Abstract
Caveolin-1 (Cav-1) is an important structural protein of caveolae and plays an oncogene-like role by influencing protein glycosylation in hepatocellular carcinoma (HCC) cells. However, the mechanism by which Cav-1 promotes invasion and metastasis capacity has not been completely clarified. In this study, we demonstrate that Pofut1 is a fucosyltransferase induced by Cav-1. Mouse Hepa1-6 HCC cells lacking Cav-1 expression exhibited low transcription levels of Pofut1, whereas strong Pofut1 expression was found in high-metastasis-potential Hca-F cells with high levels of Cav-1. Cav-1 activated MAPK signaling and promoted phosphorylation of the transcription factors CREB, Sp1, HNF4A and c-Myc, which bound to the Pofut1 promoter region to induce its transcription. As Notch signaling receptors can be modified with O-fucose by Pofut1, we further showed that Cav-1-induced upregulation of Pofut1 expression activated the Notch pathway and thus enhanced invasion and metastasis by mouse HCC cells in vitro and in vivo. Collectively, our findings reveal a novel mechanism by which Cav-1 promotes tumor metastasis by upregulating expression of Pofut1, suggesting that Cav-1 may function as a new biomarker for HCC.
Subject terms: Glycobiology, Metastasis, Cell signalling
Introduction
Caveolin-1 (Cav-1), a major structural protein of caveolae, has been implicated in many cellular processes, including lipid transport, signal transduction, and tumor progression. Recent research suggests that Cav-1 has a positive regulatory effect on tumor growth and plays a central role in tumor invasion and metastasis1–3. Moreover, there is a growing body of evidence that Cav-1 can regulate protein glycosylation4–6. It has been reported that Cav-1 modifies sugar chain biosynthesis by regulating the sublocalization of N-acetylglucosaminyltransferase III (GnT-III) in the intra-Golgi subcompartment in Huh6 cells7. Previous studies performed by our laboratory showed that Cav-1 promotes lymphatic metastasis of mouse hepatocellular carcinoma (HCC) cells by inducing CD147 glycosylation8,9. Cav-1 can also promote sialylation of HCC cells by enhancing the expression of glycosyltransferase ST6Gal-1, increasing the adhesion ability of these cells10. The above evidence indicates that Cav-1 exerts an oncogene-like effect in HCC by affecting glycosylation levels, glycosylation-related protein localization, and glycosyltransferase expression.
Fucosylation is a type of glycosylation modification that occurs via an enzyme-catalyzed reaction that covalently links fucose to oligosaccharides and proteins11. Overexpression of certain fucosyltransferases (FUTs) in tumor cells leads to an increased level of fucosylation, resulting in abnormal cell proliferation, the epithelial–mesenchymal transition (EMT), and cancer progression12. Moreover, the level of fucosylation in the serum of HCC patients is significantly elevated, and detection of alpha-fetoprotein (AFP) fucosylation has been widely used in the diagnosis of HCC13. Thirteen types of FUT catalytic fucosylation are classified into N-fucosylation and O-fucosylation based on the site of fucose addition. FUT1-11 are present in the Golgi and catalyze N-linked fucosylation; O-FUTs (Pofuts) are sublocated in the endoplasmic reticulum and catalyze O-linked fucosylation14,15. Regardless, the mechanisms regulating FUT expression and fucosylation are poorly understood.
Notch receptors are well-known substrates of O-fucosylation and are modified by Pofut1. The four Type-I transmembrane Notch receptors (Notch1-4) each contain 29–36 epidermal growth factor-like (EGF) repeats in their extracellular ligand-binding domain that can be O-fucosylated by Pofut116–20. Evidence has revealed that, in Notch signaling, >50% of EGF repeats undergo this O-fucose modification, which in turn affects the activity of the pathway in humans and other mammals21–23. Therefore, Pofut1 plays a critical regulatory role in Notch-mediated cell fate decisions in numerous biological contexts, including carcinogenesis. Despite abundant evidence regarding the core roles of Pofut1 in Notch signaling, the molecular mechanisms responsible for Pofut1- and O-fucosylation-induced metastasis in HCC have rarely been explored.
In this study, Pofut1 expression levels were found to be positively correlated with Cav-1 in mouse HCC cells with different invasion and metastasis capacities, indicating a regulatory effect on Pofut1 for Cav-1. We demonstrate that Cav-1 upregulates expression of Pofut1 by activating the mitogen-activated protein kinase (MAPK) pathway to increase the phosphorylation levels of downstream transcription factors that bind to the promoter region of Pofut1. Furthermore, Cav-1 promotes invasion and metastasis of mouse HCC cells by activating Notch signaling through Pofut1. These findings provide a new theoretical foundation and novel areas of investigation for studying the oncogene-like role of Cav-1 in HCC.
Material and methods
Cell culture
Mouse HCC Hca-F cells that highly express Cav-1 (established and stored by the Department of Pathology, Dalian Medical University, Dalian) were grown in the mouse abdominal cavity for approximately 7 days (Song et al. (2005))24. The cells were then cultured in 90% RPMI 1640 (Gibco) medium supplemented with 10% fetal bovine serum (FBS) for 24 h. Hepa1-6 mouse hepatoma cells that do not express Cav-1 (obtained from the Cell Center of Peking Union Medical College, Beijing) were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS. All cultures were maintained with 100 IU/mL penicillin and 100 μg/mL streptomycin (1% antibiotic) at 37 °C in a humidified incubator with 5% CO2.
Stable clone selection and transfection
Cultured Hca-F cells were centrifuged (800 rpm, 5 min), the supernatant was discarded, and the cells were resuspended in RPMI 1640 cell culture medium without serum. The resuspended cells were adjusted to a concentration of 106 cells/mL and placed in electroporation cuvettes with a 4-mm gap. A group transfected with an unintentional sequence plasmid was used as a negative control (NC) group, and cells mixed with the pGPU6/shCav-1 plasmid were used as the experimental group; two kinds of plasmids were purchased from GenePharma (Shanghai, China). Both groups were electroporated twice at 500 V. The Hepa1-6 cells were uniformly spread onto a 10-cm cell culture plate, and after reaching 80% confluence on the second day, the cells were transfected with the pcDNA3.1/Cav-1 plasmid, and G418 screening was used to obtain stable Cav-1-transfected Hepa1-6 cells (Hepa1-6/Cav-1) at a concentration of 800 μg/mL.
Immunohistochemical staining
Paraffin-embedded liver tissue sections from wild-type and Cav-1−/− C57BL/6J mice were dewaxed and treated with boiling citrate buffer, pH 6.0, for 3 min. The sections were incubated in hydrogen peroxide (3%) for 20 min at room temperature to block endogenous peroxidase and then incubated with anti-goat serum for 15 min at room temperature. The sections were incubated with primary antibodies specific for Cav-1 (Thermo Fisher, USA), Pofut1 (Biosynthesis Biotechnology, Beijing, China), the Notch receptor intracellular domain (NICD) (Cell Signaling Technology, USA), HES1, and HEY1 (both from Biosynthesis Biotechnology, Beijing, China) overnight at 4 °C. Immunohistochemical analyses were performed using a Histostain Kit (Beyotime, Nantong, China). The stained sections were examined using a LEICA DMI4000 B microscope.
Quantitative real-time PCR (RT-PCR)
Total mouse liver RNA was isolated from either wild-type C57BL/6J mice and Cav-1−/− mice C57BL/6J, with three mice in each group. Total cell RNA was isolated from different cells using TRIzol (Invitrogen, Carlsbad, CA, USA), and cDNA was synthesized using an RT-PCR Kit (TaKaRa, Japan) according to the manufacturer’s instructions. The cDNA was amplified by real-time PCR using primer sets specific for Pofut1 and Cav-1, with GAPDH as an internal control. The sequences of the upstream and downstream primers are shown in Supplementary Table S1. All target gene transcripts were normalized to GAPDH, and the relative fold change in the expression was calculated using the 2−ΔΔCt method.
Western blot analysis
Total protein extracts and western blot procedures were carried out as previously described25. Antibodies specific for the following targets were used: HNF4A (Abcam, USA); Sp1, c-Myc, and CREB (Cell Signaling Technology, USA); p-HNF4A (Ser313), p-Sp1 (1:500, Thr739), p-c-Myc (Ser62), p-CREB (Ser133), p-JNK (Thr183+Tyr185), and p-p38 (Tyr323) (Biosynthesis Biotechnology, Beijing, China); extracellular signal–regulated kinase (ERK; Proteintech, Wuhan, China); p-ERK (Thr202+Tyr204, Bioworld, Nanjing, China); and c-Jun N-terminal kinase (JNK), p38, and GAPDH (Proteintech, Wuhan, China). The antibodies specific for Cav-1, Pofut1, NICD, HES1, and HEY1 were identical to those used in the immunohistochemical analysis described above. Chemiluminescence detection was performed using an ECL Kit (GE Healthcare, USA). GAPDH was detected on the same membrane as a loading control.
Immunofluorescence staining
Cells were seeded onto 10-mm coverslips. After washing twice with PBS, the cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.1% Triton X-100, and incubated with primary antibodies specific for Cav-1, Pofut1, NICD, HES1, and HEY1 (all antibodies were identical to the antibodies described in the above western blot experiments) overnight at 37 °C. Nuclei were stained with DAPI. The stained cells were viewed with a microscope system.
Vector construction and luciferase reporter assays
The 3’ untranslated region of Pofut1 was amplified using genomic DNA and primers (5’-CAT GAA GAG ACC CAC AGA GGA TTA-3’ and 5’-GGA AGA GGC GCG AGT GGC TAG-3’) containing NheI and BgIII restriction sites. The PCR products were ligated to the pGL3-control luciferase expression vector using T4 ligase. Hepa1-6 cells, Hepa1-6/Cav-1 cells, Hca-F cells, and Hca-F cells transfected with pGPU6/shCav-1 were co-transfected with the reporter plasmid and the control plasmid pRL-TK. After 48 h, luciferase activities were measured using Dual-Luciferase Reporter Assay System (Promega).
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed with Hepa1-6 and Hca-F cells using EZ-Magna ChIP™ A (No. 17-408, Millipore) following the standard protocol. The protein–DNA complexes were incubated with 5 μg of antibody (specific for HNF4A, Sp1, c-Myc, and CREB, as described in the above western blot experiments) or 5 μg of normal rabbit IgG (provided in EZ-Magna ChIPTM A Kit) as the immunoprecipitating antibody. The four pairs of ChIP primers presented in Supplementary Table S1 were designed based on predicted binding site. Region 1 (from −2045 to −2145 bp) included one predicted HNF4A-binding site: −2095 to −2105. Region 2 (from −939 to −1045 bp) included one predicted c-Myc-binding site: −990 to −997. Region 3 (from −483 to −587 bp) included one predicted CREB-binding site: −493 to −498. Region 4 (from −56 to −197 bp) included two predicted Sp1-binding sites: −75 to −81 and −142 to −146. PCR was performed with ChIP products or input samples as the template. To determine whether the transcription factors were bound to the Pofut1 promoter region, the PCR products were analyzed by 2.0% agarose gel electrophoresis, followed by ethidium bromide staining.
Primer analysis of RNA-seq data and analysis of differentially expressed genes (DEGs)
After filtering RNA-seq data using the HISAT and Bowtie tools, the RSEM tool was applied for quantitative gene expression analysis; quantitative expression is expressed in fragments per kilobase million. We used the Poisson distribution method to analyze the DEGs. DEGs were plotted on a scatter plot, and colors were used to distinguish between upregulated and downregulated genes, with orange indicating upregulated genes and blue indicating downregulated genes. We then performed Gene Ontology (GO) analysis of selected DEGs and used the WEGO software to calculate GO functional classification statistics for each annotated DEG. The functional distribution characteristics of genes were recognized based on macroscopic functions: biological process, cellular component, and molecular function.
In vitro invasion and migration assays
The invasion and migration of Hepa1-6 cells, Hepa1-6/Cav-1 cells, Hepa1-6/Cav-1 cells transfected with siPofut1, Hepa1-6/Cav-1 cells treated with DAPT (a specific Notch signaling pathway inhibitor; 5 μM, 24 h), Hca-F cells, Hca-F cells transfected with pGPU6/shCav-1, Hca-F cells transfected with siPofut1, and Hca-F cells treated with DAPT (5 μM, 24 h) were assessed by seeding cells into transwell insert in medium containing 0.5% FBS. The transwell inserts (Corning; 24-well, 8-μm pore size) were left uncoated or were coated with Matrigel and used according to the manufacturer’s instructions. Forty-eight hours after seeding, non-invading cells were removed, and invading cells were fixed with ice-cold methanol and stained with 0.1% crystal violet. Independent microscopic fields were imaged using a LEICA DMI4000 B microscope at a ×10 magnification. All invasion assays were performed at least in triplicate.
In vivo tumor metastasis assays
Animal experiments were approved by the local animal ethics committee (Dalian University of Technology Medical and Physiological Ethics Committee) and performed in accordance with national guidelines and regulations. Thirty inbred 615 mice were equally divided into five groups. Hepa1-6 cells, Hepa1-6/Cav-1 cells, Hepa1-6/Cav-1 cells that had been transiently transfected with siPofut1, Hca-F cells, and Hca-F cells that had been transiently transfected with siPofut1 (5 × 106 cells for all groups) were inoculated subcutaneously into the left footpad of each mouse. After 5 weeks, the mice were sacrificed, and their left inguinal lymph nodes and left axillary lymph nodes were isolated and stained with hematoxylin and eosin. Sectioning and staining were completed with assistance from the Pathology Laboratory of Dalian Medical University.
Statistics
All data represent at least three experiments and are expressed as the mean ± S.E.M. Differences between groups were compared using Student’s t test (when comparing two groups), one-way analysis of variance (ANOVA) with Tukey’s multiple comparison test (when comparing more than two groups and considering one independent variable), or two-way ANOVA with Bonferroni post hoc test (when comparing differences between groups considering two independent variables). All statistical tests were conducted using GraphPad Prism (La Jolla, CA, USA). Differences were considered statistically significant at *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
Cav-1 upregulates Pofut1 expression
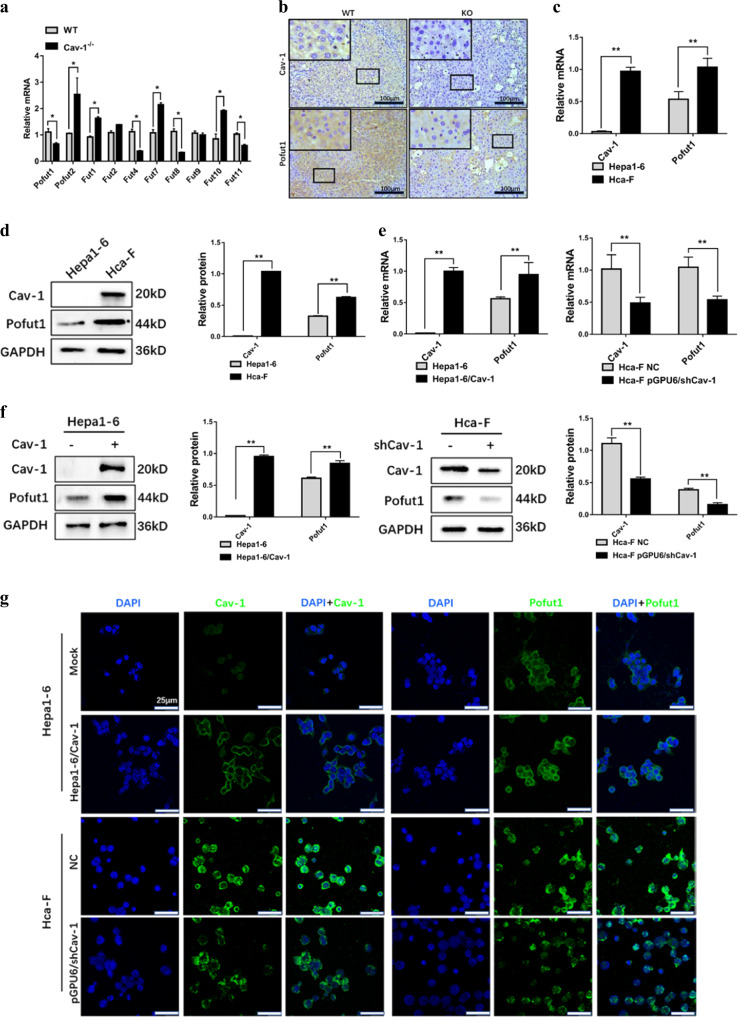
To investigate the roles of Cav-1 in the regulation of FUT expression in HCC, we first determined the expression of FUTs in Cav-1-knockout C57BL/6J mice. The results showed that transcriptional expression of FUTs in the mouse liver changed significantly following Cav-1 knockout (Fig. 1a). Among FUTs, the expression of Pofut1 was significantly reduced in the Cav-1−/− mouse liver. We then confirmed expression of Pofut1 in WT and Cav-1−/− mouse liver tissues using immunohistochemistry (Fig. 1b).
Fig. 1. Pofut1 expression is positively associated with Caveolin-1 (Cav-1).
a Analysis of the mRNA levels of different fucosyltransferases in wild-type C57BL/6J mice and Cav-1−/− C57BL/6J mice. b Expression levels of Pofut1 were examined by immunohistochemical staining. The blue staining represents the nucleus, and brown staining represents Cav-1 and Pofut1 (the secondary antibody was labeled with horseradish peroxidase, which produces a brown color with the substrate DAB). Representative sections of liver tissues from wild-type and Cav-1−/− C57BL/6 mice. Original magnification, ×100. c Endogenous Cav-1 and Pofut1 mRNA levels in Hepa1-6 and Hca-F cell lines were determined by quantitative real-time PCR (qRT-PCR). GAPDH served as a loading control. d Endogenous Cav-1 and Pofut1 protein levels in Hepa1-6 and Hca-F cell lines were determined by western blotting. GAPDH served as a loading control. e qRT-PCR assay was used to detect the mRNA expression level of Pofut1 after overexpression of Cav-1 in Hepa1-6 cells and after knockdown of Cav-1 in Hca-F cells. GAPDH served as a loading control. f Western blot assay was used to detect the protein expression level of Pofut1 after overexpression of Cav-1 in Hepa1-6 cells and after knockdown of Cav-1 in Hca-F cells. GAPDH served as a loading control. g Immunofluorescence staining was performed to further detect the protein expression of Pofut1 after overexpression of Cav-1 in Hepa1-6 cells and after knockdown of Cav-1 in Hca-F cells. Nuclei were counterstained with DAPI (×40). Statistical comparison using t test: *P < 0.05, **P < 0.01
To further analyze the relationship between Cav-1 and Pofut1 in HCC, two cell lines differing in Cav-1 expression and relative metastatic potential were selected: the poorly metastatic Cav-1-null cell line Hepa1-6 and the highly metastatic Cav-1-expressing HcaF cell line; Hca-F cells exhibit much higher metastatic potential than do Hepa1-6 cells (80% vs 0%)8. qPCR and western blotting showed that the expression of Pofut1 was significantly higher in Hca-F cells than in Hepa1-6 cells. (**P < 0.01, Fig. 1c, d). The Hepa1-6 cell line stably transfected with Cav-1 (Hepa1-6/Cav-1) was then used to characterize the impact of Cav-1 on Pofut1 expression. Notably, Cav-1 overexpression accompanied the elevation in Pofut1 mRNA and protein levels (**P < 0.01, Fig. 1e, f). Conversely, Cav-1 shRNA-transfected Hca-F cells exhibited attenuated Pofut1 expression (Fig. 1e, f). Similar results were obtained using immunofluorescence (Fig. 1g). In summary, these results implicate Cav-1 in controlling Pofut1 expression in HCC cells.
Cav-1 upregulates Pofut1 by increasing HNF4A, Sp1, c-Myc, and CREB phosphorylation
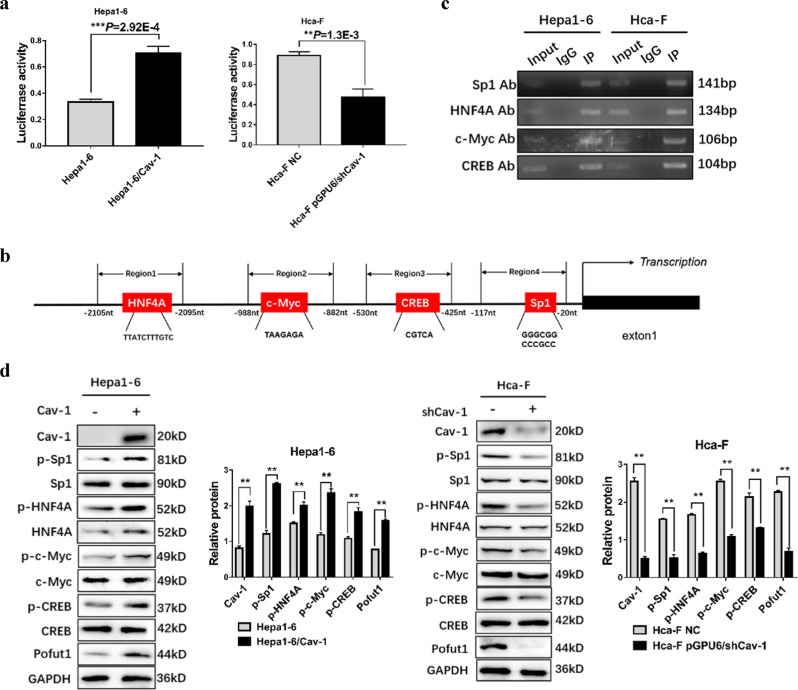
To explore the mechanism by which Cav-1 modulates Pofut1 expression, we first examined the regulation of the level of Pofut1 transcription by Cav-1. A dual-luciferase reporter gene assay revealed an obviously increased luciferase signal in Hepa1-6/Cav-1 cells and an obviously decreased luciferase signal in Hca-F cells transfected with the pGPU6/shCav-1 plasmid (**P < 0.01, Fig. 2a). Thereafter, we used the online biological software to predict transcription factor-binding sites in the Pofut1 promoter region and found sites for CREB, Sp1, HNF4A, and c-Myc within 2500 bp upstream (Fig. 2b). To corroborate this result in HCC cells, we performed ChIP analysis using Hepa1-6 and Hca-F cells. RT-PCR showed enrichment of CREB, Sp1, HNF4A, and c-Myc at the Pofut1 promoter region (Fig. 2c). Furthermore, Cav-1 overexpression significantly increased the phosphorylation but not the level of the above-mentioned transcription factors in Hepa1-6 cells (Fig. 2d), with the opposite results obtained in Hca-F cells transfected with pGPU6/shCav-1. Thus Cav-1 is able to activate CREB, Sp1, HNF4A, and c-Myc via phosphorylation. Taken together, these data suggest that Cav-1 increases Pofut1 expression by upregulating the phosphorylation of transcription factors that bind to the Pofut1 promoter region.
Fig. 2. Cav-1 upregulates Pofut1 by increasing transcription factor phosphorylation.
a Luciferase activity in Hepa1-6, Hepa1-6/Cav-1, Hca-F and Cav-1-knockout Hca-F cells was assayed at 48 h after transfection. Experiments were performed in triplicate, and the statistical significance of the difference was determined using the t-test (**P < 0.01 vs. control). b Schematic representation of predicted binding sites for HNF4A, c-Myc, CREB and Sp1. c ChIP assays confirmed that HNF4A, c-Myc, CREB, and Sp1 can bind to the following regions: region 1 (from −1088 to −1222 bp), region 2 (from −939 to −1045 bp), region 3 (from −483 to −587 bp), and region 4 (from −56 to −197 bp). d A western blot assay was used to detect the phosphorylation levels of HNF4A, Sp1, c-Myc and CREB after overexpression of Cav-1 in Hepa1-6 cells and after knockdown of Cav-1 in Hca-F cells. GAPDH served as a loading control. Statistical comparison using the t-test: **P < 0.01, **P < 0.001
Cav-1-mediated MAPK activation contributes to Pofut1 transcription
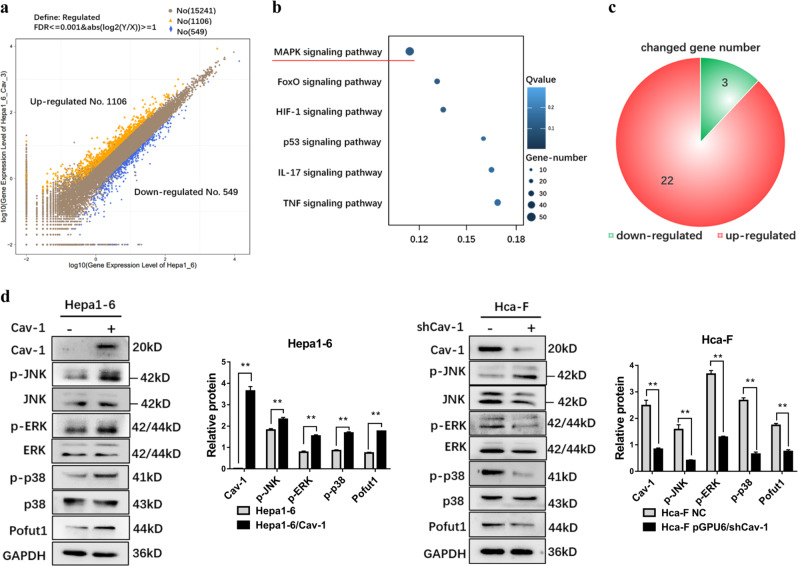
To further explore the molecular basis for the increase in Pofut1 expression, we compared transcriptomes between Hepa1-6 cells and Hepa1-6/Cav-1 cells using RNA-seq. Cluster analysis of DEGs revealed 549 genes to be downregulated and 1106 genes to be upregulated (Fig. 3a, Supplementary table S2). According to the KEGG public database, the largest number of DEGs are components of the MAPK pathway (Fig. 3b), with 22 of the 25 DEGs in pathway upregulated. These results suggest that Cav-1 overexpression activates MAPK signaling (Fig. 3c). The schematic presented in Supplementary Fig. S1 shows the specific genes in the MAPK pathway with altered expression. Western blotting showed the phosphorylation levels of ERK, JNK, and p38, three important pathways in MAPK signaling, to be significantly increased in Hepa1-6/Cav-1 cells, demonstrating that the MAPK pathway is activated by Cav-1 overexpression (Fig. 3d).
Fig. 3. Overexpression of Caveolin-1 (Cav-1) activates the mitogen-activated protein kinase (MAPK) signaling pathway.
(a) Differentially expressed genes between Hepa1-6 and Hepa1-6/Cav-1 displayed by a scatter diagram. x and y axes show the gene expression values; blue indicates downregulated genes, orange indicates upregulated genes, and brown indicates genes with nonsignificant differences; the screening condition is provided in the legend. b Statistical scatter plot of differential gene pathway enrichment for Hepa1-6 cells and Hepa1-6/Cav-1 cells. The Q value is the P value after multiple hypothesis test corrections, ranging from 0 to 1; the closer to zero the value is, the more significant is the enrichment. c There were 25 genes in the MAPK pathway with expression changes, 22 of which were upregulated in Hepa1-6/Cav-1 cells. d Western blot assay was used to detect the phosphorylation levels of extracellular signal–regulated kinase, c-Jun N-terminal kinase, and p38 after overexpression of Cav-1 in Hepa1-6 cells and after knockdown of Cav-1 in Hca-F cells. GAPDH served as a loading control. Statistical comparison using t test: **P < 0.01
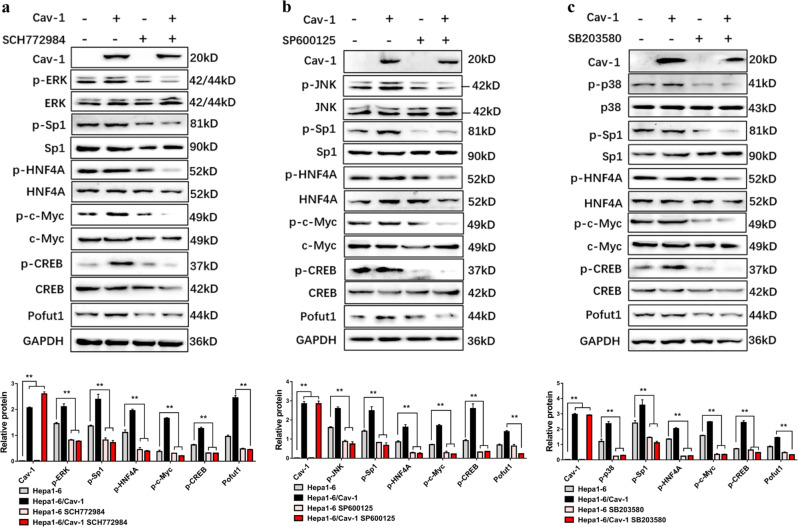
To further verify the participation of MAPK signaling in Cav-1-mediated increases in Pofut1 expression, Hepa1-6 and Hepa1-6/Cav-1 cells were treated with specific MAPK signaling inhibitors (ERK inhibitor, SCH772984, 20 nM for 6 h; JNK inhibitor, SP600125, 30 nM for 6 h; p38 inhibitor, SB203580, 20 nM for 6 h). These MAPK inhibitors were able to suppress CREB, Sp1, HNF4A, and c-Myc phosphorylation while reducing Pofut1 protein levels in both Hepa1-6 and Hepa1-6/Cav-1 cells (Fig. 4a–c). These results suggest that Cav-1 promotes phosphorylation of transcription factors via ERK, JNK, and p38 signaling pathways, thereby enhancing Pofut1 expression.
Fig. 4. Caveolin-1 (Cav-1) regulates transcription factors via mitogen-activated protein kinase and affects Pofut1 expression.
a Western blot analysis of Hepa1-6/Cav-1 or control Hepa1-6 cells treated for 6 h with 20 nM SCH772984. GAPDH served as a loading control. b Western blot analysis of Hepa1-6/Cav-1 or control Hepa1-6 cells treated for 6 h with 30 nM SP600125. GAPDH served as a loading control. c Western blot analysis of Hepa1-6/Cav-1 or control Hepa1-6 cells treated for 6 h with 20 nM SB203580. GAPDH served as a loading control. Statistical comparison using one-way analysis of variance: **P < 0.01
Cav-1-mediated increases in Pofut1 expression activates the Notch pathway
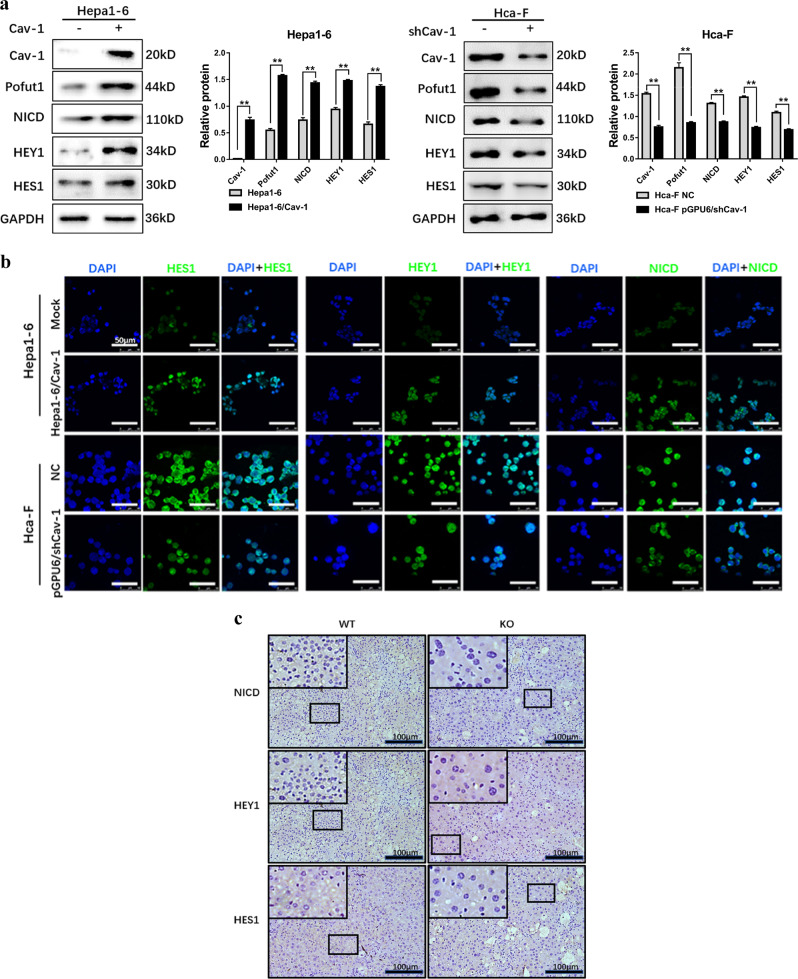
As Pofut1 has an important regulatory effect on Notch signaling, we sought to determine whether Cav-1-regulated Pofut1 expression activates this pathway. We first examined core components of the Notch pathway after Cav-1 overexpression and found that the levels of NICD and downstream key targets HES1 and HEY1 were increased in Hepa1-6/Cav-1 cells compared to those in Hepa1-6. However, downregulation of Cav-1 in Hca-F cells suppressed NICD, HES1, and HEY1 expression accompanying Pofut1 reduction (**P < 0.01, Fig. 5a). Immunofluorescence experiments confirmed this result (Fig. 5b). In addition, immunohistochemistry showed that the expression of NICD, HES1, and HEY1 in liver tissues from Cav-1−/− C57BL/6J mice was significantly lower than that in liver tissues from wild-type mice (Fig. 5c). Increased levels of NICD in the cytoplasm and nucleus were also found in Hepa1-6/Cav-1 cells (Supplementary Fig. S2a), and the results from confocal microscopy confirmed this finding (Supplementary Fig. S2a). All these data indicate that Cav-1 upregulates Pofut1 and activates the Notch pathway.
Fig. 5. Caveolin-1 (Cav-1) activates the Notch signaling pathway via Pofut1.
a Western blot analysis of Pofut1, Notch receptor intracellular domain (NICD), HES1, and HEY1 in Hepa1-6, Hepa1-6/Cav-1, Hca-F, and Hca-F-knockout Cav-1 cells. GAPDH served as a loading control. b Immunofluorescence staining was performed to detect the protein expression of Pofut1, NICD, HES1, and HEY1 after overexpression of Cav-1 in Hepa1-6 cells and after knockdown of Cav-1 in Hca-F cells. Nuclei were counterstained with DAPI (×40). c Expression levels of NICD, HEY1, and HES1 were examined by immunohistochemistry staining. The blue staining represents the nucleus, and brown staining represents Cav-1 and Pofut1 (the secondary antibody was labeled with horseradish peroxidase, which produces a brown color with the substrate DAB). Representative sections of liver tissues from wild-type C57BL/6J and Cav-1−/− C57BL/6J mice. Original magnification, ×100. Statistical comparison using t test: **P < 0.01
Cav-1-mediated increases in Pofut1 expression promote HCC cell invasion and metastasis in vitro and in vivo
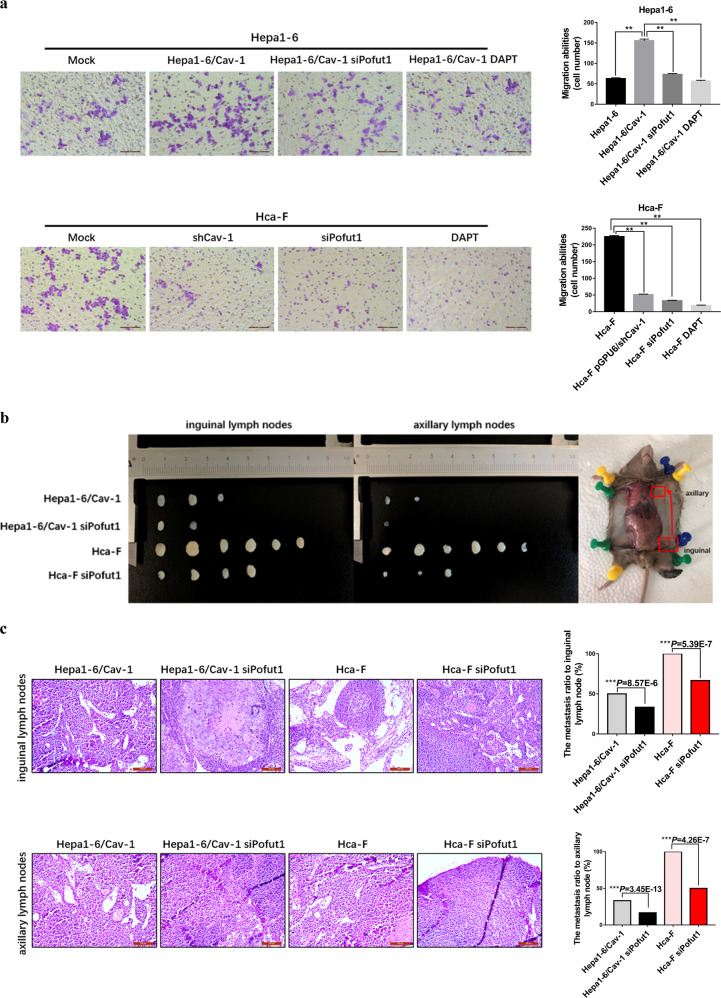
We next performed a transwell assay to determine the effect of Cav-1-mediated Pofut1 upregulation on HCC cell invasive behavior. Consistent with the oncogenic characteristics of Cav-1, the invasive ability of Hepa1-6/Cav-1 cells was significantly enhanced compared to that of Hepa1-6 cells. Nevertheless, this enhanced invasive potential was diminished by siRNA knockdown of Pofut1 in Hepa1-6/Cav-1 cells.
Reduced cell invasion potential was also observed after Pofut1 interference in Hca-F cells, suggesting the important role of Pofut1 in Cav-1-related HCC cell invasion (Fig. 6a). Moreover, a Notch signaling-specific inhibitor, DAPT, had similar effects as siPofut1, indicating that Cav-1-mediated increases in Pofut1 expression enhance HCC cell invasion through Notch signaling.
Fig. 6. Caveolin-1 (Cav-1) activates Notch by Pofut1 to enhance cell invasion and metastasis.
a A transwell assay was performed to evaluate the invasive potential of Hepa1-6 cells, Hepa1-6/Cav-1 cells, Hepa1-6/Cav-1 cells transfected with siPofut1, Hepa1-6/Cav-1 cells treated with DAPT (5 μM), Hca-F cells, Hca-F cells subjected to Cav-1 knockout, Hca-F cells transfected with siPofut1, and Hca-F cells with DAPT (5 μM). Statistical comparison using one-way analysis of variance: **P < 0.01. b Five groups (six mice in each group) of 615 mice were injected subcutaneously with Hepa1-6, Hapa1-6/Cav-1, Hepa1-6/Cav-1-siPofut1, Hca-F, and Hca-F-siPofut1 cells. After 5 weeks, the mice were sacrificed, and the inguinal lymph nodes and axillary lymph nodes were isolated and weighed. No tumor formation was observed in the inguinal and axillary lymph nodes of the mice injected with Hepa1-6 cells. Transfection of siPofut1 into Hepa1-6/Cav-1 and Hca-F cells reduced cell invasion and metastasis. c Hematoxylin and eosin (HE) staining was used to visualize the metastatic nodules in tumors from mice injected with Hapa1-6/Cav-1, Hepa1-6/Cav-1-siPofut1, Hca-F, and Hca-F-siPofut1 cells. Arrows indicate tumor foci. Scale bar = 100 μm. The lymphatic metastasis rates were significantly decreased by siRNA knockdown of Pofut1in Hepa1-6/Cav-1 and Hca-F cells. Chi-square test: ***P < 0.001
Finally, the in vivo effect on lymph node metastasis of increased Cav-1 expression mediated by Pofut1 was examined. We found that, compared to injection of Hepa1-6 cells into the footpads of 615 mice, injection of Hepa1-6/Cav-1 cells promoted lymphatic metastasis. Hepa1-6/Cav-1 cells metastasized not only to the inguinal lymph nodes near the footpad but also to the more distal axillary lymph nodes. In contrast, the lymphatic metastasis ability of Hepa1-6/Cav-1 and Hca-F cells subjected to Pofut1 knockdown was significantly decreased (Fig. 6b), and histological analysis of the lymph nodes confirmed this finding (***P < 0.001, Fig. 6c). Taken together, in vitro and in vivo results showed that Pofut1 plays an important role in Cav-1-induced mouse HCC cell invasion and lymphatic metastasis.
Discussion
Cav-1 is an essential structural and functional component of caveolae that is widely expressed in eukaryotes; this protein is mainly distributed in terminally differentiated cells, such as myocytes, endothelial cells, epithelial cells, and fibroblasts. Evidence shows that Cav-1 also participates in the signal transduction process in multiple physiological processes, including tumor metastasis26,27. Previous research has demonstrated that Cav-1 promotes the progression and metastasis of hepatocarcinoma in a variety of human hepatocarcinoma cell lines and tissues28,29. Both the amino-terminal and carboxy-terminal domains of Cav-1 are located on the cytoplasmic surface of the cell membrane, and the scaffold domain of Cav-1 can bind to downstream signaling factors; thus caveolae are key regulators of signal transduction26–29. The mature Cav-1 protein reportedly anchors a variety of signaling molecules, including EGF receptor, H-Ras, Rho-GTPases, c-Src, and endothelial nitric oxide synthase, through its scaffolding domain and plays an important role in regulating the function of various signaling molecules30–35. In this study, we found that Cav-1 influences MAPK activity to regulate Pofut1 expression and activates Notch pathway (Fig. 7). Cav-1 is also distributed in the mitochondria, endoplasmic reticulum, the late endoplasmic/lysosomal and Golgi apparatus, indicating that Cav-1 has a non-caveolae function in the membrane system36,37. It is reported that Cav-1 was found to be involved in the transport of intracellular cholesterol through the cytoplasmic surface of lysosomes38. The other report showed that Cav-1 in the Golgi was found to be involved in trans-Golgi-derived vesicle transport molecules6. In this study, we also found that Cav-1 is expressed in the cytoplasm (Fig. 1g). Since protein glycosylation occurs in the Golgi, activation of the Notch pathway by Cav-1 may not only act through upregulating Pofut1 expression but also through upregulating the glycosylation of Notch receptor by Cav-1 in the Golgi. However, the regulation of the effect on the activity of Notch pathway by Cav-1 in the cytoplasm remains to be further studied.
Fig. 7. Schematic.
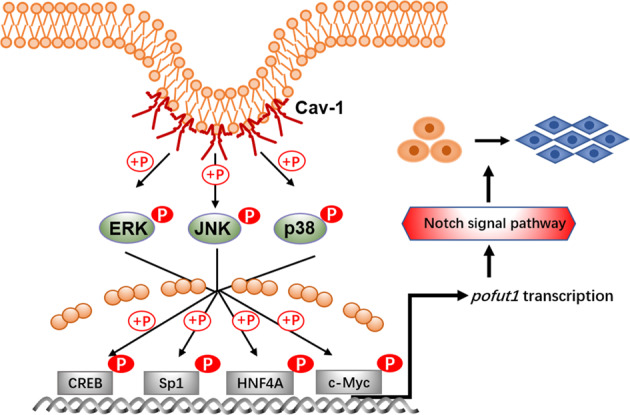
Caveolin-1 upregulates Pofut1 by increasing phosphorylation of CREB, Sp1, HNF4A, and c-Myc via the mitogen-activated protein kinase pathway and activates Notch signaling to enhance invasion and metastasis in mouse hepatocellular carcinoma cells
Abnormal protein glycosylation is closely related to the occurrence and development of many diseases, including cancer. Fucosylation is a cancer-related glycosylation, and abnormal levels of fucosylation modification are found in various tumors6,39–41. There are two main fucosylation modifications that are currently studied: core fucosylation and O-fucosylation42–45. Overall increases in core fucosylation have been associated with HCC, and these changes can be observed in serum samples15. Indeed, clinical assessment of a core fucosylated form of alpha-fetoprotein (AFP) has already been approved as a biomarker of HCC by the US Food and Drug Administration46. Although detection of AFP core-fucosylation is more specific and accurate than is detection of AFP expression alone for distinguishing between HCC and cirrhosis, little is known about the function and significance of O-fucosylation in HCC44. Here we investigated the regulation of an O-FUT, Pofut1, by Cav-1 and showed that Pofut1 plays an important role in HCC cell invasion and metastasis.
Pofut1 is a member of the O-FUT family, members of which add O-fucose monosaccharides to EGF repeats found on >100 potential targets in mammals, including Notch receptors and its ligands. The O-fucose modification catalyzed by Pofut1 on the Notch extracellular domain acts as a key regulator of Notch receptor activation, and aberrant O-fucosylation-induced dysregulation of Notch signaling disturbs cell fate decisions and other molecular events in a variety of human malignancies42,43,46. Recent studies have demonstrated that Pofut1 is highly expressed in human hepatocarcinoma cell lines (SMMC7721, Hep-3B, Hep-G2, Huh7, HCCLM3, MHCC97-H, and MHCC97-L) compared with normal hepatocytes (L-02), and Pofut1 overexpression enhances binding of the Notch ligand DLL1 to Notch receptor. In addition, the expression levels of Pofut1 are clinically correlated with the unfavorable survival and high recurrence rate in HCC, and aberrant activation of the Pofut1-Notch pathway is involved in HCC progression30.
The results presented in Figs. 5 and 6 show that Cav-1 enhances activity of the Notch pathway by upregulating Pofut1 expression. In vivo and in vitro experiments using Hepa1-6, Hepa1-6/Cav-1, and Hca-F cells indicate that Cav-1 enhances the invasion and metastasis abilities of cells by upregulating Pofut1 expression and activating Notch signaling. Hca-F and Hepa1-6 cells are mouse HCC cell lines with high and low lymphatic metastatic potential, respectively. Lymphatic metastasis ability was enhanced in Hepa1-6/Cav-1 cells, and tumors formed not only in the inguinal lymph nodes near the injection site but also in the distal axillary lymph nodes. Moreover, the lymphatic metastasis ability of Hepa1-6/Cav-1 and Hca-F cells transfected with siPofut1 was decreased, suggesting that Pofut1 plays an important role in the process by which Cav-1 enhances lymphatic metastasis in mouse HCC cells.
In summary, our results reveal the specific mechanism by which Cav-1 exerts an oncogenic effect in HCC cells, as based on enhanced glycosyltransferase expression and O-fucosylation-induced tumor metastasis, suggesting that inhibiting Pofut1 or other glycosyltransferases may constitute an optimal strategy for treating metastatic HCC.
Supplementary information
Acknowledgements
The authors would like to gratefully acknowledge support from the Natural Science Foundation of China (31570802, 31870793), the National Science and Technology Major Project of China (2018ZX10302205), and the Fundamental Research Funds for the Central Universities (DUT18ZD208, DUT18LAB09).
Competing interests
The authors declare no conflict of interest.
Footnotes
Edited by S. Tait
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1038/s41419-024-06529-5
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
7/29/2019
Since publication of this paper, the authors have noticed an error in one of the images. In Fig. 2c, the graphs of c-Myc and CREB are incorrect. During image synthesis, these two images were confused with other images from the raw data. The error does not impact the conclusions of the article. The authors would like to apologize for any inconvenience this may have caused.
This has been corrected in both the PDF and HTML versions of the Article.
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Change history
2/12/2024
This article has been retracted. Please see the Retraction Notice for more detail: 10.1038/s41419-024-06529-5
Contributor Information
Yubo Liu, Email: liuyubo@dlut.edu.cn.
Jianing Zhang, Email: jnzhang@dlut.edu.cn.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-019-1703-1).
References
- 1.Fernandez-Rojo MA, Ramm GA. Caveolin-1 function in liver physiology and disease. Trends Mol. Med. 2016;22:889–904. doi: 10.1016/j.molmed.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Nwosu ZC, Ebert MP, Dooley S, Meyer C. Caveolin-1 in the regulation of cell metabolism: a cancer perspective. Mol. Cancer. 2016;15:71. doi: 10.1186/s12943-016-0558-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ketteler J, Klein D. Caveolin-1, cancer and therapy resistance. Int. J. Cancer. 2018;143:2092–2104. doi: 10.1002/ijc.31369. [DOI] [PubMed] [Google Scholar]
- 4.Martinez-Outschoorn UE, Sotgia F, Lisanti MP. Caveolae and signalling in cancer. Nat. Rev. Cancer. 2015;15:225–237. doi: 10.1038/nrc3915. [DOI] [PubMed] [Google Scholar]
- 5.Parton RG, del Pozo MA. Caveolae as plasma membrane sensors, protectors and organizers. Nat. Rev. Mol. Cell Biol. 2013;14:98–112. doi: 10.1038/nrm3512. [DOI] [PubMed] [Google Scholar]
- 6.Kurzchalia TV, et al. VIP21, a 21-kD membrane protein is an integral component of trans-Golgi-network-derived transport vesicles. J. Cell Biol. 1992;118:1003–1014. doi: 10.1083/jcb.118.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sasai K, Ikeda Y, Ihara H, Honke K, Taniguchi N. Caveolin-1 regulates the functional localization of N-acetylglucosaminyltransferase III within the golgi apparatus. J. Biol. Chem. 2003;278:25295–25301. doi: 10.1074/jbc.M301913200. [DOI] [PubMed] [Google Scholar]
- 8.Zhou H, et al. Divergent expression and roles for caveolin-1 in mouse hepatocarcinoma cell lines with varying invasive ability. Biochem. Biophys. Res. Commun. 2006;345:486–494. doi: 10.1016/j.bbrc.2006.03.246. [DOI] [PubMed] [Google Scholar]
- 9.Jia L, et al. Caveolin-1 up-regulates CD147 glycosylation and the invasive capability of murine hepatocarcinoma cell lines. Int. J. Biochem. Cell Biol. 2006;38:1584–1593. doi: 10.1016/j.biocel.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 10.Yu S, et al. Caveolin-1 up-regulates integrinalpha2,6-sialylation to promote integrin alpha5beta1-dependent hepatocarcinoma cell adhesion. FEBS Lett. 2013;587:782–787. doi: 10.1016/j.febslet.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Holdener BC, Haltiwanger RS. Protein O-fucosylation: structure and function. Curr. Opin. Struct. Biol. 2019;56:78–86. doi: 10.1016/j.sbi.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magalhaes A, Duarte HO, Reis CA. Aberrant glycosylation in cancer: a novel molecular mechanism controlling metastasis. Cancer Cell. 2017;31:733–735. doi: 10.1016/j.ccell.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, et al. Fucosylation deficiency in mice leads to colitis and adenocarcinoma. Gastroenterology. 2017;152:193–205.e110. doi: 10.1053/j.gastro.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyoshi E, Moriwaki K, Nakagawa T. Biological function of fucosylation in cancer biology. J. Biochem. 2008;143:725–729. doi: 10.1093/jb/mvn011. [DOI] [PubMed] [Google Scholar]
- 15.Zhu, J., Warner, E., Parikh, N. D. & Lubman, D. M. Glycoproteomic markers of hepatocellular carcinoma-mass spectrometry based approaches. Mass Spectrom. Rev. 10.1002/mas21583 (2018). [DOI] [PMC free article] [PubMed]
- 16.Clausen H, Bennett EP. A family of UDP-GalNAc: polypeptide N-acetylgalactosaminyl-transferases control the initiation of mucin-type O-linked glycosylation. Glycobiology. 1996;6:635–646. doi: 10.1093/glycob/6.6.635. [DOI] [PubMed] [Google Scholar]
- 17.Brockhausen I, Yang J, Lehotay M, Ogata S, Itzkowitz S. Pathways of mucin O-glycosylation in normal and malignant rat colonic epithelial cells reveal a mechanism for cancer-associated Sialyl-Tn antigen expression. Biol. Chem. 2001;382:219–232. doi: 10.1515/BC.2001.029. [DOI] [PubMed] [Google Scholar]
- 18.Ma J, Hart GW. O-GlcNAc profiling: from proteins to proteomes. Clin. Proteomics. 2014;11:8. doi: 10.1186/1559-0275-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, et al. Modification of epidermal growth factor-like repeats with O-fucose. Molecular cloning and expression of a novel GDP-fucose protein O-fucosyltransferase. J. Biol. Chem. 2001;276:40338–40345. doi: 10.1074/jbc.M107849200. [DOI] [PubMed] [Google Scholar]
- 20.Luo Y, Haltiwanger RS. O-fucosylation of notch occurs in the endoplasmic reticulum. J. Biol. Chem. 2005;280:11289–11294. doi: 10.1074/jbc.M414574200. [DOI] [PubMed] [Google Scholar]
- 21.Haines N, Irvine KD. Glycosylation regulates Notch signalling. Nat. Rev. Mol. Cell Biol. 2003;4:786–797. doi: 10.1038/nrm1228. [DOI] [PubMed] [Google Scholar]
- 22.Rampal R, Arboleda-Velasquez JF, Nita-Lazar A, Kosik KS, Haltiwanger RS. Highly conserved O-fucose sites have distinct effects on Notch1 function. J. Biol. Chem. 2005;280:32133–32140. doi: 10.1074/jbc.M506104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okajima T, Irvine KD. Regulation of notch signaling by o-linked fucose. Cell. 2002;111:893–904. doi: 10.1016/S0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- 24.Song, B. et al. [Screening for lymphatic metastasis-associated genes in mouse hepatocarcinoma cell lines Hca-F and Hca-P using gene chip]. Ai Zheng24, 774–780 (2005). [PubMed]
- 25.Huang H, et al. MiR-23a transcriptional activated by Runx2 increases metastatic potential of mouse hepatoma cell via directly targeting Mgat3. Sci. Rep. 2018;8:7366. doi: 10.1038/s41598-018-25768-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engelman JA, et al. Reciprocal regulation of neu tyrosine kinase activity and caveolin-1 protein expression in vitro and in vivo. Implications for human breast cancer. J. Biol. Chem. 1998;273:20448–20455. doi: 10.1074/jbc.273.32.20448. [DOI] [PubMed] [Google Scholar]
- 27.Williams TM, Lisanti MP. The Caveolin genes: from cell biology to medicine. Ann. Med. 2004;36:584–595. doi: 10.1080/07853890410018899. [DOI] [PubMed] [Google Scholar]
- 28.Wang S, Jia L, Zhou H, Wang X, Zhang J. Caveolin-1 promotes the transformation and anti-apoptotic ability of mouse hepatoma cells. IUBMB Life. 2008;60:693–699. doi: 10.1002/iub.104. [DOI] [PubMed] [Google Scholar]
- 29.Tse EY, et al. Caveolin-1 overexpression is associated with hepatocellular carcinoma tumourigenesis and metastasis. J. Pathol. 2012;226:645–653. doi: 10.1002/path.3957. [DOI] [PubMed] [Google Scholar]
- 30.Ma L, et al. Overexpression of protein O-fucosyltransferase 1 accelerates hepatocellular carcinoma progression via the Notch signaling pathway. Biochem. Biophys. Res. Commun. 2016;473:503–510. doi: 10.1016/j.bbrc.2016.03.062. [DOI] [PubMed] [Google Scholar]
- 31.Cui Y, et al. Downregulation of caveolin-1 increased EGFR-TKIs sensitivity in lung adenocarcinoma cell line with EGFR mutation. Biochem. Biophys. Res. Commun. 2018;495:733–739. doi: 10.1016/j.bbrc.2017.11.075. [DOI] [PubMed] [Google Scholar]
- 32.Williams TM, et al. Combined loss of INK4a and caveolin-1 synergistically enhances cell proliferation and oncogene-induced tumorigenesis: role of INK4a/CAV-1 in mammary epithelial cell hyperplasia. J. Biol. Chem. 2004;279:24745–24756. doi: 10.1074/jbc.M402064200. [DOI] [PubMed] [Google Scholar]
- 33.Siddiqui MR, et al. Caveolin-1-eNOS signaling promotes p190RhoGAP-A nitration and endothelial permeability. J. Cell Biol. 2011;193:841–850. doi: 10.1083/jcb.201012129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cantiani L, et al. Caveolin-1 reduces osteosarcoma metastases by inhibiting c-Src activity and met signaling. Cancer Res. 2007;67:7675–7685. doi: 10.1158/0008-5472.CAN-06-4697. [DOI] [PubMed] [Google Scholar]
- 35.Jiang Y, et al. Critical role of caveolin-1 in ocular neovascularization and multitargeted antiangiogenic effects of cavtratin via JNK. Proc. Natl Acad. Sci. USA. 2017;114:10737–10742. doi: 10.1073/pnas.1706394114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi Y, et al. Critical role of CAV1/caveolin-1 in cell stress responses in human breast cancer cells via modulation of lysosomal function and autophagy. Autophagy. 2015;11:769–784. doi: 10.1080/15548627.2015.1034411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams TM, Lisanti MP. The caveolin proteins. Genome Biol. 2004;5:214. doi: 10.1186/gb-2004-5-3-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mundy DI, Li WP, Luby-Phelps K, Anderson RG. Caveolin targeting to late endosome/lysosomal membranes is induced by perturbations of lysosomal pH and cholesterol content. Mol. Biol. Cell. 2012;23:864–880. doi: 10.1091/mbc.e11-07-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dennis JW, Granovsky M, Warren CE. Protein glycosylation in development and disease. Bioessays. 1999;21:412–421. doi: 10.1002/(SICI)1521-1878(199905)21:5<412::AID-BIES8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 40.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 41.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer. 2015;15:540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 42.Li Z, et al. Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1. Nat. Chem. Biol. 2017;13:757–763. doi: 10.1038/nchembio.2381. [DOI] [PubMed] [Google Scholar]
- 43.Chen CI, et al. Structure of human POFUT2: insights into thrombospondin type 1 repeat fold and O-fucosylation. EMBO J. 2012;31:3183–3197. doi: 10.1038/emboj.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li D, Mallory T, Satomura S. AFP-L3: a new generation of tumor marker for hepatocellular carcinoma. Clin. Chim. Acta. 2001;313:15–19. doi: 10.1016/S0009-8981(01)00644-1. [DOI] [PubMed] [Google Scholar]
- 45.Tseng TH, et al. Substrate preference and interplay of fucosyltransferase 8 and N-acetylglucosaminyltransferases. J. Am. Chem. Soc. 2017;139:9431–9434. doi: 10.1021/jacs.7b03729. [DOI] [PubMed] [Google Scholar]
- 46.Yao D, et al. Protein O-fucosyltransferase 1 (Pofut1) regulates lymphoid and myeloid homeostasis through modulation of Notch receptor ligand interactions. Blood. 2011;117:5652–5662. doi: 10.1182/blood-2010-12-326074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.