Abstract
Objective: Circular RNAs (circRNAs) are involved in regulating of carcinogenesis of various cancer cells. However, the function of circRNAs in colorectal cancer (CRC) has remained largely unknown. This study investigated the characteristic expression of circRNAs in CRC and adjacent normal tissues, analyzed the miRNAs related to candidate circRNAs, and studied the correlation between circRNAs with clinical data of CRC.
Methods: Human CircRNA microarray has been applied to screen the expressions of circRNAs of the CRC tissues and adjacent normal tissues. Quantitative real-time polymerase chain reaction (qRT-PCR) verified the candidate circRNAs in CRC tissue and patients’ peripheral blood. The circRNA array data were analyzed by GeneSpring 13.0 (Agilent) software. The diseases, pathways and functional enrichment analysis of these genes were performed using the KEGG system. In addition, the circRNA-miRNA network was constructed based on the miRanda-3.3 software. Statistical analysis was performed with SPSS23.0, GraphPad Prism, and Sigmaplot software.
Results: In total, 13,198 circRNAs were identified as distinct between CRC and adjacent normal tissues, including 6,697 upregulated and 6,501 downregulated genes. Based on scores, six of them were selected for further verification in CRC tissues and peripheral blood. The hsa_circ_0004585 expression was significantly upregulated both in CRC patients tissue and peripheral blood. Hsa_circ_0004585 was positively correlated with patient’s tumor size, indicating the function of hsa_circ_0004585 in CRC carcinogenesis and metastasis.
Conclusion: Hsa_circ_0004585 could be a potential biomarker for diagnosis of CRC.
Keywords: circRNA, colorectal cancer, prognosis biomarker
Introduction
Colorectal cancer (CRC) is one of the most frequently occurring digestive tract cancers, and the leading cause of morbidity and mortality. It is estimated that over 1.3 million new CRC cases are diagnosed every year.1 The diagnosis and treatment of CRC have continuously improved, but the mortality remains high. Therefore, to screen an efficient diagnostic marker and therapeutic target is an urgent problem.
Circular RNAs (circRNAs) are a new type of non-coding RNA, which form a covalently closed continuous loop. CircRNA does not have the 3ʹ and 5ʹ ends, but has a large number of miRNAs binding sites.2 The majority of circRNAs are exonic circRNAs, which are derived from exonic regions of known protein-coding genes by back-splicing. CircRNAs are abundant and stable in exosomes and plasma, providing a more convenient method for detection. Some researchers reported that circRNA can be a new diagnostic biomarker and therapy target for cancer, due to its stable and highly specific expression in different diseases and tissues, such as breast cancer, lung cancer, colorectal cancer, liver cancer, gastric cancer, and non-small cell lung cancer.3–8 Based on the reports, circRNA plays an important role in oncogenesis and influences the cancer cells proliferation, migration, and apoptosis.
CircRNAs play their function mainly by sponging miRNA. For example, circNT5E as a “sponge” of miR-422a regulates tumorigenesis in glioblastoma. CircNT5E controlled multiple pathologic processes, including cell proliferation, migration, and invasion by directly bound miR-422a and inhibited miR-422a activity.9 Circular RNA CiRS-7 provide a promising prognostic biomarker and a potential therapeutic target of CRC. CiRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer.10 However, the potential functions and the molecular mechanism of the majority circRNAs in different cancers are still not clear.
In this study, we investigated the characteristic expressions of circRNAs in CRC and adjacent normal tissues, and analyzed the clinical value of candidate circRNA in CRC. We found that hsa_circ_0004585 could be a potential biomarker for CRC diagnosis.
Materials and methods
Tissue and plasma samples
Both plasma and tumor tissue samples were collected from the Surgery Department of the General Hospital of Ningxia Medical University (Yinchuan, China). The study obtained approval from hospital ethics committee, which was conducted in accordance with the Declaration of Helsinki. All the patients signed their consent for donating their samples for the research. A total of 50 CRC tissue samples and adjacent normal tissues and 142 CRC and 142 healthy persons’s peripheral blood samples were collected from June 2017 to August 2018. Colorectal cancer tissue samples were obtained at the site of surgery and stored directly in liquid nitrogen. RNA is usually isolated within a week and then reverse transcribed to cDNA for further research. Blood samples were collected from patients with 2 mL aseptic and RNase-free tubes and treated with Trizol reagent within half an hour to isolate the RNA. Detailed clinical, pathological, and molecular characterization of these patients were collected accordingly.
RNA isolation
Total RNA was isolated from patient tissue and blood samples using the Trizol reagent (Invitrogen) and purified with mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) according to the protocol. The purity and concentration of RNA were determined by using spectrophotometer NanoDrop2000 (Thermo Fisher Scientific), the OD260/280 ratio around 1.9 to 2.0. RNA integrity was determined by 1% formaldehyde denatured gel electrophoresis.
Microarray analysis
Human CircRNA Array v2 microarray (Beijing Capital Bio Biotechnology Corporation, China) has been used for circRNA microarray profiling. The circRNA array data was analyzed by GeneSpring 13.0 (Agilent) software. Four CRC tissues and adjacent normal tissues were used for microarray analysis. The RNA integrity was determined by Bioanalyzer 2100 (Agilent). RNA digestion, amplification, and labeling were performed according to protocol. The labeled RNAs were hybridized on the microarray (Human circRNA array, version 2.0. Beijing Capital Bio Technology) containing 162,351 human circRNA probes. Differentially expressed circRNAs were detected by the filter criteria fold-change ≥2, P-value<0.05, Fluorescence value≥100.
Reverse transcription-quantitative PCR (RT-qPCR)
The cDNA was synthesized by the Superscript Reverse Transcription System (Invitrogen). RT-qPCR was performed using TB Green qPCR Mastermix (TaKaRa, Japan) on a LightCycler® 480 real-time PCR Platform (Roche). The qRT-PCR reaction was in a total volume of 20 μL systems, including 0.8 μL/10 μM forward/reverse primers, 10 μL TB Green qPCR Mastermix, 2 μL cDNA, and 6.4 μL double-distilled water. The cycling program is 95°C for 30 seconds, followed by 40 cycles of 95°C for 8 secondss and a pre-selected annealing temperature for 30 seconds. GAPDH expression was used as control in qPCR. The primers (Table 1) were synthesized by Sangon biotech (Shanghai) Co. Ltd. The relative expression of genes was calculated using the ΔΔCT method.
Table 1.
Primers for RT-PCR
| circRNA | Primers type | 5ʹ-3’ |
|---|---|---|
| hsa_circ_0072568 | Forward | GAACTGTACGAAACAACTTTGCTG |
| Reverse | GTGCCATTGTCCACATCAAAAC | |
| hsa_circ_0072566 | Forward | AGATCACCCATGTGCAACCA |
| Reverse | TGGCCAAGACCTGTTATGGTG | |
| hsa_circ_0072567 | Forward | GCCACCATAACAGACACGGA |
| Reverse | TGGCCAAGACCTGAGCAAAT | |
| hsa_circ_0074033 | Forward | CAGCCAACAGACCTCAGGAA |
| Reverse | CAAGGAGCCTGGAAAACGCT | |
| hsa_circ_0074039 | Forward | CAGCCAACAGACCTCAGGAA |
| Reverse | AGTGGTAGGGCTAGTCGCA | |
| hsa_circ_0004585 | Forward | CAAGACTGCAAGGAGCACACTG |
| Reverse | AGAGTGAGCCAGCTGATGGT |
Abbreviation: RT-PCR, real-time polymerase chain reaction.
Bioinformatics and data analysis
The circRNA array data were analyzed for data summarization, normalization, and quality control by using the GeneSpring software V13.0 (Agilent). To select the differentially expressed genes, we used threshold values of ≥2 and ≤−2 fold change and a Benjamini-Hochberg corrected P-value<0.05. The data was Log2 transformed and median centered by genes using the Adjust Data function of CLUSTER 3.0 software. Scatterplot and Volcano Plot were analyzed by using ggPlot2 software(R). CircRNA structure was performed by circPrimer1.2. The circRNA-miRNA network was constructed based on the miRanda-3.3 (http://www.microrna.org/microrna/home.do). The miRNA-binding sites of circRNAs were predicted by the miRanda and Circ-Interactome (https://circinteractome.nia.nih.gov/index.html) bioinformatics databases. All differentially expressed circRNAs were annotated its function in terms of the diseases, pathways, and functional. Enrichment analysis of these mRNAs were performed using the KEGG orthology-based annotation system (KOBAS) 3.0. KEGG analysis was performed to determine the involvement of target genes in different biological pathways.
Statistical analysis
Statistical analyses were performed using SPSS23.0 software (IBM, USA), GraphPad Prism version 7.0 (GraphPad Software, USA) and Sigmaplot version 14.0. Data were presented as mean±SD. The fold-change of each circRNA was computed from the profile difference between the cancer and control groups, and the significance was analyzed with two independent-samples t-test or one-way ANOVA. The receiver operating characteristic curve (ROC) was built using SPSS23.0, when the AUC was equal to 0.5, the circRNA was defined as having no diagnostic value. Differences were considered significant if P<0.05 (** P<0.01; *** P<0.001).
Results
Profiling of circRNAs from the CRC patients
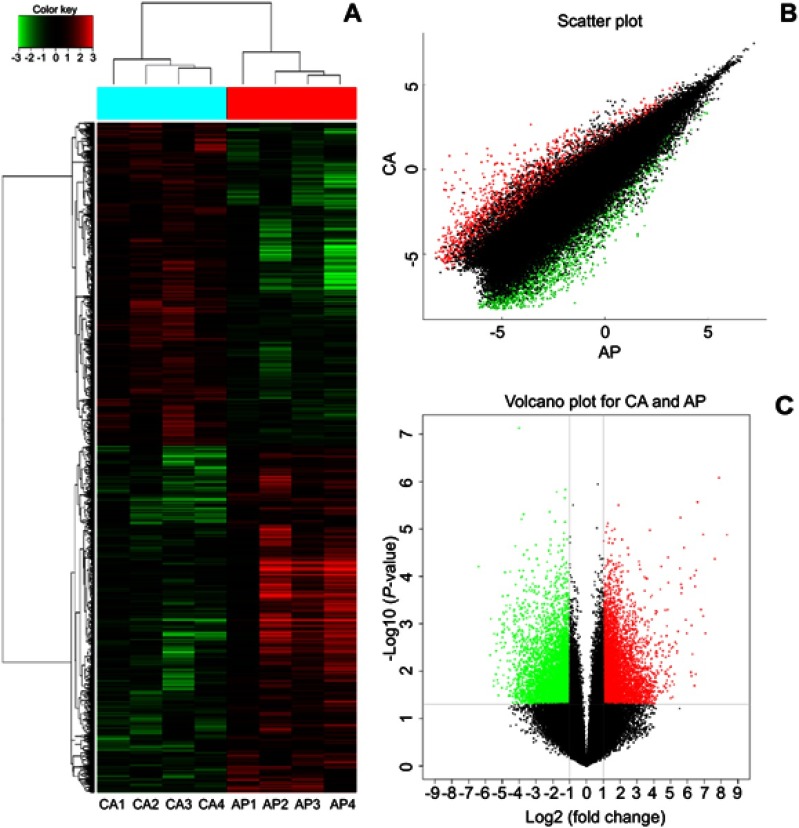
To find the specific circRNA in colorectal cancer, four CRC tissues samples (CA1–CA4) and adjacent normal samples (AP1–AP4) were selected for investigating the expression of circRNAs in CRC by microarray profiles (Figure 1). Different expression circRNAs between the two groups is displayed in the cluster analysis (Figure 1A). According to the different fluorescence signal values, scatter plots revealed that the differentially expressed circRNAs could separate CRC samples from adjacent normal tissues (Figure 1B). The Volcano plot described the variation in circRNA expression between CRC and adjacent normal tissues (Figure 1C). The figure revealed that the differentially expressed circRNAs could separate CRC samples from adjacent normal samples commendably. As is illustrated in Figure 1, 13,198 differentially expressed circRNAs were identified. Based on the screening criteria as a fold-change ≥2 or ≤-2 and a P-value<0.05, 6,697 were up-regulated and 6,501 down-regulated.
Figure 1.
Identification of the differentially expressed circRNAs between CRC tissue and adjacent. (A) The cluster graph of the samples, red is relatively high expression, blank is medium, and green is relatively low expression. CA1, CA2, CA3, and CA4 are colorectal cancer tissues, and AP1, AP2, AP3, and AP4 are corresponding adjacent normal tissue. (B) The scatter plot, the values spotted in the X and Y axes represent the normalized signals of samples in the two groups (log2-scaled). (C) The volcano plot, the Y axes is -log10 (P-values) and the X axes is log2 (fold change). Among them, the red color is the up-regulated gene, the green color is the down-regulated gene, and the black color is the no significant difference gene.
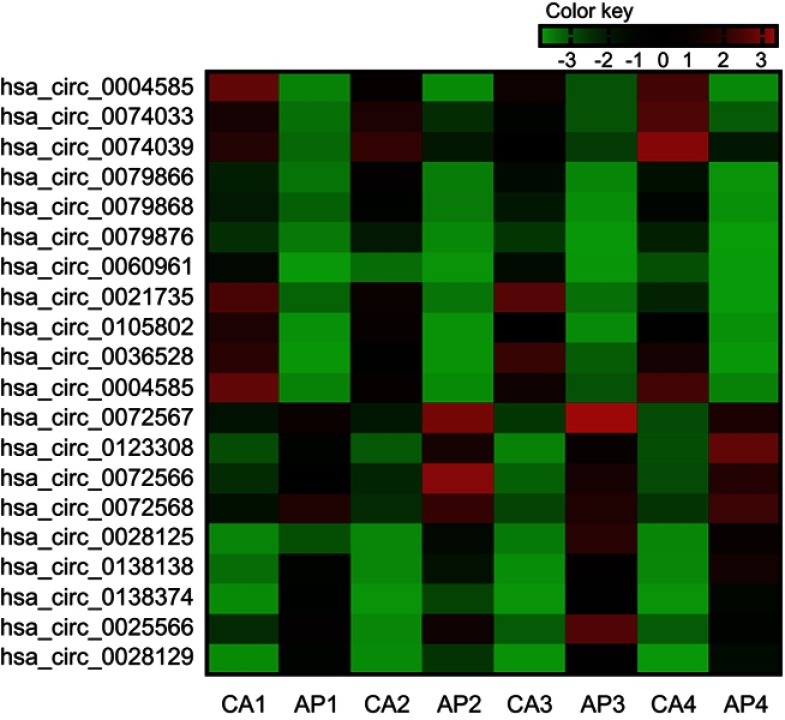
To further verify the candidate circRNA, we use fold-change ≥5 or ≤−5, P-value<0.01 and the Processed Signal ≥100 as classification criteria. The twenty candidate circRNAs selected included 11 up-regulation and ninedown-regulation. (Figure 2).
Figure 2.
Twenty differently expressed circRNAs in the colorectal cancer (the fold-change ≥5 or ≤-5, P-value<0.01 and the ProcessedSignal≥100). Red is relatively high expression, green is relatively low expression. CA1, CA2, CA3, and CA4 are colorectal cancer tissues, and AP1, AP2, AP3, and AP4 are corresponding adjacent normal tissue. The first 11 are high expression circRNAs, the rest are low expression circRNAs.
Enrichment analysis
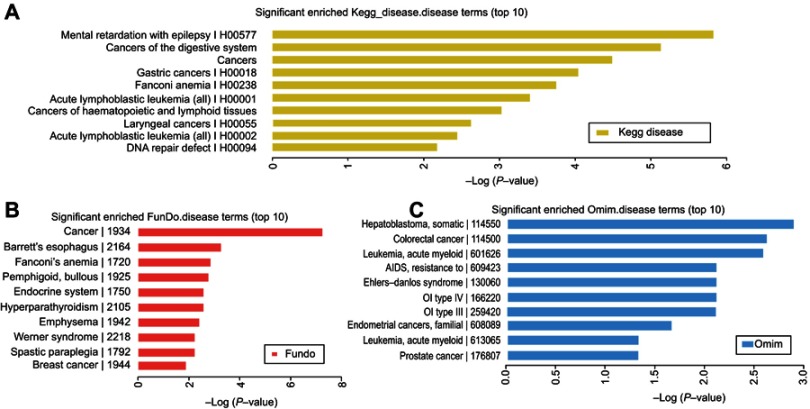
In this study, the related mRNAs (circRNA parental gene) of the differentially expressed circRNAs were annotated by KOBAS 3.0. They were enriched in some disease terms that included 433 in KEGG diseases, 795 in OMIM, and 398 in FunDO. The top 10 disease terms are shown in Figures 3A–C. The KEGG diseases contained digestive system, gastric cancer, as well as laryngeal cancer. The FunDO contained breast cancer. The OMIM contained colorectal cancer, leukemia, and prostate cancer. Based on the enrichment analysis, the differentially expressed circRNAs are involved in different tumorigenesis and development, and have in-depth research values.
Figure 3.
The enriched disease terms. The top 10 most significant enriched, (A) KEGG diseases terms, (B) FunDO terms, and (C) OMIM terms.
Validation differential expression circRNAs
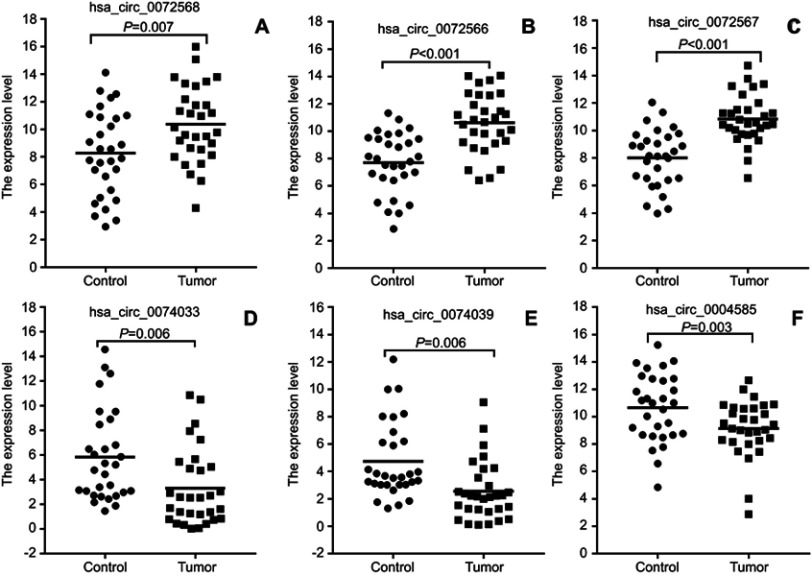
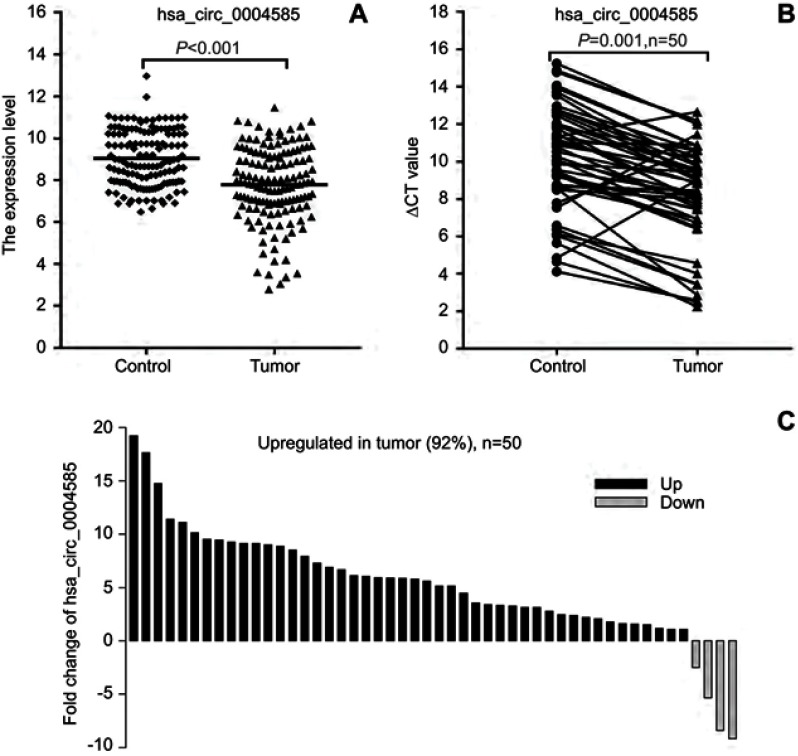
To verify the selected 20 candidate circRNAs, 30 CRC samples and adjacent normal tissues were applied in an independent cohort. Quantitative real-time reverse transcription PCR (qRT-PCR) revealed that hsa_circ_0004585, hsa_circ_0074033, and hsa_circ_0074039 were high expression in the CRC tissue, whereas hsa_circ_0072566, hsa_ circ_72567, and hsa_circ_72568 were downregulated (Figure 4). Further validation of the expression of these six candidate circRNAs, peripheral blood samples from 142 patients with CRC, and 142 healthy person were collected differently. Based on the results, the expression of hsa_circ_0004585 in CRC peripheral samples were studied, the expression of hsa_circ_0004585 was up-regulated in 121 cases and down-regulated in 21 cases (Figure 5A).
Figure 4.
Validation of differential expression circRNAs by qRT-PCR in CRC and adjacent tissues. Scatterplots display the relative expression of specific circRNAs between CRC (Tumor) and adjacent tissues (Control). Scatter plot drawing with △CT value between CRC and adjacent tissues in Graphpad Software. hsa_circ_0004585, hsa_circ_0074033, and hsa_circ_0074039 are higher expression, hsa_circ_0072566, hsa_ circ_72567, and hsa_circ_72568 are downregulated in CRC tissue.
Figure 5.
The expression level of hsa_circ_0004585. (A) The expression of hsa_circ_0004585 in 142 CRC patients’ peripheral samples compared with the normal healthy person group. (B and C) hsa_circ_0004585 was significantly upwnregulated in 92% (46/50) of 50 colorectal cancer patient tissues and the adjacent normal tissues.
Abbreviation: CT, cycle threshold.
We further added 20 cases (total 50 cases) of CRC and adjacent normal samples to verify hsa_circ_0004585 expression. The results showed that 46 cases in all were up-regulated and fourcases were down-regulated, the positive rate was 92% (Figures 5B and C). The characteristics of the 50 CRC patients are presented in Table 2. The result proved hsa_circ_0004585 expression has good consistency in the CRC tissue and peripheral samples.
Table 2.
Correlation between hsa_circ_0004585 expression and clinicopathological characteristics in 50 colorectal cancer patients
| Characteristics | No of cases | Mean±SD | P-value |
|---|---|---|---|
| Age (years) | |||
| ≤60 | 19 | 5.266±3.520 | 0.520 |
| >60 | 31 | 6.087±4.782 | |
| Gender | |||
| Male | 20 | 5.282±4.172 | 0.516 |
| Female | 30 | 6.104±4.465 | |
| Tumor size | |||
| ≤5cm | 33 | 6.655±4.725 | 0.044* |
| >5cm | 17 | 4.068±2.823 | |
| Clinical stage | |||
| I & II | 20 | 6.256±3.615 | 0.527 |
| III & IV | 30 | 5.455±4.774 | |
| CA19–9 | |||
| − | 41 | 5.412±3.996 | 0.208 |
| + | 9 | 7.419±5.570 | |
| CEA | |||
| − | 30 | 5.756±4.453 | 0.970 |
| + | 20 | 5.804±4.241 |
Note: *P<0.05.
Abbreviations: CA19-9, carbohydrate antigen 19-9; CEA, carcino-embryonic antigen.
ROC curve analysis
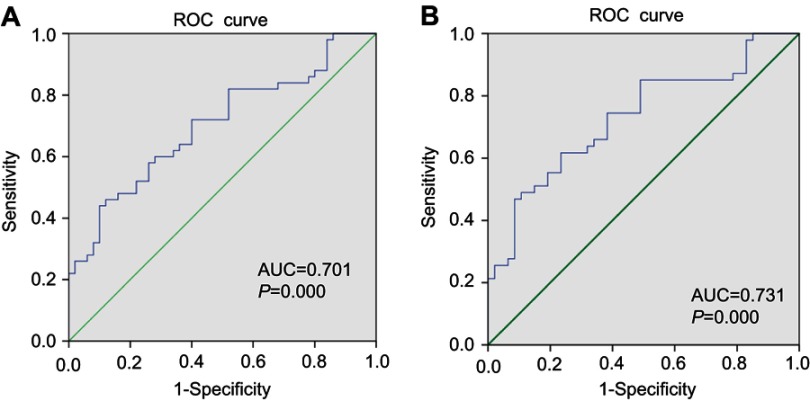
To determine the diagnostic values of circRNAs for cancer patients, Receiver Operating Characteristic (ROC) curve analysis was performed in colorectal cancer patients. The results of ROC curve analysis of six circRNAs are shown in Table 3. The results showed that hsa_circ_0004585 has higher diagnostic accuracy in 50 CRC tissues sample with an Area Under the Curve (AUC) of 0.731, P=0.000. The sensitivity and specificity are 0.851 and 0.511. In 142 peripheral samples, the AUC was 0.707, P=0.000. The sensitivity and specificity are 0.908 and 0.408 (Figure 6). The results proved that hsa_circ_0004585 have higher AUC and lower P-values in CRC.
Table 3.
Validation of the selected circRNAs by qPCR and ROC analysis
| CircRNA | Expressiona | AUC | P-value | Sensitivity | Specificity | |
|---|---|---|---|---|---|---|
| CRC tissues | Adjacent tissues | |||||
| has_circ-0004585 | 8.47±1.34 | 9.09±1.37 | 0.731 | 0.000** | 0.851 | 0.511 |
| hsa_circ_0002568 | 10.37±2.75 | 8.27±3.08 | 0.665 | 0.021* | 0.792 | 0.523 |
| hsa_circ_0002566 | 10.62±2.17 | 7.70±2.20 | 0.717 | 0.000** | 0.697 | 0.509 |
| hsa-circ-0002567 | 10.84±1.76 | 8.01±2.08 | 0.726 | 0.000** | 0.875 | 0.629 |
| hsa-circ-0074033 | 3.31±2.12 | 5.84±3.67 | 0.721 | 0.002** | 0.785 | 0.607 |
| hsa-circ-0074039 | 2.56±1.84 | 4.74±2.79 | 0.728 | 0.001** | 0.846 | 0.667 |
Notes: aThe expression of each circRNA was calculated from the ΔCt and expressed as mean±standard deviation; * P<0.05; ** P<0.01.
Abbreviations: AUC, area under the curve; CRC, colorectal cancer; qPCR, quantitative polymerase chain reaction; ROC, receiver operating characteristic.
Figure 6.
(A) ROC analysis for the expression hsa_circ_0004585 in 50 paired tissue of CRC patients. (B) ROC curves of the CRC person serum for the hsa_circ_0004585 expression.
Abbreviations: AUC, area under curve; ROC, receiver-operating characteristic.
miRNA response element (MRE) bioinformation analysis
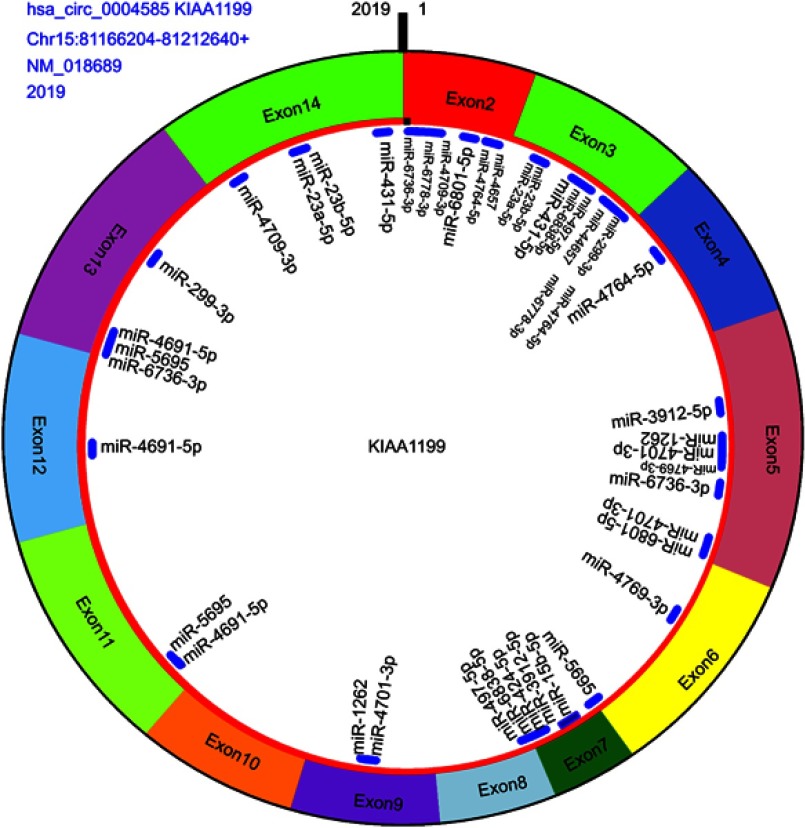
CircRNA can act as a “sponge” for miRNAs through their binding sites and modulate the activity of miRNA. In this study, we predicted the hsa_circ_0004585 binding miRNA by the circMIR software (from miRanda and RNAhybrid database). The results showed that hsa_circ_0004585 can bind with many miRNAs, some of the miRNA have multiple binding sites with hsa_circ_0004585, and some of them only have one binding site. According to the number of binding sites, the top 20 miRNA were shown in Table 4. The specific binding site location and information about the top 20 miRNAs with hsa_circ_0004585 is shown in Figure 7.
Table 4.
More binding sites of miRNA with hsa_circ_0004585 top 20
| miRNA ID | miRanda binding site (position) | Target scan binding site (positions) | Binding numbers |
|---|---|---|---|
| hsa-miR-299-3p | 1720 | 1555 1594 1736 241 99 1560 1599 1743 246 104 | 11 |
| hsa-miR-431-5p | 183 | 170 198 2008 511 977 175 205 2014 516 982 | 11 |
| hsa-miR-4657 | 222 89 | 1555 1594 1737 240 99 1560 1599 1743 246 104 | 11 |
| hsa-miR-4691-5p | 1276 | 1293 1453 1527 1873 326 1299 1458 1533 1879 331 | 11 |
| hsa-miR-15b-5p | 884 | 113 207 23 898 118 213 28 904 | 9 |
| hsa-miR-3912-5p | 869 | 1470 477 497 882 1475 483 502 888 | 9 |
| hsa-miR-424-5p | 192 | 113 207 23 898 118 213 28 904 | 9 |
| hsa-miR-497-5p | 884 | 113 207 23 898 118 213 28 904 | 9 |
| hsa-miR-6736-3p | 548 7 | 1642 20 332 561 1649 26 337 567 | 10 |
| hsa-miR-6778-3p | 15 | 253 29 458 542 260 36 463 547 | 9 |
| hsa-miR-6801-5p | 64 | 126 59 626 79 132 65 632 85 | 9 |
| hsa-miR-6838-5p | 193 884 | 113 207 23 898 118 213 28 904 | 10 |
| hsa-miR-1262 | 1048 500 | 1063 1734 520 1069 1739 526 | 8 |
| hsa-miR-23a-5p | 140 | 153 1922 300 159 1928 305 | 7 |
| hsa-miR-23b-5p | 1908 139 | 153 1922 300 159 1928 305 | 8 |
| hsa-miR-4701-3p | 508 | 1063 1734 520 1069 1739 526 | 7 |
| hsa-miR-4709-3p | 29 | 1854 44 854 1859 51 859 | 7 |
| hsa-miR-4764-5p | 229 | 100 1992 308 107 1997 314 | 7 |
| hsa-miR-4769-3p | 519 | 1422 533 699 1427 540 705 | 7 |
| hsa-miR-5695 | 806 | 1291 1630 819 1297 1636 825 | 7 |
Figure 7.
The hsa_circ_0004585 structure and miRNA binding sites, including origin gene KIAA1199, exon2–exon14 composed this circRNA, this gene from Chromosome 15, location between 81,166,204 and 81,212,640, the molecule is 2019 bp and binding miRNA top 20 (Table3).
Discussion
CircRNA, with a stable loop structure, is identified as a new type of non-coding RNA (ncRNA). It can act as competing endogenous RNAs (ceRNA) or as a miRNA “sponge” to regulate the function of mRNA expression, alternative splicing, or protein transcription.11,12 Previous research has proven that circRNA play an important role in oncogenesis and influences the cancer cells proliferation, migration, and apoptosis.13–15
Because of the stable characteristic of loop structure, many circRNAs being reported have a function in various cancers. For example, circRNA_100876 is significantly upregulated in non-small cell lung cancer (NSCLC) tissues.The survival time was significantly shorter in NSCLC patients with high circRNA_100876 expression than those patients with low circRNA_100876 expression.16 Xie et al17 found that hsa_circ_0074362 had low expression in gastric cancer tissues and gastric cancer cell lines. The expression levels of hsa_circ_0074362 were associated with gastric cancer lymphatic metastasis. Zhang et al.18 proved that hsa_circ_0001649 was down-regulated in hepatocellular carcinoma (HCC) tissues compared with adjacent normal tissues. They also demonstrated that over-expressed hsa_circ_0001649 inhibits the proliferation, migration, and invasion, and promotes the apoptosis of HCC cells. Gao et al.19 found circ_0006528 was highly expressed in breast cancer, the expression of circ_0006528 tightly related to chemotherapeutic resistance in breast cancer, and circ_0006528 could be a good biomarker for breast cancer prognostic.
Previous research reported that circRNAs have the function of promoting CRC cells growth and metastasis. CircRNA-ACAP2 and Tiam1 were shown to be highly expressed in colon cancer tissues and colon cancer SW480 cells. CircRNA-ACAP2 can act as a “sponge” to miR-21-5p and affect the proliferation, migration, and invasion of SW480 cells by regulating Tiam1 expression.20 CircHIPK3 was significantly upregulated in CRC tissues and cell lines, and promoted CRC growth and metastasis. Knockdown of circHIPK3 could inhibit CRC cells proliferation, migration, invasion, and induced apoptosis in vitro and suppressed CRC growth and metastasis in vivo.21 Hsa_circ_0020397 can enhance CRC cell viability, apoptosis, and invasion by promoting the expression of TERT and PD-L1, which was the target of the miR-138s.22 CircCCDC66 can act as the miRNA-33b and miR-93 “sponge” to promote CRC growth and matastasis.23 Those studies have revealed new insights into the pathogenicity of CRC and provide circRNA as a novel therapeutic target for the treatment of CRC.
In this study, we first screened the circRNA expression profile in four paired CRC and adjacent normal tissues following a microarray screening, 13,198 up-regulated and down-regulated circRNAs being identified. Then, 20 significantly differential expressed circRNAs, including 11 up-regulation and nine down-regulation, have been selected as candidate molecules. After verifying by RT-PCR in an independent cohort in CRC tissue, adjacent normal tissues, peripheral samples, and healthy person’s peripheral samples, hsa_circ_0004585 proved to have a good consistencyin the CRC tissue and peripheral samples.
Bioinformatics analysis found that the full-length of hsa_circ_0004585 was 2,019 bp, it was encoded by the KIAA1199 gene. Hsa_circ_0004585 was a novel circRNA derived from exons 2–14 of the KIAA1199 gene, located between 81,166,204 to 81,212,640 of human chromosome 15. Previous research has shown that the KIAA1199 gene was a high expression in CRC tissues, proving that KIAA1199 was an oncogene in CRC. Another researcher reported that the KIAA1199 protein was remarkably increased in CRC tissues and cells, which indicated the KIAA1199 protein involved in the CRC metastasis and reduced the patients’ survival.24
Although there is research which has proved that circRNA plays an important role in oncogenesis and influences the proliferation, migration, and apoptosis in different cancer, the function of most circRNAs is still unclear. CircRNA can act as a “sponge” for miRNAs through their binding sites and modulate the activity of miRNA. Different circRNAs have different miRNA binding sites, one circRNA could have several miRNA binding sites, one miRNA can bind several circRNA.25 In this study, we have predicted the hsa_circ_0004585 binding miRNA through the circMIR software (from miRanda and RNAhybrid database). According to the number of binding sites, the top 20 miRNA have been selected for in-depth analysis. We also used the enrichment to analyze the mRNAs of all differentially expressed circRNAs. We found that there were many diseases related to it.
Based on our results, hsa_circ_0004585 was up-regulated both in the CRC tissues and CRC peripheral samples compared with adjacent normal tissues and healthy person’s peripheral blood. ROC curve analysis demonstrated that hsa_circ_0004585 had high diagnostic accuracy in CRC. Our study proved that hsa_circ_0004585 has the potential to be a novel diagnostic marker and therapeutic target for CRC. However, the mechanism of hsa_circ_0004585 in CRC tumorigenesis and metastasis is still unclear, and further research is needed for validation.
Acknowledgments
This study was funded by the National Natural Science Foundation of China (No. 81560474, 81860470); the Natural Science Foundation of Ningxia (No. 2018AAC03160); the Science Research Project of Ningxia Higher Education (No. NGY2018-91); the First-Class Discipline Construction Founded Project of Ningxia Medical University and the School of Clinical Medicine (No. NXYLXK2017A05); and the Ningxia Medical University Scientific Research Project (No. XY201410, XY201413).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. Ca A Cancer J Clinicians. 2018;60:277–300. [Google Scholar]
- 2.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 3.Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi: 10.1186/s12943-017-0663-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Z, Xie L, Han L, et al. Circular RNAs: regulators of cancer-related signaling pathways and potential diagnostic biomarkers for human cancers. Theranostics. 2017;7:3106–3117. doi: 10.7150/thno.19016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang S, Zeng X, Ding T, et al. Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci Rep. 2018;8:2878. doi: 10.1038/s41598-018-21300-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen S, Zhang L, Su Y, et al. Screening potential biomarkers for colorectal cancer based on circular RNA chips. Oncol Rep. 2018;39:2499–2512. doi: 10.3892/or.2018.6372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Zhou H, Jing W, et al. The circular RNA hsa_circ_0001445 regulates the proliferation and migration of hepatocellular carcinoma and may serve as a diagnostic biomarker. Dis Markers. 2018;2018:3073467. doi: 10.1155/2018/3073467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clinica Chimica Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018 [DOI] [PubMed] [Google Scholar]
- 9.Wang R, Zhang S, Chen X, et al. CircNT5E acts as a sponge of miR-422a to promote glioblastoma tumorigenesis. Cancer Res. 2018;78:4812–4825. doi: 10.1158/0008-5472.CAN-18-0532 [DOI] [PubMed] [Google Scholar]
- 10.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568 [DOI] [PubMed] [Google Scholar]
- 11.Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. Embo J. 2014;32:923–925. doi: 10.1038/emboj.2013.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen LL. The biogenesis and emerging roles of circular RNAs. Nature Rev Mol Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32 [DOI] [PubMed] [Google Scholar]
- 13.Xu Y, Yao Y, Zhong X, et al. Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells. Biochem Biophys Res Commun. 2018;496:455–461. doi: 10.1016/j.bbrc.2018.01.077 [DOI] [PubMed] [Google Scholar]
- 14.Song YZ, Li JF. Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochem Biophys Res Commun. 2018;495:2369–2375. doi: 10.1016/j.bbrc.2017.12.050 [DOI] [PubMed] [Google Scholar]
- 15.Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11:98. doi: 10.1186/s13045-018-0643-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao JT, Zhao SH, Liu QP, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213:453–456. doi: 10.1016/j.prp.2017.02.011 [DOI] [PubMed] [Google Scholar]
- 17.Xie Y, Shao Y, Sun W, et al. Downregulated expression of hsa_circ_0074362 in gastric cancer and its potential diagnostic values. Biomark Med. 2018;12:11–20. doi: 10.2217/bmm-2017-0114 [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Qiu S, Luo P, et al. Down-regulation of hsa_circ_0001649 in hepatocellular carcinoma predicts a poor prognosis. Cancer Biomarkers. 2018;22:135–142. doi: 10.3233/CBM-171109 [DOI] [PubMed] [Google Scholar]
- 19.Gao D, Zhang X, Liu B, et al. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics. 2017;9:1175–1188. doi: 10.2217/epi-2016-0145 [DOI] [PubMed] [Google Scholar]
- 20.He JH, Li YG, Han ZP, et al. The CircRNA-ACAP2/Hsa-miR-21-5p/Tiam1 regulatory feedback circuit affects the proliferation, migration, and invasion of colon cancer SW480 cells. Cell Physiol Biochem. 2018;49:1539–1550. doi: 10.1159/000493457 [DOI] [PubMed] [Google Scholar]
- 21.Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417. doi: 10.1038/s41419-018-0454-8 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Zhang XL, Xu LL, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis, and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int. 2017;41:1056–1064. doi: 10.1002/cbin.10826 [DOI] [PubMed] [Google Scholar]
- 23.Hsiao KY, Lin YC, Gupta SK, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339. doi: 10.1158/0008-5472.CAN-16-1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J, Liu Y, Wang X, et al. Association between KIAA1199 overexpression and tumor invasion, TNM stage, and poor prognosis in colorectal cancer[J]. Int J Clin Exp Pathol. 2015;8:2909–2918. [PMC free article] [PubMed] [Google Scholar]
- 25.Grzegorz H, Małgorzata KF, Tomasz F. Relevance of microRNAs as potential diagnostic and prognostic markers in colorectal cancer. Int J Mol Sci. 2018;19:2944. doi: 10.3390/ijms19102944 [DOI] [PMC free article] [PubMed] [Google Scholar]