Version Changes
Revised. Amendments from Version 1
The article has been updated to include further rationale for its development and clarification of the open access conditions for the data and open source nature of the underlying software.
Abstract
InterMine is a data integration and analysis software system that has been used to create both inter-connected and stand-alone biological databases for the analysis of large and complex biological data sets. Together, the InterMine databases provide access to extensive data across multiple organisms. To provide more convenient access to these data from Android mobile devices, we have developed the InterMine app, an application that can be run on any Android mobile phone or tablet. The InterMine app provides a single interface for data access, search and exploration of the InterMine databases. It can be used to retrieve information on genes and gene lists, and their relatives across species. Simple searches can be used to access a range of data about a specific gene, while links to the InterMine databases provide access to more detailed report pages and gene list analysis tools. The InterMine app thus facilitates rapid exploration of genes across multiple organisms and kinds of data.
Keywords: Android app, Genomics data, Gene search, InterMine
Introduction
InterMine 1 is an open source data integration and analysis software system (license LGPL 2.1) that has been used to create a suite of both inter-connected and stand-alone biological databases for the analysis of large and complex biological data sets. InterMine databases have been developed for the major model organisms budding yeast 2, nematode worm, fruit fly 3, zebrafish, mouse 4 and rat 5, (which we will refer to as the Model Organism Database (MOD-) InterMines, together with a human database and databases for plants, bees and wasps 6, cows 7, Medicago truncatula 8, mitochondrial proteomics 9 and drug targets 10 ( Table 1; https://registry.intermine.org/). Together, the InterMine databases provide access to extensive data across multiple organisms (for full listings of data included see the website for each individual InterMine, Table 1). To provide more convenient access to these data from Android mobile devices, we have developed the InterMine app 11, an application that can be run on any Android mobile phone or tablet.
Table 1. InterMine databases available through the InterMine app by default.
The list can be configured in the app.
| Database | Data | Organisation | url |
|---|---|---|---|
| FlyMine | Fruit fly | InterMine | http://www.flymine.org |
| HumanMine | Human | InterMine | http://www.humanmine.org |
| MedicMine |
Medicago
truncatula |
JCVI | http://medicmine.jcvi.org |
| MouseMine | MGI | MGI | http://www.mousemine.org |
| PhytoMine | Multiple plant
genomes |
Phytozome | https://phytozome.jgi.doe.gov |
| RatMine | Rat | RGD | http://ratmine.mcw.edu |
| TargetMine | Support
for drug discovery |
NIBIO | http://targetmine.mizuguchilab.org |
| ThaleMine | Thale cress | Araport | https://apps.araport.org/thalemine |
| WormMine | Nematode | WormBase | http://intermine.wormbase.org/tools/wormmine |
| YeastMine | Budding
yeast |
SGD | http://yeastmine.yeastgenome.org/yeastmine |
| ZebrafishMine | Zebrafish | ZFIN | http://zebrafishmine.org/ |
| HymenopteraMine | Various
Hymenoptera |
University of
Missouri |
http://hymenopteragenome.org/hymenopteramine |
| BovineMine | Bos taurus | Bovine Genome
Database Project |
http://bovinegenome.org/bovinemine |
The InterMine app provides a single interface for data access, search and exploration of the above databases. It can be used to retrieve information on genes and gene lists, and their relatives across species. Simple searches can be used to access a range of data about a specific gene, while links to the InterMine databases provide access to more detailed report pages and gene list analysis tools. Although a number of mobile applications have been developed for the laboratory (see https://www.biocompare.com/Editorial-Articles/168745-Ten-Mobile-Apps-for-Biology-Laboratories/), only a few so far exist for biological databases. Some examples include the YeastGenome app developed by SGD 12, Molecules, for viewing 3D protein structures ( https://itunes.apple.com/us/app/molecules/id284943090?mt=8) and Pubmed on Tap ( https://itunes.apple.com/us/app/pubmed-on-tap/id301316540?mt=8) / Pubmed Mobile ( https://play.google.com/store/apps/details?id=com.bim.pubmed&hl=en) for searching Pubmed and retrieving PDFs. Thus, development of the InterMine app was largely an exploratory exercise as it was not known at the outset what sort of demand there would be for accessing data in such a way, although we were encouraged by the success of the yeastGenome app. It is intended that the app provides quick and easy access to information about Genes when researchers may be away from their main computing source, such as when attending a conference or meeting. However, in addition to providing a quick and novel way to access biological data, InterMine app also expands InterMine’s functionality in allowing all registered InterMine databases to be searched at once, thus providing a cross-organism view of the term(s) searched.
The app is available from the Google Play Store at https://play.google.com/store/apps/details?id=org.intermine.app .
Methods
Data sources
The InterMine app allows users to search a default subset of InterMine data warehouses ( Table 1). This list is configurable, and so users are able to refine or add mines to match their interests. See https://registry.intermine.org for the full list of known public InterMine instances.
InterMine databases typically integrate data from many resources. For instance BioGRID 13, IntAct 14, UniProt 15, and can include high quality curated data (from the Model Organism Databases), genome-wide high-throughput data and data from smaller more focused studies (See individual InterMine websites for more details). All the InterMine databases accessible from the app are open source (License LGPL 2.1) and the data within them is free to access and download. Some individual InterMines may have restrictions for commercial use of the data and each individual InterMine should be consulted for its policy. See Table 1 for URLs.
Search and analysis
The InterMine app provides several ways to search and explore the data available, including a keyword search, sets of pre-defined template searches and list analysis functionality. These features are described in more detail in the use case section.
MyMine accounts
InterMine databases allow users to create an account through which they can, between sessions, store lists and searches. The InterMine App therefore allows users to log in to any accounts they hold on the underlying databases, so enabling user-created lists to be accessed.
Favourite genes
Users are also able to mark genes in search results as favourite. These genes are stored on the Android device and can be accessed without needing to log in to any of the underlying databases.
Implementation
Communication
The InterMine database design and the webservices used to power the InterMine app have been previously described 1, 16. The InterMine App draws all data from the RESTful Application Programming Interfaces (APIs) that InterMine databases provide 16. RoboSpice, an Open Source (Apache 2.0 Licence) Android communications library 17, provides core network communication functionality. Data are loaded asynchronously over HTTP or HTTPS, depending on the preferred protocol of the database being accessed. For performance enhancement, most web service responses are stored on the device if headers state an appropriate cache lifetime.
When the app receives a JSON response from the web service, it transforms the data from a table-structured format to a more hierarchical view, which presents data more effectively on smaller-screened mobile devices.
Authentication
Each InterMine database is discrete and often maintained by different organisations. If a user wishes to authenticate with multiple InterMines - perhaps to view private gene lists stored on different databases - they will need to provide separate authentication details for each InterMine database. However this is only necessary once, as after a user has successfully authenticated in a given InterMine via a username/password pair, the app retrieves and stores an API authentication token. This ensures that the user can authenticate in the future without having to re-enter or store sensitive password details.
All of the user configuration settings and authentication tokens are stored locally on the device via SharedPreferences, Android’s dedicated settings storage mechanism 18.
Internal storage
Tabular data, such as favourite genes within the app, are not suited to the key/value pair storage used in SharedPreferences 19, and therefore are stored within an SQLite database on the user’s device. Data stored include the InterMine instance the data originated from, the (e.g.) gene’s identifiers, description, organism, and genomic coordinates.
Search
Keyword search is available across lists, templates and gene search results. Search results from different databases are presented to the user as a single result set, sorted by the search relevance score generated by each originating database. Search results can be shared via email, instant messaging, and other sharing media in text format, using Android’s ACTION_SEND Intent functionality 20. Further data export options are available through links to the relevant full InterMine database instance.
Advanced information
InterMine also includes advanced analysis tools - particularly data visualisations - which may not be available via the API. To access the extended information about genes or gene lists, a user can load InterMine’s advanced report pages within the app itself. This is implemented via Android’s WebView 21 functionality which allows live web pages to be embedded in an application (for example, Figure 2 shows an example of an embedded InterMine WebView).
Operation
The app is implemented in accordance with Google Material Design 22 guidelines, providing a predictable environment for the user, and also supports Android version 4.0 and above, ensuring it is able to run on over 99% of active Android phones as of November 2018 23.
Use case
The following use case introduces each of the three key features of the InterMine app; Gene search, Templates and Lists with an example of their use. Further details of these InterMine features have been fully described in previous publications from several InterMine databases 1– 3, 5, 6, 8, 9.
Cross-organism gene search
The keyword search simultaneously searches all InterMine databases selected through the settings option. Thus, a cross-organism overview of data available for further investigation is provided. Link-outs from the search results to each originating InterMine database provide access to detailed gene report pages. These pages collate information integrated for that gene and typically include functional summaries, ontology annotation, pathway, expression, interaction and disease data and links to additional related data.
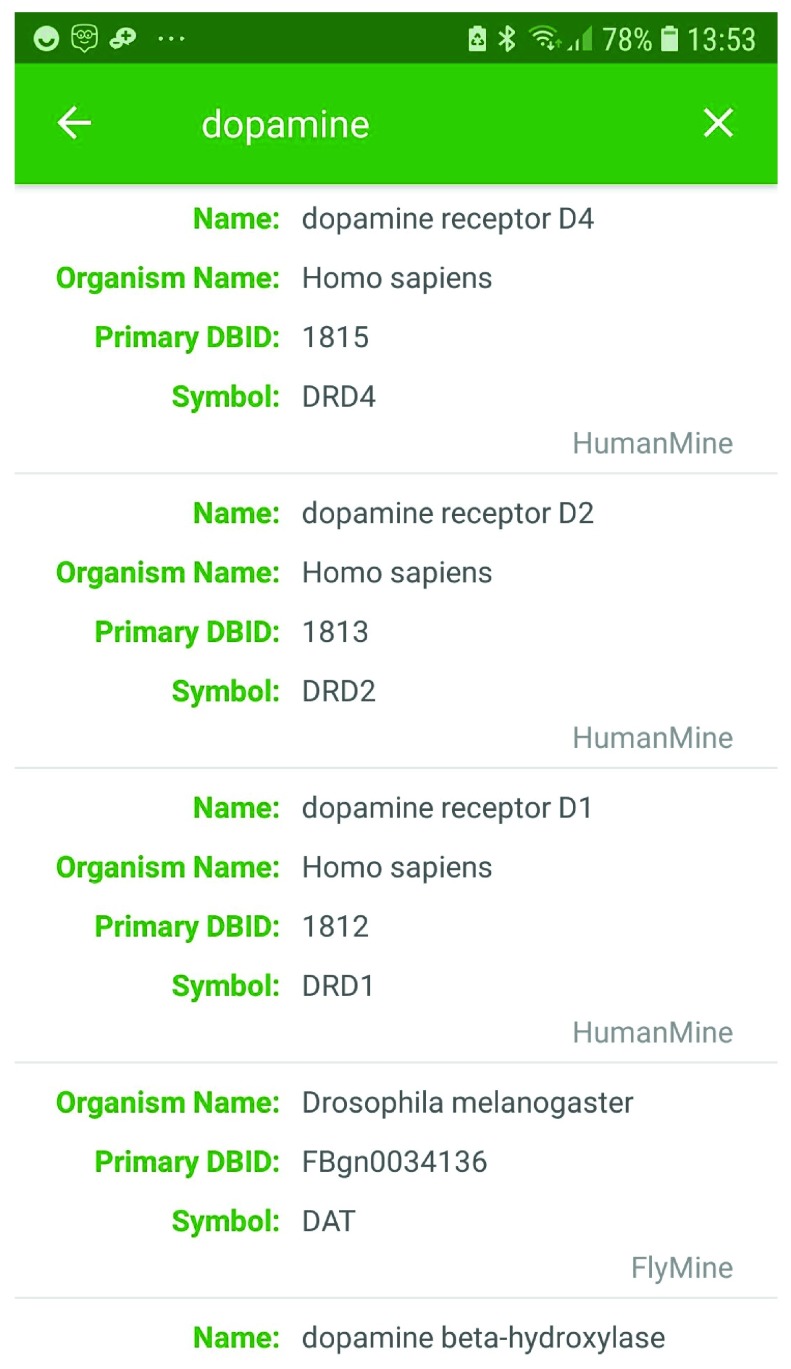
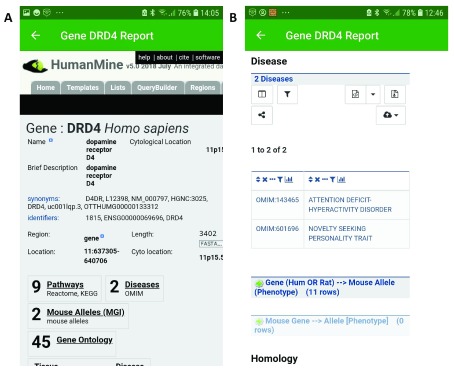
As an example, searching for ‘ dopamine’ returns dopamine-related genes from PhytoMine, MouseMine, HumanMine, TargetMine, FlyMine, RatMine, ZebrafishMine, WormMine, YeastMine, ThaleMine and HymenoperaMine ( Figure 1; see Table 1 for urls). Selecting a gene from the results, for instance the human gene DRD4 (dopamine receptor D4) displays summary information about the gene, with a link to the full gene report available from the HumanMine database. Here we learn, for example, that polymorphisms in the DRD4 gene are associated with the disorder attention deficit hyperactivity disorder (ADHD) ( Figure 2), a condition associated with low dopamine levels. The search results therefore facilitate rapid exploration across multiple organisms and kinds of data.
Figure 1. Example keyword search results.
A search for dopamine returns genes from PhytoMine, MouseMine, HumanMine, FlyMine, TargetMine, RatMine, ZebrafishMine, YeastMine and ThaleMine. A section of the results from HumanMine and FlyMine is shown.
Figure 2. The human DRD4 report page from the HumanMine webapp, displayed in the InterMine app.
( a) This view shows the first part of the report page with a summary of the Gene identifiers and data available. ( b) This view shows the Disease section of the report page.
Template searches
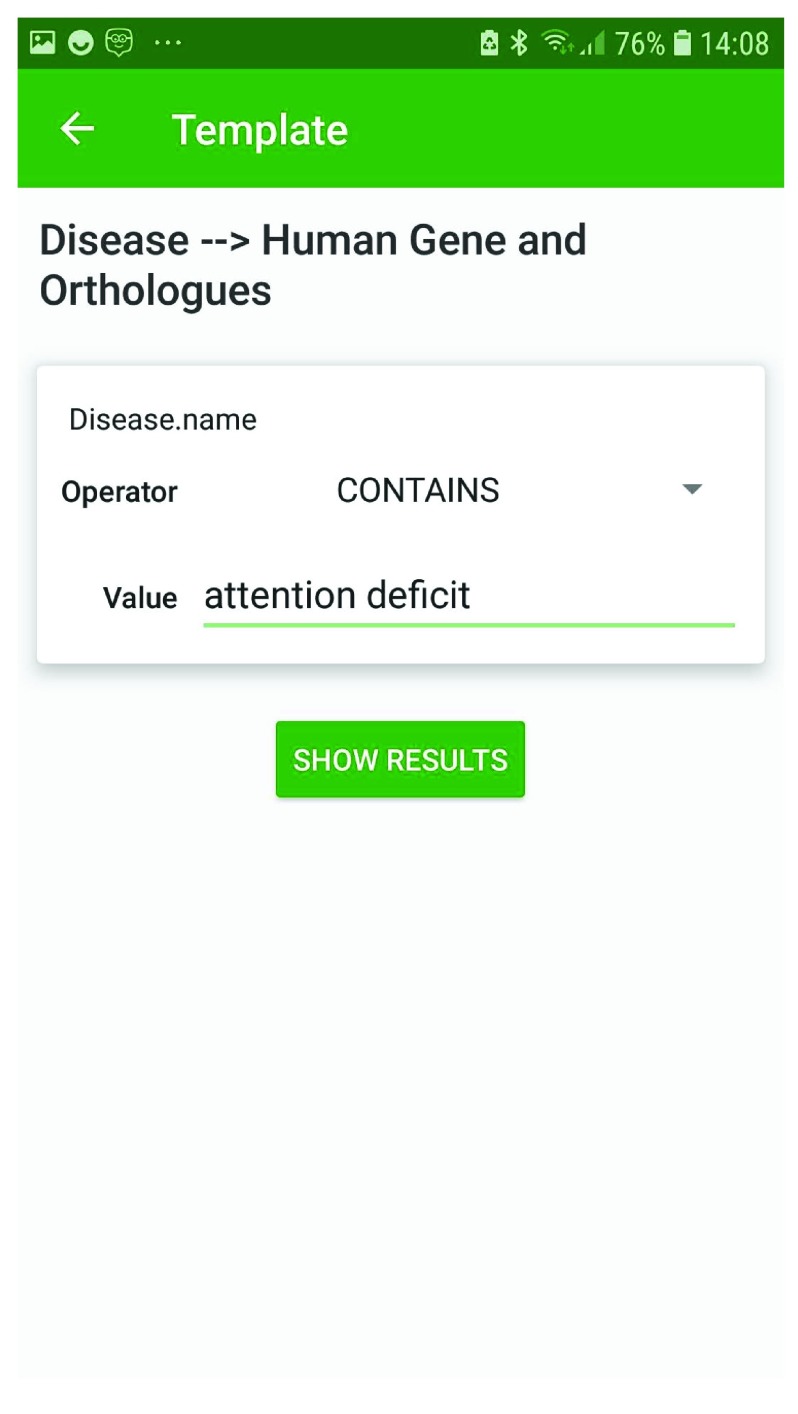
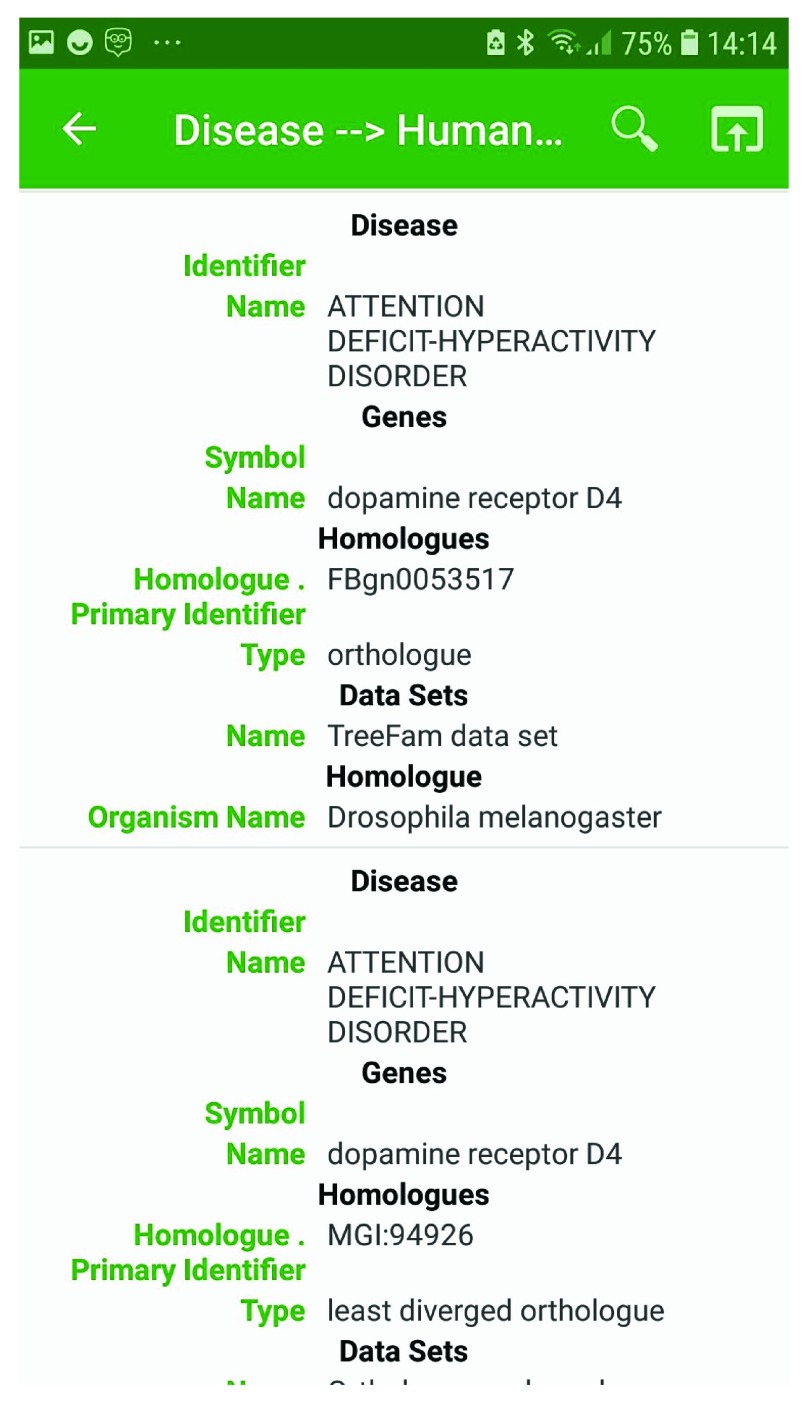
In addition to cross-organism gene search, the InterMine databases provide libraries of pre-built searches, called template searches. Such searches provide a user-friendly interface where the parameters for search filters can be specified. These templates range from simple searches, such as for a specified gene (or genes), return the corresponding Gene Ontology terms (represented as “ Gene → Gene Ontology terms”), to more complex searches combining data of more than one type, such as “ Tissue + interaction → genes”, which returns any genes expressed in the specified tissue that also interact with the product of a specified gene. Templates from each InterMine database are available within the InterMine App. The results are provided with a simple keyword search to facilitate further data browsing. For instance, continuing the above dopamine example, we can use a template search to identify genes in Drosophila associated with ADHD: on the templates page for the FlyMine database, we find the template “Disease -> Human genes and Orthologues” ( Figure 3). This template allows one to specify disease names that contains “attention deficit”, and on running the template, this returns the disease Attention deficit-hyperactivity disorder along with associated human genes and their orthologues from the available InterMine databases. Using the ability to search within the results we are able to verify that the human gene we are interested in (DRD4) is associated with this condition, and that this gene has a predicted orthologue in Drosophila melanogaster, FBgn0053517, Dop2R ( Figure 4). Through such iterative searching we can continue our investigation of this fly orthologue to identify, for instance, interacting partners, pathway and Gene Ontology annotations.
Figure 3. A FlyMine template search, “Disease -> Human genes and Orthologues”.
The disease name is an editable constraint, in this case set to search for human genes associated with diseases with a name that “contains” Attention deficit, as well as their orthologues.
Figure 4. Results from the template “Disease -> Human genes and Orthologues”, with disease constraint set to “contains” Attention deficit, available from FlyMine.
Shows the human dopamine receptor D4 associated with the disease attention deficit-hyperactivity disorder and with the D. melanogaster orthologue FBgn0053517, Dop2R.
Lists
InterMine databases are especially suited to the analysis of lists of genes or other entities. Users can create their own lists, which can be stored between sessions if the user has an account for the relevant InterMine database. Again, direct links from lists to the underlying InterMine databases provide access to list analysis tools, for instance enrichment statistics that help identify surprising properties, such as publications that cite an unexpectedly large number of the list members, or GO terms or protein domains that are associated with an unexpectedly large number of list members.
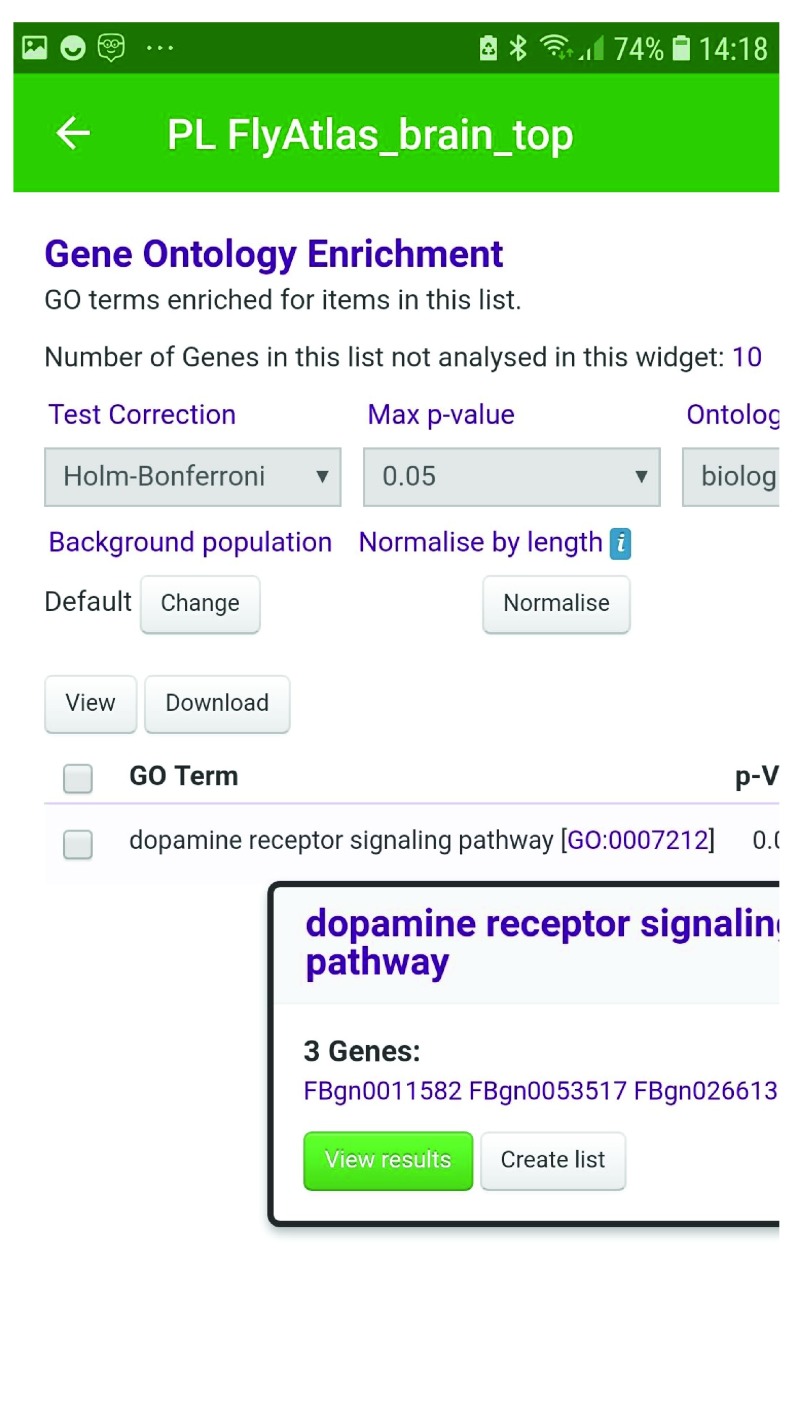
Public lists, which are typically interesting sets of genes derived from publications and other studies, are often provided by the database operators. For instance, in FlyMine, eleven of the public lists provide sets of genes whose expression increases at defined times during drosophila embryogenesis, as derived from Hooper et al. 24. Further lists show genes that are expressed at increased levels in various adult fly tissues according to data from the FlyAtlas resource 25. Within one of these sets, PL FlyAtlas_brain_top, we can identify a set of genes up-regulated in the brain. Checking within this list, we find that the dopamine receptor gene Dop2R (FBgn0053517) identified above is present. By following the link to the corresponding list analysis page on the underlying FlyMine website, and examining the enrichment statistics, we find that the Dop2R gene is part of a set in which the Gene Ontology term dopamine receptor signaling pathway (GO:0007212) occurs unexpectedly frequently (p-value 0.001303, with Holm-Bonferroni correction). It is also apparent that two other fly dopamine receptors, Dop1R1 (FBgn0011582) and Dop1R2 (FBgn0266137) are also found in this list ( Figure 5).
Figure 5. Gene Ontology enrichment analysis of the FlyMine public list PL FlyAtlas_brain_top.
A set of three genes (FBgn0011582 (Dpo1R1), FBgn0053517 (dop2R) and FBgn0266137 (Dop1R2) are enriched for the GO term dopamine receptor signaling pathway.
Conclusions
The InterMine app provides a convenient way of searching for biological information across many model organism and other databases, allowing an overview of gene function and gene relationships to be pursued. Importantly, the InterMine app reduces the effort required to obtain data available in a range of InterMine databases by removing the need to visit each one individually. Further development of the app is planned, including a single sign-in for all of the InterMine instances through oAuth2 ; further search and analysis capabilities including extending the keyword search to include all data types (instead of just genes); better cross-InterMine search result ordering; an offline mode with data cached in a local database for access when no internet connection is available, and a more sophisticated query construction capability for more advanced users.
Data availability
All data underlying the results are available as part of the article and no additional source data are required.
Software availability
The InterMine app is available from the Google Play Store: https://play.google.com/store/apps/details?id=org.intermine.app.
Source code available from: https://github.com/intermine/intermine-android .
Archived source code at time of publication: https://doi.org/10.5281/zenodo.1478646 10.
License: GNU General Public License v2.
Funding Statement
This work was supported by the Wellcome Trust (Grant number: 099133).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; peer review: 2 approved]
References
- 1. Smith RN, Aleksic J, Butano D, et al. : InterMine: a flexible data warehouse system for the integration and analysis of heterogeneous biological data. Bioinformatics. 2012;28:(23):3163–5. 10.1093/bioinformatics/bts577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balakrishnan R, Park J, Karra K, et al. : YeastMine--an integrated data warehouse for Saccharomyces cerevisiae data as a multipurpose tool-kit. Database (Oxford). 2012;2012:bar062. 10.1093/database/bar062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lyne R, Smith R, Rutherford K, et al. : FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 2007;8(7):R129. 10.1186/gb-2007-8-7-r129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Motenko H, Neuhauser SB, O’Keefe M, et al. : MouseMine: a new data warehouse for MGI. Mamm Genome. 2015;26(7–8):325–330. 10.1007/s00335-015-9573-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang SJ, Laulederkind SJ, Hayman GT, et al. : Analysis of disease-associated objects at the Rat Genome Database. Database (Oxford). 2013;2013:bat046. 10.1093/database/bat046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Elsik CG, Tayal A, Diesh CM, et al. : Hymenoptera Genome Database: integrating genome annotations in HymenopteraMine. Nucleic Acids Res. 2016;44(D1):D793–800. 10.1093/nar/gkv1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elsik CG, Unni DR, Diesh CM, et al. : Bovine Genome Database: new tools for gleaning function from the Bos taurus genome. Nucleic Acids Res. 2016;44(D1):D834–9. 10.1093/nar/gkv1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krishnakumar V, Kim M, Rosen BD, et al. : MTGD: The Medicago truncatula genome database. Plant Cell Physiol. 2015;56(1):e1. 10.1093/pcp/pcu179 [DOI] [PubMed] [Google Scholar]
- 9. Smith AC, Robinson AJ: MitoMiner v3.1, an update on the mitochondrial proteomics database. Nucleic Acids Res. 2016;44(D1):D1258–61. 10.1093/nar/gkv1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen YA, Tripathi LP, Mizuguchi K: TargetMine, an integrated data warehouse for candidate gene prioritisation and target discovery. PLoS One. 2011;6(3):e17844. 10.1371/journal.pone.0017844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Komkova D, Yehudi Y, Lyne R, et al. : Intermine-android App (Version v1.0.10). Zenodo. 2018. 10.5281/zenodo.1478646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wong ED, Karra K, Hitz BC, et al. : The YeastGenome app: the Saccharomyces Genome Database at your fingertips. Database (Oxford). 2013;2013:bat004. 10.1093/database/bat004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, et al. : The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43(Database issue):D470–8. 10.1093/nar/gku1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kerrien S, Aranda B, Breuza L, et al. : The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40(Database issue):D841–6. 10.1093/nar/gkr1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. UniProt Consortium: UniProt: a hub for protein information. Nucleic Acids Res. 2015;43(Database issue):D204–12. 10.1093/nar/gku989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kalderimis A, Lyne R, Butano D, et al. : InterMine: extensive web services for modern biology. Nucleic Acids Res. 2014;42(Web Server issue):W468–72. 10.1093/nar/gku301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. stephanenicolas: stephanenicolas/robospice. GitHub.(Accessed: 10th November 2016). Reference Source [Google Scholar]
- 18. Saving Key-Value Sets | Android Developers. (Accessed: 10th November 2016). Reference Source [Google Scholar]
- 19. Saving Data in SQL Databases | Android Developers.(Accessed: 10th November 2016). Reference Source [Google Scholar]
- 20. Sending Simple Data to Other Apps | Android Developers. (Accessed: 10th November 2016). Reference Source [Google Scholar]
- 21. Building Web Apps in WebView | Android Developers.(Accessed: 10th November 2016). Reference Source [Google Scholar]
- 22. Material Design for Android | Android Developers.(Accessed: 10th November 2016). Reference Source [Google Scholar]
- 23. Dashboards | Android Developers.(Accessed: 9th November 2016). Reference Source [Google Scholar]
- 24. Hooper SD, Boué S, Krause R, et al. : Identification of tightly regulated groups of genes during Drosophila melanogaster embryogenesis. Mol Syst Biol. 2007;3:72. 10.1038/msb4100112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Robinson SW, Herzyk P, Dow JA, et al. : FlyAtlas: database of gene expression in the tissues of Drosophila melanogaster. Nucleic Acids Res. 2013;41(Database issue):D744–50. 10.1093/nar/gks1141 [DOI] [PMC free article] [PubMed] [Google Scholar]