Abstract
The CLAVATA peptide-receptor (CLV3p-CLV1) pathway modulates a homeodomain master regulator WUSCHEL (WUS) transcription factor in the shoot apical meristem (SAM) with poorly defined signaling mechanisms. Here, we report that mitogen-activated protein kinases (MAPKs, also known as MPKs in plants) act in an intracellular signaling cascade to play an important role in the maintenance of SAM development. Interestingly, the application of exogenous CLV3p triggers rapid signaling in the SAM via dynamic activation of MPK3 and MPK6, which are positively regulated by both CLV1 and BARELY ANY MERISTEM 1 (BAM1) receptors. Surprisingly, the timing of MAPK activation is tightly correlated with the transcriptional repression of WUS expression in the SAM, indicating a fast CLV3p-CLV1/BAM1 signaling event. Furthermore, conditional mpk3,6 double mutants exhibited CLV3p insensitivity in stem cell maintenance manifested by the persistent SAM growth in the presence of exogenous CLV3p signals, as well as elevated WUS expression and repressed WUS-specific target genes. Taken together, these results suggest that MPK3 and MPK6 activated by CLV3p signals through mainly CLV1 and BAM1 receptors are key regulators controlling stem cell homeostasis in the SAM.
Keywords: Arabidopsis, CLAVATA receptor, MAPK cascade, Shoot apical meristem, Stem cell homeostasis
Introduction
Stem cells possess dual features of self-renewal and organogenesis through precisely controlled proliferation and differentiation in plants and animals (Aichinger et al. 2012; Heidstra and Sabatini 2014). Plant shoot stem cells, which are under dynamic control of the microenvironment in the primary shoot apical meristem (SAM), are the self-renewable reservoir for leaf, stem, flower and fruit organogenesis. Extensive researches have revealed a complex integration of multiple peptide-receptor and hormone signaling in modulating stem cell homeostasis in the SAM (Aichinger et al. 2012; Somssich et al. 2016). The CLAVATA-WUSCHEL (CLV-WUS) regulatory pathway, involving peptide-receptor kinase (CLV3p-CLV1) and the homeodomain WUS transcription factor, plays a central role in coordinating stem cell proliferation and differentiation in the SAM of diverse plant species (Somssich et al. 2016). The transcript of the dual repressor-activator WUS is expressed specifically in the organizing center (OC) of the SAM, but the WUS protein represses regulatory genes promoting differentiation and migrates toward overlying stem cells via plasmodesmata to directly activate CLV3 expression in the absence of WUS-interacting partner HAIRY MERISTEMs (HAMs) (Yadav et al. 2011; Yadav et al. 2013; Zhou et al. 2015; Zhou et al. 2018).
After processing, maturation and secretion, the mobile CLV3p from stem cells (Rojo et al. 2002; Kondo et al. 2006; Ni and Clark 2006; Ohyama et al. 2009) moves toward underlying cells to activate cell-surface receptor complexes and suppress WUS expression. Genetic and biochemical evidence supports the perception of CLV3p by leucine-rich repeat receptor-like kinase (LRR-RLK) complexes, including CLV1, BARELY ANY MERISTEM 1 and 2 (BAM1 and BAM2), RECEPTOR-LIKE PROTEIN KINASE 2 (RPK2), and a heterodimer with LRR receptor-like protein CLV2 and the transmembrane kinase CORYNE (CRN)/SUPPRESSOR OF LLP 1–2 (SOL2) (Müller et al. 2008; Guo et al. 2010; Kinoshita et al. 2010; Shinohara and Matsubayashi 2015). However, the intracellular signaling pathway restricting WUS expression and stem cell population via the negative feedback loop remains elusive.
Mitogen-activated protein kinase (MAPK, also called MPK in plants) cascades are evolutionarily conserved signaling modules that act as central hubs in diverse physiological and developmental processes in plants, animals and humans. Although MAPK cascades have long been proposed in SAM regulation, their molecular identities and signaling mechanisms remain unclear (Somssich et al. 2016). Over 100 Arabidopsis genes have been annotated to support putative MAPK cascades, including 20 MAPK (MPKs), 10 MAPK kinases (MKKs) and 80 MAPK kinase kinases (MAPKKKs) (Ichimura et al. 2002; Hamel et al. 2006). The best-characterized plant MAPKs are MPK3 and MPK6, which are redundantly involved in various signaling cascades related to innate immunity (Asai et al. 2002), abiotic stresses (Kovtun et al. 2000; Miles et al. 2005), ethylene response (Yoo et al. 2008), and stomatal and ovule development (Bergman et al. 2004; Wang et al. 2007; Wang et al. 2008), suggesting complexity and potentially functional redundancy of MAPK cascades in plant signal transduction networks.
In a Nicotiana benthamiana leaf system, it was reported that CLV3-triggered MAPK activation was negatively regulated by CLV1, but positively regulated by RPK2 based on the transient co-expression of CLV receptors and CLV3 ligands (Betsuyaku et al. 2011). In addition, the activated MPK6 phosphorylation was found in the clv1 mutant in the absence of the application of exogenous CLV3p (Betsuyaku et al. 2011), showing that the CLV1 receptor plays a negative role in activating MAPK signaling. However, since CLV3-induced MAPK phosphorylation was disappeared when all four receptors (CLV1, CLV2, CRN/SOL2 and RPK2) were co-expressed in N. benthamiana (Betsuyaku et al. 2011), these findings suggest not only the unique regulatory mechanism of each receptor signaling pathway, but also the antagonistic, positive roles of CLV receptors to activate MAPKs in the CLV signaling pathways. Furthermore, loss-of-function mutants of clv1, clv2 and rpk2 showed the additive phenotypes of enlarged SAMs toward higher-ordered mutants, indicating the overlapping positive functions of these receptors to promote CLV3p sensitivity restricting the stem cell population (Kinoshita et al. 2010). Because of technical limitations, there was no direct biochemical, molecular or genetic evidence to support the biological significance of the puzzling and complex MAPK function in the SAM of intact plants.
In this study, we report the rapid and dynamic MAPK activation and WUS repression in response to exogenous CLV3p treatment of the enlarged clv3 SAM exhibiting expanded CLV1-expressing cells in “quiescence” ready to perceive exogenous CLV3p signals. We also demonstrate that the CLV3p-activated MAPKs are MPK3 and MPK6 through in-gel kinase assays in WT as well as in mpk3 and mpk6 mutants. Interestingly, the phosphorylation of MPK3 and MPK6 activated by CLV3p is significantly reduced in the loss-of-function clv1 bam1 double mutant, indicating the positive role of CLV1 and BAM1 receptors in activating MAPKs. Consistently, the expression of WUS is elevated but the WUS-repressing target genes are decreased in the shoot apex tissues of inducible mpk3,6 double mutants. Moreover, the mpk3,6 mutant plants display much less sensitive phenotype of the SAM termination mediated by the application of CLV3p and exhibit enlarged SAMs in the absence of CLV3p. Therefore, our findings suggest that both MPK3 and MPK6 activated by CLV3 signals play an important role in the intracellular CLV-WUS signaling pathway for the maintenance of SAM development.
Materials and methods
Plant materials and growth conditions
Columbia-0 (Col-0) and Landsberg erecta (Ler) ecotypes were used as wild-type (WT) Arabidopsis plants in this study. The mpk3 (Salk_151594), mpk6 (Salk_062471), fls2 (Salk_141277), clv1 (WiscDsLox489–492B1), and bam1(Salk_015302) mutants and the estradiol-inducible mpk3,6 mutants (Cheng et al. 2015) had a Col-0 background, while the clv3–2 mutant had a Ler background.
For liquid culture of Arabidopsis seedlings, seeds were germinated and grown in 6-well plates containing 1 ml of liquid medium [0.5 × Murashige and Skoog (MS) and 0.5% sucrose, pH 5.8 adjusted with KOH] at 22°C–23°C and 75 μmol m−2 s−1 light intensity under a 12 h light/12 h dark photoperiod for 7 days without shaking.
RNA in situ hybridization
To amplify RNA probes, the full-length WUS cDNA was cloned into the pBluscript SK. Following PCR amplification with M13 forward/reverse primers, digoxigenin (DIG)-UTP labeled sense or antisense RNA probes were synthesized by T3 or T7 RNA polymerase, respectively (Roche). Section preparation, in situ hybridization and detection using NBT/BCIP were then performed according to the manufacturer’s protocols (Roche).
Protein in situ immunohistochemistry
The shoot apex tissues were harvested, fixed and embedded using the same procedures as for the SAM arrest assay (Kim et al. 2017). Antigen unmasking using sodium citrate was conducted according to the manufacturer’s instructions (Zymed). Sections were incubated with blocking buffer (5% [w/v] BSA in 1 × PBS) for 3 h at room temperature, then incubated with the primary rabbit α-phospho-MAPK antibody (Life technologies, #36–8800) diluted 1:100 in blocking buffer overnight at 4°C in a humidified chamber. Slides were subsequently washed three times in 1 × PBS with 0.3% [v/v] Triton X-100 with gentle shaking for 10 min. Next, slides were incubated with secondary goat α-rabbit IgG antibody diluted 1:2000 in blocking buffer overnight at 4°C (CALTAG Laboratories). Finally, slides were washed three times and detected using NBT/BCIP according to the manufacturer’s instructions (Roche).
RNA extraction and RT-qPCR assay
Total RNAs from tissues were extracted with TRIzol reagent (Invitrogen) and first-strand cDNAs were synthesized using M-MLV reverse transcriptase (Promega) according to the manufacturer’s instructions. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assays were performed with the gene specific primers listed in Table 1 using the Step One Plus™ real time PCR detection system with Power SYBR® green master mix (Applied Biosystem).
Table 1.
Primers used for RT-qPCR analysis in this study
| Gene Name | Forward primer | Reverse primer |
|---|---|---|
| WUS (At2g17950) | TGCAAGCTCAGGTACTGAATG | ATGATCCATGTTTGCCCATC |
| ARR7 (Atlgl9050) | CCTCGTATACAAGAATGTCTCAAAG | CTGCTAGCTTCACCGGTTTC |
| ARR15 (Atlg74890) | CAGCACTCAGAGAAATCCCAGT | TCATACATTGTTCTATACGAGGTTG |
| GRF6 (At2g06200) | GCCTCCTCCTACTCAATTCACTCCAAATCT | ATGATCTCTTTGTTTCAGCTTCTCCTCTAA |
| YAB3 (At4g00l80) | ACTGGGCTCATTTCCCTCACATACAC | CCAAGAGAAAATTCTAGTAGTTCCGAAA |
| ACT2 (At3gl8780) | TCCCTCAGCACATTCCAGCAGAT | AACGATTCCTGGACCTGCCTCATC |
Protein extraction and immunoblot assay
Total proteins were extracted using E buffer [135 mM Tris-HCl pH 8.8, 1% (w/v) SDS, 10% (v/v) glycerol, 50 mM Na2S2O5]. After extracts were centrifuged at maximum speed in a microfuge (13,000 g, 10 min, 4°C), the supernatant was transferred into a new tube, after which we added 1/10 volume of Z buffer [125 mM Tris-HCl pH 6.8, 12% (w/v) SDS, 10% (v/v) glycerol, 22% (v/v) β-mercaptoethanol, 0.001% (w/v) bromophenol blue]. Protein quantities were measured using a DC protein assay kit (Bio-rad). Immunoblot assays were then performed using an α-phospho-MAPK antibody (Cell signaling, #9101) or an α-TUB antibody (Sigma, T9026).
SAM termination and SAM size assays
Wild-type Col-0 and mpk3,6-es1/2 mutant seedlings were grown on MS plates (0.5 × MS, 0.5% Sucrose and 0.8% phytoagar) without estradiol for 1 week to avoid embryonic lethality, then transferred into new MS plates with or without 5 μM CLV3p in the presence of 10 μM estradiol under a 12 h light/12 h dark photoperiod for 4 weeks.
For measurement of the SAM size, seedlings were grown on MS plate for 1 week and transferred into new MS plates with 10 μM estradiol under a 12 h light/12 h dark photoperiod for 10 days. Shoot apices of seedlings were sampled and fixed in 4% paraformaldehyde/4% DMSO solution at 4°C for 16 h. Paraffin embedding and sections were performed as described previously (Kim et al. 2017). Sectioned samples were stained with 0.1% Giemsa (Sigma) for 2 min and rinsed briefly with water. The width and height of SAMs were observed using a microscope (Olympus X22LED) and measured the distance between flanking primordia using Image J program.
Statistical analysis
Statistical analyses were performed with one-way ANOVA or unpaired t test using the Prism software (version 7.0b) and significant differences were determined by Turkey’s multiple comparison test or two-tailed test, respectively.
Results
The CLV3p signal mediates rapid MAPK activation and WUS suppression in the SAM
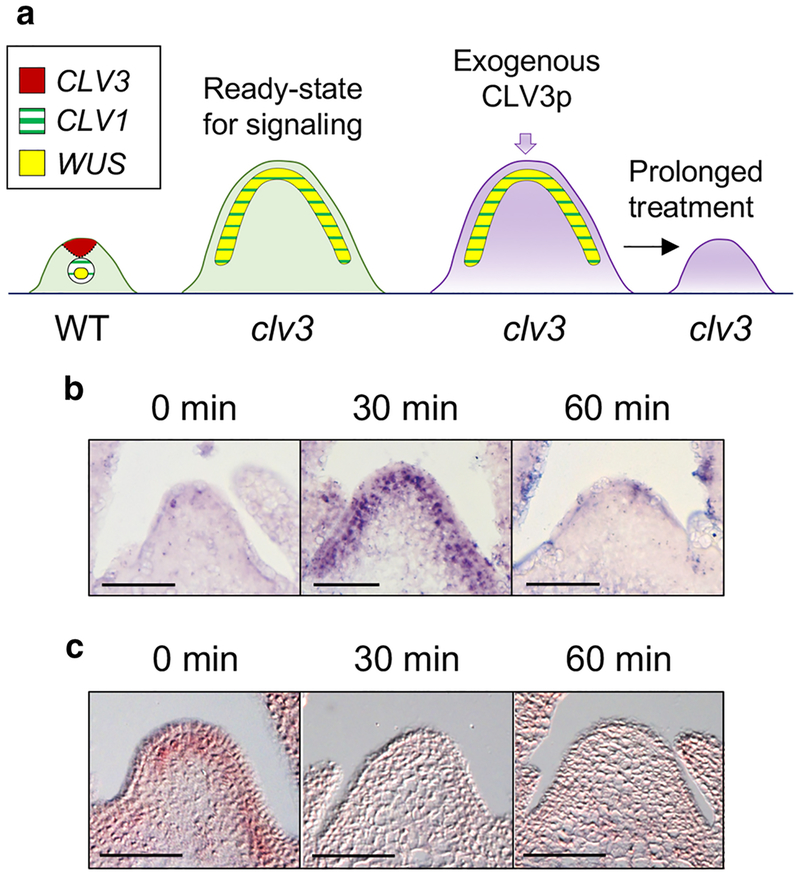
It has been challenging to investigate the primary CLV3p-CLV1 signaling events, as the key receptor CLV1 is dynamically internalized and degraded in response to endogenous CLV3 ligands in the wild-type (WT) SAM (Nimchuk et al. 2011), and CLV3p has also been shown to induce SAM immunity via the FLS2 receptor (Lee et al. 2011, 2012a, 2012b). To analyze the synchronized, dynamic and physiological stem cell signaling between the receptor and its target gene WUS in the SAM, we exploited the enlarged SAMs of the clv3 mutant, in which CLV1 receptors are stably accumulated on the plasma membrane and the expression domains of CLV1 and WUS are overlapped (Fig. 1a). Because the prolonged treatment of synthetic 12-amino acid (aa) CLV3p and 13-aa arabinosylated CLV3p was found to effectively complement the enlarged clv3 SAM growth in previous reports (Fig. 1a) (Kondo et al. 2006; Ohyama et al. 2009; Kim et al. 2017), we adopted 12-aa CLV3p as an input signal in this study. Although we previously showed that FLS2 signaling does not affect the regulation of WUS expression involved in stem cell homeostasis (Lee et al. 2011), to uncouple the CLV3p-triggered FLS2 immune response from SAM development, we used the clv3 fls2 double mutant. Importantly, the defect of the enlarged SAM in clv3–2 was fully rescued by the WT genomic CLV3 transgene, validating the key role of CLV3p signaling in the clv3–2 mutant (Song et al. 2012).
Fig. 1.
The CLV3p signal mediates rapid MAPK activation and WUS suppression in the SAM. a A schematic strategy for studying stem cell signaling using the clv3 SAM. CLV3, CLV1 and WUS genes related to stem cell signaling exhibit different expression domains in the SAM. Overlapped expression of CLV1 and WUS domains in the clv3 SAM show the ready-state for direct response triggered by CLV3p signals. b MAPK phosphorylation by CLV3p in clv3–2 fls2–24 SAMs treated with 1 μM CLV3p for 0 min, 30 min or 60 min. Protein in situ immunohistochemistry was performed using an α-phospho-MAPK antibody. c WUS repression by CLV3p in the same context with (b) through RNA in situ hybridization. Scale bar, 50 μm (b, c).
Remarkably, we detected rapid and dynamic MAPK phosphorylation activated by the application of exogenous CLV3p within 30 min in the enlarged clv3 fls2 SAM along CLV1-expressing cells (Nimchuk et al. 2015) by in situ immunolocalization using an α-phospho-MAPK antibody, and these phosphorylation signals were eventually disappeared at 60 min (Fig. 1b). Furthermore, the expanded WUS expression in the background of clv3 SAM (Schoof et al. 2000) was rapidly reduced by treatment with CLV3p within 30–60 min when MAPKs were highly activated (Fig. 1b, c). Taken together, these results suggest a correlation between CLV3p-induced MAPK activation and transcriptional repression of the downstream target WUS.
MPK3 and MPK6 are activated by treatment with CLV3p signals
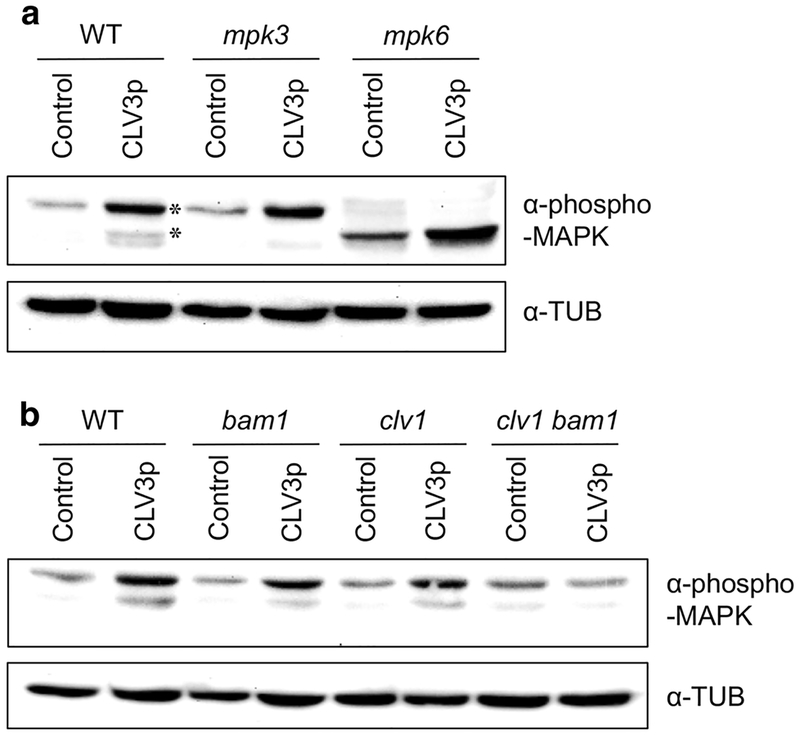
Using immunoblot assays, we further confirmed the CLV3p-triggered MAPK activation in the enriched Arabidopsis shoot apex tissues. Interestingly, we could find that MAPKs are also phosphorylated by the treatment of CLV3p in the WT Col-0 shoot apices (Fig. 2a). Importantly, because the specifically phosphorylated MAPK bands shown in WT were eliminated in mpk3 or mpk6 mutants (Fig. 2a), CLV3p-activated MAPKs were identified as MPK3 and MPK6. These findings exhibit that CLV3p signals can activate MPK3 and MPK6 to transmit intracellular signaling in the SAM.
Fig. 2.
MPK3 and MPK6 are activated by treatment with CLV3p signals. a Immunoblot assays using the shoot apex tissues harvested from WT Col-0, mpk3 and mpk6 mutant seedlings treated without (Control) or with 10 μM CLV3p for 10 min. Asterisks, endogenous MPK3 and MPK6. b Immunoblot assays using the shoot apex tissues harvested from WT Col-0, bam1, clv1 and clv1 bam1 mutant seedlings treated without (Control) or with 10 μM CLV3p for 10 min. Activated MAPKs were detected by an α-phospho-MAPK antibody and loading amounts of proteins were detected by an α-TUB antibody. These experiments were repeated at least three times with similar results
Recent findings have revealed that CLV1 and BAM1 seem to act as major receptors for CLV3p signaling, because the loss-of-function clv1 bam1 double mutant showed an almost insensitive response to the repression of WUS expression induced by CLV3p (Shinohara and Matsubayashi 2015). Interestingly, CLV3p-triggerred MAPK phosphorylation was dramatically reduced in the shoot apex tissues of the clv1 bam1 mutant compared to those of WT and each individual single mutant (Fig. 2b). Therefore, these results suggest that CLV1 and BAM1 mainly serve as corresponding receptors for CLV3p signals to induce intracellular MAPK phosphorylation.
Expression of WUS and WUS-target genes is impaired in the loss-of-function mpk3,6 mutant
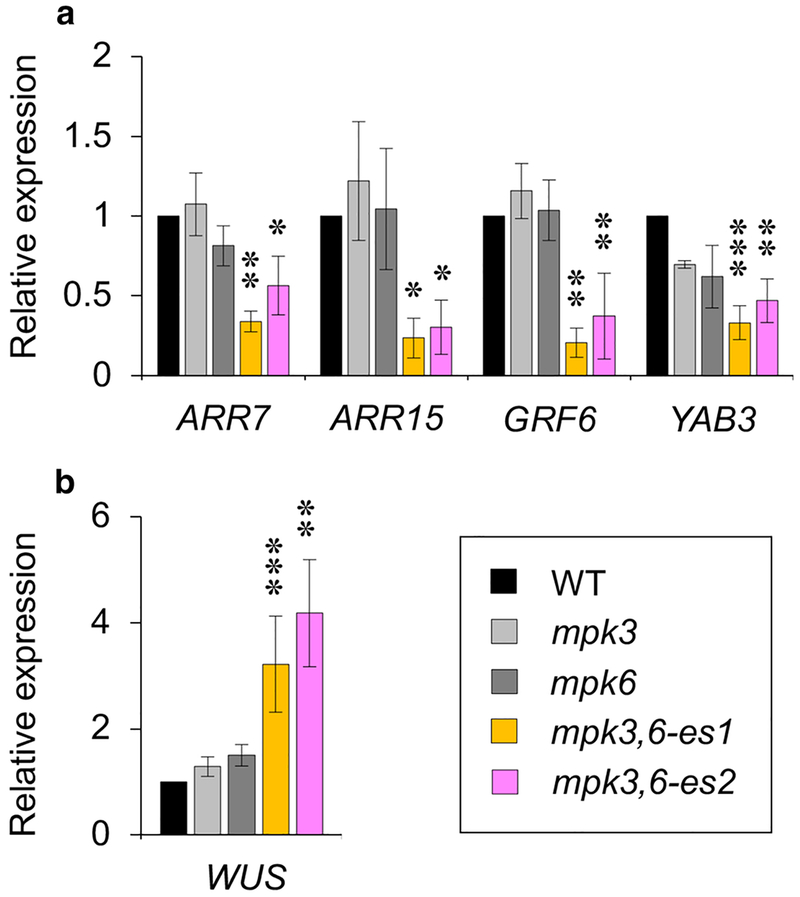
MPK3 and MPK6 have shown the functional overlap in various signaling pathways, including those associated with growth and development, abiotic stresses, hormones and pathogen defenses (Pitzschke et al. 2009; Meng et al. 2012; Lampard et al. 2014), and the mpk3,6 double homozygous mutant is known to show the embryonic lethal phenotype under normal growth conditions (Wang et al. 2007). Therefore, to further investigate the function of MPK3 and MPK6 in the shoot stem-cell homeostasis, we performed an RT-qPCR analysis to examine the expression of WUS and WUS-target genes in the enriched shoot apex tissues of WT, mpk3, mpk6 and the estradiol-inducible mpk3,6 double mutants (referred to as mpk3,6-es1 and mpk3,6-es2 for two independent lines), which bypassed the embryo lethality of mpk3,6 (Wang et al. 2007; Cheng et al. 2015). Interestingly, the RT-qPCR analysis showed that the expression of two A-type ARABIDOPSIS RESPONSE REGULATOR (ARR) genes, ARR7 and ARR15, as well as that of GROWTH-REGULATING FACTOR6 (GRF6) and YABBY3 (YAB3) were significantly decreased in the absence of MPK3/6 activities, but not MPK4 (Fig. 3a and Fig. S1), whereas the expression of WUS was significantly promoted in conditional mpk3,6-es1/2 double mutants (Fig. 3b). A-type ARR genes encoding negative regulators of cytokinin signaling are known to be directly repressed by WUS (Leibfried et al. 2005). Moreover, it has been reported that WUS directly represses differentiation-promoting regulatory genes, such as GRF6 and YAB3 (Yadav et al. 2013). Because the expression of WUS and WUS-target genes was not significantly influenced in the mpk3 or mpk6 single mutant compared to WT (Fig. 3a, b), these results indicate that MPK3 and MPK6 may redundantly affect the expression of marker genes involved in SAM development via enhanced WUS activity.
Fig. 3.
Expression of WUS and WUS-target genes is impaired in the loss-of-function mpk3,6 mutants. The expression of WUS-specific target genes (a) and WUS (b) in WT, mpk3, mpk6 and mpk3,6-es1/2 seedlings treated with 10 μM estradiol for 5 days. RNAs were isolated from the shoot apex tissues. Relative expression was normalized by ACT2. Error bars are ±S.D. (n=3). Significant differences compared to WT were determined via one-way ANOVA followed by Turkey’s test: * P<0.05, ** P<0.01 and *** P<0.001.
MPK3 and MPK6 are involved in CLV3p-mediated SAM maintenance
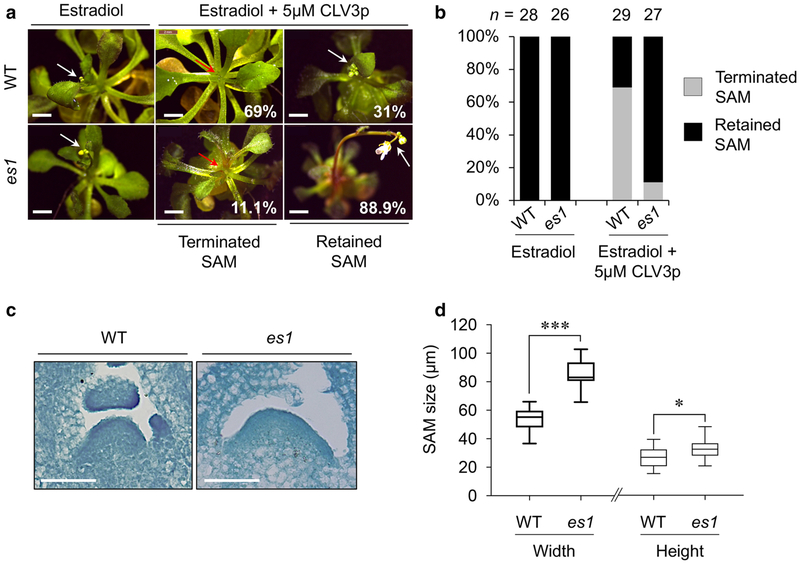
To examine the long-term effects of CLV3p treatment in the SAM development of mpk3,6-es1/2, we developed a CLV3p-dependent SAM termination assay based on the SAM transition to a visible inflorescence stem. To accomplish this, WT Col-0 and mpk3,6-es1/2 mutant seedlings were first grown on MS plates for 7 days without the application of estradiol to bypass the embryonic and seedling lethality. The majority (69%) of WT seedlings grown on MS medium containing 5 μM CLV3p for additional four weeks in the presence of 10 μM estradiol showed terminated SAMs without the inflorescence stem (Fig. 4a, b). In contrast, most estradiol-induced mpk3,6-es1/2 plants (88.9%) retained SAM growth and produced inflorescence stems in the same condition (Fig. 4a, b and Fig. S2). Moreover, the overall vegetative SAM size of mpk3,6-es1 mutants was significantly increased compared to that of WT in the presence of 10 μM estradiol (Fig. 4c, d). Taken together, these results indicate that CLV3p-triggered signaling is notably blocked in the mpk3,6 mutant, and MPK3/6 play critical roles in the regulation of stem cell homeostasis involved in SAM development.
Fig. 4.
MPK3 and MPK6 are involved in CLV3p-mediated SAM maintenance. a Significant arrest of CLV3p-mediated SAM termination in mpk3,6-es1 (es1) compared to that of WT Col-0. Red arrows, terminated SAMs. White arrows, retained SAMs. b Quantification of terminated or retained SAM growth of WT and es1 in the presence of exogenous CLV3p (n=26–29). c The phenotype of enlarged SAM shown in es1 compared to WT. Scale bar, 50 μm. d The width and height of vegetative SAMs in WT and es1 seedlings. Bands inside the boxes represent medians. Boxes represent the interquartile ranges and whiskers indicate the minimum and maximum values (n=5–8). Significant differences were determined by two-tailed t test: * P<0.05 and *** P<0.001.
Discussion
We have provided several lines of evidence to demonstrate the biological function of MPK3 and MPK6 in regulating stem cell homeostasis during SAM development. In situ immunostaining and in-gel kinase assays showed clear and dynamic MAPK activation by phosphorylation and an RNA in situ hybridization illustrated rapid WUS repression mediated by CLV3p in the enlarged clv3 SAMs (Fig 1b, c). Because CLV1 receptors perceiving CLV3p ligands are immediately diminished by endocytosis in the WT (Nimchuk et al. 2011), the loss-of-function clv3 mutant background made it feasible to analyze direct, synchronized, rapid and dynamic signaling responses triggered by exogenous CLV3p not previously possible. Interestingly, activated MAPKs in the SAM were revealed as MPK3 and MPK6, and the phosphorylation of these MPKs was induced by CLV1 and BAM1 receptors (Fig. 2b). However, since CLV3p-mediated MAPK phosphorylation was not completely eliminated in the shoot apex tissues of the clv1 bam1 double mutant, it indicates that other CLV receptors may also be involved in MAPK phosphorylation in CLV signaling. Recent findings have shown that CLV1 and BAM1 serve as direct receptors for arabinosylated CLV3p, and the clv1 bam1 double mutants reveal remarkably enlarged SAMs similar to those of the clv3 single mutant (Shinohara and Matsubyayshi 2015). Although these data represent the exclusive roles of CLV1 and BAM1 as the main receptors to perceive CLV3p signals, it is possible that other CLV receptors, including CLV1 homologues BAM2/3, CLV2 and RPK2, could be directly or indirectly involved in CLV3p-mediated MAPK phosphorylation. Moreover, it has been reported that another LRR-RLK ER-family receptor kinases, including ERECTA (ER), ERECTA-LIKE1 (ERL1) and ERL2, regulate stem cell maintenance in the SAM via a WUS-independent manner, because the loss-of-function wus er erl1 erl2 quadruple mutant restored the SAM loss found in the wus single mutant (Kimura et al. 2018). ER-family receptors are also known to function as an upstream of the YODA-MKK4/5/7/9-MPK3/6 cascade in stomatal development (Pillitteri and Torii 2012; Zoulias et al. 2018). Therefore, future researches will be required to determine how multiple receptor complexes may participate in the regulation of MAPK activation related to the complex peptide-receptor networks modulating stem cell maintenance in the SAM.
It has been reported that CLV3-mediated MAPK phosphorylation occurs in a complex manner in N. benthamiana leaves and Arabidopsis seedlings (Betsuyaku et al. 2011). However, it was shown that CLV1 receptor acts as a “negative” regulator of CLV3-dependent MPK6 phosphorylation. For example, the transient co-expression of CLV1 and CLV3 genes in N. benthamiana leaves led to the partial suppression of CLV1-dependent MAPK phosphorylation detected in the absence of CLV3 expression. In addition, elevated MPK6 activity was found in clv1 mutant seedlings, even in the absence of treatment with exogenous CLV3p. Unlike the negative role of CLV1 for MPK6 activation, our results displayed that CLV3p-mediated phosphorylation of both MPK3 and MPK6 was dramatically reduced in the clv1 bam1 mutant in the SAM, and there was no obvious MPK6 activity in the clv1 mutant without CLV3p stimulation (Fig. 2b). Because the isolated, enriched shoot apex tissues were used in this study but not whole seedlings, the use of different tissues may lead to the different results in the clv1 mutant. In addition, CLV1 has been reported to be expressed outside the SAM (Chou et al. 2016). Based on our observation of MAPK activation, these results showed that CLV3p-mediated phosphorylation of MPK3 and MPK6 is positively regulated by both CLV1 and BAM1 receptors. Moreover, since the phosphorylation of MPK3 and MPK6 was not significantly reduced in the clv1 or bam1 single mutants, it suggests that both CLV1 and BAM1 are redundantly required for the CLV3p-mediated activation of MPK3 and MPK6.
In the plant MAPK signaling pathways, well-known MAPKs such as MPK3 and MPK6 have been shown to be involved in different MAPK cascade modules induced by flg22, H2O2, phytohormones (ethylene and jasmonate), abiotic stresses (ozone and osmotic shock) and developmental signals (epidermal patterning factors 1 and 2) (Colcombet and Hirt 2008; Lee et al. 2015; Zoulias et al. 2018). Indeed, the physical interactions between MPK3/6 and upstream MKKs are not sufficient to represent the unique characteristics of each signaling pathway. For example, previous results have shown that among 10 MKKs, MPK3 interacts with six MKKs and MPK6 interacts with nine MKKs via in vitro protein microarrays (Popescu et al. 2009). One possible explanation of how the same CLV3p input signals are differently transduced via the positive role of CLV1/BAM1 on MPK3/6 activation in stem cell signaling is the regulation by scaffold proteins to specify and facilitate each signaling pathway in different MAPK cascades. Recent advances have shown that receptor for activated C kinase 1 (RACK1) functions as a scaffold protein involved in a novel plant immune pathway to link the upstream Gβ subunit and the downstream MAPK cascade module, which is distinct from the previously known flg22-induced MAPK cascade (Cheng et al. 2015). Moreover, in stomatal development, the polarity protein BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE (BASL) has also been reported to act as a scaffold protein physically interacting with MAPKKK YDA and MPK3/6 to spatially concentrate MAPK signaling for asymmetric cell division of stomatal precursor stem cells (Dong et al. 2009; Zhang et al. 2015). Since MPK3 and MPK6 have been shown to be activated by various upstream sets such as MKK4/5 or MKK7/9 in distinct stress responses and stomatal development (Colcombet and Hirt 2008; Yoo et al., 2008; Pitzschke et al. 2009; Rodriguez et al. 2010; Zoulias et al. 2018), it would be interesting to identify putative scaffold proteins and MKKs acting as an upstream of MPK3/6 activation via CLV1 and BAM1 receptors involved in stem cell homeostasis in the SAM.
Supplementary Material
Key message:
CLV3p-mediated phosphorylation of MPK3 and MPK6 occurs via CLV1 and BAM1 receptors to regulate the maintenance of SAM development.
Acknowledgements
We are grateful to Y. Xiong for the mpk3,6-es1/2 lines and HJ Kim for technical supports. HL was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2014R1A1A1006288 and 2018R1D1A1A02048850) and the Next Generation Biogreen 21 Program (PJ01109901). JS is supported by grants from the National Institute of General Medical Sciences (R01GM070567).
Abbreviations:
- MAPK (MPK)
mitogen-activated protein kinase
- SAM
shoot apical meristem
- CLV3p
CLAVATA3 peptide
- WUS
WUSCHEL
- BAM
BARELY ANY MERISTEM
- LRR-RLK
leucine-rich repeat receptor-like kinase
- ER
erecta
Footnotes
Supplementary Material
Supplementary figures are available at Plant Cell Reports Online.
Declaration of interests
The authors declare that they have no conflicts of interest.
References
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gómez-Gómez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415:977–983 [DOI] [PubMed] [Google Scholar]
- Aichinger E, Kornet N, Friedrich T, Laux T (2012) Plant stem cell niches. Annu Rev Plant Biol 63:615–636 [DOI] [PubMed] [Google Scholar]
- Bergmann DC, Lukowitz W, Somerville CR (2004) Stomatal development and pattern controlled by a MAPKK kinase. Science 304:1494–1497 [DOI] [PubMed] [Google Scholar]
- Betsuyaku S, Takahashi F, Kinoshita A, Miwa H, Shinozaki K, Fukuda H, Sawa S (2011) Mitogen-activated protein kinase regulated by the CLAVATA receptors contributes to shoot apical meristem homeostasis. Plant Cell Physiol 52:14–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Li JF, Niu Y, Zhang XC, Woody OZ, Xiong Y, Djonović S, Millet Y, Bush J, McConkey BJ, Sheen J, Ausubel FM (2015) Pathogen-secreted proteases activate a novel plant immune pathway. Nature 521:213–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J, Hirt H (2008) Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J 413:217–226 [DOI] [PubMed] [Google Scholar]
- Chou H, Zhu Y, Ma Y, Berkowitz GA (2016) The CLAVATA signaling pathway mediating stem cell fate in shoot meristems requires Ca2+ as a secondary cytosolic messenger. Plant J 85:494–506 [DOI] [PubMed] [Google Scholar]
- Dong J, MacAlister CA, Bergmann DC (2009) BASL controls asymmetric cell division in Arabidopsis. Cell 137:1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Han L, Hymes M, Denver R, Clark SE (2010) CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J 63:889–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel LP, Nicole MC, Sritubtim S, Morency MJ, Ellis M, Ehlting J, Beaudoin N, Barbazuk B, Klessig D, Lee J, Martin G, Mundy J, Ohashi Y, Scheel D, Sheen J, Xing T, Zhang S, Seguin A, Ellis BE (2006) Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci 11:192–198 [DOI] [PubMed] [Google Scholar]
- Heidstra R, Sabatini S (2014) Plant and animal stem cells: similar yet different. Nat Rev Mol Cell Biol 15:301–312 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Shinozaki K, Tena G, Sheen J, Henry Y, Champion A, Kreis M, Zhang S, Hirt H, Wilson C, Heberle-Bors E, Ellis BE, Morris PC, Innes RW, Ecker JR, Scheel D, Klessig DF, Machida Y, Mundy J, Ohashi Y, Walker JC (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7:301–308 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Wu CY, Yu HM, Sheen J, Lee H (2017) Dual CLAVATA3 peptide in Arabidopsis shoot stem cell signaling. J Plant Biol 60:506–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Tasaka M, Torii KU, Uchida N (2018) ERECTA-family genes coordinate stem cell functions between the epidermal and internal layers of the shoot apical meristem. Development 145:dev156380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita A, Betsuyku S, Osakabe Y, Mizuno S, Nagawa S, Stahl Y, Simon R, Yamaguchi-Shinozaki K, Fukuda H, Sawa S (2010) RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137:3911–3920 [DOI] [PubMed] [Google Scholar]
- Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, Sakagami Y (2006) A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313:845–848 [DOI] [PubMed] [Google Scholar]
- Kovtun Y, Chiu WL, Tena G, Sheen J (2000) Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA 97:2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard GR, Wengier DL, Bergmann DC (2014) Manipulation of mitogen-activated protein kinase kinase signaling in the Arabidopsis stomatal lineage reveals motifs that contribute to protein localization and signaling specificity. Plant Cell 26:3358–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Chah OK, Sheen J (2011) Stem-cell-triggered immunity through CLV3p-FLS2 signalling. Nature 473:376–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Chah OK, Plotnikov J, Sheen J (2012a) Stem cell signaling in immunity and development. Cold Spring Harb Symp Quant Biol 77:75–81 [DOI] [PubMed] [Google Scholar]
- Lee H, Khatri A, Plotnikov JM, Zhang XC, Sheen J (2012b) Complexity in differential peptide-receptor signaling: response to Segonzac et Al. and Mueller et Al. commentaries. Plant Cell 24:3177–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Hnilova M, Maes M, Lin YCL, Purarjunan A, Han SK, Avila J, Torii KU (2015) Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature 522:439–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU (2005) WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438:1172–1175 [DOI] [PubMed] [Google Scholar]
- Meng X, Wang H, He Y, Liu Y, Walker JC, Torii KU, Zhang S (2012) A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24:4948–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles GP, Samuel MA, Zhang Y, Ellis BE (2005) RNA interference-base (RNAi) suppression of AtMPK6, an Arabidopsis mitogen-activated protein kinase, results in hypersensitivity to ozone and misregulation of AtMPK3. Environ Pollut 138:230–237 [DOI] [PubMed] [Google Scholar]
- Müller R, Bleckmann A, Simon R (2008) The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20:934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Clark SE (2006) Evidence for functional conservation, sufficiency, and proteolytic processing of the CLAVATA3 CLE domain. Plant Physiol 140:726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk ZL, Tarr PT, Ohno C, Qu X, Meyerowitz EM (2011) Plant stem cell signaling involves ligand-dependent trafficking of the CLAVATA1 receptor kinase. Curr Biol 21:345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk ZL, Zhou Y, Tarr PT, Peterson BA, Meyerowitz EM (2015) Plant stem cell maintenance by transcriptional cross-regulation of related receptor kinases. Development 142:1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y (2009) A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol 5:578–580 [DOI] [PubMed] [Google Scholar]
- Pillitteri LJ, Torii KU (2012) Mechanisms of stomatal development. Annu Rev Plant Biol 63:591–614 [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Hirt H (2009) Disentangling the complexity of mitogen-activated protein kinases and reactive oxygen species signaling. Plant Physiol 149:606–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzschke A, Schikora A, Hirt H (2009) MAPK cascade signalling networks in plant defence. Curr Opin Plant Biol 12:421–426 [DOI] [PubMed] [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Gerstein M, Snyder M, Dinesh-Kumar SP (2009) MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev 23:80–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MCS, Petersen M, Mundy J (2010) Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61:621–649 [DOI] [PubMed] [Google Scholar]
- Rojo E, Sharma VK, Kovaleva V, Raikhel N, Fletcher JC (2002) CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 14:969–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KFX, Jürgens G, Laux T (2000) The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100:635–644 [DOI] [PubMed] [Google Scholar]
- Shinohara H, Matsubayashi Y (2015) Reevaluation of the CLV3-receptor interaction in the shoot apical meristem: dissection of the CLV3 signaling pathway from a direct ligand-binding point of view. Plant J 82:328–336 [DOI] [PubMed] [Google Scholar]
- Somssich M, Je BI, Simon R, Jackson D (2016) CLAVATA-WUSCHEL signaling in the shoot meristem. Development 143:3238–3248 [DOI] [PubMed] [Google Scholar]
- Song XF, Yu DL, Xu TT, Ren SC, Guo P, Liu CM (2012) Contributions of individual amino acid residues to the endogenous CLV3 function in shoot apical meristem maintenance in Arabidopsis. Mol Plant 5:515–523 [DOI] [PubMed] [Google Scholar]
- Wang H, Liu Y, Bruffett K, Lee J, Hause G, Walker JC, Zhang S (2008) Halo-insufficiency of MPK3 in MPK6 mutant background uncovers a novel function of these two MAPKs in Arabidopsis ovule development. Plant Cell 20:602–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S (2007) Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19:63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Girke T, Jönsson H, Reddy GV (2011) WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev 25:2025–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Ohno C, Heisler M, Girke T, Jönsson H. Reddy GV (2013) Plant stem cell maintenance involves direct transcriptional repression of differentiation program. Mol Syst Biol 9:654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Tena G, Xiong Y, Sheen J (2008) Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H2 signalling. Nature 451:789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liu X, Engstrom EM, Nimchuk ZL, Pruneda-Paz JL, Tarr PT, Yan A, Kay SA, Meyerowitz EM (2015) Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 517:377–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yan A, Han H, Li T, Geng Y, Liu X, Meyerowitz EM (2018) HAIRY MERISTEM with WUSCHEL confines CLAVATA3 expression to the outer apical meristem layers. Science 361:502–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang P, Shao W, Zhu JK, Dong J (2015) The BASL polarity protein controls a MAPK signalling feedback loop in asymmetric cell division. Dev Cell 33:136–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoulias N, Harrison EL, Casson SA, Gray JE (2018) Molecular control of stomatal development. Biochem J 475:441–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.