesity and type 2 diabetes. (a–c) Changes in IF1/β-F1-ATPase ratio. (d–h) Correlations between IF1/β-F1-ATPase ratio and BMI (d), age (e), FG (f), FI (g) and HOMA-IR (h). (i) Hierarchical clustering analyses. Rows indicate samples and columns indicate IF1/β-F1-ATPase, IF1/HSP60 and (β-F1-ATPase/HSP60 ratios. Protein expression scores: red, high; green, low. Details are provided in ESM Methods: Hierarchical clustering analyses. (j) Representative immunoprecipitation (IP) of endogenous IF1 stained with phosphoserine antibody (two samples for each condition tested). Blots of IF1 input and β-actin are shown. Histograms show quantification of three experiments. (a–c, j) Histograms show the mean ± SEM of the number of individuals analysed. **p < 0.01, ***p < 0.001 by Student’s t test and ANOVA multiple comparisons
Having observed a significantly higher increase in the IF1/β-F1-ATPase ratio in O-T2D compared with O-nT2D participants (Fig. 2c) and as the number of nO-T2D participants was relatively low, we were not convinced of the potential of these proteins of energy metabolism to serve as biomarkers of both diseases. To investigate this issue further, we carried out unsupervised hierarchical clustering of the biopsies using the expression of three ratios (IF1/β-F1-ATPase, IF1/heat shock protein 60 [HSP60] and β-F1-ATPase/HSP60) for aggregation purposes. Clustering of 80 biopsies from the four participant groups resulted in two populations with a classification sensitivity of 85% (O-T2D vs others; Fig. 2i). This suggests that a change in H+-ATP synthase activity, from either β-F1-ATPase downregulation or IF1 overexpression, could be a hallmark of both obesity and diabetes.
IF1 activity towards H+-ATP synthase can also be regulated by phosphorylation, which inactivates the protein [24]. While no significant changes in RNA expression of human IF1 between control, obese or diabetic populations were reported [26], we found that IF1 was significantly more phosphorylated in non-obese healthy individuals than in obese or type 2 diabetic individuals (Fig. 2j). This finding supports the idea of a third mechanism, phosphorylation, by which these pathologies modulate the activity of H+-ATP synthase to promote dysfunction in skeletal muscle [13].
IF1 perturbs metabolism in myotubes from obese and type 2 diabetic individuals
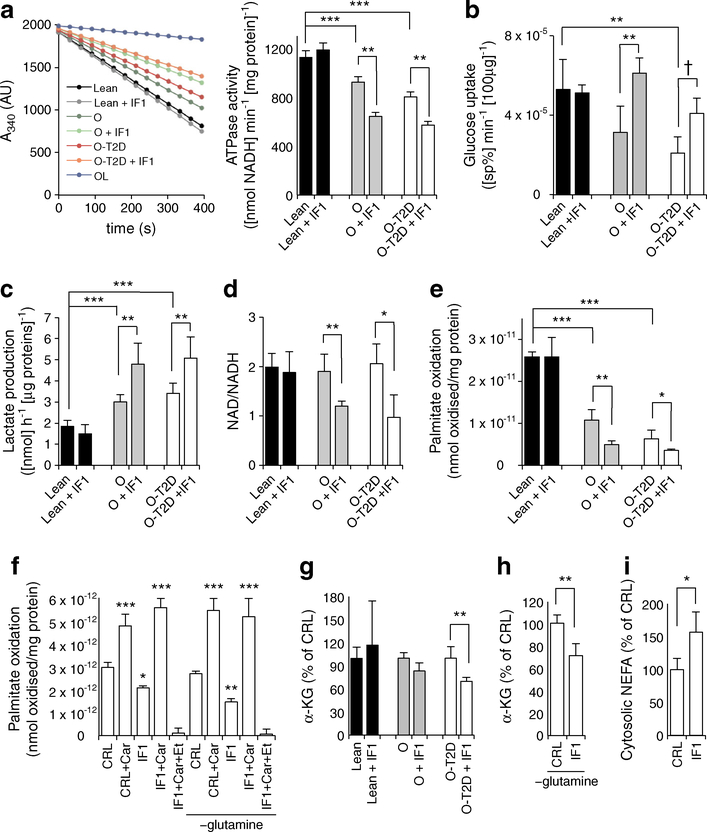
To study its role in the biology of muscle cells, we overexpressed IF1 in primary cultures of hSMCs obtained from non-obese healthy, obese and diabetic participants (Fig. 3). IF1 overexpression resulted in a 300% increase in the protein expression (ESM Fig. 4), comparable to that observed in skeletal muscle from obese and diabetic participants (Figs 1b, 2a–c). Consistent with the higher basal phosphorylation of IF1 in healthy individuals (Fig. 2j), neither endogenous nor exogenous IF1 were active (Fig. 3, black bars), as neither ATPase activity nor other assays (Figs 3, 4) were altered with IF1 overexpression (ESM Fig. 4). However, when IF1 was overexpressed in hSMCs from obese or diabetic participants, we found a significant reduction (34% and 28%, respectively) in ATPase activity in comparison to healthy individuals (Fig. 3a). As reported previously [20], glucose uptake in the absence of insulin is reduced in myotubes from obese and diabetic individuals. Yet, IF1 overexpression in these myotubes resulted in increased glucose uptake (Fig. 3b) and production of extracellular lactate (Fig. 3c). Inhibition of the rate-limiting enzyme of OXPHOS by IF1 also prompted the accumulation of ETC substrates, as indicated by a significant reduction in the NAD/NADH ratio in IF1-expressing cultures from O-nT2D and O-T2D individuals (75% and 65% of control-transfected myotubes, respectively; Fig. 3d). Moreover, in hSMCs from both obese and diabetic individuals, IF1 significantly reduced mitochondrial NEFA (palmitate) β-oxidation (60% of control, Fig. 3e). The effect of IF1 is totally abrogated by including carnitine during the oxidation assay (3 h treatment; Fig. 3f and ESM Fig. 5), suggesting a role for IF1 in carnitine metabolism.
Fig. 3.
IF1 drives a switch in hSMC metabolism. (a) ATPase activity in isolated mitochondria from hSMCs of healthy, obese and diabetic individuals. Oligomycin (OL; 5 μmol/l) was used as an external control for ATPase inhibition; the histogram shows the quantification for the line graph. Glucose uptake (b), lactate production (c), NAD/NADH ratio (d), palmitate oxidation (e, f), amounts of α-KG (g, h) in hSMCs in the presence (+IF1) or absence of IF1. Effects of glutamine-free media (−glutamine) and 3 h treatment with carnitine (+Car) or etomoxir (+Et) are shown. (i) NEFA levels in the cytosolic fraction of O-T2D hSMCs. ‘Lean’ indicates non-obese. Black bars, healthy individuals; grey bars, obese individuals; white bars, obese diabetic individuals. A, absorbance; CRL, control. Bars show the mean ± SEM of hSMCs from three healthy, four obese and ten obese diabetic individuals. *p < 0.05, **p < 0.01, ***p < 0.001, †p = 0.06 by Student’s t test and ANOVA multiple comparisons. sp%, specific percentage, representing the carrier-mediated portion of deoxyglucose uptake, after correcting for diffusion and trapping in the unstirred layer. Also see ESM Figs 4, 5
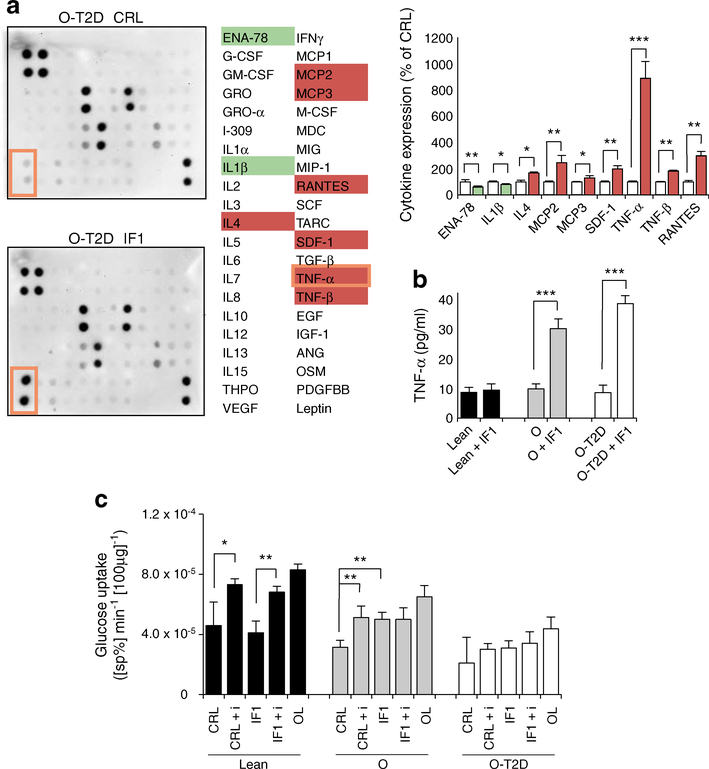
Fig. 4.
IF1 regulates myokine release and insulin sensitivity. (a) Comparison between secretomes of control (CRL, white bars) and IF1-overexpressing (IF1, coloured bars) myotubes from O-T2D participants. See ESM Fig. 6 for the array map and total myokine expression. Protein expression scores: red, high; white, normal; green, low; orange border shows TNF-α. Bars are mean ± SEM of three independent experiments. Quantification (pg/ml) of TNF-α levels (b) and rates of glucose uptake (c) in hSMCs. The effects of oligomycin (OL; 5 μmol/l), insulin (+ i, 100 nmol/l) and IF1 are shown. (b, c) Black bars, healthy individuals; grey bars, obese individuals; white bars, obese diabetic individuals. Bars show mean ± SEM for hSMCs from three healthy, four obese and ten obese diabetic individuals. *p < 0.05, **p < 0.01, ***p < 0.001 by ANOVA multiple comparisons. ANG, angiogenin; ENA-78, epithelial-derived neutrophil-activating peptide 78; G-CSF, granulocyte colony stimulating factor; GM-CSF, granulocyte–macrophage colony stimulating factor; GRO, growth-regulated protein; GROα, growth-regulated protein α; MCP, monocyte chemotactic protein; M-CSF, macrophage colony stimulating factor; MDC, macrophage-derived chemokine; MIG, monokine induced by γ interferon; MIP, macrophage inflammatory protein; OSM, oncostatin M; PDGFBB, platelet-derived growth factor-BB; RANTES, regulated on activation, normal T cell expressed and secreted; SCF, skp, cullin, F-box containing complex; SDF, stromal cell-derived factor; sp%, specific percentage; TARC, thymus and activation regulated chemokine; THPO, thrombopoietin; VEGF, vascular endothelial growth factor
Carnitine is essential for transport of NEFAs into the mitochondrial matrix [27]. Palmitate oxidation in both control and IF1 overexpressing cells was blocked by the administration of the irreversible inhibitor of carnitine palmitoyltransferase-1 (CPT-1) etomoxir (Fig. 3f, ESM Fig. 5). In line with a role for IF1 in interfering with carnitine levels or functions, IF1 overexpression led to anaplerotic perturbations of the cellular metabolome. Specifically, this was seen as a reduction in the amount of the Krebs cycle and carnitine biosynthesis intermediate α-KG in myotubes from diabetic participants (Fig. 3g). This reduction was more evident when the media was deprived of glutamine, a natural source of the metabolite (Fig. 3h). Moreover, the cytosolic content of NEFAs in myotubes from diabetic individuals were significantly increased when cells were transfected with IF1 (164% of control, Fig. 3i), highlighting the involvement of H+-ATP synthase in controlling NEFA localisation and metabolism.
IF1 prompts secretion of TNF-α, contributing to insulin resistance
pt?>Skeletal muscle has been identified as a secretory organ releasing ‘myokines’, molecules that act in an autocrine, paracrine and/or endocrine manner to regulate tissue metabolism. Overexpression of IF1 induced alterations in the secretory profile of hSMCs from diabetic individuals (Fig. 4a). In particular, IF1 stimulated the secretion of the proinflammatory myokine TNF-α from hSMCs derived from obese (10 ± 2 pg/ml vs 27 ± 5 pg/ml) and type 2 diabetic (10 ± 3 pg/ml vs 35 ± 4 pg/ml) participants (Fig. 4b). In line with its phosphorylation state and previous results (Fig. 3), the overexpression of IF1 in hSMCs from healthy participants had no effect on TNF-α secretion (7 ± 2 pg/ml vs 8 ± 3 pg/ml; Fig. 4b).
TNF-α levels have been shown to interfere with insulin stimulation of glucose uptake [28]. As has been shown previously [20], insulin effects on glucose uptake were reduced and absent in hSMCs from obese and diabetic individuals, respectively (Fig. 4c grey and white bars, respectively). However, and consistent with the effect on TNF-α, even this modest insulin effect in hSMCs from obese participants (Fig. 4c CRL vs CRL + i, grey bars) was lost after the overexpression of IF1 (Fig. 4c IF1 vs IF1 + i, grey bars), suggesting that mitochondrial OXPHOS contributes to insulin sensitivity in these myotubes. This effect is not evident in myotubes from nO-nT2D participants (Fig. 4c, black bars), where IF1 is inactive, and from O-T2D participants (Fig.4c, white bars) who are already insulin resistant. Overall, IF1 overexpression induced glucose uptake and decoupled the glucose uptake from the control of insulin, characterising an enhanced insulinindependent glucose uptake and causing metabolic inflexibility.
IF1 effects are dependent on its phosphorylation state
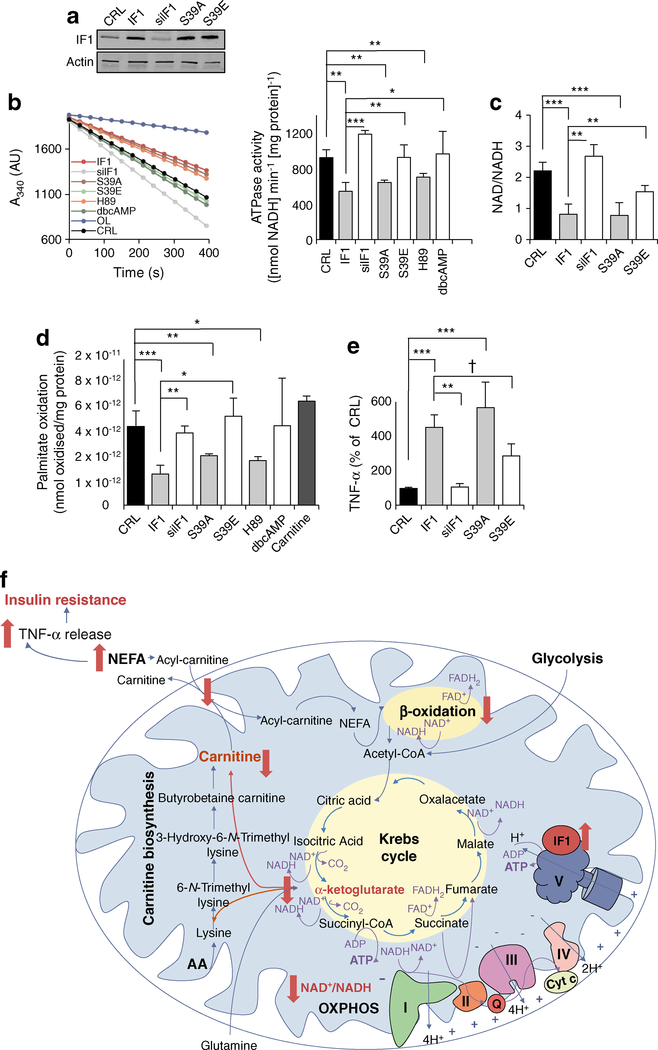
IF1 activity is regulated by its expression levels, the mitochondrial matrix pH and by phosphorylation [24]. We overexpressed constitutively active phosphodeficient (S39A) and inactive phosphomimetic (S39E) mutants of IF1 [24] in hSMCs derived from O-T2D participants (Fig. 5). Although IF1, S39A and S39E plasmids all induced an increase in the expression of IF1 protein (Fig. 5a), only IF1 and S39A led to a significant inhibition of ATPase activity (Fig. 5b) and a consequent reduction in the NAD/NADH ratio (Fig. 5c), suggesting that S39E-transfected cells respond like the IF1-insensitive hSMCs from healthy individuals.
Fig. 5.
IF1 expression and phosphorylation state regulate its activity. Effect of overexpression and dephosphorylation of IF1 (IF1, S39A, H89 [inducing the active form of IF1]) in comparison with the silencing and phosphorylation (siIF1, S39E, dbcAMP [inducing the inactive form of IF1]) or with control (CRL). (a) Representative blots in hSMCs after 36 h transfection with IF1 plasmids or siRNA (40% of IF1-expression in CRL). (b) ATPase activity in isolated mitochondria from hSMCs. 5 μmol/l oligomycin was used to inhibit H+-ATP synthase. The histogram shows the quantification for the line chart. (c) hSMC NAD/NADH ratio, (d) palmitate oxidation rates and (e) TNF-α release. (b–e) Bars are the mean ± SEM of at least four sets of myotubes from O-T2D individuals. *p < 0.05, **p < 0.01, ***p < 0.001, †p = 0.07 by ANOVA multiple comparisons. (f) Schematic representation of the effects of the obesity- and diabetes-driven overexpression and dephosphorylation of IF1 on the biology of a skeletal muscle cell. AA, amino acids; cyt, cytochrome; OXPHOS, oxidative phosphorylation
It has been demonstrated that a mitochondrial protein kinase A (PKA)-like kinase is responsible for IF1 phosphorylation [24]. Accordingly, administration of the competitive PKA inhibitor H89 (6 h) significantly reduced H+-ATP synthase activity, while 6 h treatment with the PKA activator dbcAMP did not induce significant changes in ATPase activity in myotubes from type 2 diabetic individuals (Fig. 5b and [24]). As might be expected, palmitate oxidation was significantly modified by IF1, S39A and H89, but not by S39E, dbcAMP or by the silencing of IF1 (Fig. 5d). Conversely, myotube TNF-α secretion was significantly increased by the expression of IF1 (420%) and S39A (530%), while phosphorylation of IF1 significantly halved its effect (Fig. 5e). Also, silencing of IF1 did not alter ATPase activity (Fig. 5b), NADH pool (Fig. 5c) and TNF-α secretion (Fig. 5e), highlighting dephosphorylation as one of the principle mechanisms of IF1 activation during obesity and diabetes.
Discussion
Proteomic analysis of skeletal muscle biopsies, and the study of the biology of hSMCs from a cohort of participants with obesity and/or type 2 diabetes, offer valuable tools for the investigation of the relevance of mitochondrial impairment in these metabolic disorders. We demonstrate that mitochondrial OXPHOS, ROS and dynamic machineries, involved in both the pathogenesis and long-term complications of type 2 diabetes [29, 30], are modified differently in obesity and diabetes, with the downregulation of H+-ATP synthase activity being a common feature. The overexpression of the dephosphorylated active form of the H+-ATP synthase inhibitor, IF1, in myotubes from obese and diabetic individuals boosts aerobic glycolysis, altering the NADH and α-KG pools, thus attenuating lipid metabolism and insulin sensitivity while stimulating TNF-α release. Conversely, the phosphorylated deactivated forms of IF1, either occurring naturally in healthy individuals or from the inactive phosphomimetic (S39E) mutant, lack these effects. This paper provides the first account highlighting the relevance of inhibiting the mitochondrial H+-ATP synthase activity in glucose and lipid metabolism in human muscle, and stresses the potential value of IF1 as a target for therapeutic intervention.
We have identified different processes by which muscle cells from obese or diabetic individuals can repress H+-ATP synthase activity. Type 2 diabetes preferentially induces downregulation of the catalytic subunit β-F1-ATPase (this study and [11]), while obesity causes the upregulation of IF1 both ex vivo and in vitro. Moreover, in line with recent reports about the dephosphorylation and activation of IF1 during hypoxic conditions, carcinogenesis and lack of β-adrenergic stimulation [24], the dephosphorylated active form of IF1 is prevalent in both obesity and type 2 diabetes. These findings provide further support for the idea that H+-ATP synthase repression represents one of the principal strategies of the cell to alter energy metabolism. Indeed, the impaired capacity to switch between fatty acid and glucose oxidation that we have reported during IF1 overexpression has been described in insulin resistance [31], as well as in obesity, in the heart with impaired glucose tolerance [32] and in diabetic nephropathy [2, 12].
Whether mitochondrial dysfunction is a cause or consequence of insulin resistance remains controversial. Insulin has a stimulatory effect on ATP production and mRNA transcripts of OXPHOS enzymes in skeletal muscle [33]. Moreover, it can activate PGC1α and mitogen-activated protein kinase pathways [34], favouring cell growth and mitogenesis, which could become impaired during insulin resistance [33]. Mitochondrial dysfunction might impair insulin signalling by altering fatty acid metabolism, resulting in increased cytosolic diacylglycerol content [1] and blocking insulin signalling cascades. In line with this, PGC1α-stimulated mitogenesis has been shown to increase the energetic state of the cell, upregulating the expression of subunits of H+-ATP synthase, thus counteracting chronic pathologies such as diabetes [35]. Now, we report that IF1-dependent modulation of H+-ATP synthase activity in hSMCs from both obese and diabetic individuals is sufficient to induce alterations in fatty acid oxidation and insulin sensitivity. While IF1 has a significant effect on lipid metabolism in hSMCs, the effect on insulin sensitivity is not evident in myotubes derived from participants with type 2 diabetes who are already insulin resistant [21] or from healthy individuals where IF1 is phosphorylated and inactive [24, 36].
We have shown that active IF1 OXPHOS inhibition indirectly alters the mitochondrial NADH pool. This event has been linked with the inhibition of anaplerotic reactions of the Krebs cycle, leading to perturbations in the mitochondrial metabolome [37, 38]. Consistent with this, we demonstrated that levels of the Krebs cycle metabolite α-KG are reduced during IF1 overexpression. Interestingly, although complete loss of mitochondrial function is detrimental, partial suppression of H+-ATP synthesis by increased α-KG or IF1 levels has been shown to extend Caenorhabditis elegans lifespan [39] and protect neurons from glutamate excitotoxicity [7]. This suggests that modulation of H+-ATP synthase activity is a double-edged sword, dependent on substrate availability and metabolic conditions. Accordingly, IF1 overexpression has no effect in non-obese healthy participants and α-KG does not extend the lifespan of dietary-restricted animals [39].
Since α-KG is also a cofactor in L-carnitine biosynthesis, IF1 could cause an alteration in L-carnitine levels, leading to an impairment of efficient carnitine-dependent entry of NEFAs into the mitochondrial matrix (Fig. 5f). These events led to the cytosolic accumulation of NEFAs and a subsequent reduction of mitochondrial β-oxidation of NEFAs, in line with the decreased skeletal muscle lipid oxidation found in obesity and type 2 diabetes [22, 40]. A high level of NEFAs coordinates a mitochondrial-to-cytosolic stress response [41] and is associated with an increase in the release of TNF-α from the liver, adipocytes and skeletal muscle cells [42, 43] that correlates with insulin resistance [28]. Given that skeletal muscle is the largest organ in the human body, myokines have the potential to influence crosstalk between different organs, playing a role in metabolic disorders such as obesity and type 2 diabetes [44, 45]. We have demonstrated that the overexpression of an active form of IF1 ultimately prompts myotube TNF-α secretion (Fig. 5f) in a concentration sufficient to alter insulin sensitivity [28].
While the focus of the current work is on the role(s) of IF1 and H+-ATP synthase in line with the smaller size and density of mitochondria in skeletal muscle biopsies from obese and diabetic participants [46, 47], we also found differential effects of obesity and type 2 diabetes on components of the mitochondrial dynamics network that define mitochondrial morphology. These data, together with our findings regarding regulation of IF1 and H+-ATP synthase, further highlight that obesity and type 2 diabetes contribute distinctly to skeletal muscle mitochondrial dysfunction observed in both disorders. Indeed, whether a shared disease process underlies obesity and diabetes is an ongoing question [48]. Improving the understanding of how these diseases differ with respect to molecular and functional mechanisms could help to develop more effective approaches to treatment.
Limitations of the current study include the incomplete metabolic phenotyping of the participants, as substrate oxidation and mitochondrial activity were not measured in vivo, as well as the smaller number of nO-T2D individuals available; although this lower proportion reflects their prevalence in the general type 2 diabetes population. Further type 2 diabetes proteomics and metabolomics profiling could provide additional insight. Strengths include the number of participants and the ability to directly manipulate IF1 expression and phosphorylation in myotubes derived from many of the same individuals, allowing us to advance from observations of correlations to the investigation of potential mechanisms.
Overall, the results point to IF1 as an intrinsic player within the muscle cell that can alter lipid metabolism during obesity and type 2 diabetes, and thus alter retrograde signalling to the nucleus for programmes of myokine secretion and insulin resistance. Hence, we propose H+-ATP synthase activity as a pivotal regulator of skeletal muscle endocrine signalling.
Supplementary Material
Acknowledgements
We acknowledge the contribution of A. Gaspar (VA San Diego Healthcare System, San Diego, CA, USA) in performing the muscle biopsies, and D. Armstrong and P. Burke (VA San Diego Healthcare System, San Diego, CA, USA) for patient care and assisting in the biopsy procedures. Parts of this work were presented in an abstract at the 75th Annual Scientific Sessions of the ADA, 5–9 June 2015, Boston, MA, USA; Diabetes 64(Suppl 1): 269-LB. The content of this paper does not represent the views of the US Department of Veterans Affairs or the US Government.
Funding This work was supported by grants from: Merit Review Award IO1CX000635-01A1 from the United States Department of Veterans Affairs Clinical Sciences Research and Development Service; HHS/NIH/NIDDK (DK098717-01A1); Spanish Ministry of Economy, Industry and Competitiveness (Ministerio de Economía, Industria y Competitividad, MINECO), Spain (SAF2013-41945-R and SAF2016-76028R); Comunidad de Madrid, Spain (S2011/BMD-2402); Centro de Investigación Biomédica en Red (CIBER) de Enfermedades Raras, Spain (CB06/07/0017). LF is supported by the Ramón y Cajal Spanish programme (call of 2013). FV is a Cardero Therapeutics co-founder and stockholder.
Abbreviations
- dbcAMP
Dibutyryl cAMP
- DRP-1
Dynamin-related protein 1
- ETC
Electron transport chain
- FG
Fasting plasma glucose
- FI
Fasting plasma insulin
- hSMC
Human primary culture of skeletal muscle cells
- HSP60
Heat shock protein 60
- IF1
ATPase inhibitory factor 1
- α-KG
α-ketoglutarate
- nO-nT2D
Non-obese non-diabetic participants
- nO-T2D
Non-obese diabetic participants
- OPA-1
Mitochondrial dynamin-like GTPase
- O-nT2D
Obese non-diabetic participants
- O-T2D
Obese diabetic participants
- OXPHOS
Oxidative phosphorylation
- PKA
Protein kinase A
- ROS
Reactive oxygen species
- SOD2
Superoxide dismutase 2
Footnotes
Duality of interest The authors declare that there is no duality of interest associated with this manuscript.
Data availability The data sets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material The online version of this article (doi:10.1007/s00125-017-4379-z) contains peer-reviewed but unedited supplementary material, which is available to authorised users.
References
- 1.Lowell BB, Shulman GI (2005) Mitochondrial dysfunction and type 2 diabetes. Science 307:384–387 [DOI] [PubMed] [Google Scholar]
- 2.Sivitz WI, Yorek MA (2010) Mitochondrial dysfunction in diabetes: from molecular mechanisms to functional significance and therapeutic opportunities. Antioxid Redox Signal 12:537–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asmann YW, Stump CS, Short KR et al. (2006) Skeletal muscle mitochondrial functions, mitochondrial DNA copy numbers, and gene transcript profiles in type 2 diabetic and nondiabetic subjects at equal levels of low or high insulin and euglycemia. Diabetes 55: 3309–3319 [DOI] [PubMed] [Google Scholar]
- 4.Sanchez-Arago M, Formentini L, Cuezva JM (2013) Mitochondria-mediated energy adaption in cancer: the H(+)-ATP synthase-geared switch of metabolism in human tumors. Antioxid Redox Signal 19: 285–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman JR, Nunnari J (2014) Mitochondrial form and function. Nature 505:335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshida M, Muneyuki E, Hisabori T (2001) ATP synthase—a marvellous rotary engine of the cell. Nat Rev Mol Cell Biol 2:669–677 [DOI] [PubMed] [Google Scholar]
- 7.Formentini L, Pereira MP, Sanchez-Cenizo L et al. (2014) In vivo inhibition of the mitochondrial H+-ATP synthase in neurons promotes metabolic preconditioning. EMBO J 33:762–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strauss M, Hofhaus G, Schroder RR, Kuhlbrandt W (2008) Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J 27:1154–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Formentini L, Sanchez-Arago M, Sanchez-Cenizo L, Cuezva JM (2012) The mitochondrial ATPase inhibitory factor 1 triggers a ROS-mediated retrograde prosurvival and proliferative response. Mol Cell 45:731–742 [DOI] [PubMed] [Google Scholar]
- 10.Martinez-Reyes I, Cuezva JM (2014) The H(+)-ATP synthase: a gate to ROS-mediated cell death or cell survival. Biochim Biophys Acta 1837:1099–1112 [DOI] [PubMed] [Google Scholar]
- 11.Hojlund K, Wrzesinski K, Larsen PM et al. (2003) Proteome analysis reveals phosphorylation of ATP synthase beta-subunit in human skeletal muscle and proteins with potential roles in type 2 diabetes. J Biol Chem 278:10436–10442 [DOI] [PubMed] [Google Scholar]
- 12.Guan SS, Sheu ML, Wu CT, Chiang CK, Liu SH (2015) ATP synthase subunit-beta down-regulation aggravates diabetic nephropathy. Sci Rep 5:14561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hojlund K, Yi Z, Lefort N et al. (2010) Human ATP synthase beta is phosphorylated at multiple sites and shows abnormal phosphorylation at specific sites in insulin-resistant muscle. Diabetologia 53: 541–551 [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Bermudez J, Cuezva JM (2016) The ATPase inhibitory factor 1 (IF1): a master regulator of energy metabolism and of cell survival. Biochim Biophys Acta 1857:1167–1182 [DOI] [PubMed] [Google Scholar]
- 15.Gledhill JR, Montgomery MG, Leslie AG, Walker JE (2007) How the regulatory protein, IF(1), inhibits F(1)-ATPase from bovine mitochondria. Proc Natl Acad Sci U S A 104:15671–15676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez-Arago M, Formentini L, Martinez-Reyes I et al. (2013) Expression, regulation and clinical relevance of the ATPase inhibitory factor 1 in human cancers. Oncogenesis 2:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez-Arago M, Garcia-Bermudez J, Martinez-Reyes I, Santacatterina F, Cuezva JM (2013) Degradation of IF1 controls energy metabolism during osteogenic differentiation of stem cells. EMBO Rep 14:638–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vazquez-Martin A, Corominas-Faja B, Cufi S et al. (2013) The mitochondrial H(+)-ATP synthase and the lipogenic switch: new core components of metabolic reprogramming in induced pluripotent stem (iPS) cells. Cell Cycle 12:207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henry RR, Abrams L, Nikoulina S, Ciaraldi TP (1995) Insulin action and glucose metabolism in nondiabetic control and NIDDM subjects. Comparison using human skeletal muscle cell cultures. Diabetes 44:936–946 [DOI] [PubMed] [Google Scholar]
- 20.Ciaraldi TP, Abrams L, Nikoulina S, Mudaliar S, Henry RR (1995) Glucose transport in cultured human skeletal muscle cells. Regulation by insulin and glucose in nondiabetic and non-insulindependent diabetes mellitus subjects. J Clin Invest 96:2820–2827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santacatterina F, Sanchez-Cenizo L, Formentini L et al. (2016) Down-regulation of oxidative phosphorylation in the liver by expression of the ATPase inhibitory factor 1 induces a tumorpromoter metabolic state. Oncotarget 7:490–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cha BS, Ciaraldi TP, Carter L et al. (2001) Peroxisome proliferator-activated receptor (PPAR) gamma and retinoid X receptor (RXR) agonists have complementary effects on glucose and lipid metabolism in human skeletal muscle. Diabetologia 44:444–452 [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Yang F, Gritsenko MA et al. (2011) Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics 11:2019–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Bermudez J, Sanchez-Arago M, Soldevilla B, del Arco A, Nuevo-Tapioles C, Cuezva JM (2015) PKA phosphorylates the ATPase inhibitory factor 1 and inactivates its capacity to bind and inhibit the mitochondrial H(+)-ATP synthase. Cell Rep 12:2143–2155 [DOI] [PubMed] [Google Scholar]
- 25.Pernas L, Scorrano L (2016) Mito-morphosis: mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu Rev Physiol 78:505–531 [DOI] [PubMed] [Google Scholar]
- 26.Yang X, Pratley RE, Tokraks S, Bogardus C, Permana PA (2002) Microarray profiling of skeletal muscle tissues from equally obese, non-diabetic insulin-sensitive and insulin-resistant Pima Indians. Diabetologia 45:1584–1593 [DOI] [PubMed] [Google Scholar]
- 27.Reuter SE, Evans AM (2012) Carnitine and acylcarnitines: pharmacokinetic, pharmacological and clinical aspects. Clin Pharmacokinet 51:553–572 [DOI] [PubMed] [Google Scholar]
- 28.Lorenzo M, Fernandez-Veledo S, Vila-Bedmar R, Garcia-Guerra L, De Alvaro C, Nieto-Vazquez I (2008) Insulin resistance induced by tumor necrosis factor-alpha in myocytes and brown adipocytes. J Anim Sci 86:E94–E104 [DOI] [PubMed] [Google Scholar]
- 29.Chouchani ET, Kazak L, Jedrychowski MP et al. (2016) Mitochondrial ROS regulate thermogenic energy expenditure and sulfenylation of UCP1. Nature 532:112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sena LA, Chandel NS (2012) Physiological roles of mitochondrial reactive oxygen species. Mol Cell 48:158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muoio DM, Newgard CB (2008) Mechanisms of disease: molecular and metabolic mechanisms of insulin resistance and beta-cell failure in type 2 diabetes. Nat Rev Mol Cell Biol 9:193–205 [DOI] [PubMed] [Google Scholar]
- 32.Crnz-Topete D, List EO, Okada S, Kelder B, Kopchick JJ (2011) Proteomic changes in the heart of diet-induced pre-diabetic mice. J Proteome 74:716–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stump CS, Short KR, Bigelow ML, Schimke JM, Nair KS (2003) Effect of insulin on human skeletal muscle mitochondrial ATP production, protein synthesis, and mRNA transcripts. Proc Natl Acad Sci US A 100:7996–8001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pagel-Langenickel I, Bao J, Joseph JJ et al. (2008) PGC-1alpha integrates insulin signaling, mitochondrial regulation, and bioenergetic function in skeletal muscle. J Biol Chem 283:22464–22472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Handschin C, Spiegelman BM (2008) The role of exercise and PGC1alpha in inflammation and chronic disease. Nature 454: 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shinoda K, Ohyama K, Hasegawa Y et al. (2015) Phosphoproteomics identifies CK2 as a negative regulator of beige adipocyte thermogenesis and energy expenditure. Cell Metab 22:997–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG (2015) Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 162:552–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Titov DV, Cracan V, Goodman RP, Peng J, Grabarek Z, Mootha VK (2016) Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio. Science 352:231–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chin RM, Fu X, Pai MY et al. (2014) The metabolite alpha-ketoglutarate extends lifespan by inhibiting ATP synthase and TOR Nature 510:397–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaster M, Rustan AC, Aas V, Beck-Nielsen H (2004) Reduced lipid oxidation in skeletal muscle from type 2 diabetic subjects may be of genetic origin: evidence from culturedmyotubes. Diabetes 53:542–548 [DOI] [PubMed] [Google Scholar]
- 41.Kim HE, Grant AR, Simic MS et al. (2016) Lipid biosynthesis coordinates a mitochondrial-to-cytosolic stress response. Cell 166: 1539–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Febbraio MA, Steensberg A, Starkie RL, McConell GK, Kingwell BA (2003) Skeletal muscle interleukin-6 and tumor necrosis factor-α release in healthy subjects and patients with type 2 diabetes at rest and during exercise. Metabolism 52:939–944 [DOI] [PubMed] [Google Scholar]
- 43.Feldstein AE, Werneburg NW, Canbay A et al. (2004) Free fatty acids promote hepatic lipotoxicity by stimulating TNF-alpha expression via a lysosomal pathway. Hepatology 40:185–194 [DOI] [PubMed] [Google Scholar]
- 44.Pedersen BK, Febbraio MA (2012) Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nat Rev Endocrinol 8:457–465 [DOI] [PubMed] [Google Scholar]
- 45.Amir Levy Y, Ciaraldi TP, Mudaliar SR, Phillips SA, Henry RR (2015) Excessive secretion of IL-8 by skeletal muscle in type 2 diabetes impairs tube growth: potential role of PI3K and the Tie2 receptor. Am J Physiol Endocrinol Metab 309:E22–E34 [DOI] [PubMed] [Google Scholar]
- 46.Kelley DE, He J, Menshikova EV, Ritov VB (2002) Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 51: 2944–2950 [DOI] [PubMed] [Google Scholar]
- 47.Morino K, Petersen KF, Dufour S et al. (2005) Reduced mitochondrial density and increased IRS-1 serine phosphorylation in muscle of insulin-resistant offspring of type 2 diabetic parents. J Clin Invest 115:3587–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eckel RH, Kahn SE, Ferrannini E et al. (2011) Obesity and type 2 diabetes: what can be unified and what needs to be individualized? Diabetes Care 34:1424–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.